Abstract
Tissue factor pathway inhibitor 2 (TFPI2) is a Kunitz-type serine proteinase inhibitor, which plays an important role in the etiology of human malignancies. DNA methylation is a common epigenetic modification of the genome that is involved in regulating many cellular processes. In addition to human papilloma virus (HPV) infection, DNA methylation may play a role in the carcinogenesis of cervical cancer. Methylation of 22 CpG sites in the promoter region of the TFPI2 gene was detected by MassARRAY spectrometry and a gene mass spectrogram was drawn using MALDI-TOF MS. HPV16 was detected by PCR. We show that aberrant methylation of TFPI2 is present in a higher proportion of invasive cervical carcinoma (ICC) clinical samples as compared to normal cervical samples in Uygur and Han. Across the four pathologic lesions of the progression of cervical cancer, ICC showed the highest level of aberrant methylation, and with a stronger correlation between CpG site and lesion grade in Uygur than in Han. Moreover, a difference in TFPI2 methylation between Uygur patients positive and negative for HPV16 infection was observed at CpG_6 (P = 0.028) and CpG_15 (P = 0.007). Altogether, these results indicate that DNA methylation of TFPI2 may play an important role in the carcinogenesis of cervical cancer and that the differential methylation of TFPI2 may at least partially explain the disparity in cervical cancer incidence between Uygur and Han women.
Keywords: TFPI2, HPV16, cervical cancer, Uygur, Han, methylation
Introduction
Cervical cancer remains a major cause of cancer mortality in women worldwide. According to GLOBOCAN estimates, approximately 528,000 patients were diagnosed with cervical cancer in 2012 (GLOBOCAN 2012). In China, there are approximately 130,000 new cases are diagnosed every year, accounting for one third of the total new cases worldwide, with 53,000 deaths attributed to the cervical cancer annually, one fourth of the total deaths worldwide [1]. Kashi, Xinjiang, inhabited by a large number of Uygur, is an area of high cervical cancer incidence in China. According to a recent report, the incidence of cervical cancer in Uygur was 622/100,000 in Xinjiang in 2008 [2], much higher than the 126.94/100,000 incidence in the Han [3].
Testing for high-risk human papilloma virus (HPV) may help to triage patients with pre-invasive disease and determine appropriate clinical intervention. However, HPV presence/absence does not solely dictate the molecular status of the cervical epithelial cell. Persistent infection with high-risk types of human papillomavirus (HR-HPV) is known to cause cervical cancer; however, additional genetic alterations are also required for disease progression [4]. DNA methylation is an important epigenetic modification of the genome that is involved in regulating many cellular processes, including embryonic development, transcription, chromatin structure, X chromosome inactivation, genomic imprinting, and chromosome stability. Consistent with these important roles, an increasing number of human cancers have been found to be associated with aberrant DNA methylation [5]. Tissue factor pathway inhibitor 2 (TFPI2), a Kunitz-type serine proteinase inhibitor associated with the extracellular matrix, has been shown to reduce tumor invasion [6]. Downregulation of TFPI2 may thus enhance the invasive potential of neoplastic cells in several cancers, including cervical cancer. The human TFPI2 gene is located on chromosome 7q22 [7] and its promoter contains a CpG island region of at least 220 bp that spans exon 1 and the three transcription initiation sites. The methylation state of TFPI2 CpG islands was first determined in a cervical cancer cell line by Sove et al., who found that the methylation detection rate of TFPI2 reached 82% while the normal control group had a rate of only 38% [6]. Kahn et al. proposed that TFPI2 may be an effective indicator to distinguish cervical cancer and cervical intraepithelial neoplasia (CIN) [8]. We hypothesize that detection of TFPI2 gene methylation to predict the development of CIN and enable early diagnosis of cervical cancer is of great significance.
As DNA methylation is an epigenetic phenomenon, populations with different genetic backgrounds may have different gene methylation [9]. Minority Uygur women residing in Xinjiang in the northwest region of China have a high incidence of cervical cancer [10]. Few reports on DNA methylation in cervical cancers of the Uygur have been published and there has been no comparison between Uygur and Han patients to date. The aim of the present study was to investigate the epigenetic status of TFPI2 in cervical cancers from patients with invasive cervical carcinomas (ICC), cervical intraepithelial neoplasm 2/3 (CIN2/3), cervical intraepithelial neoplasm 1 (CIN1) and healthy controls of both the Uygur and Han populations. In this study, Sequenom MassARRAY was utilized to evaluate the CpG methylation status of TFPI2. PCR and next-generation amplicon deep sequencing was performed to test for HPV16 infection. Our goal was to investigate the relationship between TFPI2 methylation and HPV16 infection as a means to explain the high incidence of ICC in the Uygur and identify molecular markers of ICC to enable early diagnosis.
Materials and methods
Patients
Multistage cluster sampling was utilized to randomly select 302 cases of Han and Uygur patients diagnosed with histologically confirmed ICC (55 cases of Uygur, 58 cases of Han), CIN (CIN2/3: 36 cases of Uygur, 57 cases of Han; CIN1: 11 cases of Uygur, 36 cases of Han) and normal cervical tissues (21 cases of Uygur, 28 cases of Han). All patients were referred from 2001 to 2009 and obtained from the 12th Army Hospital in Kashi, Xinjiang, Kashi District People’s Hospital, and Shihezi University School of Medicine. The samples from the normal group in these two nations were diagnosed with no cervical lesions, obtaining after total hysterectomy in patients with uterine fibroids. Surgical excision was performed according to routine protocols of clinical care. Hematoxylin-eosin and immunohistochemistry staining of slides prepared from paraffin-embedded tissue was used to confirm the diagnosis of each case. Each sample was confirmed by two experienced pathologists according to the WHO Pathology & Genetics Tumours of the Breast and Female Genital Organs (seventh edition).
DNA extraction, purification, and bisulfate modification
An EZ DNA methylation process kit was purchased from Zymo Research, Irvine, CA; a PCR MassCLEAVE Reagent kit was purchased from Sequenom, San Diego, CA. 5×TBE dNTPs (10 mM), and the PUC19DNA/Msp (Hpa II) marker were obtained from Sangon Biotech Company, Shanghai. Genomic DNA was previously extracted from paraffin-embedded tissue using a phenolchloroform protocol and resuspended. Quantification of total DNA was performed using a NanoDrop spectrophotometer (NanoDrop Products, Wilmington, DE). DNA was bisulfite treated using the EZ DNA Methylation Kit (Zymo Research) and eluted in buffer [11]. One microliter of bisulfite-treated DNA was used as a template for methylation quantification with PCR. Primers for each marker were designed to target the bisulfite-modified methylated sequences of each gene. The primer sequence for the TFPI2 gene was designed using EpiDesigner (http://www.epidesigner.com; Sequenom) while primers for β-globin and HPV16 were obtained from literature [12]. The primer sequences for the β-globin cDNA were as follows: β-globin-RT-forward: 5’-CAACTTCATCCACGTTCACC-3’, and β-globin-RT-reverse: 5’-GAAGAGCCAAGGACAGGTAC-3’, generating a 268-bp PCR product. The primer sequences for the TFPI2 cDNA were: TFPI2-RT-forward: 5’-aggaagagagAGGGGTAGGGGAGATTAGATAAGTT-3’, and TFPI2-RT-reverse: 5’-cagtaatacgactcactatagggagaaggctCAAAAATAACTACACCCACACCTTC-3’, generating a 354-bp PCR product. The primer sequences for the HPV16 cDNA were: HPV16-RT-forward: 5’-GACCCAGAAAGTTACCACAG-3’, and HPV16-RT-reverse: 5’-CACAACGGTTTGTTGTATTG-3’, generating a 268-bp PCR product. PCR conditions were 94°C for 4 minutes, 35 cycles of 94°C for 45 seconds/58°C for 45 seconds/72°C for 1 minute, and 72°C for 10 minutes. PCR products were resolved through 2% agarose gels, visualized using a transilluminator, and then analyzed by pyrosequencing using the Biotage Sample Prep kit and the forward primer for sequencing.
Quantitative MassARRAY analysis of gene methylation status
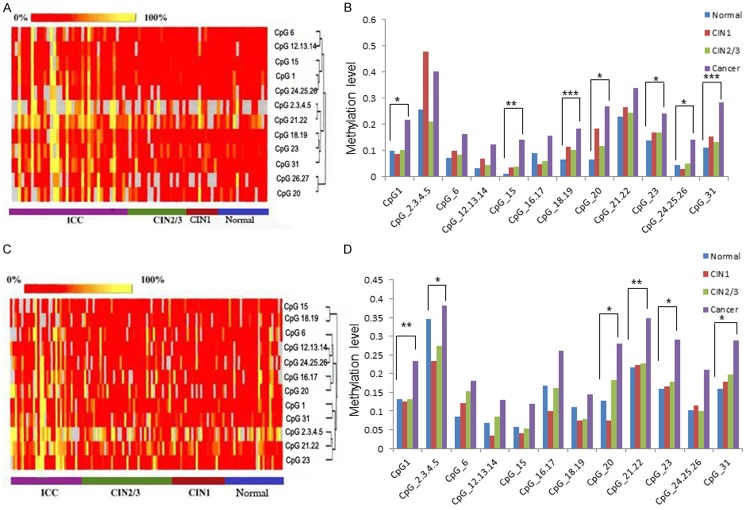
A nanodispenser was used to distribute 22 nL of the pyrolysis reaction liquid to a SpectroCHIP (Sequenom), which was loaded with the reaction substrate. Gene mass spectrograms were acquired by MassARRAY (Bruker-Sequenom) using MALDI-TOF MS and analyzed using EpiTYER® software (v1.05). Methylation detection provides the intensity of methylation of every CpG site in the TFPI2 gene, which appears as different colors. A red-to-yellow gradient equals sample methylation intensity from 0% to 100%, as in Figure 1A. CpG island sequences were obtained from the UCSC website (http://www.genome.ucsc.edu).
Figure 1.
DNA methylation status of TFPI2 in Uygur patients. A. Clustering analysis diagram of ICC, CIN2/3, CIN1 and normal lesion grades in Uygur. Yellow shows that the methylation rate is 100%, red shows 0%, gray indicates areas that have not been analyzed. Each line represents one CpG site, and each column represents one lesion grade. Clustering analysis diagram showing positive correlation of methylation level with lesion grade. B. A histogram depicting the methylation level of every CpG site with differences between the four lesion grades and the trend of the methylation indicated. C. Clustering analysis diagram in Han. Clustering analysis diagram showing positive correlation of methylation level with lesion grade. D. The histogram indicates the difference between the four lesion grades in Han. *P < 0.05, **P < 0.01, ***P < 0.001.
Statistical analysis
The TFPI2 methylation data were analyzed using EpiTYERv1.05 software, which reports a P value for each CpG site based on a comparison of the mean methylation level in each of the four groups. A Kruskal-Wallis H test was used to compare the DNA methylation status of TFPI2 between the four groups in Uygur or Han patients. A Student-Newman-Keuls analysis was used to analyze the difference between two groups in Uygur or Han patients. A student’s t-test was used to compare the TFPI2 methylation level between the Uygur and Han patients. We assessed associations between TFPI2 methylation at each CpG site and lesion grade using the Spearman rank correlation. Descriptive data about HPV16 infection were analyzed using Chi-square (X2) and t-tests were used for comparisons of DNA methylation levels between patients positive and negative for HPV16 infection. Observed differences with a P value of < 0.05 were considered statistically significant.
Results
DNA methylation status of TFPI2 in uygur patients
Based on the clustering analysis of ICC (n = 36), CIN2/3 (n = 21), CIN1 (n = 8), and normal pathology (n = 16) samples from Uygur patients, we generated a heat map, which demonstrates an increased rate of methylation in the ICC samples as compared to the other lesion classifications (Figure 1A). To determine if there was a trend in the methylation present in these lesion grades, we compared methylation at each CpG site via a histogram. As shown in Figure 1B, the methylation level is significantly different between ICC samples and the other lesion classifications at CpG_1, CpG_15, CpG_18.19, CpG_20, CpG_23, CpG_24.25.26, and CpG_31. P values of each comparison are provided in the supplementary material, Table S1. Moreover, methylation of the TFPI2 gene in ICC is higher than in CIN2/3, CIN1, and the normal pathology groups (Figure 1B).
Next, we performed a pairwise comparison of the four lesion classifications in the Uygur cohort. Striking differences were observed between ICC and the other lesion classifications at CpG_1, CpG_24.25.26, and CpG_31. Differential TFPI2 methylation levels were also found between ICC and either the normal pathology samples or CIN2/3 at CpG_15, CpG_18.19, CpG_20 and CpG_23 (Table 1).
Table 1.
Pairwise comparison of lesion grades in the Uygur cohort
| Sites | Lesion Grades | Normal | CIN1 | CIN2/3 | ICC |
|---|---|---|---|---|---|
| CpG_1 | Normal | 1 | |||
| CIN1 | 0.873 | 1 | |||
| CIN2/3 | 0.946 | 0.826 | 1 | ||
| ICC | 0.019* | 0.047* | 0.013* | 1 | |
| CpG_15 | Normal | 1 | |||
| CIN1 | 0.696 | 1 | |||
| CIN2/3 | 0.57 | 0.963 | 1 | ||
| ICC | 0.003** | 0.052 | 0.008** | 1 | |
| CpG_18.19 | Normal | 1 | |||
| CIN1 | 0.244 | 1 | |||
| CIN2/3 | 0.274 | 0.75 | 1 | ||
| ICC | 0.000*** | 0.068 | 0.004** | 1 | |
| CpG_20 | Normal | 1 | |||
| CIN1 | 0.267 | 1 | |||
| CIN2/3 | 0.527 | 0.523 | 1 | ||
| ICC | 0.008** | 0.379 | 0.034* | 1 | |
| CpG_23 | Normal | 1 | |||
| CIN1 | 0.586 | 1 | |||
| CIN2/3 | 0.497 | 0.971 | 1 | ||
| ICC | 0.009** | 0.178 | 0.046* | 1 | |
| CpG_24.25.26 | Normal | 1 | |||
| CIN11 | 0.79 | 1 | |||
| CIN2/3 | 0.935 | 0.732 | 1 | ||
| ICC | 0.024* | 0.041* | 0.019* | 1 | |
| CpG_31 | Normal | 1 | |||
| CIN12 | 0.488 | 1 | |||
| CIN2/14 | 0.665 | 0.706 | 1 | ||
| ICC | 0.000*** | 0.023* | 0.000*** | 1 |
Note: There is a significant difference between ICC and other lesion grades in these CpG sites in the Uygur cohort. Student-Newman-Keuls analysis of variance between different lesion grades in Uygur.
P < 0.05;
P < 0.01;
P < 0.001.
DNA methylation status of TFPI2 in Han patients
Based on the clustering analysis of ICC (n = 33), CIN2/3 (n = 38), CIN1 (n = 26), and normal pathology (n = 25) samples from Han patients, the TFPI2 methylation level was found to be correlated with cervical lesion grade (Figure 1C), with significant differences between the four lesion classifications at CpG_1, CpG_2.3.4.5, CpG_20, CpG_21.22, CpG_23, and CpG_31. (Figure 1D). P values are reported in the supplementary material, Table S2. Methylation of the TFPI2 gene was greater in the ICC group as compared to CIN1, CIN2/3, and normal pathology samples. Further, at CpG_21.22, CpG_23, and CpG_31, the methylation level increased with advancing pathology (Figure 1D).
Similar to the study in Uygur patients, we compared TFPI2 methylation between lesion grades in Han patients in a pairwise fashion. Interestingly, no difference was observed in the DNA methylation of TFPI2 between the normal pathology samples and CIN1 or CIN2/3, or between CIN1 and CIN2/3. At CpG_1, CpG_21.22, CpG_23, and CpG_31, a statistically significant difference (P < 0.05) was observed between ICC and the other lesion classifications. Increased methylation was observed in ICC relative to CIN1 and CIN2/3 at CpG_2.3.4.5 (P = 0.009 and 0.045, respectively). Between ICC and normal pathology samples as well as ICC and CIN1, differences in methylation were also present at CpG20 (P = 0.013 and 0.007, respectively) (Table 2).
Table 2.
Pairwise comparison of lesion grades in the Han cohort
| Sites | Lesion Grades | Normal | CIN1 | CIN2/3 | ICC |
|---|---|---|---|---|---|
| CpG_1 | Normal | 1 | |||
| CIN1 | 0.913 | 1 | |||
| CIN2/3 | 0.973 | 0.877 | 1 | ||
| ICC | 0.005** | 0.003** | 0.002** | 1 | |
| CpG_2.3.4.5 | Normal | 1 | |||
| CIN1 | 0.115 | 1 | |||
| CIN2/3 | 0.369 | 0.397 | 1 | ||
| ICC | 0.364 | 0.009** | 0.045* | 1 | |
| CpG_20 | Normal | 1 | |||
| CIN1 | 0.733 | 1 | |||
| CIN2/3 | 0.377 | 0.23 | 1 | ||
| ICC | 0.013* | 0.007** | 0.066 | 1 | |
| CpG_21.22 | Normal | 1 | |||
| CIN1 | l0.815 | 1 | |||
| CIN2/3 | 0.759 | 0.955 | 1 | ||
| ICC | 0.003** | 0.004** | 0.003** | 1 | |
| CpG_23 | Normal | 1 | |||
| CIN1 | 0.852 | 1 | |||
| CIN2/3 | 0.659 | 0.807 | 1 | ||
| ICC | 0.012* | 0.018* | 0.019* | 1 | |
| CpG_31 | Normal | 1 | |||
| CIN1 | 0.683 | 1 | |||
| CIN2/3 | 0.415 | 0.704 | 1 | ||
| ICC | 0.003** | 0.01* | 0.015* | 1 |
Note: There is a significant difference between ICC and other lesion grades for most CpG sites in the Han cohort. Student-Newman-Keuls analysis of variance between different lesion grades in the Han Chinese.
P < 0.05;
P < 0.01.
Comparison of TFPI2 methylation between Han and Uygur patients amongst four lesion classifications
Although we observed similar TFPI2 methylation profiles between Uygur and Han patients, the Uygur have an increased incidence of cervical cancer as compared to the Han [13]. We hypothesized that Uygur women may have different genetic factors accounting for their increased susceptibility to ICC compared to Han women living in the same region. To address this question, we compared the differences between the TFPI2 DNA methylation profiles of the Uygur and Han patients. Of the CpG sites compared, only CpG_31 in the normal pathology samples and CpG_24.25.26 in the CIN2/3 samples were different between the two cohorts (P = 0.044 and 0.043, respectively) (Table 3 and Figure 2C). In CIN1 and ICC groups, the differences in CpG methylation were not statistically significant (Figure 2C and 2D).
Table 3.
Differences in the methylation levels of the TFPI2 gene
| Lesion grade | CpG_1 | CpG_2.3.4.5 | CpG_6 | CpG_12.13.14 | CpG_15 | CpG_16.17 | CpG_18.19 | CpG_20 | CpG_21.22 | CpG_23 | CpG_24.25.26 | CpG_31 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Normal | t | 1.026 | 0.88 | 0.471 | 1.322 | 1.85 | 1.177 | 1.511 | 1.034 | -0.271 | 0.541 | 1.58 | 2.087 |
| P | 0.311 | 0.387 | 0.641 | 0.195 | 0.072 | 0.247 | 0.14 | 0.308 | 0.788 | 0.592 | 0.124 | 0.044* | |
| CIN1 | t | 0.944 | -1.94 | 0.473 | -1.234 | 0.521 | 0.542 | -0.861 | -0.896 | -0.646 | -0.044 | 1.469 | 0.693 |
| P | 0.352 | 0.066 | 0.64 | 0.227 | 0.606 | 0.592 | 0.396 | 0.379 | 0.523 | 0.965 | 0.153 | 0.494 | |
| CIN2/3 | t | 1.24 | 0.856 | 1.319 | 1.42 | 0.346 | 1.657 | -1.16 | 0.986 | -0.485 | 0.258 | 2.078 | 1.455 |
| P | 0.22 | 0.398 | 0.193 | 0.161 | 0.731 | 0.105 | 0.251 | 0.329 | 0.63 | 0.798 | 0.043* | 0.151 | |
| ICC | T | 0.354 | 0.039 | 0.346 | 0.225 | -0.421 | 1.695 | -1.309 | 0.354 | 0.328 | 0.931 | 1.101 | 0.18 |
| P | 0.725 | 0.969 | 0.731 | 0.823 | 0.675 | 0.095 | 0.196 | 0.725 | 0.744 | 0.355 | 0.276 | 0.858 |
Note: Comparison of methylation levels of the TFPI2 gene between Han and Uygur cohorts in different lesion grades. There is a statistical difference only for CpG_31 of normal pathology samples and CpG_24.25.26 of CIN2/3 samples. A t-test was used to analyze the differences amongst CpG sites. P values were obtained from comparisons between Han and Uygur cohorts for the methylation levels of each CpG site in the four lesion grades.
P < 0.05.
Figure 2.
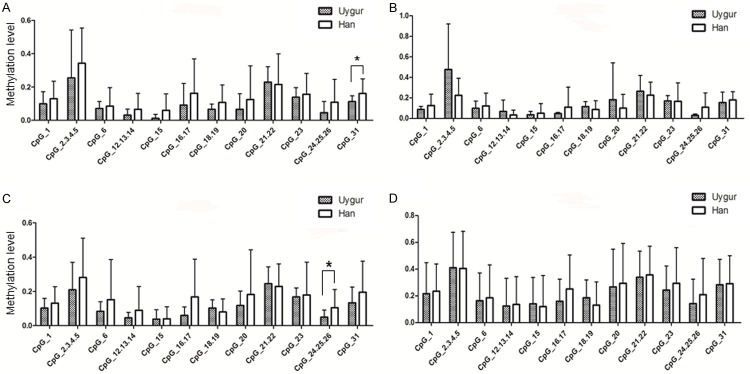
Comparison of methylation in Han and Uygur across the four lesion grades. The X axis represents every CpG site; the Y axis represents methylation level. Presented are two examples of such a contrast as shown in Table 3 using a histogram. *P < 0.05.
In order to confirm whether there is a correlation between TFPI2 methylation at every CpG site amongst the four lesion classifications, a Spearman analysis was performed. In the Uygur cohort, a correlation was found for CpG_1, CpG_12.13.14, CpG_15, CpG_18.19, CpG_20, CpG_21.22, CpG_23, and CpG_31, with strong statistical significance (P < 0.001) observed for CpG_1, CpG_15, CpG_18.19, and CpG_31 (Table 4). In contrast, in the Han cohort, such a correlation was only found for CpG_1, CpG_16.17, CpG_20, CpG_21.22, CpG_23, and CpG_31 (Table 4).
Table 4.
Correlation of methylation of each CpG site and lesion grade in Uygur and Han cohorts
| CpG_1 | CpG_2.3.4.5 | CpG_6 | CpG_12.13.14 | CpG_15 | CpG_16.17 | CpG_18.19 | CpG_20 | CpG_21.22 | CpG_23 | CpG_24.25.26 | CpG_31 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lesion grades (r) | Uygur | 0.288*** | 0.304 | 0.108 | 0.266* | 0.404*** | 0.247 | 0.413*** | 0.348** | 0.301* | 0.306** | 0.231 | 0.456*** |
| Han | 0.204* | 0.118 | 0.059 | 0.164 | 0.058 | 0.239* | -0.007 | 0.281** | 0.299** | 0.191* | 0.09 | 0.249** | |
Note: Uygur cohort has a stronger correlation between CpG site and lesion grade. Spearman rank correlation was used to analyze the relationship between CpG site and lesion grade. R value is correlation coefficient.
P < 0.05;
P < 0.01;
P < 0.001.
Association of cervical lesion grade with HPV16 infection
Based on a Chi-square test in the Uygur cohort, cervical lesion grade was associated with HPV16 infection, with an increased rate of positive HPV16 infections according to disease progression (Table S3). In contrast, the rate of positive HPV16 infections did not increase with disease progression in the Han cohort (Table S4), indicating that HPV16 infection may not be involved in the progression from CIN2/3 to cancer in Han patients. No further differences were observed between the Uygur and Han cohorts regarding HPV16 infection rate in cervical cancer (Table 5).
Table 5.
Difference in the presence of HPV16 infection between Uygur and Han cervical cancer patients
| HPV16 n (%) | X2 | P | ||
|---|---|---|---|---|
|
| ||||
| + | - | |||
| Uygur | 42 (76.4%) | 13 (23.6%) | 0.475 | 0.491 |
| Han | 23 (69.7%) | 10 (30.3%) | ||
Note: No difference was observed between the two cohorts. “+” is HPV16-positive infection, “-” is HPV16-negative infection. Chi-square test was used in this analysis.
We also tested the difference in methylation levels between ICC patients positive and negative for HPV16 infection. In the Uygur samples, three CpG sites (CpG_6, CpG_15, and CpG_23) were found have increased methylation correlated with HPV16 infection (Table 6), while, in the Han samples, there was no significant association between any of the CpG sites and HPV16 infection (Table 6). Our observations indicate an association between TFPI2 gene methylation and HPV16 infection in Uygur cervical cancer patients.
Table 6.
Differential methylation levels between the four lesion grades at each CpG site relative HPV16 infection in cervical cancer
| CpG_1 | CpG_2.3.4.5 | CpG_6 | CpG_12.13.14 | CpG_15 | CpG_16.17 | CpG_18.19 | CpG_20 | CpG_21.22 | CpG_23 | CpG_24.25.26 | CpG_31 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| t | Uygur | -1.27 | -1.81 | -2.32* | -1.65 | -2.89** | -0.05 | -0.90 | 0.01 | -1.15 | -3.12** | -0.77 | -1.50 |
| Han | -1.04 | -0.06 | 0.14 | 0.15 | -0.95 | -0.12 | -0.41 | 0.08 | -0.41 | -0.41 | 0.07 | 0.15 |
Note: Only three CpG sites are differentially methylated according to HPV16 infection in Uygur cervical cancer patients. A t-test was used to analyze difference among CpG sites.
P < 0.05;
P < 0.01.
Discussion
TFPI2 was previously identified as a tumor suppressor [14-16]; however, some studies have found TFPI2 hypermethylation in several cancer types [17], suggesting an important role for TFPI2 in the etiology of human malignancies. Accordingly, we also found a high level of TFPI2 methylation in cervical lesions with a trend of increased TFPI2 methylation from normal pathology to ICC. In the ICC group, almost every CpG site had a high level of methylation. We speculated that TFPI2 methylation is involved in cervical carcinogenesis.
We analyzed the methylation level of the TFPI2 gene in different lesion grades in Uygur and Han patient samples by a clustering analysis and observed a higher TFPI2 methylation level in ICC compared to the other lesion grades. Based on the results of the Kruskal-Wallis H test of ICC, CIN2/3, CIN1, and normal pathology samples, a statistically significant difference in TFPI2 methylation was found between the four lesion grades. A previous study reported that gene hypermethylation is associated with cervical cancer in Uygur women in Xinjiang [18]. Sova et al. found that the TFPI2 gene had a high frequency of aberrant methylation in ICC specimens [19]. The present study confirms these results by demonstrating TFPI2 methylation in cervical cancer. As we found a trend in increased TFPI2 methylation from normal pathology to ICC, we wanted to further elucidate the differences between lesion grades on the progression from normal pathology to ICC.
Next, we performed pairwise comparisons of TFPI2 methylation between the four lesion grades separately in the Uygur and Han cohorts, with a statistically significant difference (P < 0.05) observed between ICC and the other groups, especially at CpG_1, CpG_20, CpG_23, and CpG_31, which also were statistically significant between the two cohorts. These results demonstrate that DNA methylation of the TFPI2 gene may be involved in the tumorigenesis of cervical cancer.
We specifically focused on an area of high cervical cancer incidence in Xinjiang, China inhabited by a large number of Uygur people, of whom the incidence of ICC was 622/100,000 in 2008 [20], much higher than the incidence in the Han [2]. We speculated that TFPI2 methylation in different lesion grades was likely relative to ethnic differences. There have been few reports of the differences in the epigenetics of these populations. We analyzed the difference in TFPI2 methylation between the Uygur and Han cohorts across the four lesion classifications. Statistical significance was thus demonstrated only for CpG_31 of normal pathology samples and CpG_24.25.26 of CIN2/3 samples. We first employed a Student’s t-test, a rapid assay that allowed us to evaluate the differential methylation level of each CpG site amongst all Uygur and Han patients. The results suggest that there is a weak difference in TFPI2 methylation between Uygur and Han patients in the process of tumor formation. This weak difference may play a role in the ethnic disparity of cervical cancer incidence.
In order to elucidate the pathogenesis of cervical cancer, we analyzed the correlation of TFPI2 methylation between each CpG site and lesion grade in the Uygur and Han cohorts. This was further supported by a Spearman analysis, which revealed a stronger correlation between CpG site and lesion grade in Uygur patients than in Han patients. This may be particularly of importance in explaining the high incidence of cervical cancer in the Uygur people.
It is widely accepted that HPV is the main etiological factor determining cervical cancer development [21]. In the present study, we found no difference in HPV16 infection rates between the ICC patients of both cohorts. We also evaluated the difference between the four lesion grades in each cohort and found a significant difference in HPV16 infection rate across the four lesion grades in the Uygur cohort, further verifying the previous conclusion that HPV16 infection and its integration in host genome is a key event in the malignant transformation of cervical cells [22]. Earlier studies, both semi-quantitative [23] and quantitative [12,24-29], also demonstrated a correlation between HPV viral load and disease progression. In some studies, however, a synergistic effect between HPV16 infection and other factors was not examined. We compared the presence of HPV16 infection and TFPI2 DNA methylation using a Student’s t-test. The results revealed that three CpG sites (CpG_6, CpG_15, and CpG_23) were differentially methylated in Uygur; however, there was no difference in the Han patients (Table 6). To our knowledge, this is the first report of an association between HPV infection and DNA methylation. Our data indicate a strong association between HPV16 infection in cervical cancer and TFPI2 DNA methylation, and we speculate that there is a synergistic effect of HPV16 infection and TFPI2 methylation in the progression of cervical cancer in Uygur but not Han patients.
Our results give insight into a potential mechanism responsible for the high incidence of cervical cancer in Uygur, while the reason for the differential morbidity associated with the disease amongst groups living in the same region in the northwest of China is not yet clear. Regarding breast cancer, Shan et al. [30] suggested that management strategies should be implemented to improve patient outcomes due to the differential characteristics of Uygur breast cancer patients as compared to Han breast cancer patients, including their lower survival rates. Similarly, further research into whether different ethnic groups have varying treatment strategies regarding cervical cancer is also necessary.
Thus far, most studies on cancer have been performed in urban or high-income regions and few have examined low-income populations. It is worth paying more attention to the remote western China, especially the city-Kashi in western Xinjiang where Uygur is the main resident. In addition, the majority of Kashi residents have low incomes. More than 92% of low-income women lived on US$1.00 per day or less [31]. We should focus on the higher incidence of cervical cancer in this region, because whether the high incidence of cervical cancer in Xinjiang results from geographic, economic, or genetic/epigenetic factors - or a combination thereof - remains to be fully determined.
In conclusion, TFPI2 hypermethylation may play an important role in the tumorigenesis of cervical cancer. Moreover, the difference in TFPI2 methylation between Uygur and Han patients may contribute to high incidence of cervical cancer in Uygur and the disparity between the two groups. A synergistic effect of HPV16 infection and TFPI2 methylation in the progression of cervical cancer in Uygur, but not Han, patients was also determined. These results are useful in understanding the etiology of cervical cancer in the Uygur people and will aid in future studies of Uygur women’s health.
Acknowledgements
This work was funded by the Preeminent Youth Foundation of Shihezi University School of Medicine (No. 2013ZRKXYQ-YD18). The funding agencies had no role in the design, execution, and analysis of the study, or the decision to submit the paper for publication.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Mayinuer N, Liu F, Zhu KC, Dai JX, Ayeti S, Wang L, Kunduozi U. [Relationship between human papillomavirus 16 infection and the expression of p33(ING1b), human telomerase reverse transcriptase in cervical squamous cell carcinoma of Uygur female in Xinjiang Uygur Autonomous Region] . Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2007;29:592–596. [PubMed] [Google Scholar]
- 2.Chen YX, Ma CL, Chen ZF, Zhang W. [Clinical significance of miR-143 expression in women with cervical cancer of Uyghur and Han ethnicities, in Xinjiang] . Zhonghua Liu Xing Bing Xue Za Zhi. 2013;34:1120–1124. [PubMed] [Google Scholar]
- 3.Hu JM, Sun Q, Li L, Liu CX, Chen YZ, Zou H, Pang LJ, Zhao J, Yang L, Cao YW, Cui XB, Qi Y, Liang WH, Zhang WJ, Li F. Human leukocyte antigen-DRB1*1501 and DQB1*0602 alleles are cervical cancer protective factors among Uighur and Han people in Xinjiang, China. Int J Clin Exp Pathol. 2014;7:6165–6171. [PMC free article] [PubMed] [Google Scholar]
- 4.Yang HJ. Aberrant DNA methylation in cervical carcinogenesis. Chinese Journal of Cancer. 2013;32:42. doi: 10.5732/cjc.012.10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Eijk KR, de Jong S, Strengman E, Buizer-Voskamp JE, Kahn RS, Boks MP, Horvath S, Ophoff RA. Identification of schizophrenia-associated loci by combining DNA methylation and gene expression data from whole blood. Eur J Hum Genet. 2014 doi: 10.1038/ejhg.2014.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hubé F, Reverdiau P, Iochmann S, Rollin J, Cherpi-Antar C, Gruel Y. Transcriptional silencing of the TFPI-2 gene by promoter hypermethylation in choriocarcinoma cells. Biol Chem. 2003;384:1029–1034. doi: 10.1515/BC.2003.115. [DOI] [PubMed] [Google Scholar]
- 7.Rollin J, Iochmann S, Blechet C, Hube F, Regina S, Guyetant S, Lemarie E, Reverdiau P, Gruel Y. Expression and methylation status of tissue factor pathway inhibitor-2 gene in non-small-cell lung cancer. Br J Cancer. 2005;92:775–783. doi: 10.1038/sj.bjc.6602298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kahn SL, Ronnett BM, Gravitt PE, Gustafson KS. Quantitative methylation-specific PCR for the detection of aberrant DNA methylation in liquid-based Pap tests. Cancer. 2008;114:57–64. doi: 10.1002/cncr.23258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qu Y, Dang S, Hou P. Gene methylation in gastric cancer. Clin Chim Acta. 2013;424:53–65. doi: 10.1016/j.cca.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Zheng XZ, Yang AQ, Pan XL, Zheng LL, Wang XL, Zhou QY, Li XM, Yan LH, Zhang B, Li HA. Ethnicity determines association of p53Arg72Pro alleles with cervical cancer in China. European Journal of Cancer Prevention. 2008;17:460–466. doi: 10.1097/CEJ.0b013e3282f75f3e. [DOI] [PubMed] [Google Scholar]
- 11.Kisiel JB, Yab TC, Taylor WR, Chari ST, Petersen GM, Mahoney DW, Ahlquist DA. Stool DNA testing for the detection of pancreatic cancer. Cancer. 2012;118:2623–2631. doi: 10.1002/cncr.26558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andersson S, Safari H, Mints M, Lewensohn-Fuchs I, Gyllensten U, Johansson B. Type distribution, viral load and integration status of high-risk human papillomaviruses in pre-stages of cervical cancer (CIN) Br J Cancer. 2005;92:2195–2200. doi: 10.1038/sj.bjc.6602648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng XZ, Yang AQ, Pan XL, Zheng LL, Wang XL, Zhou QY, Li XM, Yan LH, Zhang B, Li HA, Jiang JF. Ethnicity determines association of p53Arg72Pro alleles with cervical cancer in China. Eur J Cancer Prev. 2008;17:460–466. doi: 10.1097/CEJ.0b013e3282f75f3e. [DOI] [PubMed] [Google Scholar]
- 14.Kempaiah P, Kisiel W. Human tissue factor pathway inhibitor-2 induces caspase-mediated apoptosis in a human fibrosarcoma cell line. Apoptosis. 2008;13:702–715. doi: 10.1007/s10495-008-0207-8. [DOI] [PubMed] [Google Scholar]
- 15.Bretz N, Noske A, Keller S, Erbe-Hofmann N, Schlange T, Salnikov AV, Moldenhauer G, Kristiansen G, Altevogt P. CD24 promotes tumor cell invasion by suppressing tissue factor pathway inhibitor-2 (TFPI-2) in a c-Src-dependent fashion. Clin Exp Metastasis. 2012;29:27–38. doi: 10.1007/s10585-011-9426-4. [DOI] [PubMed] [Google Scholar]
- 16.Gessler F, Voss V, Seifert V, Gerlach R, Kogel D. Knockdown of TFPI-2 promotes migration and invasion of glioma cells. Neurosci Lett. 2011;497:49–54. doi: 10.1016/j.neulet.2011.04.027. [DOI] [PubMed] [Google Scholar]
- 17.Glöckner SC, Dhir M, Yi JM, McGarvey KE, Van Neste L, Louwagie J, Chan TA, Kleeberger W, de Bruïne AP, Smits KM. Methylation of TFPI2 in stool DNA: a potential novel biomarker for the detection of colorectal cancer. Cancer Res. 2009;69:4691–4699. doi: 10.1158/0008-5472.CAN-08-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zuo Q, Zheng W, Zhang J, Pan Z, Liu Y, Long H, Fan P, Guo C, Li F, Shao R. Methylation in the promoters of HS3ST2 and CCNA1 genes is associated with cervical cancer in Uygur women in Xinjiang. Int J Biol Markers. 2014;29:e354–62. doi: 10.5301/jbm.5000107. [DOI] [PubMed] [Google Scholar]
- 19.Sova P, Feng Q, Geiss G, Wood T, Strauss R, Rudolf V, Lieber A, Kiviat N. Discovery of novel methylation biomarkers in cervical carcinoma by global demethylation and microarray analysis. Cancer Epidemiol Biomarkers Prev. 2006;15:114–123. doi: 10.1158/1055-9965.EPI-05-0323. [DOI] [PubMed] [Google Scholar]
- 20.Rountree MR, Bachman KE, Herman JG, Baylin SB. DNA methylation, chromatin inheritance, and cancer. Oncogene. 2001;20:3156–3165. doi: 10.1038/sj.onc.1204339. [DOI] [PubMed] [Google Scholar]
- 21.Stone SC, Rossetti RA, Lima AM, Lepique AP. HPV associated tumor cells control tumor microenvironment and leukocytosis in experimental models. Immun Inflamm Dis. 2014;2:63–75. doi: 10.1002/iid3.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shukla S, Mahata S, Shishodia G, Pande S, Verma G, Hedau S, Bhambhani S, Kumari A, Batra S, Basir SF. Physical state & copy number of high risk human papillomavirus type 16 DNA in progression of cervical cancer. Indian J Med Res. 2014;139:531–543. [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang H, Wu J, Wang B, Lu Z, Yang G. [Regulation of SOCS-3, OB, GLUT4 and PP- ARgamma gene expression by insulin and dexamethasone in porcine primary adipocyte] . Sheng Wu Gong Cheng Xue Bao. 2008;24:1354–1360. [PubMed] [Google Scholar]
- 24.Briolat J, Dalstein V, Saunier M, Joseph K, Caudroy S, Prétet JL, Birembaut P, Clavel C. HPV prevalence, viral load and physical state of HPV-16 in cervical smears of patients with different grades of CIN. Int J Cancer. 2007;121:2198–2204. doi: 10.1002/ijc.22959. [DOI] [PubMed] [Google Scholar]
- 25.Cricca M, Morselli-Labate AM, Venturoli S, Ambretti S, Gentilomi GA, Gallinella G, Costa S, Musiani M, Zerbini M. Viral DNA load, physical status and E2/E6 ratio as markers to grade HPV16 positive women for high-grade cervical lesions. Gynecol Oncol. 2007;106:549–557. doi: 10.1016/j.ygyno.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 26.Saunier M, Monnier-Benoit S, Mauny F, Dalstein V, Briolat J, Riethmuller D, Kantelip B, Schwarz E, Mougin C, Prétet JL. Analysis of human papillomavirus type 16 (HPV16) DNA load and physical state for identification of HPV16-infected women with high-grade lesions or cervical carcinoma. J Clin Microbiol. 2008;46:3678–3685. doi: 10.1128/JCM.01212-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lefevre J, Hankins C, Pourreaux K, Voyer H, Coutlée F. Real-time PCR assays using internal controls for quantitation of HPV-16 and β-globin DNA in cervicovaginal lavages. J Virol Methods. 2003;114:135–144. doi: 10.1016/j.jviromet.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 28.Flores R, Papenfuss M, Klimecki WT, Giuliano AR. Cross-sectional analysis of oncogenic HPV viral load and cervical intraepithelial neoplasia. Int J Cancer. 2006;118:1187–1193. doi: 10.1002/ijc.21477. [DOI] [PubMed] [Google Scholar]
- 29.Fiander A, Hart K, Hibbitts S, Rieck G, Tristram A, Beukenholdt R, Powell N. Variation in human papillomavirus type-16 viral load within different histological grades of cervical neoplasia. J Med Virol. 2007;79:1366–1369. doi: 10.1002/jmv.20875. [DOI] [PubMed] [Google Scholar]
- 30.Shan M, Wang X, Sun G, Ma B, Yao X, Ainy A, Ma J, Dong C, Li H, Abudukeremu M. A retrospective study of the clinical differences of Uygur breast cancer patients compared to Han breast cancer patients in the Xinjiang region of China. Int J Clin Exp Med. 2014;7:3482–3490. [PMC free article] [PubMed] [Google Scholar]
- 31.Cong L, Zhan JQ, Yang L, Zhang W, Li SG, Chen C, Zhang HY, Ma ZP, Hao XL, Simayi D, Tao L, Zhao J, Amanguli A, Mohemaiti M, Jing MX, Wang W, Saimaiti A, Zou XG, Gu Y, Li L, Wang YH, Li F, Zhang WJ. Overweight and obesity among low-income Muslim Uyghur women in far western China: correlations of body mass index with blood lipids and implications in preventive public health. PLoS One. 2014;9:e90262. doi: 10.1371/journal.pone.0090262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.