Abstract
Kank1, which was first described as a potential tumor suppressor for renal cell carcinoma (RCC), mapped to 9p24.3 and encoded an ankyrin-repeat domain-containing protein. Its frequent deletion was found to be associated with several human malignant tumors, cerebral palsy, and neuronal and developmental diseases. However, its functional role in nasopharyngeal cancer (NPC) was still unknown. In the present study, we found that Kank1 expression was down-regulated in NPC cells than in human nasopharyngeal epithelial cell line NP69 and demethylating agent 5-aza-2’-deoxycytidine (5-aza-CdR) could improve its mRNA and protein expression level. Further studies demonstrated that DNA methylation might be the mainly cause for Kank1 decreased expression and restored Kank1 expression mediated by 5-aza-CdR played a key role in suppressing NPC cells growth and inducing its apoptosis. Our primary results revealed new function of Kank1 for NPC and implied that epigenetic regulation especially demethylation may have a potential value for NPC treatment.
Keywords: Kank1, nasopharyngeal carcinoma, 5-aza-CdR, methylation, proliferation, apoptosis
Introduction
Nasopharyngeal cancer (NPC) is a common malignant tumor in southern China and Southeast Asia [1]. According to the statistics, there were 41503 new diagnosed cases and 20058 deaths of NPC patients in China in 2010 [2]. In addition to environmental factors (e.g. Early-age Epstein-Barr virus (EBV) infection) [3,4], genetic/epigenetic modification including proto-oncogenes activation, tumor suppressor genes inactivation, gene deletion, point mutation, as well as DNA methylation all contribute to the initiation and progression of NPC [5,6].
The human Kank1 gene (ANKRD15), located at 9p24.3, was first described as a potential tumor suppressor for renal cell carcinoma (RCC) [7]. It belongs to Kank family which consists of other three members, Kank2, Kank3 and Kank4 [8]. The Kank family proteins share a similar structure containing a coiled-coil domain in the N-terminal region, an ankyrin-repeats domain in the C-terminal region and a KN motif at the N-terminus [8,9]. It has been proven that there were at least two types of Kank1 protein due to alternative splicing of Kank gene at the first exon [10]. Kank1 has multiple biological functions in physiological and pathological processes. It plays an important role in cytoskeleton formation and cell motility by regulating actin polymerization [11,12]. Meanwhile, the deletions of Kank1 have been reported to be involved in many malignant tumors, such as RCC [13,14], bladder cancer [15], hepatocellular carcinoma [16], pancreatic carcinoma [17], lung cancer [18], cervical carcinoma [19], breast cancer and acute lymphocytic leukemia [20,21]. However, the roles of Kank1 in NPC have not been reported yet.
In this study, we firstly evaluated Kank1 mRNA expression level in human nasopharyngeal epithelial cell line NP69 and in four human NPC cell lines (5-8F, 6-10B, CNE1, CNE2) and found that Kank1 was significantly downregulated in NPC cells. Loss of heterozygosity (LOH) and aberrant hypermethylation of CpG islands in promoters are known as the two leading causes of tumor suppressor genes inactivation [22]. There are already some researches about LOH of Kank1 in many malignant tumors, but whether DNA methylation plays a vital role in deletion of Kank1 expression in NPC is still unknown. Here, by treating NPC cells with 5-aza-2-deoxycytidine (5-aza-CdR), a DNA methyltransferase enzyme (DNMT) inhibitor, the expression level of Kank1 was significantly elevated. Meanwhile, the reexpression of Kank1 induced by 5-aza-CdR suppressed NPC cells proliferation and facilitated its apoptosis. Therefore, we demonstrated that the loss of function of Kank1 in NPC mainly attributed to DNA methylation. And it shed lights on us that using demethylating agents to improve Kank1 expression may have a potential value in NPC treatment.
Materials and methods
Cell lines and drugs
NPC cell lines CNE-1, CNE-2, 5-8F, 6-10B and non-cancerous human nasopharyngeal epithelial cell line NP69 were provided by Key Laboratory of Cancer Proteomics of Chinese Ministry of Health. 5-aza-CdR (Sigma) was dissolved in dimethylsulfoxide (DMSO) to the concentration of 5 mol/L, the final concentration in the culture medium was 0, 1, 2, 5, 10 μmol/L.
Quantitative real time fluorescence polymerase chain reaction (qRT-PCR)
The relative Kank1 mRNA expression was determined by qRT-PCR. The total RNA was extracted from cells with trizol (Invitrogen). cDNA was synthesized by reverse-transcription using the All-in-One First-Strand Kit (GeneCopoeia). The qRT-PCR was subsequently performed with SYBR Green PCR Master Mix (GeneCopoeia) using an ABI ViiA 7 instrument (Applied Biosystems) at 95°C for 10 min, followed by 40 cycles of 95°C for 10 s, 60°C for 20 s and 72°C for 20 s. Reactions were performed in triplicate with human β-actin as an internal control. The primers used for qRT-PCR are as follows: Kank1, forward 5’-GCAAGAAGAGAAAAGGCAGTTG-3’, reverse 5’-TCCTCACACCACAGACATTGAT-3’; β-actin: forward 5’-s TGACGTGGACATCCGCAAAG-3’, reverse 5’-CTGGAAGGTGGACAGCGAGG-3’. 2-ΔΔCT method was used to calculate the relative gene expression.
Western blotting
The Kank1 protein expression was determined with western blotting. The cells were rinsed with cold PBS before treated with RIPA lysis solution at 4°C for 30 min. Then the mixture was centrifuged at 12000 rpm for 30 min under 4°C, and protein concentration was assayed using BCA method. Total proteins were separated by electrophoresis on SDS-PAGE gel (6% separating gel, 5% stacking gel). Immunodetection of Kank1 and β-actin were carried out by using anti-Kank1 antibody at a dilution of 1:1500 and anti-β-actin (1:1000). The ECL kit (Millipore) and X-ray films were used for visualization.
Bisulfite sequencing PCR
Bisulfite sequencing PCR (BS-PCR) was used to examine the methylation status of Kank1 gene promoter. Genomic DNA from cells was extracted for methylation status screening. Approximately 1 μg of genomic DNA was bisulfite-modified using the EN-EpiTect-Bisulfile (QianGen) according to the manufacturer’s recommendations. Based on the functional promoter sequence of the Kank1 gene, the primers (F: 5’-AGTGTATTTTTTTGGAAGGTAAATT-3’; R: 5’-CAACTACCAAATCCTCTCTATCTTC-3’) used for bisulfite-specific PCR (BSP) detection were designed using MethPrimer (http://www.urogene.org/methprimer/index.html), and the amplified fragment was 173 bp which contains 9 CG sites. The PCR reaction was performed in a 50 μl reaction system, the reaction procedure was as follows: 95°C 4 min, (95°C 30 s, 60°C 34s, 72°C 30 s) × 42 cycles, 95°C 4 min. The BSP products were then cloned into a pGEM-T-vector (Promega), and DH5α competent cells were used for transformation. Positive clones were selected by EcoR Ⅰrestriction endonuclease and sequenced by Shanghai Sangon Biotechnology.
MTT assay
Proliferation of the cell lines were detected by MTT. CNE1 and 6-10B cells were seeded in 96-well culture plates with 100 μl of growth medium, cells were treated with different concentrations (0, 1, 2, 5, 10 μmol/L) of 5-aza-CdR and cultured for 3 days. Subsequently, 20 μl MTT was added to each well and incubated in CO2 at 37°C for 4 h. DMSO was added to terminate the reaction and the absorbance value was detected at 490 nm. Cellular growth curves were constructed based on the results.
FCM analysis
FCM analysis was used with annexin V-FITC and PI (ABI) combined staining to demonstrate the apoptosisinduced by 5-Aza-CdR. Apoptosis was observed after treatment with 10 μmol/L 5-aza-CdR for 72 hours. The samples were washed with PBS two times and adjusted to 1 × 106 cells /mL. Then the cells were stained with annexin V-FITC (10 μL) and PI (10 μL). After incubation for 30 minutes in the dark at room temperature, 400 μL 1× Binding buffer was added to each tube and analyzed under FCM (BD Biosciences) within 30 minutes.
Statistics
The SPSS 17.0 software was used for statistical analyses. The data were presented as mean ± SD and comparison between two groups used pair-t test. The statistical significance was defined as a P value less than 0.05.
Results
5-aza-CdR increases the expression of Kank1 in NPC cells
To evaluate the kank1 mRNA expression level, we performed qRT-PCR in NPC cells and human nasopharyngeal epithelial cell line NP69 and the results showed that kank1 mRNA expression significantly decreased in the NPC cell lines (5-8F, 6-10B, CNE1, CNE2) comparing with the control NP69 cell, especially for CNE1 and 6-10B cells (Figure 1A). Thus, we next examined the impact of 5-aza-CdR on the expression of Kank1 in 6-10B and CNE1 cells and found that Kank1 mRNA expression gradually enhanced by treating with increasing concentration of 5-aza-CdR (Figure 1B and 1C). When the concentration of 5-aza-CdR reached 10 μM, the Kank1 protein level in 6-10B and CNE1 cells were as well significantly evaluated (Figure 1D). Therefore, we speculated that the decreased expression of Kank1 in NPC cells was primarily mediated by DNA methylation.
Figure 1.
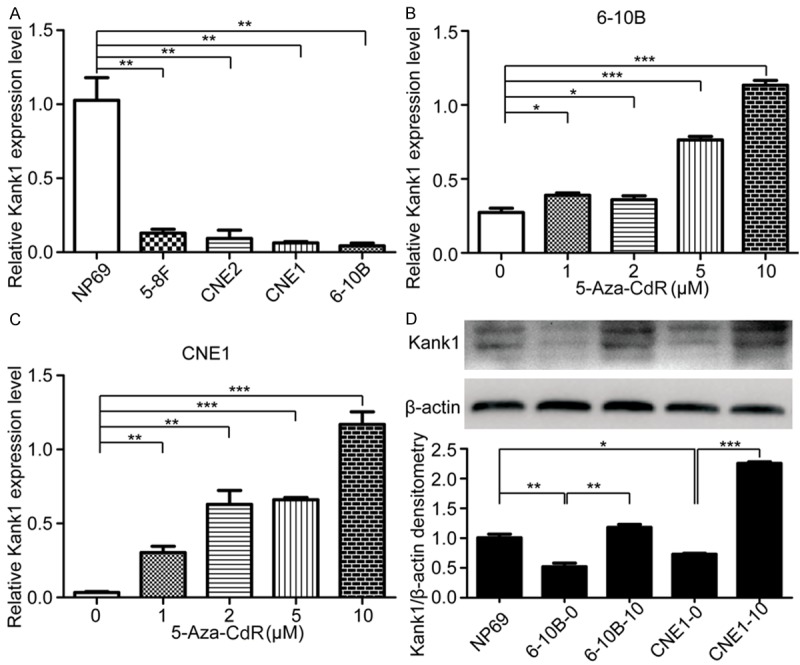
Kank1 is down-regulated in NPC cells and 5-Aza-CdR can increase its expression. A. Relative mRNA expression of Kank1 was examined by qPCR in NPC cells and human nasopharyngeal epithelial cell line NP69. Values are means ± SD (**P < 0.01). B. Different concentration of 5-Aza-CdR elevated Kank1 mRNA expression in 6-10B cell. Values are means ± SD (*P < 0.05, ***P < 0.001). C. Kank1 mRNA expression in CNE1 cell can be enhanced by treating with 1, 2, 5, 10 μM 5-Aza-CdR. Values are means ± SD (**P < 0.01, ***P < 0.001). D. Western blotting analysis of Kank1 expression in NP69, 6-10B, CNE1 cells and in 6-10B, CNE1 cells which were treated with 10 μM 5-Aza-CdR. 6-10B-0 and 6-10B-10 respectively represent 6-10B cell treated with no 5-Aza-CdR or 10 μM 5-Aza-CdR. Similarly, CNE1-0 and CNE1-10 represent CNE1 cell treated with no 5-Aza-CdR or 10 μM 5-Aza-CdR. Values are means ± SD (*P < 0.05, **P < 0.01, ***P < 0.001).
5-aza-CdR ameliorates the methylation of Kank1 promoter
To further detect the methylation status of Kank1 in NPC, we performed BS-PCR in 6-10B and CNE1 cells. The results showed that all the 9 CG sites in the Kank1 promoter were hypermethylated in the two NPC cells. However, after treated with 10 μM 5-aza-CdR, the methylation rates of Kank1 in 6-10B and CNE1 cells were obvious reduced (Figure 2A and 2B). All these suggest that promoter methylation might be one of the principal reasons of Kank1 inactivation in NPC.
Figure 2.
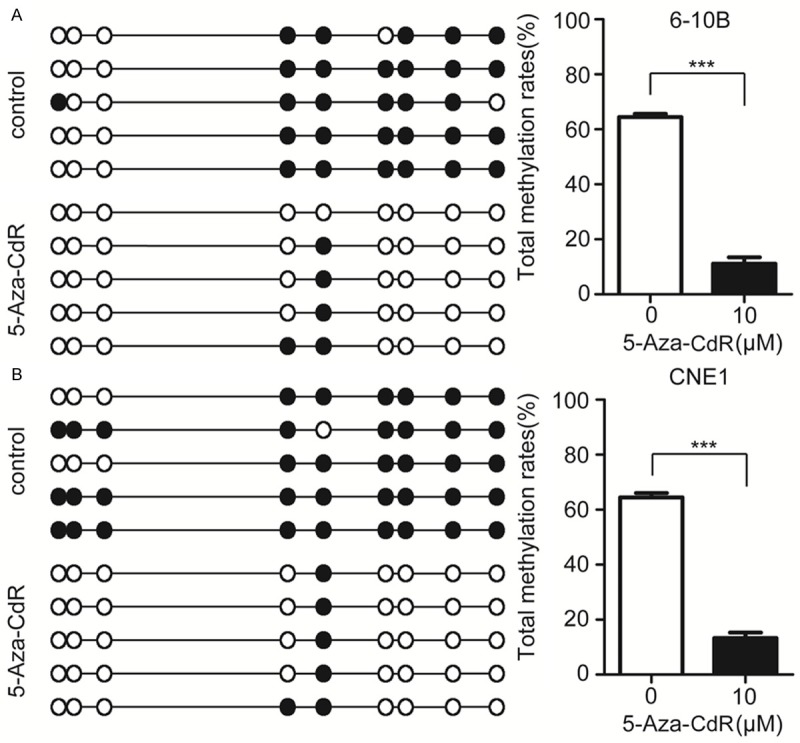
5-Aza-CdR decreases the methylation of Kank1 promoter, A. BS-PCR results of 6-10B cell treated with or without 10 μM 5-Aza-CdR. Open circles indicate unmethylated and solid circles represent methylated CpG dinucleotides. The total methylation rates of Kank1 promoter was shown by histogram. Values are means ± SD (***P < 0.001). B. The methylation status of Kank1 promoter in CNE1 cell treated with 10 μM 5-Aza-CdR compared to no drug. Values are means ± SD (***P < 0.001).
5-aza-CdR suppresses the proliferation of NPC cells
To check the effect of 5-aza-CdR on the growth of NPC cells, MTT assay was used after treating 6-10B and CNE1 cells with different concentration of 5-aza-CdR for 72 hours. The results in Figure 3A and 3B exhibited that 10 μM 5-aza-CdR strikingly restrained the proliferation of 6-10B and CNE1 cells.
Figure 3.
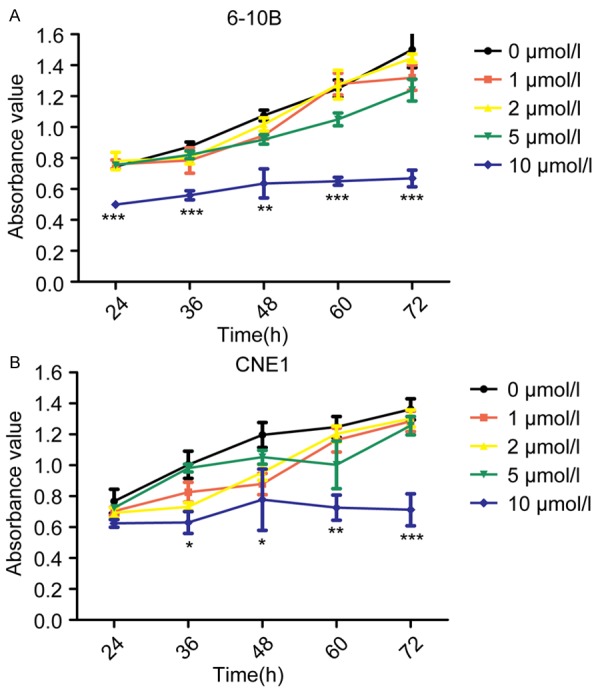
5-Aza-CdR suppresses 6-10B and CNE1 cells proliferation. A. MTT assays were used to investigate the effects of different concentration of 5-Aza-CdR on 6-10B cell proliferation. Values are means ± SD (**P < 0.01, ***P < 0.001). B. Cell proliferation analysis of different concentration of 5-Aza-CdR onCNE1 cell. The results shown are means ± SD (*P < 0.05, **P < 0.01, ***P < 0.001).
5-aza-CdR induces the apoptosis of 6-10B and CNE1 cells
As an important physiological phenomenon, apoptosis plays a key role during tumor progression. Therefore, we investigated whether 5-aza-CdR had any effects on NPC cells apoptosis. After treated with 10 μM 5-aza-CdR for 72 hours, 6-10B and CNE1 cells all appeared cell shrinkage, nuclear fragmentation etc. apoptotic phenotype (Figure 4A and 4B). To further confirm the promoting role of 5-aza-CdR to NPC cells, we stained 6-10B and CNE1 cells with annexin V-FITC and PI after treating with 10 μM 5-aza-CdR, followed by analyzing the cells apoptosis via flow cytometry. Consistent with the primary observations, 5-aza-CdR significantly facilitated 6-10B and CNE1 cells apoptosis compared with control (Figure 4C and 4D).
Figure 4.
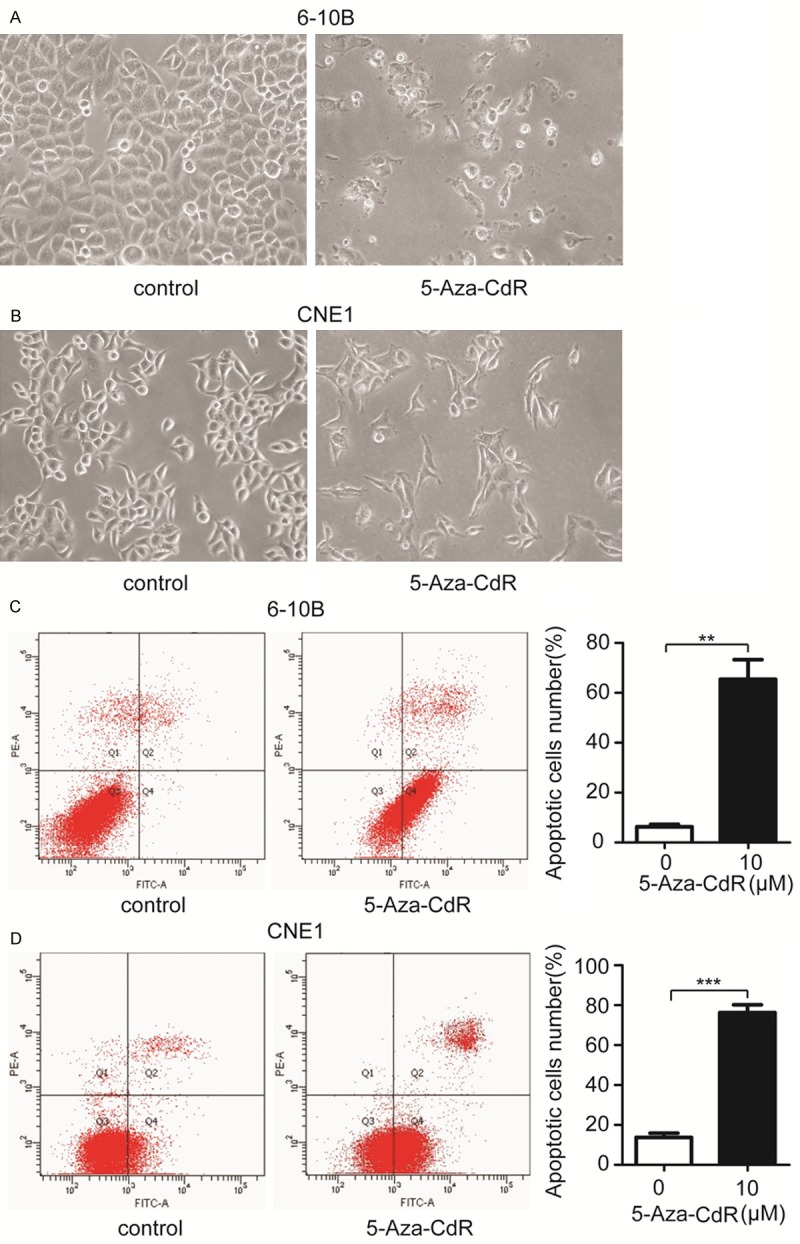
5-Aza-CdR promotes 6-10B and CNE1 cells apoptosis. A. Representative images of 6-10B cells treated with 10 μM 5-Aza-CdR for 72 hours or DMSO as control. B. The survival status of CNE1 cells which were treating with 10 μM 5-Aza-CdR or not. C. FCM analysis of the effects of 5-Aza-CdR on 6-10B cells apoptosis. Apoptotic cells rates were shown on the right panel. Values are means ± SD (**P < 0.01). D. 10 μM 5-Aza-CdR significantly facilitated CNE1 cells apoptosis, as measured by FCM. Values are means ± SD (***P < 0.001).
Discussion
NPC is a high-incidence malignancy and tends to be one of the most serious public health problems in southern China and Southeast Asia [23]. Though great advancement has been reached in diagnosing and treatment of NPC, the 5-year survival rate remains unsatisfactory due to the complicated etiology and high metastasis [24]. Therefore, it is extremely crucial to explore the key gene involved in NPC initiation and progression.
Kank1 initially was identified as a tumor suppressor in RCC [7] and afterward its deletion was found to have a close relationship with several human cancers such as bladder cancer, hepatocellular cancer, lung cancer et al [15-18]. However, there is no report about its function in NPC. In the present study, we found that Kank1 was significantly down-regulated in NPC cells compared to human nasopharyngeal epithelial cell line NP69 and its expression can be elevated in 6-10B and CNE1 cells by treating with a DNA methyltransferase enzyme (DNMT) inhibitor 5-aza-CdR. Through BS-PCR assays, we primarily confirmed that the methylation of CpG island in promoter was the leading cause for Kank1 decreased expression in NPC cells. Furthermore, we demonstrated that 5-aza-CdR can suppresses 6-10B and CNE1 cells proliferation and promote them apoptosis. All these results suggest us Kank1 may act as a suppressor in NPC and enhanced Kank1 expression by 5-aza-CdR may have a potential value for NPC treatment.
It is demonstrated that dysregulated gene expression caused by aberrant DNA methylation contributes to the progression of several human cancers [25]. The human Kank1 gene was firstly found by a LOH analysis based on microsatellite markers using candidate loci obtained by a genomic subtraction method [7]. Sarkar et al. also revealed the LOH of Kank1 occurred in 25% of RCC tissues [14] and methylation might be the most likely mechanism of inactivation of Kank1 gene expression [7]. Though there are many reports about LOH of Kank1 gene in several human cancers and other diseases [26,27], there have rare researches about the methylation of Kank1. Through BS-PCR method we first demonstrated the Kank1 was hypermethylated in NPC cells. Therefore, Kank1 reexpression in NPC induced by 5-aza-CdR could have an inhibition effect for NPC progression. Recent studies shed light on the epigenetic therapy including numerous drugs targeted specific enzymes involved in the epigenetic regulation of gene expression for cancers [28]. 5-aza-CdR, as a DNMT inhibitor, whether can be used for NPC treatment needs a further study.
Recent studies revealed Kank1 protein not only localized in the cytoplasm but also acts as a nucleo-cytoplasmic shuttling molecule responsible for relocalization of β-catenin to the nucleus to activate β-catenin-dependent transcription [29]. In addition, Kank1 plays an important role in inhibiting the activation of RhoA through PI3K/Akt/14-3-3 and Rac1 signaling via its binding to IRSp53, resulting in the inhibition of actin polymerization at the cell periphery and cell migration [11,12,30]. Thus, more functional analysis of Kank1 is ongoing to confirm the importance of it in NPC development.
Acknowledgements
The authors acknowledge the grants from Science and Technology Project of Hunan Province (2013FJ4112) and the National Natural Science Foundation of China (81372516, 81102046).
Disclosure of conflict of interest
None.
References
- 1.Chang ET, Adami HO. The enigmatic epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol Biomarkers Prev. 2006;15:1765–1777. doi: 10.1158/1055-9965.EPI-06-0353. [DOI] [PubMed] [Google Scholar]
- 2.Wei KR, Zheng RS, Zhang SW, Liang ZH, Ou ZX, Chen WQ. Nasopharyngeal carcinoma incidence and mortality in China in 2010. Chin J Cancer. 2014;33:381–387. doi: 10.5732/cjc.014.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith C. EBV and nasopharyngeal carcinoma: a target for cellular therapies. Immunotherapy. 2013;5:821–824. doi: 10.2217/imt.13.68. [DOI] [PubMed] [Google Scholar]
- 4.Lin CT. Relationship between Epstein-Barr virus infection and nasopharyngeal carcinoma pathogenesis. Ai Zheng. 2009;28:791–804. doi: 10.5732/cjc.009.10107. [DOI] [PubMed] [Google Scholar]
- 5.Yu MC, Yuan JM. Epidemiology of nasopharyngeal carcinoma. Semin Cancer Biol. 2002;12:421–429. doi: 10.1016/s1044579x02000858. [DOI] [PubMed] [Google Scholar]
- 6.Bakos A, Banati F, Koroknai A, Takacs M, Salamon D, Minarovits-Kormuta S, Schwarzmann F, Wolf H, Niller HH, Minarovits J. High-resolution analysis of CpG methylation and in vivo protein-DNA interactions at the alternative Epstein-Barr virus latency promoters Qp and Cp in the nasopharyngeal carcinoma cell line C666-1. Virus Genes. 2007;35:195–202. doi: 10.1007/s11262-007-0095-y. [DOI] [PubMed] [Google Scholar]
- 7.Sarkar S, Roy BC, Hatano N, Aoyagi T, Gohji K, Kiyama R. A novel ankyrin repeat-containing gene (Kank) located at 9p24 is a growth suppressor of renal cell carcinoma. J Biol Chem. 2002;277:36585–36591. doi: 10.1074/jbc.M204244200. [DOI] [PubMed] [Google Scholar]
- 8.Zhu Y, Kakinuma N, Wang Y, Kiyama R. Kank proteins: a new family of ankyrin-repeat domain-containing proteins. Biochim Biophys Acta. 2008;1780:128–133. doi: 10.1016/j.bbagen.2007.09.017. [DOI] [PubMed] [Google Scholar]
- 9.Kakinuma N, Zhu Y, Wang Y, Roy BC, Kiyama R. Kank proteins: structure, functions and diseases. Cell Mol Life Sci. 2009;66:2651–2659. doi: 10.1007/s00018-009-0038-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Onishi Y, Kakinuma N, Roy BC, Aoyagi T, Kiyama R. Alternative splicing of the human Kank gene produces two types of Kank protein. Biochem Biophys Res Commun. 2005;330:1247–1253. doi: 10.1016/j.bbrc.2005.03.106. [DOI] [PubMed] [Google Scholar]
- 11.Kakinuma N, Roy BC, Zhu Y, Wang Y, Kiyama R. Kank regulates RhoA-dependent formation of actin stress fibers and cell migration via 14-3-3 in PI3K-Akt signaling. J Cell Biol. 2008;181:537–549. doi: 10.1083/jcb.200707022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roy BC, Kakinuma N, Kiyama R. Kank attenuates actin remodeling by preventing interaction between IRSp53 and Rac1. J Cell Biol. 2009;184:253–267. doi: 10.1083/jcb.200805147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roy BC, Aoyagi T, Sarkar S, Nomura K, Kanda H, Iwaya K, Tachibana M, Kiyama R. Pathological characterization of Kank in renal cell carcinoma. Exp Mol Pathol. 2005;78:41–48. doi: 10.1016/j.yexmp.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 14.Hatano N, Nishikawa NS, McElgunn C, Sarkar S, Ozawa K, Shibanaka Y, Nakajima M, Gohiji K, Kiyama R. A comprehensive analysis of loss of heterozygosity caused by hemizygous deletions in renal cell carcinoma using a subtraction library. Mol Carcinog. 2001;31:161–170. doi: 10.1002/mc.1051. [DOI] [PubMed] [Google Scholar]
- 15.Simon R, Burger H, Semjonow A, Hertle L, Terpe HJ, Bocker W. Patterns of chromosomal imbalances in muscle invasive bladder cancer. Int J Oncol. 2000;17:1025–1029. doi: 10.3892/ijo.17.5.1025. [DOI] [PubMed] [Google Scholar]
- 16.Shao J, Li Y, Li H, Wu Q, Hou J, Liew C. Deletion of chromosomes 9p and 17 associated with abnormal expression of p53, p16/MTS1 and p15/MTS2 gene protein in hepatocellular carcinomas. Chin Med J (Engl) 2000;113:817–822. [PubMed] [Google Scholar]
- 17.Heidenblad M, Schoenmakers EF, Jonson T, Gorunova L, Veltman JA, van Kessel AG, Hoglund M. Genome-wide array-based comparative genomic hybridization reveals multiple amplification targets and novel homozygous deletions in pancreatic carcinoma cell lines. Cancer Res. 2004;64:3052–3059. doi: 10.1158/0008-5472.can-03-3159. [DOI] [PubMed] [Google Scholar]
- 18.Sato M, Takahashi K, Nagayama K, Arai Y, Ito N, Okada M, Minna JD, Yokota J, Kohno T. Identification of chromosome arm 9p as the most frequent target of homozygous deletions in lung cancer. Genes Chromosomes Cancer. 2005;44:405–414. doi: 10.1002/gcc.20253. [DOI] [PubMed] [Google Scholar]
- 19.Zimonjic DB, Simpson S, Popescu NC, DiPaolo JA. Molecular cytogenetics of human papillomavirus-negative cervical carcinoma cell lines. Cancer Genet Cytogenet. 1995;82:1–8. doi: 10.1016/0165-4608(95)91129-7. [DOI] [PubMed] [Google Scholar]
- 20.An HX, Claas A, Savelyeva L, Seitz S, Schlag P, Scherneck S, Schwab M. Two regions of deletion in 9p23-24 in sporadic breast cancer. Cancer Res. 1999;59:3941–3943. [PubMed] [Google Scholar]
- 21.Heyman M, Grander D, Brondum-Nielsen K, Liu Y, Soderhall S, Einhorn S. Deletions of the short arm of chromosome 9, including the interferon-alpha/-beta genes, in acute lymphocytic leukemia. Studies on loss of heterozygosity, parental origin of deleted genes and prognosis. Int J Cancer. 1993;54:748–753. doi: 10.1002/ijc.2910540507. [DOI] [PubMed] [Google Scholar]
- 22.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vokes EE, Liebowitz DN, Weichselbaum RR. Nasopharyngeal carcinoma. Lancet. 1997;350:1087–1091. doi: 10.1016/S0140-6736(97)07269-3. [DOI] [PubMed] [Google Scholar]
- 24.Lin S, Tham IW, Pan J, Han L, Chen Q, Lu JJ. Combined high-dose radiation therapy and systemic chemotherapy improves survival in patients with newly diagnosed metastatic nasopharyngeal cancer. Am J Clin Oncol. 2012;35:474–479. doi: 10.1097/COC.0b013e31821a9452. [DOI] [PubMed] [Google Scholar]
- 25.Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005;2(Suppl 1):S4–11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 26.Vanzo RJ, Martin MM, Sdano MR, South ST. Familial KANK1 deletion that does not follow expected imprinting pattern. Eur J Med Genet. 2013;56:256–259. doi: 10.1016/j.ejmg.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 27.Lerer I, Sagi M, Meiner V, Cohen T, Zlotogora J, Abeliovich D. Deletion of the ANKRD15 gene at 9p24.3 causes parent-of-origin-dependent inheritance of familial cerebral palsy. Hum Mol Genet. 2005;14:3911–3920. doi: 10.1093/hmg/ddi415. [DOI] [PubMed] [Google Scholar]
- 28.Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006;5:37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Kakinuma N, Zhu Y, Kiyama R. Nucleo-cytoplasmic shuttling of human Kank protein accompanies intracellular translocation of beta-catenin. J Cell Sci. 2006;119:4002–4010. doi: 10.1242/jcs.03169. [DOI] [PubMed] [Google Scholar]
- 30.Li CC, Kuo JC, Waterman CM, Kiyama R, Moss J, Vaughan M. Effects of brefeldin A-inhibited guanine nucleotide-exchange (BIG) 1 and KANK1 proteins on cell polarity and directed migration during wound healing. Proc Natl Acad Sci U S A. 2011;108:19228–19233. doi: 10.1073/pnas.1117011108. [DOI] [PMC free article] [PubMed] [Google Scholar]


