Abstract
Recent clinical trials on patients with glioblastoma revealed that O6-Methylguanine-DNA methyltransferase (MGMT) methylation status significantly predicts patient’s response to alkylating agents. In this study, we sought to develop and validate a quantitative MGMT methylation assay using pyrosequencing on glioblastoma. We quantified promoter methylation of MGMT using pyrosequencing on paraffin-embedded fine needle aspiration biopsy tissues from 43 glioblastoma. Using a 10% cutoff, MGMT methylation was identified in 37% cases of glioblastoma and 0% of the non-neoplastic epileptic tissue. Methylation of any individual CpG island in MGMT promoter ranged between 33% and 95%, with a mean of 65%. By a serial dilution of genomic DNA of a homogenously methylated cancer cell line with an unmethylated cell line, the analytical sensitivity is at 5% for pyrosequencing to detect MGMT methylation. The minimal amount of genomic DNA required is 100 ng (approximately 3,000 cells) in small fine needle biopsy specimens. Compared with methylation-specific PCR, pyrosequencing is comparably sensitive, relatively specific, and also provides quantitative information for each CpG methylation.
Keywords: MGMT, methylation, glioblastoma, epigenetics, pyrosequencing
Introduction
Glioblastoma multiforme (GBM), or high grade glioblastoma, is the most common malignant brain tumor in adults. It has a very poor prognosis with a median overall survival of only 12-15 months. Current treatments of GBM include surgical resection, radiotherapy, and administration of alkylating agents such as temozolomide [1]. Alkylating agents inhibit DNA replication and induce tumor cell apoptosis by producing cross-links between adjacent DNA strands. The most common site for alkylation is the O6 position of guanine [2]. MGMT as a DNA repair enzyme specifically removes promutagenic alkyl groups from the O6 position of guanine in DNA. Repair of O6-alkylguanine adducts in tumor DNA reduces the cytotoxicity of alkylating chemotherapeutic agents and thus confers tumor chemoresistance [3].
Both preclinical and clinical evidence showed that the expression of MGMT is mainly regulated at the epigenetic level by promoter methylation [4]. Under normal circumstances, the CpG sites spanning a large portion of the promoter and exon 1 of MGMT gene are unmethylated. In some tumor cells, however, the cytosines at certain CpG sites are methylated. This prevents transcriptional factors from binding and eventually leads to the reduced expression of MGMT protein, which sensitizes the cancer cells to alkylating agents.
Many studies have established a significant correlation between methylation of MGMT and a positive clinical outcome for patients with glioblastoma treated with alkylating agents such as temozolomide, carmustine, and procarbazine with or without radiotherapy [2,5-7]. Furthermore, significant differences were demonstrated among varying levels of methylation, with higher MGMT methylation level associated with longer overall survival [8]. These studies suggested that MGMT methylation can predict glioblastoma chemosensitivity to alkylating agents and help determine the patient population who likely benefit from such treatment.
Several methods have been used to identify MGMT methylation status [3,9-12]. Methylation-specific PCR (MSP) with its high sensitivity is widely used for assessing MGMT methylation in the research setting. But it suffers from increased false-positivity and its qualitative nature. Its mediocre compatibility with formalin-fixed paraffin-embedded (FFPE) tissue specimens and requirement of high quality DNA limited its application in the clinical setting [11]. Pyrosequencing is a highly reproducible and quantitative method for confirming methylation status. It has been shown to be the best approach for assessing MGMT methylation status in GBM patients and correlating with clinical outcomes [3]. However, no studies have been done to validate this method in the clinical setting for its compatibility with FFPE tissue specimens, analytical quality, and clinical feasibility. In this study, we aimed to validate a quantitative MGMT methylation assay using pyrosequencing on FFPE biopsy tissue of glioblastoma and determine its analytical sensitivity and precision as well as clinical feasibility for pathologic diagnosis and prediction of glioblastoma chemosensitivity.
Materials and methods
A total of 53 cases of brain tissue for quantitative analysis of MGMT promoter methylation status were selected for the study. These tissue specimens came from 10 patients with epilepsy as non-neoplastic controls, 43 patients with resected GBM or stereotaxic biopsy for GBM. All specimens were FFPE tissue transported and stored at room temperature. No special preparation was needed.
The genomic DNA was extracted from 10 μm tissue sections of FFPE tissue samples using the Gentra Puregene tissue kit (Qiagen Inc., Valencia, CA). DNA was further cleaned and purified by running through the QIAamp MiniElute column (Qiagen Inc., Valencia, CA) according to the manufacture’s manual. The DNA concentration, protein to nucleic acid ratio, and DNA to RNA ratio for purity were assessed by spectrophotometer (NanoDrop products, Wilmington, DE). Approximately 100 to 200 ng total DNA was subjected to bisulfite conversion using EZ DNA Methylation Gold kit (Zymo Research, Orange, CA).
A total of 10-20 ng bisulfite-treated DNA was carried on for PCR using the PyroMark PCR kit (Qiagen Inc., Valencia, CA). The PCR control was reaction mixture with water in place of DNA. The PCR conditions for MGMT gene were 95°C for 15 minutes; 45 cycles of 95°C for 20 seconds, 53°C for 20 seconds, and 72°C for 20 seconds; 72°C for 5 minutes, and then on hold at 4°C. The PCR products were immobilized to beads and strand separation. We then conducted pyrosequencing methylation assay using the sequencing primer provided in the PyroMark CpG MGMT kit on the PyroMark Q96ID pyrosequencer (Qiagen Inc., Valencia, CA). The Pyromark CpG MGMT kit detected the level of methylation on 5 CpG sites located in exon 1 of MGMT gene. A cytosine not followed by a guanine served as an internal control for completion of bisulfite conversion. The percent methylation (% of C’s present) was reported for each CpG site. A sample with methylation above 10% of any single CpG island or the average of all 5 CpG islands was interpreted as positive. A sample below 10% methylation was interpreted as negative.
Two cell lines were used in analytical sensitivity tests. SW40, a colon cancer cell line with a mean MGMT methylation of 95% was used as the positive control; MRC5, a fibroblast cell line with 0% MGMT methylation was used as the negative control. We diluted SW40 DNA with MRC5 DNA from a ratio of 1:10 to 1:1000. The most diluted concentration with detectable methylation was recorded as the analytical sensitivity, which was compared with the standard MSP method. In addition, we tested 50, 100, 150, and 200 ng DNA obtained from FFPE tissues in MGMT methylation pyrosequencing for the minimal requirement of DNA volume. To measure the biopsy tissue size needed for MGMT methylation pyrosequencing, we reviewed 50 consecutive GBM biopsy specimens in 2010 at our institution and documented the size of these specimens. We chose 16 biopsy specimens that represent the biopsy size below 30 percentile for testing. Genomic DNA was extracted from 8 sections of 15 μm thick FFPE blocks. The concentration of DNA was measured in duplicate in each case.
Statistical analysis for this study was descriptive in nature. Thus we did not perform any sample size calculation or power estimation. We summarized categorical data as frequency counts and percentages, and continuous measures as means, standard deviations (s.d.), medians, and ranges. All the statistical analyses were performed using VassarStats (Richard Lowry, PhD, Vassar College, NY).
Results
Optimizing the DNA extraction and purification procedure for pyrosequencing assay
We optimized the experimental conditions for MGMT methylation pyrosequencing assay. After extraction, genomic DNA was further purified to generate highly purified DNA for pyrosequencing. The amount of DNA sample loss was less than 15%. We used EZ DNA methylation gold kit for bisulfite conversion. This method converts greater than 95% of unmethylated cytosines in the genome which satisfies the requirement of PyroMark Q96ID pyrosequencer for consistent results.
MGMT methylation in GBM and epilepsy brain tissues
We determined the level of methylation on 5 CpG sites located in exon 1 of MGMT gene from the brain tissues of both patients with epilepsy as non-neoplastic control and those with glioblastoma. Typical examples of pyrograms are shown in Figure 1. Shaded boxes encompass T/C pairs with methylation of 5 CpG sites measured quantitatively. In a typical tissue specimen from an epilepsy patient, most of the CpG sites are unmethylated with occasional sites bearing less than 10% methylation. In contrast, a typical tumor specimen from a glioma patient often has more than 25% methylation in all the CpG sites. As shown in Figure 2, we measured MGMT gene methylation in 10 non-neoplastic epilepsy patients. None of the epilepsy brain tissues harbors > 10% MGMT methylation. We only observed low level of MGMT methylation in two cases (4% and 8%, respectively). In contrast, among the glioblastoma tissues from 43 patients, the range of methylation percentage of any CpG island in MGMT gene is between 33% and 95%, with a mean of 65%. We observed > 10% MGMT methylation in 37% (16/43) patients and > 5% MGMT methylation in 42% (18/43) patients. Ten percent of methylation is the best cutoff to distinguish MGMT methylation positive cases from negative cases.
Figure 1.
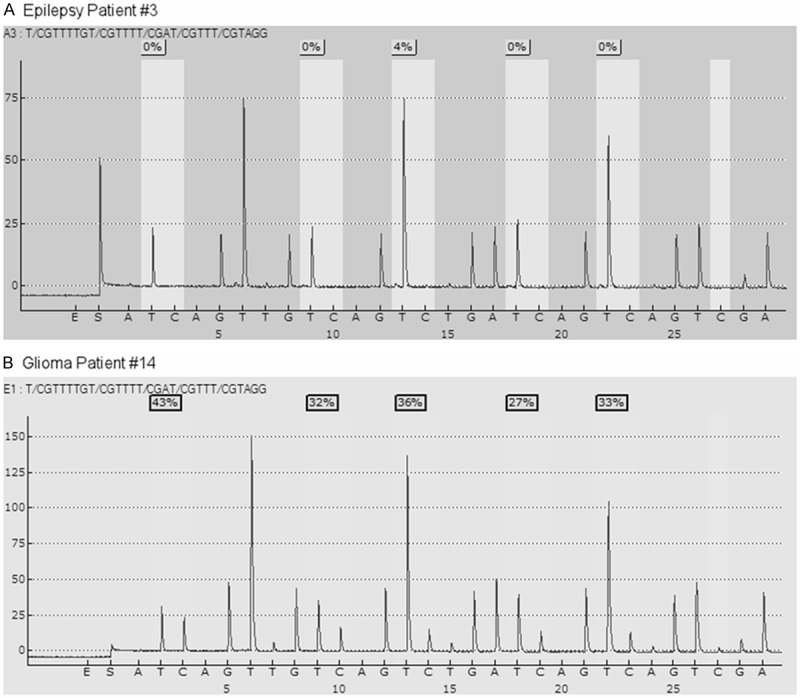
Pyrograms for MGMT methylation pyrosequencing test from the brain tissues of an epilepsy patient (Panel A) and a glioma patient (Panel B).
Figure 2.
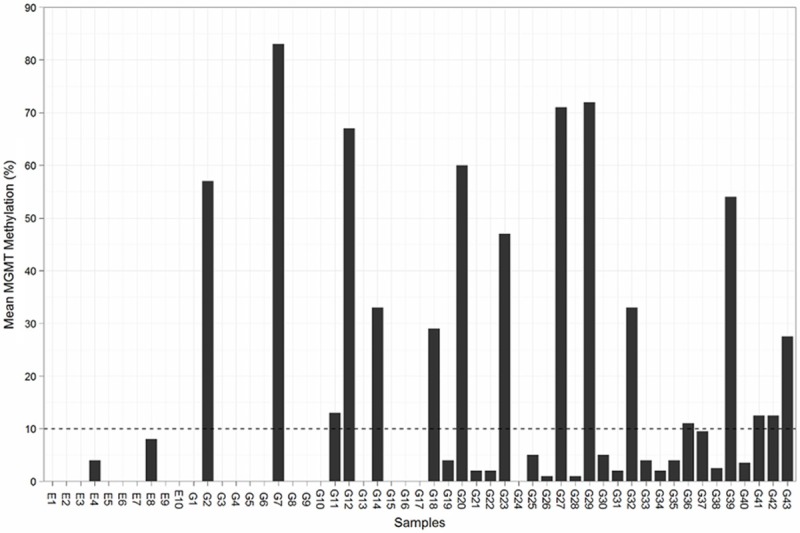
Mean MGMT methylation of tissue specimens from epilepsy patients (E1-E10) and glioma patients (G1-G43). The dash line represents a 10% cutoff.
Comparing the analytical sensitivity between pyrosequencing and MSP assays
We determined the analytical sensitivity of this methylation test by pyrosequencing using a series of DNA dilutional tests. By a serial dilution of a methylated cancer cell line with an unmethylated normal cell line, pyrosequencing can detect 1 tumor cell harboring MGMT methylation out of 80-100 cells without MGMT methylation. For clinical application, in comparison with MSP, pyrosequencing is comparably sensitive with less false-positive cases and also provide quantitative methylation value of each CpG island.
Minimal DNA amount and minimal tissue volume needed for pyrosequencing assay
Our measurement of minimal DNA requirement revealed that the minimal amount of genomic DNA required for successful detection of MGMT methylation by pyrosequencing is 100 ng (approximately 3,000 cells). Our efforts to determine the tissue size that is needed for MGMT methylation pyrosequencing test demonstrated that most of the clinical GBM biopsy specimens range from 0.1 to 3.0 cm in size with a median of 1.0 cm. We chose the cases with biopsy size less than 1.0 cm. to test the minimal tissue needed for obtaining enough genomic for conducting pyrosequencing-based MGMT methylation assay. As shown in Figure 3, the mean DNA yield increased in line with the increase of biopsy tissue size. Four of 15 μm thick tissue sections should yield approximately > 300 ng genomic DNA, which corresponds to 0.3 cm biopsy size with less than 15%. We could use 150 ng DNA samples for each run and fulfill the clinical requirement of duplicate tests. After testing protocol has been finalized, we run a precision test with a known MGMT methylation case along with positive and negative controls to polish up the procedure. Results of duplicated tests were consistent and compatible with expected results.
Figure 3.
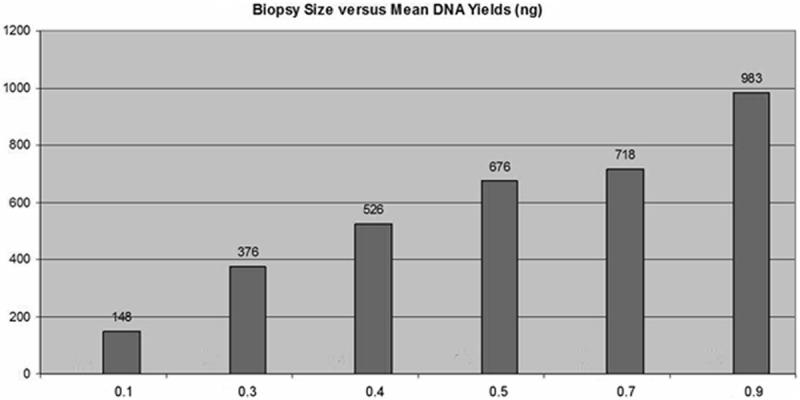
The relationship between biopsy size and mean DNA yields.
Discussion
Since the establishment of MGMT methylation as a favorable predictor of positive clinical outcome for patients treated for glioblastoma [2], several methods have been developed to determine its methylation status including the widely used MSP method in the research setting [9,11]. However, various limitations of these methods such as qualitative result report, low specificity, and high tissue quality requirement hindered their further utilization in the clinical setting [3,11]. This fact was clearly demonstrated by Karayan-Tapon and coworkers [3] during their assessment of prognostic value of MGMT status in glioblastoma patients with five different methods. It turned out that pyrosequencing is the best approach to stratify patients treated with temozolomide into different prognostic groups based on their measured MGMT methylation status.
The reliability and reproducibility of most of the previously developed methods require high-quality DNA. These methods such as MSP often require fresh frozen specimen [7]. However, in most clinical settings, FFPE specimens are needed for easy transport and storage. It has been reported that MSP can only provide reliable MGMT methylation status on two thirds of the FFPE tumor specimens [7]. To solve this problem, we further purified the genomic DNA extract from FFPE tissue samples using Qiagen MiniElute column. After elution with 100 μl AE buffer, we were able to generate highly purified genomic DNA for the following steps with less than 15% DNA loss.
Bisulfite conversion of cytosine is a crucial step toward success. Complete conversion of unmethylated cytosine to uracil is the basis of methylation quantification. Incomplete conversion leads to overestimation of MGMT gene methylation. It also severely compromises the robustness of methods for clinical samples often with low DNA quantity. In our study, we compared two bisulfite treatment methods: EZ DNA methylation gold kit and Qiagen DNA methylation kit. The former method converts greater than 95% of unmethylated cytosines in the genome which satisfies the requirement of PyroMark Q96ID pyrosequencer for consistent results. However, the latter method used in some of the previous studies converts only about 90% of unmethylated cytosines which does not fulfill the requirement of pyrosequencer.
The 10% cutoff of methylation we chose to stratify patients was based on the maximum level of methylation (8%) in the control group. No formal analysis of receiver operating characteristics was conducted due to small sample size. However, both the 10% cutoff and the prevalence of MGMT hypermethylation in the tumor tissues of glioblastoma patients thereof were consistent with the previous reports [4,11].
The analytical sensitivity of our method for MGMT methylation detection is from 1:80 to 1:100, which is approximately five times less than MSP method (1:500). Although MSP is more sensitive than pyrosequencing, it often generates approximately 10% false positive results [10]. This concern with false-positivity using MSP was mentioned in the literature due to its “super-sensitivity” and mispriming (PCR bias), which can be further aggravated by high cycle number and nested primers used for decreased sample quality of PPFE specimen [13]. Since most testing samples will be GBM lesional materials by FNAB and are less diluted with normal brain tissue, sensitivity of 1:80-1:100 should be good enough for clinical usage. Further analysis in this study demonstrated approximately 95% of cases matched between pyrosequencing and MSP assays. On the other hand, this non-inferior sensitivity of pyrosequencing is accompanied by its higher specificity with quantitative value of both average and individual CpG island methylation status. In contrast, MSP is not quantitative and cannot tell methylation status of individual CpG island which may be more predictive to clinical outcomes as previously reported [11].
In our study, 100 ng DNA is adequate to repeatedly detect MGMT methylation by pyrosequencing, which is much less than the recommendation of Qiagen kit using > 300 ng genomic DNA for research purposes. The biopsy size requirement (0.3 cm) of our method is a lot smaller than the current median biopsy size of 1.0 cm (range: 0.1-3.0 cm). The DNA yielded from even the smallest GBM biopsy size (0.1 cm) at our institution in 2010 was enough for duplicated assays. Thereby our tissue requirement is reasonable clinically and is significantly less demanding than the currently commercially available kits of MSP method, which requires > 1.0 cm tissue with minimal 500 ng input for their assays. Last, we created both positive and negative report templates compatible with CAP MP reporting guidelines.
In summary, we described the first validated pyrosequencing-based MGMT methylation test on clinical FFPE biopsy tissue from 33 patients with glioblastoma and 10 patients with epilepsy as control. We demonstrated that pyrosequencing detection of MGMT methylation has an analytical sensitivity, feasible DNA and biopsy tissue requirement suitable for routine clinical application. In addition to pathologic diagnosis, MGMT methylation assay shall provide a useful molecular biomarker for prediction of chemosensitivity in patients with glioblastoma multiforme.
Disclosure of conflict of interest
None.
References
- 1.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 2.Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, Baylin SB, Herman JG. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–4. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 3.Karayan-Tapon L, Quillien V, Guilhot J, Wager M, Fromont G, Saikali S, Etcheverry A, Hamlat A, Loussouarn D, Campion L, Campone M, Vallette FM, Gratas-Rabbia-Ré C. Prognostic value of O6-methylguanine-DNA methyltransferase status in glioblastoma patients, assessed by five different methods. J Neurooncol. 2010;97:311–22. doi: 10.1007/s11060-009-0031-1. [DOI] [PubMed] [Google Scholar]
- 4.Esteller M, Herman JG. Generating mutations but providing chemosensitivity: the role of O6-methylguanine DNA methyltransferase in human cancer. Oncogene. 2004;23:1–8. doi: 10.1038/sj.onc.1207316. [DOI] [PubMed] [Google Scholar]
- 5.Paz MF, Yaya-Tur R, Rojas-Marcos I, Reynes G, Pollan M, Aguirre-Cruz L, García-Lopez JL, Piquer J, Safont MJ, Balaña C, Sanchez-Cespedes M, García-Villanueva M, Arribas L, Esteller M. CpG island hypermethylation of the DNA repair enzyme methyltransferase predicts response to temozolomide in primary gliomas. Clin Cancer Res. 2004;10:4933–8. doi: 10.1158/1078-0432.CCR-04-0392. [DOI] [PubMed] [Google Scholar]
- 6.Hegi ME. Clinical Trial Substantiates the Predictive Value of O-6-Methylguanine-DNA Methyltransferase Promoter Methylation in Glioblastoma Patients Treated with Temozolomide. Clin Cancer Res. 2004;10:1871–1874. doi: 10.1158/1078-0432.ccr-03-0384. [DOI] [PubMed] [Google Scholar]
- 7.Hegi ME, Diserens AC, Gorlia T. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 8.Dunn J, BaboriE A, Alam F, Alam F, Joyce K, Moxham M, Sibson R, Crooks D, Husband D, Shenoy A, Brodbelt A, Wong H, Liloglou T, Haylock B, Walker C. Extent of MGMT promoter methylation correlates with outcome in glioblastomas given temozolomide and radiotherapy. Brit J Cancer. 2009;101:124–31. doi: 10.1038/sj.bjc.6605127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Nati Acad Sci U S A. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaw RJ, Akufo-Tetteh EK, Risk JM, Field JK, Liloglou T. Methylation enrichment pyrosequencing: combining the specificity of MSP with validation by pyrosequencing. Nucleic Acids Res. 2006;34:e78. doi: 10.1093/nar/gkl424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mikeska T, Bock C, El-Maarri O, Hübner A, Ehrentraut D, Schramm J, Felsberg J, Kahl P, Büttner R, Pietsch T, Waha A. Optimization of quantitative MGMT promoter methylation analysis using pyrosequencing and combined bisulfite restriction analysis. J Mol Diagn. 2007;9:368–81. doi: 10.2353/jmoldx.2007.060167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jha P, Suri V, Jain A, Sharma MC, Pathak P, Jha P, Srivastava A, Suri A, Gupta D, Chosdol K, Chattopadhyay P, Sarkar C. O6-methylguanine DNA methyltransferase gene promoter methylation status in gliomas and its correlation with other molecular alterations: first Indian report with review of challenges for use in customized treatment. Neurosurgery. 2010;67:1681–91. doi: 10.1227/NEU.0b013e3181f743f5. [DOI] [PubMed] [Google Scholar]
- 13.Derks S, Lentjes MHFM, Hellebrekers DMEI. Methylation-specific PCR unraveled. Cellular Oncologyy. 2004;26:291–9. doi: 10.1155/2004/370301. [DOI] [PMC free article] [PubMed] [Google Scholar]


