Abstract
TMSG1, as a novel tumor metastasis suppressor gene, has been demonstrated to closely relate to the metastasis and drug-resistant of breast cancer. However, its molecular mechanism is still unclear. In this study, we explored the effect of small interference RNA (siRNA) targeting TMSG1 on the invasion of human breast carcinoma cell line MCF-7 and its molecular mechanisms associated with the extracellular pH. qRT-PCR and Western blot analysis revealed dramatic reduction of the levels of TMSG1 mRNA and protein after transfection of siRNA in MCF-7 cells. Cell migration and invasion were obviously increased by TMSG1 siRNA treatment. The activity of vacuolar ATPase (V-ATPase) and MMP-2 was significantly increased in MCF-7 cells transfected with the TMSG1 siRNA compared with the controls. Furthermore, acidic intracellular environment significantly increased the MMP-2 activity and the capacity of cell migration and invasion. In conclusion, silencing of TMSG1 increased V-ATPase activity, decreased extracellular pH and in turn the activation of secreted MMP-2, which ultimately promoted metastasis capacity of breast cancer cell.
Keywords: Breast cancer, TMSG1, V-ATPase, MMP-2, extracellular pH
Introduction
Breast cancer is the most common malignancy, threatening the health of women around the world. Although diagnostic and therapeutic methods have greatly improved over the last decade, more than 1.3 million women worldwide are diagnosed with breast cancer and about half-a-million women still die because of breast cancer each year [1,2]. The high figures of incidences and mortality, even with the advancement of primary screening and diagnosis, increase the urgency to study the metastasis mechanism of breast cancer and explore the genes and treatment target associated with the metastasis of breast cancer.
Accumulating evidence suggests a key role of tumor acidic microenvironment in cancer development, progression, and metastasis [3-5]. V-ATPase, as a newly identified ATP-dependent proton pump, pumps H+ out of the cell membrane and thus play an important role in formation and maintenance of the extracellular acidic microenvironment [6-8]. V-ATPases are overexpressed in many types of metastatic cancers and positively correlated to their invasion and metastasis by decreasing extracellular pH and increasing secretion and activation of degradative enzymes, such as matrix metalloproteinases (MMPs) [6,9]. Furthermore, low extracellular pH may promote the degradation and remolding of ECM through proteolytic enzyme activation, thus contributing to cancer invasion and metastasis [10]. In breast cancer cells, the abundance of V-ATPase on the plasma membrane correlates with an invasive phenotype [11], and V-ATPase inhibitors reduce cell migration in cancer cells with high levels of plasma membrane V-ATPase [12,13].
TMSG1 gene (tumor metastasis suppressor gene 1) is a novel tumor metastasis suppressor gene, which plays a prominent role in suppression of tumor invasion and metastasis [14]. It has been found that TMSG1 is negative correlation with tumor metastatic potential. Silencing of TMSG1 gene in PC-3M-2B4 cells enhanced invasion and metastasis of PCA cell [15]. The roles played by TMSG-1 in suppression of tumor invasion and metastasis have been reported to be achieved through various mechanisms. Recent studies showed that TMSG1 could regulate V-ATPase activity and extracellular pH by targeting the C subunit of V-ATPase, thereby induce apoptosis of tumor cells [16,17]. In addition, TMSG1 also promotes cell apoptosis through the synthesis of ceramide, which in turn induces cell cycle arrest and affects the activity of telomerase as well as cell life span [18,19]. Recently, it has been reported that low expression of LASS2 was associated with poor prognosis in patients with breast cancer [20]. However, the molecular mechanism by which TMSG1 on breast cancer metastasis is unclear at present.
In this study, small interfering RNAs (siRNAs) targeting TMSG1 will be adopted to silence the gene expression in human breast cancer cell line MCF-7. Meanwhile, we further elucidated the molecular mechanism of TMSG1 involved in the regulation of tumor metastasis by investigating the activity of V-ATPase, the expression and activity of matrix metalloproteinase 2 (MMP-2) in MCF-7 cells, then further revealed the key role of V-ATPase induced extracellular acidic microenvironment in metastasis of breast cancer.
Materials and methods
Cells and cell culture
MCF-7 breast cancer cells (American Type Culture Collection, Manassas, VA, USA; HTB-22) was cultured at 37°C in a humidified 5% CO2 atmosphere in RPMI-1640 medium with 10% fetal calf serum (Gibco, Invitrogen, Carlsbad, CA, USA), 100 IU/ml penicillin G, and 100 mg/ml streptomycin sulfate (Sigma-Aldrich, St Louis, MO, USA).
Construction of siRNAs
We designed and purchased four different siRNA duplexes of TMSG1 from GenePharma company (Shanghai, China). siRNA-1 primers sense is 5’-CUGCCUUCUUUGGCUAUUATT, and reverse is, 5’-UAAUAGCCAAAGAAGGCAGTT-3’; siRNA-2 primers sense is 5’-GCUGCCCUCUUGAACAUAATT, and reverse is, 5’-UUAUGUUCAAGAGGGCAGCTT-3’; siRNA-3 primers sense is 5’-GAGUCAGCCAAGAUGUUUATT, and reverse is, 5’-UAAACAUCUUGGCUGACUCTT-3’; siRNA- negative control primers sense is 5’-UUCUCCGAACGUGUCACGUTT, and reverse is, 5’-ACGUGACACGUUCGGAGAATT-3’;
Transfection
Twenty-four hours before transfection cells were seeded, TMSG1 siRNA and Negative Control siRNA were transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Twenty-four or forty-eight hours later, MCF-7 cells were collected and subjected to further analysis. Total RNA or protein was extracted from the indicated cells for analysis. The experiment was done in triplicates. More than nine wells were treated with the same kind of siRNA.
RNA extraction and qRT-PCR
Total RNA was isolated from cells with the TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. Real-time polymerase chain reaction was performed with the StepOne Plus sequence detection system (Applied Biosystems, Foster City, CA). To ensure the reproducibility of the results, all the genes were tested in triplicate. ITGB1 generated using sense (5’-gaagccagctggagattcac-3’) and antisense (5’-gacatcagaggcaatgctga-3’) primers. β-action sense primers: 5’-cattaaggagaagctgtgct-3’ and antisense primers: 5’-gttgaaggtagtttcgtgga’.
Western blot analysis
Whole cell extracts were prepared with a cell lysis reagent (Sigma-Aldrich, St. Louis, MO, USA) according to the manual, and then, the protein was quantified by a BCA assay (Pierce, Rockford, IL, USA). Then, the protein samples were separated by SDS-PAGE (10%) and detected by Western blot using polyclonal goat anti-TMSG1 antibody (Santa Cruz Bio-technology) or rabbit anti-MMP-2 antibody (Santa Cruz Bio-technology) and monoclonal mouse anti-β-actin (Santa Cruz Bio-technology) as a control. Goat anti-rabbit IgG (Pierce, Rockford, IL, USA) secondary antibody conjugated to horseradish peroxidase and ECL detection systems (SuperSignal West Femto, Pierce) were used for detection.
Activity of V-ATPase
Assays V-ATPase activity was performed as described previously by Fan et al [21].
Gelatin zymography
MMP Zymography assay kit (for MMP-2 and MMP-9) (Applygen Technologie, China) was used to detect the activity of MMP2 and MMP9. Protein extracts and positive mixture were mixed with an equal volume of 2X SDS-PAGE non-reducing buffer, and electrophoresed on 8% SDS polyacrylamide gels containing 2 mg/ml of gelatin. Gels were then washed twice for 30 min in buffer A at room temperature, and incubated for 4 hours at room temperature in incubation buffer B. Gels were then stained for 2 hour with 0.25% Coomassie brilliant blue and then destained in destaining buffer (10% acetic acid and 20% methanol) for 60 min.
Invasion assay
Cells were cultivated to 80% confluence on the 12-well plates. Then, we observed the procedures of cellular growth at 24 h. All the experiments were repeated in triplicate. The transwell invasion chambers were used to evaluate cell invasion. Then cells invading across the membrane were counted under a light microscope.
Wound healing assay
For the wound healing assay, cells were seeded in 12-well plates and grown to 90% confluence. Monolayers in the center of the wells were scraped with pipette tips and washed with PBS. Cell movement into the wound area was monitored and photographed at 0 and 24 h using a light microscope. The migration distance between the leading edge of the migrating cells and the edge of the wound was compared as previous work [22].
Statistical analysis
Each experiment was repeated at least three times. Data were shown as mean ± s.d and analyzed using SPSS 18.0. Statistical comparisons between groups were analyzed using Student’s t-test and a two-tailed P < 0.05 was considered to indicate statistical significance.
Results
Effects of TMSG1 siRNAs on the expression of TMSG1 in MCF-7 cells
The MCF-7 cells were transfected with TMSG1 siRNAs and with negative control siRNA containing scrambled random sequence, respectively. We examined four siRNAs to target human TMSG1 as described above. qRT-PCR analysis showed that siRNA-2 effectively blocked the expression of TMSG1 mRNA. qRT-PCR revealed dramatic reduction of 88.1% with siRNA-2, 60.1% with siRNA-3, and 12.2% with siRNA-1 in the levels of TMSG1 mRNA after transfection of siRNA in MCF-7 cells, compared with TMSG1 negative control siRNA or the control (untransfected) group (P < 0.05; Figure 1). Thus, the cells transfected with TMSG1 shRNA2 were used for further experiments.
Figure 1.
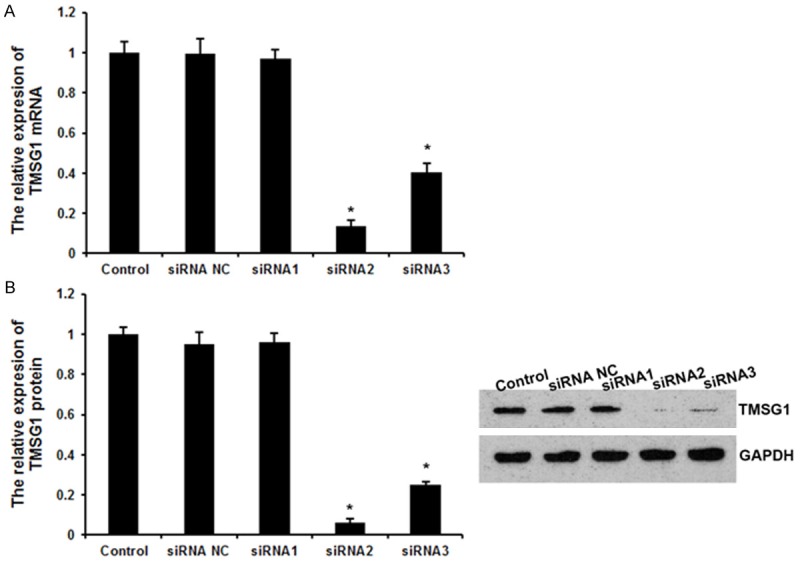
TMSG1 siRNAs silenced the expression of TMSG1 in MCF-7 cells. A. qRT-PCR analyzed the expression of TMSG1 after transfection with different siRNA fragments in MCF-7 cells. B. The western blot analysis showed the expression of TMSG1 protein after interference. Control, untreated cells. siRNA NC, transfection with negative control siRNA. siRNA1, transfection with TMGS1 siRNA 1. siRNA2, transfection with TMGS1 siRNA 2. siRNA3, transfection with TMGS1 siRNA 3. Data are the mean ± SD of duplicates from a representative experiment of three independent experiments. *P < 0.01 vs. control and siRNA NC group.
TMSG1 siRNA promotes the migration and invasion of MCF-7 cells
As shown in Figure 2A, transwell assays without Matrigel demonstrated that TMSG1 siRNA could significantly promoted migration of MCR-7 cells when compared with siRNA NC group and control (untreated) group. To further confirm this observation, we also determined the migration ability of breast cancer cells in the condition of TMSG1 silencing using a wound migration assay. The results showed that the cell-free wound gaps of MCF-7 cells monolayers healed slowly in the untreated cells or Negative Control siRNA transfected cells. However, in TMSG1 siRNA-2 transfected cells, the closure of the wounded area was significantly accelerated (Figure 2B).
Figure 2.
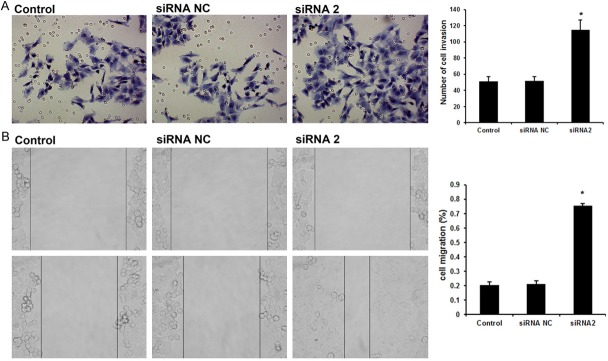
TMSG1 siRNA affects the migration and invasion of MCF-7 cells. A. Transwell assays showed that the TMSG1 siRNA transfected cells displayed dramatically increased invasion ability compared with the untreated cells or Negative Control siRNA transfected cells. B. In wound migration assay, the cell-free wound gaps of MCF-7 cells monolayers healed slowly in the untreated cells or Negative Control siRNA transfected cells. Data are the mean ± SD of duplicates from a representative experiment of three independent experiments. *P < 0.01 vs. control and siRNA NC group.
TMSG1 siRNA increase the activity of V-ATPase in MCF-7 cells
To explore whether TMSG1 siRNA suppress breast cancer cell migration and invasion by inhibiting the V-ATPase activity through binding to ATP6V1H, we examined the influence of TMSG1 siRNA on the V-ATPase activity by GEMEND’s V-ATPase activity assay kit. The activity of V-ATPase = OD of sample/(concentration × 31.1) × 103 U/mg. As shown in Figure 3, the V-ATPase activity was greatly increased in TMSG1 siRNA-2 treated MCF-7 cells (2.53 ± 0.031) compared with untreated cells (1.43 ± 0.022) and siRNA-NC treated cells (1.39 ± 0.014).
Figure 3.
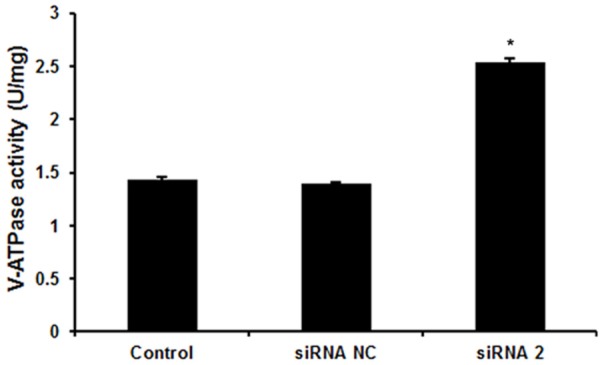
Effects of TMSG1 siRNA on V-ATPase activity. Data are the mean ± SD of duplicates from a representative experiment of three independent experiments. *P < 0.01 vs. control and siRNA NC group.
Effects of TMSG1 siRNA on the expression and activity of MMP-2 protein
V-ATPases pumps proton out of the cell membrane and thus plays an important role in formation and maintenance of the extracellular acidic microenvironment, which positively are positively correlated to secretion and activation of degradative enzymes, such as matrix metalloproteinases (MMPs) [6,9]. Here, we determined the expression levels and activities of MMP-2, which are closed related to cancer invasion and metastasis [23]. Western blot showed that TMSG1 siRNA didn’t have obvious effect on the expression of total MMP-2 protein in the MCF-7 cells (Figure 4A). Furthermore, the supernatant of cultured cells was collected and the gelatinase activity was assayed with gelatin zymography. As shown in Figure 4B, the activity of MMP-2 was significantly increased by TMSG1 siRNA. These results indicated that TMSG1 didn’t have obvious effects on the expression levels of MMP-2 protein, but have significant effects on the activity of MMP-2 protein in the MCF-7 cells.
Figure 4.
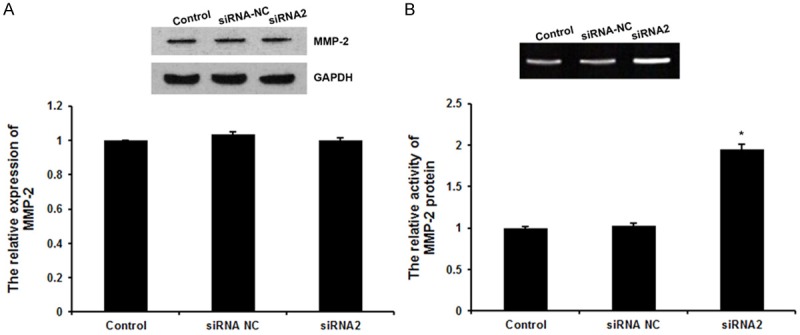
Effects of TMSG1 siRNA on MMP-2 protein. A. Western blot analysis was used to detect the expression levels of MMP-2 protein. B. The activity of MMP-2 was examined by Gelatin zymography. Data are the mean ± SD of duplicates from a representative experiment of three independent experiments. *P < 0.01 vs. control and siRNA NC group.
Extracellular pH affects the activity of MMP-2 protein and the migration and invasion of MCF-7 cells
Accumulating studies show a key role of tumor acidic microenvironment in cancer development, progression, and metastasis. The acidic tumor extracellular microenvironment induces the increased secretion and activation of proteases, and promotes the degradation and remolding of extracellular matrix (ECM) through proteolytic enzyme activation, thus contributing to cancer invasion and metastasis. In this study, we further analyzed the effects of acidic extracellular microenvironment on MMP-2 protein in MCF-7 cells. As shown in Figure 5A, extracellular pH didn’t affect obviously the expression of total MMP-2 protein in the MCF-7 cells using western blot analysis. Furthermore, the gelatinase activity assayed the revealed the positive correlation between extracellular pH and MMP-2 activity. As shown in Figure 5B, the activity of MMP-2 was significantly increased in a PH-dependent manner. These results indicated that extracellular pH didn’t have obvious effects on the expression levels of MMP-2 protein, but have significant effects on the activity of MMP-2 protein in the MCF-7 cells.
Figure 5.
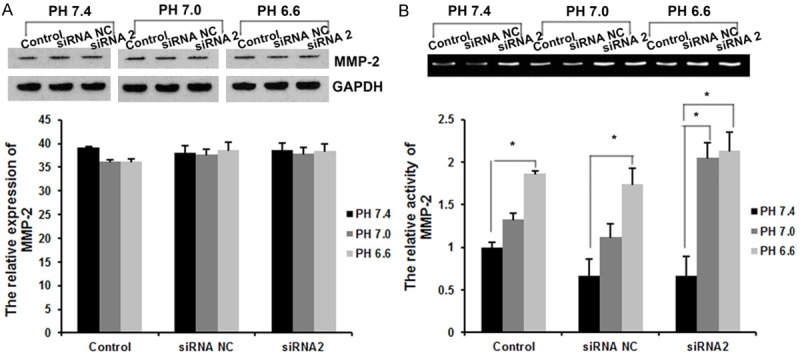
Effects of extracellular pH on MMP-2 protein. A. Western blot analysis was used to detect the expression levels of MMP-2 protein. B. The activity of MMP-2 was examined by Gelatin zymography. Data are the mean ± SD of duplicates from a representative experiment of three independent experiments.
Furthermore, we demonstrated the effects of extracellular PH on invasion and migration of MCF-7 cells. As shown in Figure 6A, acidic extracellular microenvironment (PH 6.6) dramatically increased invasion ability compared with the PH 7.0 and PH 7.4 extracellular microenvironment. The wound migration assay further confirm this observation that acidic extracellular microenvironment greatly increased migration ability compared with the PH 7.0 and PH 7.4 extracellular microenvironment (Figure 6B).
Figure 6.
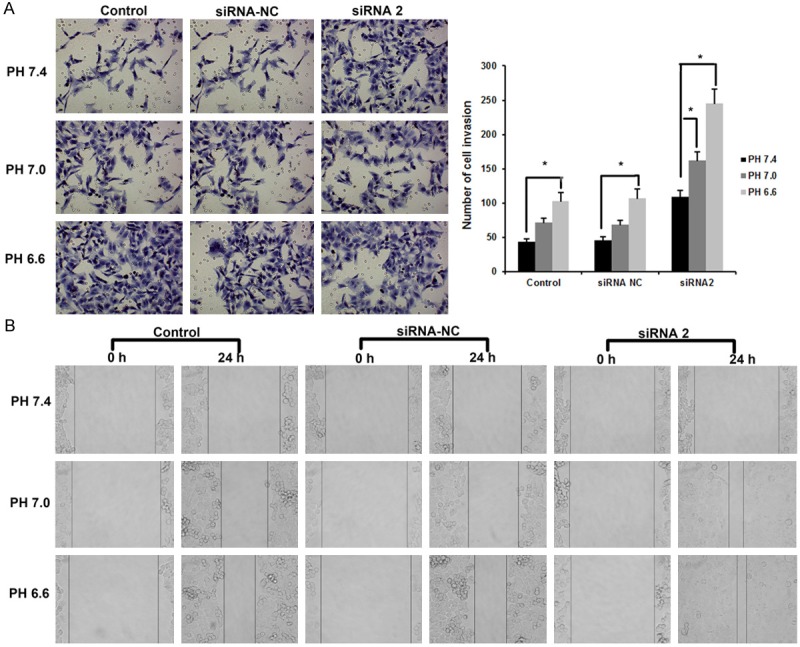
Extracellular pH affects the invasion and migration of MCF-7 cells. A. Transwell assays showed the invasion ability at extracellular PH 7.4, PH 7.0 and PH 6.6 respectively. B. Wound migration assay showed the migration ability at extracellular PH 7.4, PH 7.0 and PH 6.6 respectively. Data are the mean ± SD of duplicates from a representative experiment of three independent experiments.
Therefore, acidic extracellular microenvironment significantly increased the migration and invasion ability of MCF-7 cells.
Discussion
Tumor metastasis suppressor gene 1 (TMSG1), also known as LASS2, is a newly found tumor metastasis suppressor gene. Accumulating evidences showed that it not only suppressed tumor growth but was closely related to tumor metastasis. Recently, it has been reported that expression levels of TMSG-1 was altered in breast cancer and TMSG-1 overexpression inhibited the proliferation, invasion and apoptosis of metastatic breast cancer cells significantly [20,24]. However, as a novel TMSG, the precise molecular mechanisms of TMSG1 on breast cancer metastasis are unclear.
Therefore, to further investigate the molecular mechanisms of the antitumor effect of TMSG-1 on metastasis of breast cancer, small interference RNA (siRNA) was adopted to silence the gene expression of TMSG1 in human breast cancer cell line MCF-7. Furthermore, we confirmed that siRNA2 could significantly decreased the TMSG1 expression at the mRNA level and protein level, reduced 88.1% TMSG1 mRNA and 93.9% TMSG-1 protein. Then, we further analyzed the effects of LASS2 silencing on invasion and migration of breast cancer. Our results showed that silencing of TMSG1 could promote the migration and invasion of MCF-7 cells, which in turn suggested that TMSG1 gene function as tumor metastasis suppressor.
It is well-known that extracellular pH is usually low in solid tumors, in contrast to the approximately neutral intracellular pH. VATPase, as ATPdependent hydrogen ion pump, pumps out H+ through transmembrane transportation and plays important roles in formation and maintenance of extracellular acidic microenvironment and metastasis [9]. Recently, Sennoune et al. showed the correlation of V-ATPases located at the plasma membrane with the metastatic potential of breast cancer cells [12]. The preferential expression of V-ATPase at the cell surface is important for the acquisition of invasiveness and metastasis of the breast cancer cells [12]. Moreover, TMSG1, as a suppressor of V-ATPase, inhibited the V-ATPase activity by interacting with ATP6V0C, the c subunit of V-ATPase proton pump [16]. Here, in order to investigate the breast tumor metastasis suppression mechanism of TMSG1, we studied the interaction between TMSG1 and V-ATPase. As shown in Figure 3, the activity of V-ATPase was greatly increased byTMSG1 siRNA in MCF-7 cells. The findings were also confirmed in previous work that TMSG1 siRNA and shRNA could enhance the activity of V-ATPase and increase extracellular H+ concentration in prostate cancer cells [15,25]. These results suggested that TMSG-1 could inhibit the activity of V-ATPase, suppress the proton transmembrane secretion and thus decrease extracellular H+ concentration in breast cancer cells. The promoting effect of V-ATPase on cancer invasion and metastasis mainly relies on its maintaining acidic pH of extracellular microenvironment, which is related to the activation, secretion, and cellular distribution of many proteases involved in the digestion of ECM [11]. The pH-sensitive proteases include cathepsin and MMPs (MMP-2, MMP-9, MMP-3, etc.). Here, we demonstrated that TMSG1 siRNA could increase the activity of MMP-2 in the supernatant of the MCF-7 cells, but had no effect on the expression and secretion of MMP-2 protein, which was consistent with previous studies by Xu et al [15,25]. These results indicated that migration of tumor cells might be regulated by MMP activity, rather than by MMP expression.
To reveal the relationship among V-ATPase-induced acidic extracellular microenvironment, tumor metastasis and MMP2 protein, we further analyzed invasion and migration capacity of MCF-7 cells at PH 6.6, PH 7.0 and PH 7.4 extracellular microenvironment. As shown in Figure 5, MMP2 expression level in MCF-7 cells was not affected by different PH cultured conditions, however, the MMP2 activity was significantly upregulated from pH 7.4 and pH 6.6 cultured conditions. Furthermore, acidic extracellular microenvironment (PH 6.6) dramatically increased invasion and migration ability compared with the PH 7.0 and PH 7.4 extracellular microenvironment. TGMS1 silencing- induced invasion and migration were reversed by high extracellular pH (PH 7.4). These data also indicated that V-ATPase played an important role in breast cancer metastasis.
In conclusion, silencing of TMSG1 promoted the formation of the extracellular acidic microenvironment by targeting V-ATPase and promoting the V-ATPase activity in breast cancer. The acidic extracellular microenvironment activated degradative enzymes, such as matrix metalloproteinases (MMPs) and degradation of ECM, ultimately accelerated tumor’s invasion and metastasis. Our findings suggest that TGMS1, as a novel tumor metastasis suppressor gene, is a promising therapeutic target for advanced breast cancer.
Acknowledgements
This work was supported by a grant of Youth Foundation of Education Bureau of Hunan Province, China (NO 13B111) and a grant of Innovation Center Foundation of the Higher Education Institutions of Hunan Province, China (NO 13K113).
Disclosure of conflict of interest
None.
References
- 1.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jabbarzadeh Kaboli P, Rahmat A, Ismail P, Ling KH. Targets and mechanisms of berberine, a natural drug with potential to treat cancer with special focus on breast cancer. Eur J Pharmacol. 2014;740:584–595. doi: 10.1016/j.ejphar.2014.06.025. [DOI] [PubMed] [Google Scholar]
- 3.Navratilova J, Hankeova T, Benes P, Smarda J. Acidic pH of tumor microenvironment enhances cytotoxicity of the disulfi ram/Cu2+ complex to breast and colon cancer cells. Chemotherapy. 2013;59:112–120. doi: 10.1159/000353915. [DOI] [PubMed] [Google Scholar]
- 4.Tong Z, Luo W, Wang Y, Yang F, Han Y, Li H, Luo H, Duan B, Xu T, Maoying Q, Tan H, Wang J, Zhao H, Liu F, Wan Y. Tumor tissue-derived formaldehyde and acidic microenvironment synergistically induce bone cancer pain. PLoS One. 2010;5:e10234. doi: 10.1371/journal.pone.0010234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liotta LA, Kohn EC. The microenvironment of the tumour-host interface. Nature. 2001;411:375–379. doi: 10.1038/35077241. [DOI] [PubMed] [Google Scholar]
- 6.Sennoune SR, Luo D, Martinez-Zaguilan R. Plasmalemmal vacuolar-type H+-ATPase in cancer biology. Cell Biochem Biophys. 2004;40:185–206. doi: 10.1385/CBB:40:2:185. [DOI] [PubMed] [Google Scholar]
- 7.Rojas JD, Sennoune SR, Maiti D, Bakunts K, Reuveni M, Sanka SC, Martinez GM, Seftor EA, Meininger CJ, Wu G, Wesson DE, Hendrix MJ, Martinez-Zaguilan R. Vacuolar-type H+-ATPases at the plasma membrane regulate pH and cell migration in microvascular endothelial cells. Am J Physiol Heart Circ Physiol. 2006;291:H1147–1157. doi: 10.1152/ajpheart.00166.2006. [DOI] [PubMed] [Google Scholar]
- 8.Torigoe T, Izumi H, Ishiguchi H, Uramoto H, Murakami T, Ise T, Yoshida Y, Tanabe M, Nomoto M, Itoh H, Kohno K. Enhanced expression of the human vacuolar H+-ATPase c subunit gene (ATP6L) in response to anticancer agents. J Biol Chem. 2002;277:36534–36543. doi: 10.1074/jbc.M202605200. [DOI] [PubMed] [Google Scholar]
- 9.Fais S, De Milito A, You H, Qin W. Targeting vacuolar H+-ATPases as a new strategy against cancer. Cancer Res. 2007;67:10627–10630. doi: 10.1158/0008-5472.CAN-07-1805. [DOI] [PubMed] [Google Scholar]
- 10.Rofstad EK, Mathiesen B, Kindem K, Galappathi K. Acidic extracellular pH promotes experimental metastasis of human melanoma cells in athymic nude mice. Cancer Res. 2006;66:6699–6707. doi: 10.1158/0008-5472.CAN-06-0983. [DOI] [PubMed] [Google Scholar]
- 11.Chung C, Mader CC, Schmitz JC, Atladottir J, Fitchev P, Cornwell ML, Koleske AJ, Crawford SE, Gorelick F. The vacuolar-ATPase modulates matrix metalloproteinase isoforms in human pancreatic cancer. Lab Invest. 2011;91:732–743. doi: 10.1038/labinvest.2011.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sennoune SR, Bakunts K, Martinez GM, Chua-Tuan JL, Kebir Y, Attaya MN, Martinez-Zaguilan R. Vacuolar H+-ATPase in human breast cancer cells with distinct metastatic potential: distribution and functional activity. Am J Physiol Cell Physiol. 2004;286:C1443–1452. doi: 10.1152/ajpcell.00407.2003. [DOI] [PubMed] [Google Scholar]
- 13.Wiedmann RM, von Schwarzenberg K, Palamidessi A, Schreiner L, Kubisch R, Liebl J, Schempp C, Trauner D, Vereb G, Zahler S, Wagner E, Muller R, Scita G, Vollmar AM. The V-ATPase-inhibitor archazolid abrogates tumor metastasis via inhibition of endocytic activation of the Rho-GTPase Rac1. Cancer Res. 2012;72:5976–5987. doi: 10.1158/0008-5472.CAN-12-1772. [DOI] [PubMed] [Google Scholar]
- 14.Mizutani Y, Kihara A, Igarashi Y. Mammalian Lass6 and its related family members regulate synthesis of specifi c ceramides. Biochem J. 2005;390:263–271. doi: 10.1042/BJ20050291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu X, Liu B, Zou P, Zhang Y, You J, Pei F. Silencing of LASS2/TMSG1 enhances invasion and metastasis capacity of prostate cancer cell. J Cell Biochem. 2014;115:731–743. doi: 10.1002/jcb.24716. [DOI] [PubMed] [Google Scholar]
- 16.Yu W, Wang L, Wang Y, Xu X, Zou P, Gong M, Zheng J, You J, Wang H, Mei F, Pei F. A novel tumor metastasis suppressor gene LASS2/TMSG1 interacts with vacuolar ATPase through its homeodomain. J Cell Biochem. 2013;114:570–583. doi: 10.1002/jcb.24400. [DOI] [PubMed] [Google Scholar]
- 17.Xu XY, Pei F, You JF. TMSG-1 and its roles in tumor biology. Chin J Cancer. 2010;29:697–702. doi: 10.5732/cjc.009.10745. [DOI] [PubMed] [Google Scholar]
- 18.Rossi MJ, Sundararaj K, Koybasi S, Phillips MS, Szulc ZM, Bielawska A, Day TA, Obeid LM, Hannun YA, Ogretmen B. Inhibition of growth and telomerase activity by novel cationic ceramide analogs with high solubility in human head and neck squamous cell carcinoma cells. Otolaryngol Head Neck Surg. 2005;132:55–62. doi: 10.1016/j.otohns.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 19.Ma C, Liu Y, Zheng J, Fang W, You J, Wang J, Cui X, Wu B. Identifi cation of tumor metastasisrelated gene TMSG-1 by mRNA differential display. Sci China C Life Sci. 2002;45:553–560. doi: 10.1360/02yc9061. [DOI] [PubMed] [Google Scholar]
- 20.Fan S, Niu Y, Tan N, Wu Z, Wang Y, You H, Ke R, Song J, Shen Q, Wang W, Yao G, Shu H, Lin H, Yao M, Zhang Z, Gu J, Qin W. LASS2 enhances chemosensitivity of breast cancer by counteracting acidic tumor microenvironment through inhibiting activity of V-ATPase proton pump. Oncogene. 2013;32:1682–1690. doi: 10.1038/onc.2012.183. [DOI] [PubMed] [Google Scholar]
- 21.Fan SH, Wang YY, Lu J, Zheng YL, Wu DM, Zhang ZF, Shan Q, Hu B, Li MQ, Cheng W. CERS2 Suppresses Tumor Cell Invasion and Is Associated with Decreased V-ATPase and MMP-2/MMP-9 activities in Breast Cancer. J Cell Biochem. 2014;116:502–513. doi: 10.1002/jcb.24978. [DOI] [PubMed] [Google Scholar]
- 22.Liang CC, Park AY, Guan JL. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc. 2007;2:329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 23.Fagan-Solis KD, Schneider SS, Pentecost BT, Bentley BA, Otis CN, Gierthy JF, Arcaro KF. The RhoA pathway mediates MMP-2 and MMP-9-independent invasive behavior in a triple-negative breast cancer cell line. J Cell Biochem. 2013;114:1385–1394. doi: 10.1002/jcb.24480. [DOI] [PubMed] [Google Scholar]
- 24.Su J, You JF, Wang JL, Cui XL, Fang WG, Zheng J. [Overexpression of tumor metastasis suppressor gene 1 suppresses proliferation and invasion, but enhances apoptosis of human breast cancer cells MDA-MB-231 cells] . Zhonghua Bing Li Xue Za Zhi. 2007;36:672–676. [PubMed] [Google Scholar]
- 25.Xu X, You J, Pei F. Silencing of a novel tumor metastasis suppressor gene LASS2/TMSG1 promotes invasion of prostate cancer cell in vitro through increase of vacuolar ATPase activity. J Cell Biochem. 2012;113:2356–2363. doi: 10.1002/jcb.24106. [DOI] [PubMed] [Google Scholar]


