Abstract
Fusobacterium species are part of the gut microbiome in humans. Recent studies have identified over-representation of Fusobacterium in colorectal cancer (CRC) tissues but it is not yet clear whether this is pathogenic or simply an epiphenomenon. In this study, we evaluated the relationship between Fusobacterium status and molecular features in CRCs through quantitative real-time PCR in 149 CRC tissues, 89 adjacent normal appearing mucosae and 72 colonic mucosae from cancer-free individuals. Results were correlated with CpG island methylator phenotype (CIMP) status, microsatellite instability (MSI) and mutations in BRAF, KRAS, TP53, CHD7 and CHD8. Whole exome capture sequencing data were also available in 11 cases. Fusobacterium was detectable in 111/149 (74%) CRC tissues and heavily enriched in 9% (14/149) of the cases. As expected, Fusobacterium was also detected in normal appearing mucosae from both cancer and cancer-free individuals but the amount of bacteria was much lower compared to CRC tissues (a mean of 250-fold lower for Pan-fusobacterium). We found the Fusobacterium-high CRC group (FB-high) to be associated with CIMP positivity (p=0.001), TP53 wild type (p=0.015), hMLH1 methylation positivity (p=0.0028), MSI (p=0.018) and CHD7/8 mutation positivity (p=0.002). Among the 11 cases where whole exome sequencing data was available, two that were FB-high cases also had the highest number of somatic mutations (a mean of 736 per case in FB-high vs. 225 per case in all others). Taken together, our findings show that Fusobacterium enrichment is associated with specific molecular subsets of CRCs, offering support for a pathogenic role in CRC for this gut microbiome component
Keywords: Fusobacterium, colorectal cancer, DNA methylation, CpG island methylator phenotype, exome sequencing
Introduction
The non-spore-forming, anaerobic Gram-negative bacteria, Fusobacterium is part of the normal flora in the human mouth and gut mucosa. Fusobacterium species are highly heterogeneous and some species have been recognized as opportunistic pathogens implicated in inflammatory diseases of both the mouth, such as periodontitis, and the gut, such as appendicitis and inflammatory bowel diseases1-5. Two recent studies have linked Fusobacterium species with colorectal cancer (CRC). These studies demonstrated that Fusobacterium nucleatum (F. nucleatum) and whole Fusobacterium species (Pan-fusobacterium) were abundant in CRC tissues compared to adjacent normal mucosa6,7. Several infectious bacteria and viruses were previously associated with neoplasia such as human papillomavirus in cervical cancer8, Kaposi’s sarcoma-associated herpesvirus in Kaposi’s sarcoma9 and Epstein–Barr virus in lymphomas and gastric cancer10. Fusobacterium in CRC provided a novel concept in that a part of the normal intestinal microflora may be relevant to tumorigenesis. However, the previous studies could not exclude the possibility that the presence of Fusobacterium in CRC is an epiphenomenon, related to local changes triggered by the neoplastic process.
CRCs are characterized by specific genetic and epigenetic lesions. Besides common mutations in TP53, KRAS and APC genes11,12, epigenetic alterations in CRCs are frequent, particularly gene promoter DNA methylation. Classification of CRCs according to mutation and DNA methylation status has identified distinct subtypes based on the CpG Island Methylator Phenotype (CIMP)13. Typical high-level CIMP (CIMP-high, CIMP1) CRCs are associated with microsatellite instability (MSI) through epigenetic silencing of a mismatch repair gene MLH1, as well as BRAF mutation. Frequent mutation in chromatin regulator genes, notably, CHD7 and CHD8, members of the chromodomain helicase/ ATP-dependent chromatin remodeling family were recently also discovered in CIMP1 CRCs14. Low-level CIMP (CIMP-low, CIMP2) is characterized by methylation of a limited group of genes and mutation in KRAS. CIMP-negative cases have less frequent methylation changes and very frequent TP53 mutation and chromosomal instability15,16.
Since CRCs have heterogeneous molecular and clinical features15-19, we investigated whether Fusobacterium status is associated with different subtypes of CRCs. We found that Fusobacterium-high cases have a unique genetic and epigenetic profile, supporting potential links between the gut microbiome and molecular features of CRC.
Materials and Methods
Tissue samples
We used genomic DNA samples of 149 primary CRCs and 89 normal-appearing adjacent tissues from patients undergoing surgery or colonoscopy at the Johns Hopkins Hospital, MD Anderson Cancer Center, Sapporo medical University, Akita Red Cross Hospital and Aichi Cancer Center Research Institute. All CRCs used in this study were characterized previously for CIMP (all cases), MSI (n=113), BRAF mutation (n=144), KRAS mutation (n=148) and TP53 mutation status (n=143) 15,20-23. CHD7 and CHD8 mutation were also characterized in 100 out of 149 cases14. Genomic DNA was also obtained from 72 colonic biopsies in 65 cancer free subjects undergoing colonoscopy at the MD Anderson Cancer Center and Fujita Health University Hospital. 52 out of 72 these samples were from distal colon (descending and sigmoid colon, and rectum) and the remaining 20 were from the proximal colon (cecum, ascending and transverse colon). Samples were collected in accordance with institutional policies and written informed consent for tissue collection was provided by all the participants.
Quantitative PCR analysis for Fusobacterium
Quantitative real-time PCR was performed using the Universal PCR Master Mix (Bio-Rad) and StepOnePlus™ Real-Time PCR System (Applied Biosystems). F. nucleatum and pan-fusobacterium TaqMan primer/probe sets used in this study were described previously6,24. The cycle threshold (Ct) values for F. nucleatum and pan-fusobacterium were normalized to the amount of human DNA in each reaction by using a primer/probe set for the reference gene, prostaglandin transporter (PGT), as described previously25. All assays were done in duplicate and we averaged the results.
DNA methylation analysis for cancer free subjects
Bisulfite-treated genomic DNA from cancer free subjects was used to evaluate the methylation status of 7 CpG islands (ER, SFRP1, MYOD1, MGMT, SLC16A2, SPOCK2 and N33) using the primers listed in supplementary Table 1. Bisulfite treatment of DNA was performed with an EpiTect bisulfite kit (Qiagen) according to the manufacturer’s protocol. Pyrosequencing was carried out using a Pyro Mark Q96 MD system with a Pyro-Gold reagent Kit (QIAGEN), and the results were analyzed using PyroMark Q96 ID software version 1.0 (QIAGEN).
Whole exome capture sequencing and Gene Ontology analysis
Genomic DNA specimens from 11 colorectal tumors and their adjacent normal tissues were submitted to Otogenetics Corporation (Norcross, GA USA) for exome capture and sequencing. Genomic DNAs were fragmented and then tested for size distribution and concentration. Illumina libraries were made using Next reagents (New England Biolabs, Ipswich, MA USA), and the resulting libraries were subjected to exome enrichment using NimbleGen SeqCap EZ Human Exome Library v2.0 (Roche NimbleGen, Inc., Madison, WI USA). The samples were then sequenced on an Illumina HiSeq2000 (Illumina, Inc., San Diego, CA USA), which generated paired-end reads of 90 or 100 nucleotides. All paired samples (tumor and normal) were sequenced on the same run, using same depth and coverage. Read results from both replicates were combined in the final analysis. Data were analyzed for quality, exome coverage, and exome-wide single nucleotide polymorphism (SNP)/InDel using the platform provided by DNAnexus (Mountain View, CA USA). We excluded all variants with a PHRED-encoded probability score lower than 35, those that were present in the DNA of the corresponding normal samples (thus excluding germline events), and those that were not in coding regions, as well as silent changes and known SNPs (except for clinically associated SNPs). DNAnexus Genome Browser was used for visual validation of all potential somatic mutations to ensure that they were present in forward and reverse strands. The clinicopathological data for the studied cases, a detailed protocol of data analysis, summary of sequencing statistics and somatic mutations list for all samples can be found in this manuscript 14. Functional enrichment of mutated genes was determined by the gene ontology analysis using DAVID Bioinformatics Resources 6.7 (http://david.abcc.ncifcrf.gov/). P-values were corrected for multiple hypothesis testing using the Benjamini method.
Statistical analysis
Continuous variables among matched samples (cancer and normal tissues) were examined using the Wilcoxon signed-rank test. Continuous variables among two and three different groups were examined using the Student’s t-test and One-way ANOVA, respectively. Categorical variables among two or three different groups were examined using two-sided Fisher’s exact test. Two sided P value < .05 was considered statistically significant.
Results
Clinicopathologic characteristics of CRCs
We studied 104 CRCs selected based on sample availability and subsequently added 26 CIMP1, 18 CIMP2 and 1 CIMP-negative cases to expand this cohort. In total, these cases consisted of 60 CIMP-negative, 42 CIMP1 and 47 CIMP2 tumors. Clinicopathologic characteristics are shown in Table 1. As expected, CIMP1 cases presented at a higher age and were principally located in the proximal colon. CIMP1 cases were characterized by a higher incidence of mutations in BRAF and MSI and rare mutations in KRAS and TP53. The CIMP2 cases were characterized by a higher incidence of mutations in KRAS and rare MSI. The CIMP-negative cases were characterized by a higher incidence of mutations in TP53 and rare MSI.
Table1.
Clinicopathological characteristics of 149 CRCs studied
| CIMP-negative | CIMP1 | CIMP2 | |
|---|---|---|---|
| Total number | 60 | 42 | 47 |
| Age: mean +/− SEM& | 64.0+/−1.9 | 71.8+/−1.3 | 66.7+/−1.6 |
| Female | 21 (35.0%) | 21 (50.0%) | 18 (38.3%) |
| Proximal location# | 26 (52.0%) | 26 (86.7%) | 22 (75.9%) |
| BRAF mutant$ | 2 (3.6%) | 31 (73.8%) | 0 (0%) |
| KRAS mutant* | 23 (40.0%) | 0 (0%) | 38 (80.9%) |
| TP53 mutant$$ | 37 (66.1%) | 3 (7.1%) | 18 (40.0%) |
| MSI## | 6 (13.0%) | 36 (97.3%) | 0 (0%) |
CIMP1 vs. CIMP-negative, p=0.002, CIMP1 vs.CIMP2, p=0.01.
CIMP1 vs. CIMP-negative, p=0.002. Data were missing in 28 cases.
CIMP1 vs. CIMP-negative, p<0.0001, CIMP1 vs. CIMP2, p<0.0001. Data were missing in 5 cases.
CIMP2 vs. CIMP-negative, p=0.0001, CIMP2 vs. CIMP1, p<0.0001, CIMP-negative vs. CIMP2, p<0.0001. Data was missing in one case.
CIMP-negative vs. CIMP1, p<0.0001, CIMP-negative vs. CIMP2, p=0.02, CIMP2 vs. CIMP1, p=0.0004. Data were missing in 6 cases.
CIMP1 vs. CIMP-negative, p<0.0001, CIMP1 vs. CIMP2, p<0.000. Data were missing in 36 cases.
Note: Proximal, cecum, and ascending and transverse colon; distal, descending and sigmoid colon, and rectum ND, not determined.
Detection of Fusobacterium in CRC tissues and their adjacent mucosa
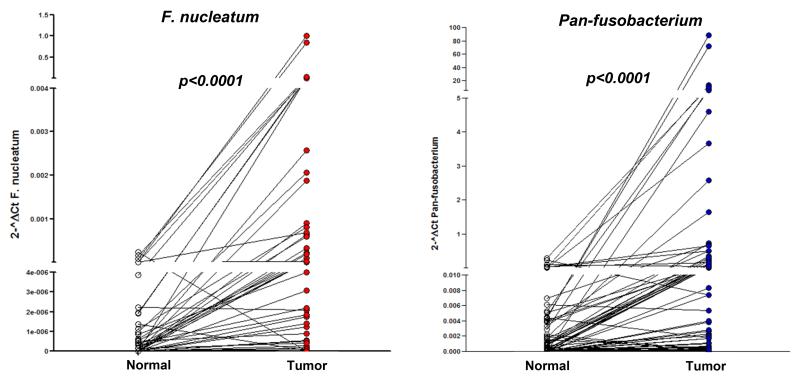
Among 149 CRC tumor tissues, F. nucleatum and pan-fusobacterium were detectable in 78 (52.3%) and 110 (73.8%) cases, respectively and 111 patients (74.4%) had either F. nucleatum or pan-fusobacterium detectable. Among 89 adjacent normal colonic mucosae, F. nucleatum and pan-fusobacterium were detectable in 27 (30.3%) and 47 (52.8%) cases, respectively (Supplementary Fig. 1). To determine the abundance of Fusobacterium in CRC tissues, we initially compared the amount of bacteria in 89 matched tumor tissues and normal mucosae. In agreement with previous studies6,7, we found significant enrichment of both F. nucleatum and pan-fusobacterium in CRC tissues compared to adjacent normal mucosae (approximate enrichment of F. nucleatum, 3600 fold and pan-fusobacterium, 250 fold, both p values <0.0001 by the Wilcoxon signed-rank test, Fig.1). Over-representation of both F. nucleatum and pan-fusobacterium in tumor versus matched normal specimens was found in more than half of the cases (51%, 45/89 for F. nucleatum and 62%, 55/89 for pan-fusobacterium).
Fig.1.
Over-representation of F. nucleatum (left) and pan-fusobacterium (right) in CRC tissues relative to adjacent normal colonic mucosa in 89 paired cases. Statistical analysis was performed using the Wilcoxon signed-rank test.
Association between fusobacterium high and clinical and molecular characteristics of CRC
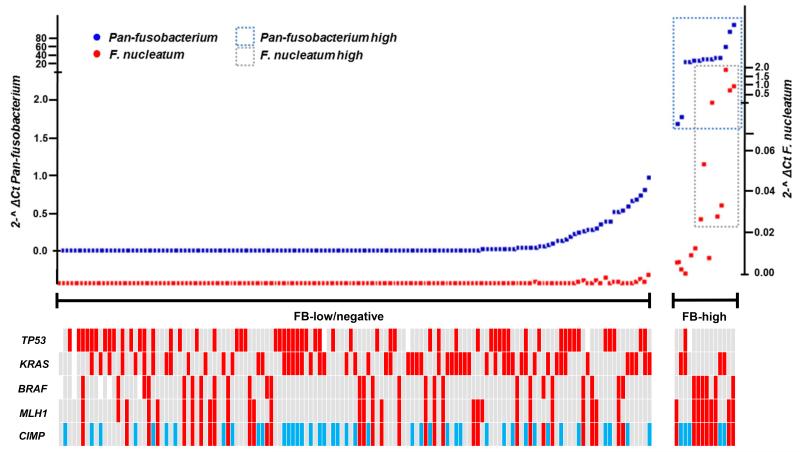
The amount of F. nucleatum and pan-fusobacterium in detectable cases varied considerably among the samples. Pan-fusobacterium was more commonly detected, being measurable in 74%. For both F. nucleatum and pan-fusobacterium, the amount of bacteria in measurable cases had an approximately Gaussian distribution, with over representation of bacteria-high cases. Based on this, we set cut-off values of 0.01 and 1 (2-^ ΔCt) for F. nucleatum and pan-fusobacterium and identified 8 (5.4%) and 14 (9.4%) cases as having a high amount of F. nucleatum and pan-fusobacterium, respectively (Supplementary Fig. 2). Since F. nucleatum and pan-Fusobacterium status was highly correlated in both cancer and normal tissues (p <0.0001, Supplementary Table 2), we defined a high amount of Fusobacterium (FB-high) as those cases with either high F. nucleatum or pan-fusobacterium or both. In cancer tissues, all 8 cases with high F. nucleatum were included in high pan-fusobacterium cases. Therefore, all FB-high cases (n=14) corresponded to high pan-fusobacterium cases. (Supplementary Table 2, Fig. 2). On average, these cases had 250 fold enrichment of pan-fusobacterium when compared to the overall average of the other cancer cases. We next analyzed clinico-pathologic correlations of FB-high status.
Fig. 2.
Distribution of Fusobacterium in CRC patients (n=149). The cases were ranked according to the amount of pan-fusobacterium (right=high amount, left=low amount). Note that all F. nucleatum high cases (n=8) were included in pan-fusobacterium high cases (n=14) and there is clear separation of FB high group (n=14, 9.4%) and FB low/negative group (n=135, 90.6%). Red, CIMP1, MLH1 methylated, BRAF, KRAS and TP53 mutated; blue, CIMP2; grey CIMP-negative, MLH1 unmethylated, BRAF, KRAS and TP53 wild type; white, not determined;
The prevalence of FB-high was significantly elevated in CIMP-positive CRCs including CIMP1 (9/42, 21.4%) and CIMP2 CRCs (5/47, 10.6%) compared to CIMP-negative cases (0/64, 0%, p=0.001). Consistent with this, FB-high was significantly associated with molecular features that are common in CIMP CRCs, such as TP53 wild type (p=0.015), hMLH1 methylation positivity (p=0.0028) and MSI (p=0.018) (Table 2). On the other hand, prevalence of fusobacterium measurable cases were similar among CIMP1, CIMP2 and CIMP-negative cases for both F. nucleatum and pan-fusobacterium (all p values >0.05, data not shown). We also found a significant association between FB-high and CHD7/8 mutation positivity (CHD7: p=0.025, CHD8: p=0.035 and CHD7/8 mutation: p=0.002). CHD7 and CDH8 are members of the chromodomain helicase/ ATP-dependent chromatin remodeling family and both are commonly mutated in CIMP-positive CRCs in our recent study14. Since CIMP-positive CRCs are more common in proximal colon and it is conceivable that the gut microbiome differs by site, we next assessed whether FB-high is associated with CIMP-positive CRCs in the proximal colon. Among 72 proximal CRCs, FB-high was significantly associated with CIMP (p=0.047). FB-high was also associated with CHD7/8 mutation (p=0.046) and older age (p=0.01), while weak associations were also found between FB-high and TP53 wild type status (p=0.05), hMLH1 methylation positivity (p=0.05) and CHD7 mutation (p=0.06) (Supplementary Table 4). We also investigated whether FB-high is associated with any clinical or molecular features within CIMP1 CRCs but found no significant correlations (Supplementary Table 5).
Table 2.
Association between high amount of Fusobacterium and clinical and molecular subtypes of CRCs
| Variables: n (%) | FB-high | (%) | FB-low/negative | (%) | p value |
|---|---|---|---|---|---|
| CIMP status | |||||
| CIMP-negative | 0 | 0.0 | 60 | 100.0 | |
| CIMP-1 | 9 | 21.4 | 33 | 78.6 | |
| CIMP-2 | 5 | 10.6 | 42 | 89.4 | 0.001 |
| BRAF | |||||
| Wild type | 8 | 7.2 | 103 | 92.8 | |
| Mutated | 6 | 18.2 | 27 | 81.8 | 0.09 |
| KRAS | |||||
| Wild type | 10 | 11.5 | 77 | 88.5 | |
| Mutated | 4 | 6.6 | 57 | 93.4 | 0.4 |
| P53 | |||||
| Wild type | 12 | 14.1 | 73 | 85.9 | |
| Mutated | 1 | 1.7 | 57 | 98.3 | 0.015 |
| hMLH1 | |||||
| Unmethylated | 5 | 4.6 | 103 | 95.4 | |
| Methylated | 9 | 22.0 | 32 | 78.0 | 0.0028 |
| MSI | |||||
| MSS | 3 | 4.2 | 68 | 95.8 | |
| MSI | 8 | 19.0 | 34 | 81.0 | 0.018 |
| CHD7 | |||||
| Wild type | 7 | 8.0 | 81 | 92.0 | |
| Mutated | 4 | 33.3 | 8 | 66.7 | 0.025 |
| CHD8 | |||||
| Wild type | 7 | 8.0 | 80 | 92.0 | |
| Mutated | 4 | 30.8 | 9 | 69.2 | 0.035 |
| CHD7 or 8 | |||||
| Wild type | 4 | 5.1 | 74 | 94.9 | |
| Mutated | 7 | 31.8 | 15 | 68.2 | 0.002 |
| Location | |||||
| Distal colon | 2 | 4.1 | 47 | 95.9 | |
| Proximal colon | 9 | 12.5 | 63 | 87.5 | 0.2 |
| Gender | |||||
| Male | 7 | 7.9 | 82 | 92.1 | |
| Female | 7 | 11.7 | 53 | 88.3 | 0.57 |
| Age | |||||
| ~70y | 5 | 5.8 | 81 | 94.2 | |
| 70y< | 9 | 14.5 | 53 | 85.5 | 0.09 |
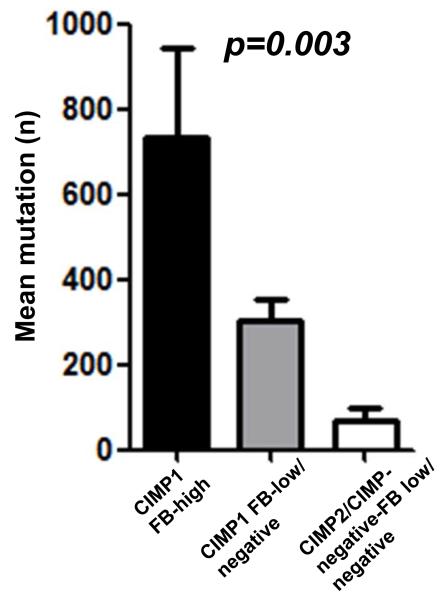
Whole exome capture and sequencing data was available for 11 CRCs and their matched normal colonic tissues14. The 11 CRCs consisted of 8 CIMP1, 1 CIMP2 and 2 CIMP-negatives, and 2 of CIMP1 CRCs were classified as FB-high. This technology determines the sequence of ~30,000 coding genes, based on RefSeq, CCDS and miR base. There were 3495 non-silent somatic mutations in 2913 genes. The somatic mutations in the two FB-high (mean 736) was higher than that seen in CIMP1 with low/undetectable FB (mean 302, range 94 to 436) and CIMP2/CIMP-negative with low/undetectable FB presented the lowest somatic mutation rate (mean 71, range 24 to 122). These differences were statistically significant (p=0.003) (Fig.3). We also compared the distribution of different types of mutations (non-synonymous, stop codon and frame shift) and the context of the single base substitution mutations. Although CIMP-1 CRCs had increased mutations in polynucleotide tracts, there was no difference in the types of mutations or the context of the single base substitution mutations across the different CIMP and Fusobacterium status. Non-synonymous, C to T and G to A transitions within the CpG sites were the most frequent in all the samples14.
Fig. 3.
Number of mutated genes determined by whole exome sequencing analysis in 11 CRCs. (2 FB-high, 6 FB-low/negative CIMP1 and 3 FB-low/negative CIMP2/CIMP-negative). Statistical analysis was performed using One-way ANOVA.
To further evaluate functional differences of gene mutations among FB-high cases, we next performed Gene Ontology analysis to determine whether there was an enrichment for specific functional categories among the mutated genes in FB-high cases. This analysis showed that mutated genes in FB-high cases frequently encoded genes related to nervous system development. Interestingly, this functional category is not represented among the genes exclusively mutated in CIMP1 with low/undetectable FB and CIMP2/CIMP-negative with low/undetectable FB nor among the genes mutated in both tumor categories (Supplementary Table 6, 7). However, the number of cases available for analysis is small and these conclusions need confirmation in other datasets.
Detection of Fusobacterium in non-neoplastic colonic mucosa
Although the amount was much lower than that of cancer tissues (Fig.1), the amount of F. nucleatum and pan-fusobacterium in adjacent normal mucosae also showed a Gaussian distribution with an excess of bacteria-high cases. Based on this, we set a cut-off value of 3*10^(−6) and 0.1 (2-^ ΔCt) for F. nucleatum and pan-fusobacterium, respectively. Among the 89 samples analyzed, 9 (10.1%) and 8 (9.9%) were classified as having a high amount of F. nucleatum and pan-fusobacterium, respectively (Supplementary Fig. 3). F. nucleatum and pan-Fusobacterium status was highly correlated in normal tissues (p <0.0001, Supplementary Table 3). We then classified 13 out of 89 cases (14.6%) as FB-high, having either a high amount of F. nucleatum or pan-fusobacterium in the normal adjacent mucosa. FB-high status in normal appearing mucosae was associated with a 15-fold increased likelihood of FB-high status in cancer tissues (p=0.0005, Table 3).
Table 3.
Association between Fusobacterium status in adjacent tissues and cancer tissues
| FB-high | FB low/negative | |
|---|---|---|
| Adjacent tissues | ||
| FB-low/negative (n=76) | 4 (5.3%) | 72 (94.7%) |
| FB-high (n=13) | 6 (46.2%) | 7 (53.8%) |
Odds ratio=15.4, 95% confidence intervals=3.5-68.1, p=0.0005.
We next examined 72 non-neoplastic colonic biopsies from 65 cancer free subjects. 14 biopsies from 12 subjects (18.4%) were classified as FB-high using the same cut off value used in cancer cases. The prevalence of FB-high was not significantly different between patients with CRCs and cancer free subjects (14.6% vs. 18.4%, p=0.66, Table 4). Patients with CIMP1 CRC were more likely to be FB-high in their adjacent tissues than patients with CIMP-negative CRC (29.2% vs. 6.8%, p=0.03, Table 4). FB-high state in the cancer free subjects was not associated with any clinical characteristics including gender, location and age (Supplementary Table 8). We also found no significant difference of FB-high state among samples from the United States (7/37, 18.9%) or from Japan (7/35, 20%) (p=0.92). Finally, we investigated the association between FB-high and DNA methylation status in non-neoplastic colonic mucosa using 7 different markers (ER, SFRP1, MYOD1, MGMT, SLC16A2, SPOCK2 and N33). No significant association was found between FB-high and methylation status of any marker (Supplementary Fig.4).
Table 4.
Fusobacterium status in non-neoplastic colonic mucosa in cancer free and CRC patients
| FB-high | FB low/negative | |
|---|---|---|
| Cancer free (n=65) | 12 (18.4%) | 53 (80.6%) |
| CRC cases (n=89) | 13 (14.6%) | 76 (85.4%) |
| CIMP-negative (n=44) | 3 (6.8%) | 41 (93.2%) |
| CIMP1$ (n=24) | 7 (29.2%) | 17 (70.8%) |
| CIMP2 (n=21) | 3 (14.3%) | 18 (85.7%) |
| All CIMP (n=45) | 10 (22.2%) | 35 (77.8%) |
CIMP1 vs. CIMP-negative, p=0.03
Discussion
Our data show that CRC patients with a high level of Fusobacterium in their cancer tissues have a molecularly distinct type of cancer, with a high degree of CpG island methylation and a high rate of mutations overall (though not of the TP53 gene). These data provide evidence for a pathogenic rather than passenger role for these bacteria. In favor of this argument are the facts that (1) a high level of bacteria can detected in both cancer, uninvolved adjacent mucosa and unaffected controls, (2) that the FB-high state in normal mucosa is strongly predictive of the specific molecular subtype of CRC patients and (3) that FB-high CRC have a distinct molecular profile; all these points suggest that bacteria were not simply an epiphenomenon of the cancer state. Although the data imply a contributory role of Fusobacterium, they fall short of proving causation. Clearly, not all people with high levels of Fusobacterium have colon cancer. Thus, the interaction of this normal flora bacterium with cancer is best viewed in the light of emerging data on a pathogenic link between neoplastic cells and a permissive microenvironment. Our data are consistent with previous studies linking high-relative-abundance of Fusobacterium in tumor with regional lymph node metastases6, which are also more likely to be CIMP positive cancers 26. Fusobacterium was also detected in a subset of resected CRC metastases7, suggesting that Fusobacterium may be also required for the survival or maintenance of colorectal cancer cells. In fact, all FB-high CRCs were CIMP-1 or CIMP2 and none were CIMP-negative; however, only a small fraction of the total CIMP tumors are in this high FB-group.
Prevalence of fusobacterium measurable cases did not significantly differ across the different molecular subtypes of CRCs (data not shown). This suggests that bacteria high cases rather than simply detectable cases are important for the development of CIMP-positive CRCs. FB-high status may contribute to the development of a subset of CIMP-positive CRCs, affecting different molecular pathways. For example, we found that somatic mutations in the FB-high cases were significantly more frequent compared to CIMP1 and CIMP2/CIMP-negative with low/undetectable FB, and affected pathways seemed to be different though the small number of cases analyzed makes this conclusion tentative. Whether the different molecular pathways targeted affect patient prognosis should also be evaluated.
Although F. nucleatum and other Fusobacterium species are part of the gut microbiome in human, their invasive3,27, adherent28,29, and pro-inflammatory30-32 features have been noted. Fusobacterium have been associated with inflammatory disorders such as periodontitis1, cerebral abscesses33, acute appendicitis2 and inflammatory bowel diseases3-5. It is interesting to note that the tumor subtype most associated with Fusobacterium (CIMP1 cases) have a distinct immune response with abundant tumor infiltrating lymphocytes 26, 34. This inflammatory reaction has been thought to be a host immune response to the tumor cells and is associated with a better prognosis and longer survival26, 34. Our data suggest that it could also be linked to an immune response to the high levels of bacteria in the peritumoral tissues. More broadly, inflammation may provide the pathogenic link between infections and cancer. Increased CpG island methylation is a noted feature of chronic inflammation, whether in the context of normal tissues (e.g. Ulcerative Colitis35, 36) or cancer (e.g. EBV positive gastric cancer 37). Fusobacterium has a reported association with inflammatory bowel diseases (IBD), including both ulcerative colitis (UC) and Crohn’s diseases4, 5, and IBD is one of the highest risk factors for CRC. Thus, the high rate of aberrant DNA methylation and somatic mutations in FB-high CRCs may reflect the fact that these cancers arise on a background of immune response triggered (or contributed to) by high levels of Fusobacterium.
One of the interesting implications of this work is the potential of Fusobacterium as a biomarker of cancer risk. In our studies, Fusobacterium levels in normal colonic mucosa were higher in CIMP1 compared to CIMP-negative cases, but were also prevalent in cancer free subjects (and not associated with DNA methylation there). Thus, Fusobacterium levels alone would not be useful as a biomarker of risk. Still, the hypothesis that Fusobacterium contributes to neoplasia as a co-factor through tumor-microenvironment interactions suggest that it should be tested as a risk modifier, e.g. in patients with genetic and/or environmental predisposition to cancer. Also, the mean age of cancer free subjects analyzed in this study was younger than that in CRC cases, and we could not exclude the possibility that a considerable percentage of the FB-High cancer free subjects may be at increased risk of developing CRC in the future. Whether the Fusobacterium levels in normal colonic mucosa would increase the risk of specific subtypes of CRC needs to be confirmed by prospective clinical studies. The hypothesis also deserves to be tested in animal models, where one could specifically explore the possibility of therapeutic intervention targeting Fusobacterium in the prevention or treatment of colorectal cancer.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health grants CA098006 and CA158112 to JPJI and the G.S. Hogan Gastrointestinal Research Fund at MD Anderson Cancer Center to M.R.H.E. JPI is an American Cancer Society Clinical Research professor supported by a generous gift from the F. M. Kirby Foundation.
Footnotes
Author contributions
T.T., M.R.H.E. and J.-P.J.I conceived and designed the study.
T.T. designed and performed experiments, analyzed data. Experiments were also designed by M.R.H.E., J.-P.J.I., Y.K. and M.T.
M.R.H.E. performed bioinformatics analysis.
Data were additionally analyzed and interpreted by W.C., J.G., J.J., and J.-P.J.I.
Clinical samples were collected by E.Y., H.S., R.M., Y.K., B.A., S.I., and H.Y.
Histopathological examination was performed by T.S.
Molecular analysis was performed by E.Y., H.S., R.M., Y.K., B.A., and M.T.
T.T., M.R.H.E and J.-P.J.I. wrote the manuscript.
All authors reviewed and commented on the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Signat B, Roques C, Poulet P, Duffaut D. Fusobacterium nucleatum in periodontal health and disease. Curr Issues Mol Biol. 2011;13:25–36. [PubMed] [Google Scholar]
- 2.Swidsinski A, Dörffel Y, Loening-Baucke V, et al. Acute appendicitis is characterised by local invasion with Fusobacterium nucleatum/necrophorum. Gut. 2011;60:34–40. doi: 10.1136/gut.2009.191320. [DOI] [PubMed] [Google Scholar]
- 3.Strauss J, Kaplan GG, Beck PL, et al. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis. 2011;17:1971–1978. doi: 10.1002/ibd.21606. [DOI] [PubMed] [Google Scholar]
- 4.Neut C, Bulois P, Desreumaux P, et al. Changes in the bacterial flora of the neoterminal ileum after ileocolonic resection for Crohn’s disease. Am J Gastroenterol. 2002;97:939–946. doi: 10.1111/j.1572-0241.2002.05613.x. [DOI] [PubMed] [Google Scholar]
- 5.Ohkusa T, Sato N, Ogihara T, Morita K, Ogawa M, Okayasu I. Fusobacterium varium localized in the colonic mucosa of patients with ulcerative colitis stimulates species-specific antibody. J Gastroenterol Hepatol. 2002;17:849–853. doi: 10.1046/j.1440-1746.2002.02834.x. [DOI] [PubMed] [Google Scholar]
- 6.Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet. 2007;370:890–907. doi: 10.1016/S0140-6736(07)61416-0. [DOI] [PubMed] [Google Scholar]
- 9.Chang Y, Cesarman E, Pessin MS, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 10.Fukayama M, Hino R, Uozaki H. Epstein-Barr virus and gastric carcinoma: virus-host interactions leading to carcinoma. Cancer Sci. 2008;99:1726–1733. doi: 10.1111/j.1349-7006.2008.00888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rustgi AK. The genetics of hereditary colon cancer. Genes Dev. 2007;21:2525–2538. doi: 10.1101/gad.1593107. [DOI] [PubMed] [Google Scholar]
- 12.Walther A, Johnstone E, Swanton C, Midgley R, Tomlinson I, Kerr D. Genetic prognostic and predictive markers in colorectal cancer. Nat Rev Cancer. 2009;9:489–499. doi: 10.1038/nrc2645. [DOI] [PubMed] [Google Scholar]
- 13.Toyota M, Ohe-Toyota M, Ahuja N, Issa JP. Distinct genetic profiles in colorectal tumors with or without the CpG island methylator phenotype. Proc Natl Acad Sci U S A. 2000;97:710–715. doi: 10.1073/pnas.97.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tahara T, Yamamoto E, Madireddi P, et al. Colorectal Carcinomas with CpG Island Methylator Phenotype 1 Frequently Contain Mutations in Chromatin Regulators. Gastroenterology. doi: 10.1053/j.gastro.2013.10.060. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen L, Toyota M, Kondo Y, et al. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc Natl Acad Sci U S A. 2007;104:18654–18659. doi: 10.1073/pnas.0704652104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahn JB, Chung WB, Maeda O, et al. DNA methylation predicts recurrence from resected stage III proximal colon cancer. Cancer. 2011;117:1847–1854. doi: 10.1002/cncr.25737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23:609–618. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 18.Ribic CM, Sargent DJ, Moore MJ, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jover R, Nguyen TP, Pérez-Carbonell L, et al. 5-Fluorouracil adjuvant chemotherapy does not increase survival in patients with CpG island methylator phenotype colorectal cancer. Gastroenterology. 2011;140:1174–1181. doi: 10.1053/j.gastro.2010.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki H, Igarashi S, Nojima M, et al. IGFBP7 is a p53-responsive gene specifically silenced in colorectal cancer with CpG island methylator phenotype. Carcinogenesis. 2010;31:342–349. doi: 10.1093/carcin/bgp179. [DOI] [PubMed] [Google Scholar]
- 21.Kimura T, Yamamoto E, Yamano HO, et al. A novel pit pattern identifies the precursor of colorectal cancer derived from sessile serrated adenoma. Am J Gastroenterol. 2012;107:460–469. doi: 10.1038/ajg.2011.457. [DOI] [PubMed] [Google Scholar]
- 22.Konishi K, Watanabe Y, Shen L, et al. DNA methylation profiles of primary colorectal carcinoma and matched liver metastasis. PLoS One. 2011;6:e27889. doi: 10.1371/journal.pone.0027889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.An B, Kondo Y, Okamoto Y, et al. Characteristic methylation profile in CpG island methylator phenotype-negative distal colorectal cancers. Int J Cancer. 2010;127:2095–2105. doi: 10.1002/ijc.25225. [DOI] [PubMed] [Google Scholar]
- 24.Boutaga K, van Winkelhoff AJ, Vandenbroucke-Grauls CM, Savelkoul PH. Periodontal pathogens: a quantitative comparison of anaerobic culture and real-time PCR. FEMS Immunol Med Microbiol. 2005;45:191–199. doi: 10.1016/j.femsim.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 25.Wilson GM, Flibotte S, Chopra V, Melnyk BL, Honer WG, Holt RA. DNA copy-number analysis in bipolar disorder and schizophrenia reveals aberrations in genes involved in glutamate signaling. Hum Mol Genet. 2006;15:743–749. doi: 10.1093/hmg/ddi489. [DOI] [PubMed] [Google Scholar]
- 26.Ogino S, Nosho K, Irahara N, et al. Lymphocytic reaction to colorectal cancer is associated with longer survival, independent of lymph node count, microsatellite instability, and CpG island methylator phenotype. Clin Cancer Res. 2009;15:6412–6420. doi: 10.1158/1078-0432.CCR-09-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han YW, Shi W, Huang GT, et al. Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect Immun. 2000;68:3140–3146. doi: 10.1128/iai.68.6.3140-3146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bachrach G, Ianculovici C, Naor R, Weiss EI. Fluorescence based measurements of Fusobacterium nucleatum coaggregation and of fusobacterial attachment to mammalian cells. FEMS Microbiol Lett. 2005;248:235–240. doi: 10.1016/j.femsle.2005.05.055. [DOI] [PubMed] [Google Scholar]
- 29.Uitto VJ, Baillie D, Wu Q, et al. Fusobacterium nucleatum increases collagenase 3 production and migration of epithelial cells. Infect Immun. 2005;73:1171–1179. doi: 10.1128/IAI.73.2.1171-1179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krisanaprakornkit S, Kimball JR, Weinberg A, Darveau RP, Bainbridge BW, Dale BA. Inducible expression of human beta-defensin 2 by Fusobacterium nucleatum in oral epithelial cells: multiple signaling pathways and role of commensal bacteria in innate immunity and the epithelial barrier. Infect Immun. 2000;68:2907–2915. doi: 10.1128/iai.68.5.2907-2915.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peyret-Lacombe A, Brunel G, Watts M, Charveron M, Duplan H. TLR2 sensing of F. nucleatum and S. sanguinis distinctly triggered gingival innate response. Cytokine. 2009;46:201–210. doi: 10.1016/j.cyto.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 32.Moore RA, Warren RL, Freeman JD, et al. The sensitivity of massively parallel sequencing for detecting candidate infectious agents associated with human tissue. PLoS One. 2011;6:e19838. doi: 10.1371/journal.pone.0019838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kai A, Cooke F, Antoun N, Siddharthan C, Sule O. A rare presentation of ventriculitis and brain abscess caused by Fusobacterium nucleatum. J Med Microbiol. 2008;57:668–671. doi: 10.1099/jmm.0.47710-0. [DOI] [PubMed] [Google Scholar]
- 34.Ogino S, Odze RD, Kawasaki T. Correlation of pathologic features with CpG island methylator phenotype (CIMP) by quantitative DNA methylation analysis in colorectal carcinoma. Am J Surg Pathol. 2006;30:1175–1183. doi: 10.1097/01.pas.0000213266.84725.d0. J Med Microbiol. [DOI] [PubMed] [Google Scholar]
- 35.Konishi K, Shen L, Wang S, Meltzer SJ, Harpaz N, Issa JP. Rare CpG island methylator phenotype in ulcerative colitis-associated neoplasias. Gastroenterology. 2007;132:1254–1260. doi: 10.1053/j.gastro.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 36.Issa JP, Ahuja N, Toyota M, Bronner MP, Brentnall TA. Accelerated age-related CpG island methylation in ulcerative colitis. Cancer Res. 2001;61:3573–3577. [PubMed] [Google Scholar]
- 37.Kusano M, Toyota M, Suzuki H, et al. Genetic, epigenetic, and clinicopathologic features of gastric carcinomas with the CpG island methylator phenotype and an association with Epstein-Barr virus. Cancer. 2006;106:1467–14. doi: 10.1002/cncr.21789. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.