Abstract
Human leukocyte antigen B27 (HLA-B27) is associated with protection in human immunodeficiency virus (HIV) and hepatitis C virus (HCV) infection. This protective role is linked to single immunodominant HLA-B27-restricted CD8+ T-cell epitopes in both infections. In order to define the relative contribution of a specific HLA-B27-restricted epitope to the natural course of HCV infection, we compared the biological impact of the highly conserved HCV genotype 1 epitope, for which the protective role has been described, with the corresponding region in genotype 3 that differs in its sequence by three amino acid residues. The genotype 3a peptide was not recognized by CD8+ T cells specific for the genotype 1 peptide. Furthermore, patients with acute or chronic infection with HCV genotype 3a did not mount T-cell responses to this epitope region, and their autologous viral sequences showed no evidence of T-cell pressure. Finally, we found a significantly higher frequency of HLA-B27 positivity in patients with chronic HCV genotype 3a infection compared to genotype 1 infection, indicating that there is no protection by HLA-B27 in HCV genotype 3 infection.
Conclusion
Our data indicate that the protective effect of HLA-B27 is limited to HCV genotype 1 infection and does not expand to other genotypes such as genotype 3a. This can most likely be explained by intergenotype sequence diversity leading to the loss of the immunodominant HLA-B27 epitope in viral strains other than genotype 1. Our results underline the central role of a single HLA-B27-restricted epitope-specific CD8+ T-cell response in mediating protection in HCV genotype 1 infection.
Human leukocyte antigen B27 (HLA-B27) is a prominent major histocompatability complex (MHC) class I-allele in human immune biology. It is associated with autoimmune diseases such as ankylosing spondylitis and related spondyloarthritis, but also confers protection in viral infections. Indeed, it is associated with slow disease progression in human immunodeficiency virus (HIV) infection1,2 and a high rate of spontaneous viral clearance in hepatitis C virus (HCV) infection.3 The mechanisms by which HLA-B27 mediates susceptibility to rheumatic disease and at the same time protection in infectious diseases are thought to be related, with several theories being currently discussed. This includes theories suggesting that the role of HLAB27 is independent from epitope presentation or that HLA-B27 might even be in gene linkage with another, decisive factor. It has also been suggested that the strong immunogenicity of HLA-B27 is linked to its unusual cell biology, including its tendency to misfold or to build noncanonical forms such as heavy chain homodimers, or the failure of B27 ligands to engage KIR3DL1, leading to an increased natural killer cell activation.4,5
A putative arthritogenic peptide has not been identified so far, further complicating the analysis of mechanisms contributing to the association between HLA-B27 and spondyloarthritis. In contrast, the protective role of HLA-B27 in HIV and HCV infection has been linked to single, immunodominant CD8+ T-cell epitopes.1,6 In both infections, escape from the CD8+ T-cell response targeting this epitope is difficult to achieve. In HIV, one mutation is typically selected during the early phase of infection. However, this single mutation is not sufficient for immune escape, as the variant is still targeted. Full immune escape is only achieved when a second mutation develops at the main HLA-B27 binding anchor, arginine at position 2 of the epitope. Of note, this second mutation in the epitope can only occur together with a compensatory mutation outside the epitope.7-12 This complicated pathway may explain why escape from this immunodominant HIV epitope occurs only late in infection. In HCV we have previously shown that fitness constraints limit the ability to mutate at the main HLA-B27 binding anchor of the immunodominant HLA-B27 epitope that is located within a conserved region of the RNA-dependent RNA polymerase (NS5B2841-2849). Instead, a mosaic of several mutations at the T-cell receptor contact residues within the epitope needs to evolve in order to allow significant escape from the HLA-B27-restricted CD8+ T-cell response.6,13 Similar to HIV, these results suggest that viral escape cannot be achieved easily, giving the T-cell response sufficient time to clear the virus. These virological factors presumably contribute to the protective effect of HLA-B27.
Although there is strong immunological and virological evidence that the protective effect of HLA-B27 in HIV and HCV infection, respectively, is indeed linked to these particular immunodominant epitopes, this has not been conclusively demonstrated, as this would require a prospective analysis of a large number of B27-positive subjects with acute infection. In order to determine the contribution of the viral epitope on protection by HLAB27, we took advantage of the fact that the immunodominant viral region targeted by HLA-B27-restricted CD8+ T cells is conserved in HCV genotype 1 only, in which the protective effect of HLA-B27 has been described.6 In other HCV genotypes, e.g., genotype 3a (the most frequent genotype after genotype 1 in most countries) the epitope sequence differs by three out of nine amino acid residues from genotype 1. We therefore hypothesized that lack of recognition of the epitope in patients infected with HCV genotypes other than 1 might lead to a loss of protection by HLA-B27. In order to test this hypothesis, in this study we analyzed the CD8+ T-cell response and autologous viral sequences in a new cohort of HLA-B27+ patients acutely or chronically infected with HCV geno-type 3a and determined the frequency of HLA-B27 in a large cohort of patients chronically infected with HCV genotype 1 or 3a. Our results suggest that HLA-B27 is indeed protective in patients with HCV genotype 1 infection but not in patients infected with HCV genotype 3a. This lack of protection is most likely caused by intergenotypic sequence differences leading to the loss of the immunodominant HLA-B27 epitope in infection with HCV genotypes other than 1. Our results further support the important role of a single immunodominant NS5B epitope in mediating the protective and genotype-specific effect of HLA-B27 in HCV infection.
Patients and Methods
Study Subjects
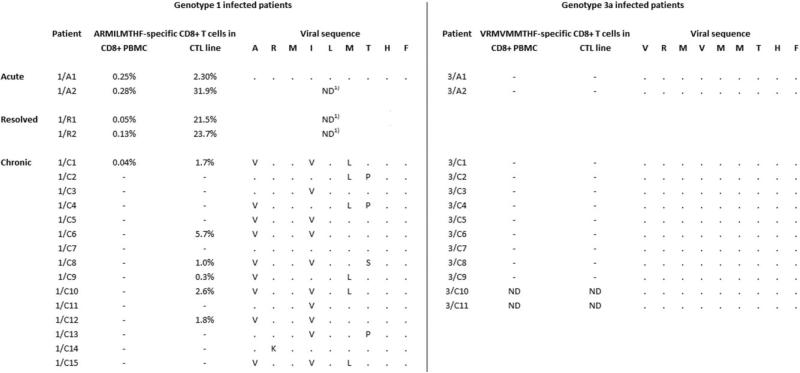
Eleven HLA-B27+ individuals with chronic HCV genotype 3a infection who presented to the University Hospital of Freiburg were included (patient codes 3/C1 to 3/C11, Fig. 3). In addition, two patients with acute-persistent HCV genotype 3a infection who presented to the University Hospital of Freiburg (3/A1) or the Massachusetts General Hospital (3/A2), respectively, were included and followed over time. In addition, the following patients infected with HCV genotype 1 were studied: One patient with acute-resolving HCV genotype 1a infection who presented to the University Hospital Freiburg (1/A1),6 one patient who developed acute-resolving HCV genotype 1a infection after receiving a contaminated patellar ligament graft (1/A2),14 two patients who had resolved HCV genotype 1b infection from a contaminated anti-D immunoglobulin preparation in 1977 (1/R1 and 1/R2),3 one patient from the same cohort who developed chronic genotype 1b infection (1/C1), and 14 further patients with chronic HCV genotype 1 infection who presented to the University Hospital Freiburg (1/C2 to 1/C15). Data from most of the geno-type 1-infected patients have been published previously6,13 and are shown for comparison only. As negative controls, nine HLA-B27+ individuals that are HCV antibody-negative and have no history of HCV infection were included. For HLA allele frequency, a cohort of 265 patients with chronic HCV genotype 1 infection and 98 patients with chronic HCV genotype 3a infection, respectively, who presented to the University Hospital of Freiburg was analyzed. After written informed consent and in agreement with the 1975 Declaration of Helsinki, federal guidelines, and the local ethics committee, blood was obtained from the patients. EDTA anticoagulated blood was used for the isolation of peripheral blood mononuclear cells (PBMCs) by using lymphocyte separation medium-density gradients (PAA Laboratories, Pasching, Austria).
Fig. 3.
HLA-B27+ individuals with acute or chronic HCV genotype 3a infection do not target the HLA-B27 NS5B2841-2849 epitope region and show no sequence variation within the epitope region. CD8+ T cells from HLA-B27+ patients with acute (3/A1 and 3/A2) or chronic (3/C1-C9) HCV genotype 3a infection were tested for INF-γ production after 5 hours of stimulation with the genotype-specific HLA-B27 NS5B2841-2849 peptide. In addition, PBMC were stimulated for 2 weeks with the genotype-specific peptide, and the resulting CTL lines were tested for INF-γ production after 5 hours of peptide restimulation. Autologous viral sequences corresponding to the epitope region were determined and are aligned to genotypespecific consensus. For comparison, data from genotype 1-infected patients that have in part been published6,13 are also shown. Percent values are percent INF-γ+ per CD8+ T cells. “−” indicates below the cutoff of 0.02%. “·” indicates homologous to consensus. ND: not done. 1Infecting virus is known to harbor the genotype 1 consensus sequence within the epitope region.
Peptides and Antibodies
Peptides were synthesized with a free amino and carboxy terminus by standard Fmoc chemistry by Genaxxon Bioscience (Biberach, Germany). The peptides were dissolved and diluted as described.15 Anti-CD8 PE and anti-interferon-γ (IFN-γ) fluorescein isothiocyanate (FITC) antibodies as well as isotype phycoerythrin (PE) and FITC (all BD PharMingen, San Jose, CA) were used according to the manufacturer's instructions.
Generation of Peptide-Specific T-Cell Lines
Four × 106 PBMC were resuspended in 1 mL complete medium (RPMI 1640 containing 10% fetal calf serum, 1% streptomycin/penicillin, and 1.5% HEPES buffer 1 mol/L) and stimulated with peptide at a final concentration of 10 μg/mL and anti-CD28 (BD PharMingen) at a final concentration of 0.5 μg/mL. On days 3 and 10, 1 mL complete medium (see above) and recombinant interleukin 2 (rIL-2; Hoffmann-La Roche, Basel Switzerland) at a final concentration of 20 U/mL was added to each well. On day 7 the cultures were restimulated with the corresponding peptide (10 μg/mL) and 106 irradiated autologous feeder cells (for some of the samples, no feeder cells were used due to limitation of available cell number). On day 14 the cells were tested for peptide-specific IFN-γ production.
Intracellular IFN-γ Staining
Procedures were performed essentially as described.15 Briefly, CD8+ PBMC or peptide-specific T-cell lines (0.2 × 106 per well, 96-well plate) were stimulated with peptides in the concentrations indicated in the figure panels in the presence of 50 U/mL human rIL-2 and 1 μL/mL brefeldin A (BD PharMingen). After 5 hours incubation (37°C, 5% CO2), cells from each well were blocked with immunoglobulin G1 (IgG1) antibodies and stained with antibodies against CD8. After permeabilization with Cytofix/cytoperm (BD PharMingen), cells were stained with antibodies against IFN-γ and fixed in 100 μL CellFIX (BD PharMingen) per well before FACS analysis. When indicated in the figure legends, cells were not directly restimulated with peptide, but with autologous or partially HLA-matched Epstein-Barr virus (EBV) immortalized B-cell lines that had been loaded with peptide overnight and extensively washed (6×) prior to the 5-hour coculture with the target cells.16
Additional experimental procedures can be found in the Supporting Materials.
Results
Immunodominant HLA-B27-Restricted Epitope NS5B2841-2849 Is Highly Conserved Within but Not Between Different HCV Genotypes
First, we performed a database analysis and compared the consensus sequence of the NS5B2841-2849 epitope region between the different HCV genotypes. Although the consensus sequences from subtypes 1a and 1b were identical to the described HLA-B27-restricted CD8+ T-cell epitope, the consensus sequences of genotypes other than 1 differed by one, two, or three amino acid residues within the nine amino acid-long epitope region (Fig. 1A). In Fig. 1B, available sequence data are shown for genotypes 1a (178 sequences), 1b (242 sequences), and 3a (163 sequences), which are the most frequent subtypes world-wide and for which sufficient sequence data are currently available. Although the epitope region was conserved within a given genotype, genotype 3a sequences differed by three amino acids from the genotype 1 consensus sequence. Interestingly, the consensus sequence of genotype 3a reflects the possible escape variants observed in genotype 1. The HLA-binding positions are identical in both genotypes; however, different combinations of the A284lV, I2844V, and L2845M substitutions are frequently observed in sequences from HLA-B27-positive genotype 1-infected subjects.6,13,17
Fig. 1.
The HLA-B27 NS5B2841-2849 epitope region is highly conserved within but not between different HCV genotypes. (A) Genotype-specific consensus sequences corresponding to the HLA-B27 NS5B2841-2849 epitope are displayed. The numbers present sequences identical to consensus/total sequences in the Los Alamos HCV Sequence Database. (B) Genotype 1a, 1b, and 3a sequences available at the Los Alamos HCV Sequence Database have been aligned to the respective genotypespecific consensus sequence. Amino acids differing between genotype 1a/1b and genotype 3a are underlined.
Genotype 3a Epitope Peptide Is Not Cross-Recognized by CD8+ T-Cell Lines Generated by Stimulation with the Genotype 1 Epitope Peptide
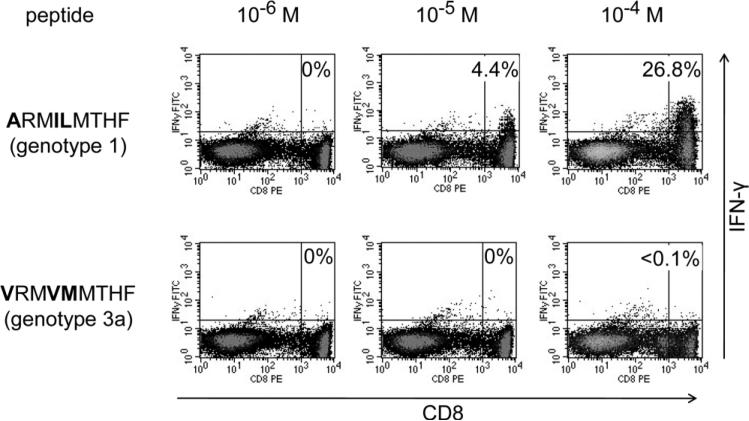
The above results raised the possibility that sequences from genotype 3a (and probably also the remaining HCV genotypes other than 1) may not be recognized by CD8+ T cells specific for the genotype 1 epitope region. To address this issue, we generated cytotoxic T lymphocyte (CTL) lines from nine HLA-B27+ patients infected with HCV geno-type 1 (two patients with acute-resolving infection, two patients with spontaneously resolved infection, and five patients with chronic infection) through two rounds of peptide stimulation with the genotype 1 consensus NS5B2841-2849 peptide (ARMILMTHF). Indeed, these CTL lines produced significant amounts of IFN-γ in response to the genotype 1 consensus peptide only, but no or only little IFN-γ in response to the genotype 3a consensus peptide (V RM VM MTHF) (see Fig. 2 for a representative staining and Supporting Fig. 1 for all nine cell lines). Although genotype 3a is the genotype found next most often in patients after genotype 1, other geno-types occur rarely in patients in Europe and North America. However, we took advantage of the fact that the genotype 2a strain JFH-1 that is used in the HCV replicon model also differs from genotype 1 consensus by two amino acids (V RM V LMTHF). We constructed a hepatoma cell line (Huh7-Lunet) expressing HLA-B27 and transfected it with a subgenomic JFH-1 replicon. Importantly, coculture of these replicon cells with a T-cell line specific for the genotype 1 epitope peptide (ARMILMTHF) did not induce significant IFN-γ production unless the replicon sequence was mutated to the genotype 1 consensus sequence (Supporting Fig. 2). In sum, these findings suggest that a CD8+ T-cell response targeting the immunodominant NS5B2841-2849 epitope can only recognize the HCV genotype 1 but not the genotype 2a or 3a sequence.
Fig. 2.
CD8+ T cells specific for the genotype 1 HLA-B27 NS5B2841-2849 epitope do not cross-recognize the corresponding genotype 3a peptide. PBMC from an HLA-B27+ patient chronically infected with HCV genotype 1 were stimulated for 2 weeks with the genotype 1 HLA-B27 NS5B2841-2849 epitope peptide (ARMILMTHF). The resulting CTL line was restimulated for 5 hours prior to intracellular INF-γ staining either with the genotype 1 peptide at different concentrations as indicated (upper panel) or with the corresponding genotype 3a peptide (V RM VM MTHF) (lower panel). Percent values indicate the subset of IFN-γ+ CD8+ T cells after subtracting the background in the absence of peptide.
Patients Infected with HCV Genotype 3a Do Not Mount T-Cell Responses Against the Genotype 3a Epitope Peptide
Because the HLA-B27 binding positions are identical in the genotype 1 and the genotype 3a variant, it is possible that the epitope is still presented and a genotype 3-speficic response is primed in genotype 3 infected patients. To address this, we next analyzed whether genotype 3a-infected patients mount a response against the genotype 3a epitope peptide. For this reason, we tested nine HLA-B27+ patients with chronic HCV genotype 3a infection and two HLA-B27+ patients with acute HCV genotype 3a infection for CD8+ T-cell responses against the genotype 3a consensus peptide V RM VM MTHF. There was no significant CD8+ T-cell response to this peptide in any of the subjects, either in CD8+ PBMC, or in peptide-stimulated CTL lines (Fig. 3, right). This is in clear contrast to our previous findings in genotype 1-infected patients, where two out of two acutely infected patients and six out of 15 chronically infected patients showed a response (Fig. 3, left; Supporting Fig. 1). It is important to point out that the procedure used here to expand peptide-specific CD8+ T cells is not capable of priming a peptide-specific responses in HCV-naïve individuals. After stimulation of PBMC from nine HLA-B27+ HCV-naïve individuals in the presence of the genotype 1 or genotype 3a peptide, respectively, no peptide-specific T cells became detectable (data not shown). In addition, we also tried to expand genotype 3a-specific CD8+ T cells from genotype 1-infected patients. The majority of patients (six out of nine) with a response after expansion with the genotype 1 peptide did not display any response after expansion with the geno-type 3a peptide (Supporting Fig. 1). From three patients (1/A2, 1/R1, and 1/C8) specific T cells could be expanded with the genotype 3a-derived peptide; however, these responses were substantially weaker compared to responses observed after expansion with the genotype 1 peptide. Moreover, T cells expanded in the presence of the genotype 3a peptide still preferentially targeted the genotype 1 peptide (Supporting Fig. 1), indicating that these responses were indeed genotype 1-specific, with some degree of cross-reactivity with the genotype 3a peptide. Taken together, these data indicate that in contrast to the genotype 1 epitope region, the corresponding genotype 3a sequence does not prime virus-specific CD8+ T cells in vivo.
Genotype 1-Specific CD8+T-Cell Response in a Genotype 3a-Infected Patient Suggests Previous Exposure to HCV Genotype 1
Next, we tested PBMC from patients infected with HCV genotype 3a for CD8+ T-cell responses against the genotype 1 epitope peptide. As expected, most patients also showed no response against the genotype 1 peptide (data not shown). Of note, however, one patient (patient 3/C3) showed a significant CD8+ T-cell response against the genotype 1 peptide in both CD8+ PBMC and a CTL line stimulated with the genotype 1 peptide (Fig. 4A, upper). However, this cell line did not produce IFN-γ after restimulation with the genotype 3a peptide (Fig. 4A, lower). These results suggested that the CD8+ T-cell response detected in this patient did not target the current HCV genotype 3a infection, but rather may represent an immunological “scar” from a previously resolved HCV genotype 1 infection, as has been previously reported.18,19 To further analyze this hypothesis, we screened this patient (3/C3) for additional responses to other HCV genotype 1-specific CD8+ T-cell responses. Of note, the patient targeted three additional genotype 1-specific CD8+ T-cell epitopes (two restricted by HLA-A3 and one restricted by HLA-B35). Importantly, similar to the HLA-B27-restricted epitope, the two HLA-A3-restricted CD8+ T-cell responses showed no cross-recognition with the corresponding genotype 3a peptides (Fig. 4B). The T-cell line generated by stimulation with the genotype 1 derived HLA-B35 epitope peptide displayed cross-recognition with the corresponding genotype 3a peptide (Fig. 4B); however, titration experiments performed in the presence of peptide-loaded antigen-presenting cells revealed preferential targeting of the genotype 1a peptide (Fig. 4C). This is in line with the much stronger predicted HLAB35 binding of the genotype 1a peptide (genotype 1a peptide: median inhibitory concentration [IC50] 55 nM; genotype 3a peptide: IC50 773 nM; www.iedb.org).20 In sum, these data support the hypothesis that the CD8+ T-cell responses detected in this patient might be remainders of a previous genotype 1 infection.
Fig. 4.
Genotype 1 epitope-specific CD8+ T cell response suggest a previous genotype 1 infection. (A) PBMC of patient 3/C3 were stimulated for 2 weeks with the genotype 1 HLA-B27 NS5B2841-2849 epitope peptide and then tested for INF-γ production after 5 hours of restimulation with the genotype 1 peptide (ARMILMTHF) (upper panel) or the corresponding genotype 3a peptide (V RM VM MTHF) (lower panel) in the indicated concentration. (B) PBMCs of the same patient were stimulated for 2 weeks with three additional genotype 1a-specific epitope peptides and then tested for IFN-γ production after 5 hours of restimulation with the genotype 1a-specific peptide as well as the corresponding genotype 3a specific peptide at a concentration of 10−5 M. (C) For the HLA-B35 restricted epitope, restimulation was also performed with autologous B-cell lines that had been loaded with peptide overnight in the indicated concentrations and washed extensively prior to the 5-hour coculture with the CTL line.
Patients Infected with HCV Genotype 3a Show No Evidence of Sequence Variation Within the Epitope Region
The few HLA-B27+ patients infected with HCV genotype 1 who progress to chronic infection develop clustered escape mutations within the immuno-dominant HLA-B27 epitope (Fig. 3, left).6,13,17 Because CD8+ T cells in patients with acute or chronic HCV genotype 3a infection did not target this region, we hypothesized that in contrast to genotype 1, in genotype 3a infection no HLA-B27-driven sequence polymorphisms should evolve. To address this point, we analyzed the autologous sequences in sera from 11 patients with chronic HCV genotype 3a infection. Of note, no patient showed any sequence variation from the consensus sequence within this region (Fig. 3, right). In addition, sequential sequence analyses in two patients with acute-persistent HCV genotype 3a infection showed no evidence of viral escape mutations over a time of one or two years, respectively (data not shown). In line with the results from the cellular assays these data strongly suggest that there is no CD8+ T-cell pressure within the NS5B2841-2849 region in HCV genotype 3a-infected HLA B27+ patients.
Frequency of HLA-B27 Is Significantly Higher in Patients Chronically Infected with Genotype 3a Compared to Patients Infected with Genotype 1
Based on our immunological findings, it is tempting to speculate that in contrast to HCV genotype 1 infection, lack of the immunodominant target of the HLA-B27-restricted CD8+ T-cell responses is associated with loss of the protective effect of HLA-B27 in genotype 3a infection. To address this question, we determined the frequency of HLA-B27 positivity in a large cohort of patients chronically infected with either HCV genotype 1 (265 patients) or 3a (98 patients). The frequency of HLA-B27 positivity was significantly higher in patients infected with genotype 3a (12/98 patients, 12.5%) compared with patients infected with genotype 1 (14/265 patients, 5.3%; P = 0.0363) (Fig. 5). In this context, it is important to note that the frequency of HLA-B27 positivity in the general German population is ≈8.5% based on the analysis of 11,407 individuals in the German bone marrow registry (compare www.allelefrequencies.net).21 Thus, the protective effect of HLA-B27 in HCV genotype 1 infection is reflected by the low prevalence of HLA-B27 positivity in patients chronically infected with HCV genotype 1. In contrast, HLA-B27 has no protective effect (or may even have a disadvantageous effect) in HCV genotype 3a infection, reflected by the relatively high frequency of HLA-B27 positivity in patients infected with HCV genotype 3a.
Fig. 5.
The frequency of HLA-B27 is significantly higher in patients chronically infected with genotype 3a compared to patients infected with genotype 1. The frequency of HLA-B27 was determined in a cohort of 265 patients chronically infected with HCV genotype 1 and 98 patients chronically infected with HCV genotype 3a. The frequency of HLA-B27 in the general population includes 11,407 individuals from the German bone marrow registry. P-value was calculated by two-tailed Fisher's exact test.
Importantly, we did not observe a significant difference between the frequencies in genotype 1 or 3a-infected patients for any other HLA-A or HLA-B allele (data not shown). Thus, the different frequency of HLA-B27 in HCV genotype 1 versus 3a infection is unique and consistent with the central role of the immunodominant NS5B2841-2849 epitope in mediating the protective effect of HLA-B27 in HCV genotype 1 infection.
Discussion
We have recently linked the protective effect of HLAB27 in the outcome of HCV infection to an immuno-dominant HLA-B27-restricted HCV epitope.6 This epitope is located in a highly conserved and functionally constrained region within NS5B, the RNA-dependent RNA polymerase. In our previous study we showed that viral escape from this epitope is limited by viral fitness costs as well as cross-recognition by CD8+ T cells, thus requiring a complex mosaic of two or more mutations for significant viral escape.13 During the acute phase of infection, the strong HLA-B27-restricted CD8+ T-cell response is most likely able to prevent the evolution of this complex mosaic of viral mutations, best explaining the protective effect of HLA-B27. The protective effect of HLA-B27 and the strong immune pressure mediated by the immunodominant HLA-B27 epitope have been identified in HCV genotype 1 infection only. Although HCV genotype 1 is the most prevalent genotype in North America (subtype 1a > 1b) and Europe (subtype 1b > 1a), other genotypes may become more relevant in the near future, because they predominate in many developing countries with a high incidence of HCV infection as well as in defined cohorts such as injection drug users, whereHCVgenotype3afrequencyisincreasing.22Intriguingly, the immunodominant HLA-B27 NS5B2841-2849 epitope region is highly conserved within but not between different HCV genotypes. Based on our previous findings in patients infected with HCV genotype 1,6 we therefore hypothesized that this epitope region may not be targeted by HCV-specific CD8+ T cells in patients infected with HCV genotypes other than genotype 1, abrogating the protective effect of HLA-B27 in these patients. This was addressed in the current study by analyzing a new cohort of patients with acute or chronic HCV genotype 3a infection.
Indeed, we could demonstrate that CD8+ T cells specific for the HCV genotype 1 NS5B2841-2849 epitope (ARMILMTHF) do not recognize the HCV genotype 3a peptide (V RM VM MTHF). In addition, patients with genotype 3a infection do not target the region corresponding to the B27-epitope. Accordingly, there is no evidence of mutational escape in genotype 3 isolates supporting lack of HLA-B27-associated T-cell pressure on this region in genotype 3a-infected patients. Collectively, these data suggest that HCV genotype 3a lacks the HLAB27 epitope, which is immunodominant in HCV geno-type 1 infection and most likely significantly contributes to the protective effect of HLA-B27. It is not clear why patients infected with genotype 3a are unable to target the genotype 3a epitope region, especially because the main HLA-B27 binding anchors (arginine at position 2 and phenylalanine at the C-terminus) are conserved between the genotypes. The variant sequence might have an impact on binding to HLA-B27 despite intact primary anchor residues. Alternatively, it has been proposed previously that the failure of the immune system to target certain epitope variants that are efficiently presented by the restricting HLA allele is caused by a hole in the T-cell repertoire.23 In this context, it is intriguing to note that both the variant described in the previous study as well as the genotype 3a epitope variant described here contain a leucine to methionine (L → M) substitution at amino acid residue four.
We also compared the frequency of HLA-B27 positivity in patients chronically infected with genotypes 1 and 3a, respectively. Interestingly, the frequency of HLA-B27 positivity was significantly higher in patients infected with genotype 3a than in patients infected with genotype 1, indicating that HLA-B27+ patients had a lower risk of developing chronic HCV infection when infected with HCV genotype 1 compared to genotype 3a. These data demonstrate that the protective role of HLA-B27 is indeed limited to HCV genotype 1 infection, and does not extend to HCV genotype 3a, which does not share the protective NS5B2841-2849 epitope. This finding may also explain why in some cohorts infected with divers HCV genotypes24 or infected exclusively with other genotypes25 a protective role of HLA-B27 has not been shown. At the same time, the protective effect of HLA-B27 has been reproduced in the largest study performed on this issue so far, including primarily patients infected with HCV genotype 1.26 In this study the prevalence of certain HLA-class I alleles in 5,901 North American patients with chronic HCV infection undergoing liver transplantation and in 11,728 individuals undergoing liver transplantation for other liver diseases was compared. HLA-B27 and HLA-B39 positivity, respectively, was associated with the greatest level of protection from chronic HCV infection within the different HLA class I alleles in that study. Thus, it is important to point out that the differences in the CD8+ T-cell responses to different genotypes of HBV27 and HCV19 or to different clades of HIV28 indeed might translate clinically into different outcomes of infection, also underlining the notion that HLA-driven footprints might have a significant contribution to intergenotype/interclade variability.28
In conclusion, we show that intergenotype sequence diversity is associated with the absence of an immuno-dominant and protective HLA-B27 epitope in HCV genotypes other than 1. At the same time, this is a possible explanation why HLA-B27 is protective in HCV geno-type 1 infection only, but not in infection with other HCV genotypes. Our findings support the hypothesis that the protective effect of HLA-B27 is indeed mediated by HLA-B27-restricted CD8+ T cells and not by other indirect effects such as gene linkage. In addition, our findings highlight the importance to consider biological differences between HCV genotypes in molecular, immunological, as well as clinical terms. Clearly, a precise definition of immunodominant and protective HCV epitopes in different HCV genotypes is an important prerequisite for the development of strategies to prevent or treat HCV infection by vaccination.
Supplementary Material
Acknowledgment
We thank all the study subjects. We thank Natalie Wischniowski for excellent technical assistance. HLA-B27 tetramers were kindly supplied by the National Institutes of Health (NIH) tetramer core facility at Emory University, Atlanta, GA. Recombinant human IL-2 was kindly supplied by the NIH AIDS Research and Reference Reagent Program, Germantown, MD.
Supported by the Bundesministerium für Bildung und Forschung (BMBF 01KI0793 and 01EO0803) and the Deutsche Forschungsgemeinschaft (Heisenberg Professorship TH 719/3-1, FOR 1202 “Mechanisms of persistence of hepatotropic viruses”, and TI323/3-1).
Abbreviations
- CTL
cytotoxic T lymphocyte
- HCV
hepatitis C virus
- HIV
human immunodeficiency virus
- HLA-B27
human leukocyte antigen B27
- INF-γ
interferon-γ
- NS5B
nonstructural protein 5B
- PBMC
peripheral blood mononuclear cell
Footnotes
Potential conflict of interest: Nothing to report.
Additional Supporting Information may be found in the online version of this article.
The authors have no conflicting financial interests.
References
- 1.Goulder PJ, Watkins DI. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat Rev Immunol. 2008;8:619–630. doi: 10.1038/nri2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaslow RA, Carrington M, Apple R, Park L, Munoz A, Saah AJ, et al. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med. 1996;2:405–411. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- 3.McKiernan SM, Hagan R, Curry M, McDonald GS, Kelly A, Nolan N, et al. Distinct MHC class I and II alleles are associated with hepatitis C viral clearance, originating from a single source. Hepatology. 2004;40:108–114. doi: 10.1002/hep.20261. [DOI] [PubMed] [Google Scholar]
- 4.Marcilla M, Lopez de Castro JA. Peptides: the cornerstone of HLA-B27 biology and pathogenetic role in spondyloarthritis. Tissue Antigens. 2008;71:495–506. doi: 10.1111/j.1399-0039.2008.01051.x. [DOI] [PubMed] [Google Scholar]
- 5.Stewart-Jones GB, di Gleria K, Kollnberger S, McMichael AJ, Jones EY, Bowness P. Crystal structures and KIR3DL1 recognition of three immunodominant viral peptides complexed to HLA-B*2705. Eur J Immunol. 2005;35:341–351. doi: 10.1002/eji.200425724. [DOI] [PubMed] [Google Scholar]
- 6.Neumann-Haefelin C, McKiernan S, Ward S, Viazov S, Spangenberg HC, Killinger T, et al. Dominant influence of an HLA-B27 restricted CD8+ T cell response in mediating HCV clearance and evolution. Hepatology. 2006;43:563–572. doi: 10.1002/hep.21049. [DOI] [PubMed] [Google Scholar]
- 7.Goulder PJ, Brander C, Tang Y, Tremblay C, Colbert RA, Addo MM, et al. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature. 2001;412:334–338. doi: 10.1038/35085576. [DOI] [PubMed] [Google Scholar]
- 8.Goulder PJ, Phillips RE, Colbert RA, McAdam S, Ogg G, Nowak MA, et al. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat Med. 1997;3:212–217. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- 9.Kelleher AD, Long C, Holmes EC, Allen RL, Wilson J, Conlon C, et al. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J Exp Med. 2001;193:375–386. doi: 10.1084/jem.193.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lichterfeld M, Kavanagh DG, Williams KL, Moza B, Mui SK, Miura T, et al. A viral CTL escape mutation leading to immunoglobulin-like transcript 4-mediated functional inhibition of myelomonocytic cells. J Exp Med. 2007;204:2813–2824. doi: 10.1084/jem.20061865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneidewind A, Brockman MA, Sidney J, Wang YE, Chen H, Suscovich TJ, et al. Structural and functional constraints limit options for CTL escape in the Immunodominant HLA-B27 restricted epitope in HIV-1 capsid. J Virol. 2008;82:5594–5605. doi: 10.1128/JVI.02356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneidewind A, Brockman MA, Yang R, Adam RI, Li B, Le Gall S, et al. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J Virol. 2007;81:12382–12393. doi: 10.1128/JVI.01543-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dazert E, Neumann-Haefelin C, Bressanelli S, Fitzmaurice K, Kort J, Timm J, et al. Loss of viral fitness and cross-recognition by CD8+ T cells limit HCV escape from a protective HLA-B27-restricted human immune response. J Clin Invest. 2009;119:376–386. doi: 10.1172/JCI36587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tester I, Smyk-Pearson S, Wang P, Wertheimer A, Yao E, Lewinsohn DM, et al. Immune evasion versus recovery after acute hepatitis C virus infection from a shared source. J Exp Med. 2005;201:1725–1731. doi: 10.1084/jem.20042284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thimme R, Oldach D, Chang KM, Steiger C, Ray SC, Chisari FV. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J Exp Med. 2001;194:1395–1406. doi: 10.1084/jem.194.10.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neumann-Haefelin C, Timm J, Spangenberg HC, Wischniowski N, Nazarova N, Kersting N, et al. Virological and immunological determinants of intrahepatic virus-specific CD8+ T-cell failure in chronic hepatitis C virus infection. Hepatology. 2008;47:1824–1836. doi: 10.1002/hep.22242. [DOI] [PubMed] [Google Scholar]
- 17.Timm J, Li B, Daniels MG, Bhattacharya T, Reyor LL, Allgaier R, et al. Human leukocyte antigen-associated sequence polymorphisms in hepatitis C virus reveal reproducible immune responses and constraints on viral evolution. Hepatology. 2007;46:339–349. doi: 10.1002/hep.21702. [DOI] [PubMed] [Google Scholar]
- 18.Schulze Zur Wiesch J, Lauer GM, Timm J, Kuntzen T, Neukamm M, Berical A, et al. Immunologic evidence for lack of heterologous protection following resolution of HCV in patients with non-genotype 1 infection. Blood. 2007;110:1559–1569. doi: 10.1182/blood-2007-01-069583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giugliano S, Oezkan F, Bedrejowski M, Kudla M, Reiser M, Viazov S, et al. Degree of cross-genotype reactivity of hepatitis C virus-specific CD8+ T cells directed against NS3. Hepatology. 2009;50:707–716. doi: 10.1002/hep.23096. [DOI] [PubMed] [Google Scholar]
- 20.Lundegaard C, Lamberth K, Harndahl M, Buus S, Lund O, Nielsen M. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8-11. Nucleic Acids Res. 2008;36:W509–512. doi: 10.1093/nar/gkn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Middleton D, Menchaca L, Rood H, Komerofsky R. New allele frequency database. Tissue Antigens. 2003;61:403–407. doi: 10.1034/j.1399-0039.2003.00062.x. http://www.allelefrequencies.net. [DOI] [PubMed] [Google Scholar]
- 22.Esteban JI, Sauleda S, Quer J. The changing epidemiology of hepatitis C virus infection in Europe. J Hepatol. 2008;48:148–162. doi: 10.1016/j.jhep.2007.07.033. [DOI] [PubMed] [Google Scholar]
- 23.Wolfl M, Rutebemberwa A, Mosbruger T, Mao Q, Li HM, Netski D, et al. Hepatitis C virus immune escape via exploitation of a hole in the T cell repertoire. J Immunol. 2008;181:6435–6446. doi: 10.4049/jimmunol.181.9.6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thio CL, Gao X, Goedert JJ, Vlahov D, Nelson KE, Hilgartner MW, et al. HLA-Cw*04 and hepatitis C virus persistence. J Virol. 2002;76:4792–4797. doi: 10.1128/JVI.76.10.4792-4797.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chuang WC, Sarkodie F, Brown CJ, Owusu-Ofori S, Brown J, Li C, et al. Protective effect of HLA-B57 on HCV genotype 2 infection in a West African population. J Med Virol. 2007;79:724–733. doi: 10.1002/jmv.20848. [DOI] [PubMed] [Google Scholar]
- 26.Hraber P, Kuiken C, Yusim K. Evidence for human leukocyte antigen heterozygote advantage against hepatitis C virus infection. Hepatology. 2007;46:1713–1721. doi: 10.1002/hep.21889. [DOI] [PubMed] [Google Scholar]
- 27.Tan AT, Loggi E, Boni C, Chia A, Gehring AJ, Sastry KS, et al. Host ethnicity and virus genotype shape the hepatitis B virus-specific T-cell repertoire. J Virol. 2008;82:10986–10997. doi: 10.1128/JVI.01124-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matthews PC, Leslie AJ, Katzourakis A, Crawford H, Payne R, Prendergast A, et al. HLA footprints on human immunodeficiency virus type 1 are associated with interclade polymorphisms and intraclade phylogenetic clustering. J Virol. 2009;83:4605–4615. doi: 10.1128/JVI.02017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.