Abstract
Purpose: To reveal the potential microRNAs (miRNAs), genes, pathways and regulatory network involved in the process of nasopharyngeal carcinoma (NPC) by using the method of bioinformatics. Methods: Gene expression profiles GSE12452 (31 NPC and 10 normal samples) and GSE53819 (18 NPC and 18 normal samples), as well as miRNA expression profiles GSE32960 (312 NPC and 18 normal samples) and GSE36682 (62 NPC and 6 normal samples) were obtained from Gene Expression Omnibus database. The differentially expressed genes (DEGs) and miRNAs (DEmiRNAs) between NPC and normal samples were identified by using t-test based on MATLAB software (FDR < 0.01), followed by pathway enrichment analysis based on DAVID software (P-value < 0.1). Then, DEmiRNA-DEG regulatory network was constructed. Results: A total of 1254 DEGs and 107 DEmiRNAs were identified, respectively. Then, 16 pathways (including cell cycle) and 32 pathways (including pathways in cancer) were enriched by DEGs and target genes of DEmiRNAs, respectively. Furthermore, DEmiRNA-DEG regulatory network was constructed, containing 12 DEmiRNAs (including has-miR-615-3P) and 180 DEGs (including MCM4 and CCNE2). Conclusion: has-miR-615-3p might take part in the pathogenetic process of NPC through regulating MCM4 which is enriched in cell cycle. The DEmiRNAs identified in the present study might serve as new biomarkers for NPC.
Keywords: Nasopharyngeal carcinoma, differentially expressed genes, microRNAs, pathway enrichment, regulatory network
Introduction
Nasopharyngeal carcinoma (NPC), one of the most common cancers originating in nasopharynx, is caused by various factors like virus, environmental influences, and heredity [1]. Previous studies indicate that NPC is associated with the infection of Epstein-Barr virus (EBV) [2], consumption of salted food [3], smoking, and alcohol consumption [4]. Although NPC can be treated by surgery, chemotherapy or radiotherapy [5], the morbidity and risk of NPC is increasing, causing a significant decline in health-related life quality. In 2010, NPC resulted in 65,000 deaths globally [6], and NPC is extremely common in China [3,7]. However, the detailed biological mechanism in the development of NPC is still unclear [8].
Available data have suggested that polymorphisms of genes, including CYP2E1, XRCC1, and hOGG1, are involved in DNA damage or repair, which further participate in the process of NPC [9,10]. Studies of families at high risk of NPC have suggested that there is a linkage between DNA in chromosomal 4 and NPC [11]. Besides of variation in DNA, the dysregulation of microRNAs (miRNAs) is also implicated in the development and progression of NPC: miR-18a promotes the malignant progression by impairing microRNA biogenesis in NPC [12]; miRNA-125a-5p increased p53 protein expression in HNE-1 cells and decreased Her2 protein expression in HNE-1 and HK-1 cells [13]; miR-BART7 is highly expressed and regularly secreted into the extracellular environment of NPC cells, which is also proved to be a biomarker for the diagnosis and treatment of NPC [14]. Therefore, miRNAs and target genes might play important roles in the process of NPC, requiring further studies.
Herein, microarray data of genes and miRNAs expression from GEO (Gene Expression Omnibus) database were used in the present study. The differentially expressed genes (DEGs) and miRNAs (DEmiRNAs) were identified, followed by the KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment analysis and DEmiRNA-DEG regulatory network analysis. This study might provide evidence for the candidate genes and miRNAs involved in NPC.
Materials and methods
Microarray data
Gene expression profiles GSE12452 [15] (platform: GPL570, Affymetrix Human Genome U133 Plus 2.0 Array) and GSE53819 (platform: GPL6480, Agilent-014850 Whole Human Genome Microarray 4 × 44K G4112F) were obtained from NCBI (National Center for Biotechnology Information) GEO database (http://www.ncbi.nlm.nih.gov/geo/). A total of 31 NPC and 10 normal nasopharyngeal specimens were included in GSE12452, while 18 NPC and 18 normal nasopharyngeal specimens were included in GSE53819.
MiRNA expression profiles GSE32960 [16] (platform: GPL14722, microRNA array) and GSE36682 (platform: GPL15311, Human miRNA 1K) were obtained from NCBI GEO database as well. A total of 312 NPC and 18 normal samples were included in GSE32960, while 62 NPC and 6 normal samples were included in GSE36682. For all expression profiles, data after normalization were downloaded.
Data of miRNA-target and protein-protein interaction (PPI)
The miRTarBase (http://mirtarbase.mbc.nctu.edu.tw) database has accumulated more than fifty thousand miRNA-target interactions (MTIs), which are collected by systematically manual literature mining [17]. The Human Protein Reference Database (HPRD) [18] is a protein database accessible through the internet, storing a huge amount of PPIs. Totally, 37443 MTIs (including 596 miRNAs and 12104 target genes) were downloaded from miRTarBase, and 37080 PPIs were downloaded from HPRD.
Identification of DEGs and DEmiRNAs
DEGs and DEmiRNAs were identified by using t-test based on MATLAB software [19]. The criterion for this analysis was false discovery rate (FDR) < 0.01. In this study, DEGs represent the genes differentially expressed between NPC and normal specimens in both of GSE12452 and GSE53819, and DEGs must have same change direction (up or down) in GSE12452 and GSE53819. Similarly, DEmiRNAs represent the miRNAs differentially expressed between NPC and normal specimens in both of GSE32960 and GSE36682, and DEmiRNAs must have same change direction (up or down) in GSE32960 and GSE36682.
Pathway enrichment analysis
The KEGG database [20] contains information of how molecules or genes are networked, which is complementary to most of the existing molecular biology databases that contain the information of individual molecules or individual genes. Online software DAVID [21] consists of biological knowledgebase and analytic tools aimed at systematically extracting biological meaning from large gene lists. We used DAVID to identify significant KEGG pathways with P-value < 0.1.
Construction of DEmiRNA-DEG regulatory network
Based on the data of MTIs and PPIs, MTI-PPI network was constructed, and the identified DEGs and DEmiRNAs were mapped to MTI-PPI network. In this study, both direct regulation (miRNA-target) and indirect regulation (miRNA-target-gene with PPI) between DEmiRNAs and DEGs were remained. In doing so, DEmiRNA-DEG regulatory network was constructed, and then visualized by Cytoscape software [22].
Results
Identification of DEGs and DEmiRNAs
After DEGs screening, 1254 significant DEGs (FDR < 0.01) were found to exist in both of GSE12452 and GSE53819 and have same change direction in GSE12452 and GSE53819. Among these DEGs, 503 DEGs were significantly up-regulated, and 751 DEGs were significantly down-regulated in NPC specimens, compared with normal specimens. Furthermore, 107 significant DEmiRNAs (FDR < 0.01) were identified to exist in both of GSE32960 and GSE36682 and have same change direction in GSE32960 and GSE36682. Among these DEmiRNAs, 45 DEmiRNAs were significantly up-regulated, and 62 DEmiRNAs were significantly down-regulated. The top 10 up-regulated and down-regulated DEGs (or DEmiRNAs) were listed in Table 1.
Table 1.
Top 10 up-regulated and down-regulated DEGs (or DEmiRNAs)
| Gene ID | Gene symbol | miRNA ID | miRNA symbol | |
|---|---|---|---|---|
| Up-regulated | 100 | ADA | hsa-miR-34c-5p | hsa-miR-34c-5p |
| 128 | ADH5 | hsa-miR-145 | hsa-miR-145 | |
| 140 | ADORA3 | hsa-miR-768-3p | hsa-miR-768-3p | |
| 191 | AHCY | hsa-miR-200a | hsa-miR-200a | |
| 204 | AK2 | hsa-miR-199a-3p | hsa-miR-199a-3p | |
| 377 | ARF3 | hsa-let-7e | hsa-let-7e | |
| 468 | ATF4 | hsa-miR-34b | hsa-miR-34b | |
| 518 | ATP5G3 | hsa-miR-363 | hsa-miR-363 | |
| 526 | ATP6V1B2 | hsa-miR-26a | hsa-miR-26a | |
| 637 | BID | hsa-miR-203 | hsa-miR-203 | |
| Down-regulated | 18 | ABAT | hsa-miR-125b | hsa-miR-125b |
| 124 | ADH1A | hsa-miR-100 | hsa-miR-100 | |
| 125 | ADH1B | hsa-miR-191 | hsa-miR-191 | |
| 126 | ADH1C | hsa-miR-143 | hsa-miR-143 | |
| 131 | ADH7 | hsa-miR-451 | hsa-miR-451 | |
| 150 | ADRA2A | hsa-let-7d | hsa-let-7d | |
| 203 | AK1 | hsa-miR-421 | hsa-miR-421 | |
| 246 | ALOX15 | hsa-miR-29c | hsa-miR-29c | |
| 267 | AMFR | hsa-miR-140-3p | hsa-miR-140-3p | |
| 311 | ANXA11 | hsa-miR-26b | hsa-miR-26b |
DEGs: differentially expressed genes; DEmiRNAs: differentially expressed microRNAs.
KEGG pathways involved in NPC
The online software DAVID was used to identify significant KEGG pathways with P-value < 0.1. As a result, a total of 16 pathways were enriched by DEGs, e.g., cell cycle, p53 signaling pathway, and DNA replication. The top 10 pathways enriched by DEGs were listed in Table 2. According to MTIs, a total of 32 KEGG pathways were enriched by the target genes of DEmiRNAs, e.g., cell cycle, pathways in cancer, p53 signaling pathway, and focal adhesion. The top 10 KEGG pathways enriched by the target genes of DEmiRNAs were listed in Table 3.
Table 2.
Top 10 pathways enriched by differentially expressed genes
| Pathway ID | Pathway name | Total gene | P-value | Genes |
|---|---|---|---|---|
| hsa04110 | Cell cycle | 125 | 0.000758 | MCM4, CCNE2, CDC6, CCND2, HDAC2, etc. |
| hsa00982 | Drug metabolism | 62 | 0.001483 | GSTA1, GSTA3, CYP2B6, CYP2C8, MAOB, etc. |
| hsa05222 | Small cell lung cancer | 84 | 0.006000 | CKS1B, COL4A2, E2F3, COL4A1, PTGS2, etc. |
| hsa00450 | Selenoamino acid metabolism | 26 | 0.021307 | AHCY, GGT7, MAT2A, MARS2, PAPSS2, etc. |
| hsa00980 | Metabolism of xenobiotics by cytochrome P450 | 60 | 0.033427 | GSTA1, GSTA3, CYP2F1, CYP2B6, CYP2C8, etc. |
| hsa04640 | Hematopoietic cell lineage | 86 | 0.043065 | CR1, CD19, TFRC, FCER2, MS4A1, etc. |
| hsa00230 | Purine metabolism | 153 | 0.057698 | POLR2H, GDA, AK1, AK2, AK7, etc. |
| hsa04115 | p53 signaling pathway | 68 | 0.062786 | CCNE2, BID, CDK1, TNFRSF10B, CCND2, etc. |
| hsa03410 | Base excision repair | 35 | 0.066446 | POLD4, UNG, TDG, NEIL1, PCNA, etc. |
| hsa03050 | Proteasome | 47 | 0.073129 | PSMA2, PSMA1, PSMD14, PSMA4, PSMB3, etc. |
Table 3.
Top 10 pathways enriched by the target genes of differentially expressed microRNAs
| Pathway name | Total gene | P-value | Genes |
|---|---|---|---|
| Ribosome | 87 | 2.70E-11 | RPL13, RPL15, RPL27A, RPL36, RPS2, etc. |
| Spliceosome | 126 | 3.44E-09 | ISY1, SNRPD2, SF3A1, SF3B2, HSPA8, etc. |
| Lysine degradation | 44 | 3.74E-05 | SETDB1, DLST, EHMT1, SETD1A, SUV39H1, etc. |
| Cell cycle | 125 | 1.09E-04 | MCM4, CCNE2, CDK4, CCND2, HDAC2, etc. |
| Glycolysis/Gluconeogenesis | 60 | 0.0013266 | HK1, PGAM1, ALDOA, ALDOC, ALDH1B1, etc. |
| Pathways in cancer | 328 | 0.0017572 | HSP90AB1, E2F3, HRAS, FGF9, SPI1, etc. |
| Pancreatic cancer | 72 | 0.0030168 | E2F3, RALBP1, ERBB2, TGFBR1, SMAD4, etc. |
| Huntington’s disease | 180 | 0.0035936 | ATP5E, ATP5B, CYC1, NDUFAB1, CYTB, etc. |
| Parkinson’s disease | 128 | 0.0048767 | ATP5E, ND4, SLC25A5, ND5, ATP5B, etc. |
| Adherens junction | 77 | 0.0058749 | FGFR1, PARD3, ACTN4, ERBB2, TGFBR1, etc. |
Construction of DEmiRNA-DEG regulatory network
The 1254 DEGs and 107 DEmiRNAs were mapped to MTI-PPI network, resulting in the construction of DEmiRNA-DEG regulatory network. This network contained 253 regulatory relationships, 41 PPIs, 180 DEGs, and 12 DEmiRNAs (Figure 1). The DEGs like ADRA2A and CTPS, as well as the DEmiRNAs like hsa-miR-615-3p, hsa-miR-296-3p, and hsa-miR-342-3p had high degree in this network. Furthermore, DEGs in this network were mainly enriched in KEGG pathways like cell cycle, p53 signaling pathway, and pathways in cancer. Especially, MCM4, CCNE2, CDC6, CCND2, HDAC2, CDK4, PCNA, MAD2L1, and E2F3 were significantly enriched in cell cycle. Among these genes, MCM4, CDC6, PCNA, and MAD2L1 were regulated by hsa-miR-615-3p, CCNE2, CDK4, and E2F3 were regulated by hsa-miR-34c-5p, and CCND2 and HDAC2 were regulated by hsa-miR-342-3p.
Figure 1.
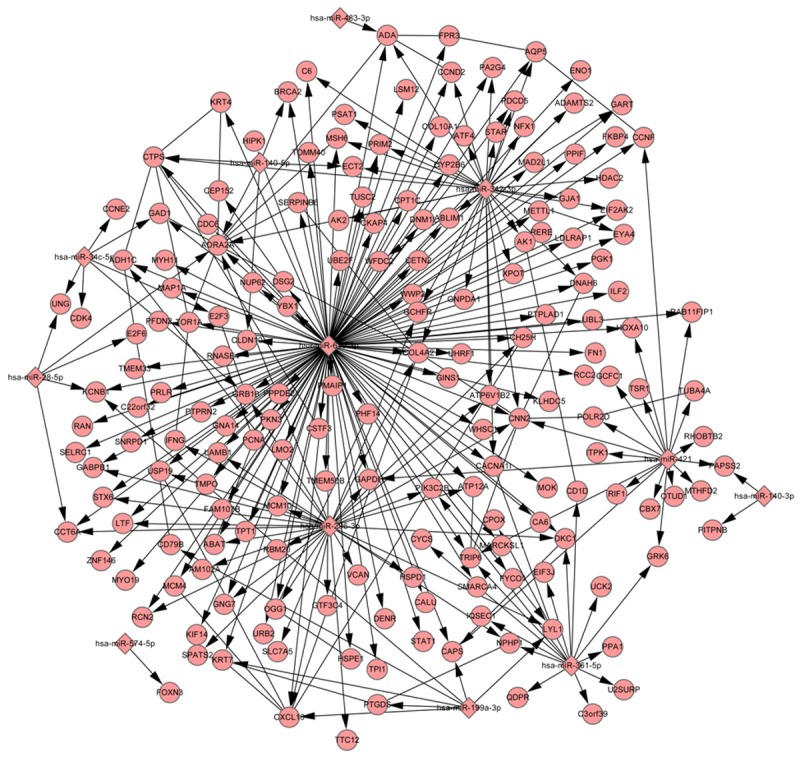
DEmiRNA-DEG regulatory network. DEmiRNA: differentially expressed microRNA; DEG: differentially expressed gene; diamond node represents DEmiRNA; circular node represents DEG; line with arrow represents the regulatory interaction between DEmiRNA and DEG; line without arrow represents protein-protein interaction between DEGs.
Discussion
NPC is one of the most common cancers originating in nasopharynx worldwide. Previous studies indicate that some genes and miRNAs play important roles in the process of NPC. In the present research, a series of bioinformatics analyses were performed based on two human nasopharyngeal gene expression profiles and two human nasopharyngeal miRNAs expression profiles. Consequently, 1254 DEGs were both existed in two gene expression profiles, and significantly enriched in 16 pathways. A total of 107 DEmiRNAs were both existed in two miRNAs expression profiles, and their target genes were significantly enriched in 32 pathways. Furthermore, the DEmiRNA-DEG regulatory network was constructed, involving 180 DEGs and 12 DEmiRNAs. Especially, MCM4, CCNE2, CDC6, CCND2, HDAC2, CDK4, PCNA, MAD2L1, and E2F3 in the regulatory network were significantly enriched in cell cycle.
MCM4 codes a member of highly conserved mini-chromosome maintenance proteins (MCM) that are essential for the initiation of eukaryotic genome replication [23]. Watanabe et al. have reported that MCM4 mutation can cause tumors in mouse through affecting the formation of MCM4/6/7 complex [24]. The partial MCM4 deficiency can result in natural killer cell deficiency and cancer [25-27]. CCNE2 (cyclin E2), which is encoded by the CCNE2 gene in humans, plays a critical role in the G1/S portion of cell cycle [28]. CCNE2 increased proportion of abnormal mitoses, micronuclei and chromosomal aberrations in cancer setting [29]. In the present study, the potential NPC-related genes including MCM4 and CCNE2 were enriched in cell cycle and p53 signaling pathway which are both associated with the pathogenesis of NPC [30-33]. Thus, the results of pathway enrichment analysis in the present study were consistent with previous studies, and we speculated that the regulation of DEGs involved in these pathways might have a positive effect on NPC inhibition and treatment.
MiRNAs are post-transcriptional regulators of gene expression with critical functions in health and disease [34]. Genome-wide analyses of radio resistance-associated miRNAs expression profile in NPC have shown the important relationship between miRNAs and NPC [35,36]. In the present study, 9 DEGs (including MCM4, CCNE2, CDC6, CCND2, HDAC2, CDK4, PCNA, MAD2L1, and E2F3) involved in cell cycle were regulated by has-miR-615-3p, hsa-miR-34c-5p and has-miR-342-3p. These DEmiRNAs were estimated to regulate the process of NPC through targeting these DEGs. Thus, the DEmiRNAs identified in the present study might serve as new biomarkers for NPC.
In conclusion, the DEGs (including MCM4 and CCNE2) enriched in the biological pathways like cell cycle and p53 signaling pathway were found to be related with NPC. Furthermore, DEmiRNAs including has-miR-615-3p, hsa-miR-34c-5p and has-miR-342-3p might take part in the process of NPC. However, further studies were required to validate these predictions.
Disclosure of conflict of interest
None.
References
- 1.Zhang F, Zhang J. Clinical hereditary characteristics in nasopharyngeal carcinoma through Ye-Liang’s family cluster. Chin Med J (Engl) 1999;112:2. [PubMed] [Google Scholar]
- 2.Lo K, Chung G, To K. Deciphering the molecular genetic basis of NPC through molecular, cytogenetic, and epigenetic approaches. Semin Cancer Biol. 2012;22:7. doi: 10.1016/j.semcancer.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 3.Chang E, Adami H. The enigmatic epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol Biomarkers Prev. 2006;15:12. doi: 10.1158/1055-9965.EPI-06-0353. [DOI] [PubMed] [Google Scholar]
- 4.Cheng Y, Hildesheim A, Hsu M, Chen I, Brinton L, Levine P, Chen C, Yang C. Cigarette smoking, alcohol consumption and risk of nasopharyngeal carcinoma in Taiwan. Cancer Causes Control. 1999;10:6. doi: 10.1023/a:1008893109257. [DOI] [PubMed] [Google Scholar]
- 5.Brennan B. Nasopharyngeal carcinoma. Orphanet J Rare Dis. 2006;1:5. doi: 10.1186/1750-1172-1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn S, Alvarado M, Anderson H, Anderson L, Andrews K, Atkinson C, Baddour L, Barker-Collo S, Bartels D, Bell M, Benjamin E, Bennett D, Bhalla K, Bikbov B, Bin Abdulhak A, Birbeck G, Blyth F, Bolliger I, Boufous S, Bucello C, Burch M, Burney P, Carapetis J, Chen H, Chou D, Chugh S, Coffeng L, Colan S, Colquhoun S, Colson K, Condon J, Connor M, Cooper L, Corriere M, Cortinovis M, De Vaccaro K, Couser W, Cowie B, Criqui M, Cross M, Dabhadkar K, Dahodwala N, De Leo D, Degenhardt L, Delossantos A, Denenberg J, Des Jarlais D, Dharmaratne S, Dorsey E, Driscoll T, Duber H, Ebel B, Erwin P, Espindola P, Ezzati M, Feigin V, Flaxman A, Forouzanfar M, Fowkes F, Franklin R, Fransen M, Freeman M, Gabriel S, Gakidou E, Gaspari F, Gillum R, Gonzalez-Medina D, Halasa Y, Haring D, Harrison J, Havmoeller R, Hay R, Hoen B, Hotez P, Hoy D, Jacobsen K, James S, Jasrasaria R, Jayaraman S, Johns N, Karthikeyan G, Kassebaum N, Keren A, Khoo J, Knowlton L, Kobusingye O, Koranteng A, Krishnamurthi R, Lipnick M, Lipshultz S, Ohno S, Mabweijano J, Macintyre M, Mallinger L, March L, Marks G, Marks R, Matsumori A, Matzopoulos R, Mayosi B, Mcanulty J, Mcdermott M, Mcgrath J, Mensah G, Merriman T, Michaud C, Miller M, Miller T, Mock C, Mocumbi A, Mokdad A, Moran A, Mulholland K, Nair M, Naldi L, Narayan K, Nasseri K, Norman P, O’donnell M, Omer S, Ortblad K, Osborne R, Ozgediz D, Pahari B, Pandian J, Rivero A, Padill R, Perez-Ruiz F, Perico N, Phillips D, Pierce K, Pope C, Porrini E, Pourmalek F, Raju M, Ranganathan D, Rehm J, Rein D, Remuzzi G, Rivara F, Roberts T, De León F, Rosenfeld L, Rushton L, Sacco R, Salomon J, Sampson U, Sanman E, Schwebel D, Segui-Gomez M, Shepard D, Singh D, Singleton J, Sliwa K, Smith E, Steer A, Taylor J, Thomas B, Tleyjeh I, Towbin J, Truelsen T, Undurraga E, Venketasubramanian N, Vijayakumar L, Vos T, Wagner G, Wang M, Wang W, Watt K, Weinstock M, Weintraub R, Wilkinson J, Woolf A, Wulf S, Yeh P, Yip P, Zabetian A, Zheng Z, Lopez A, Murray C, Almazroa M, Memish Z. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;30:33. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang W, Li X, Jiang Q, Liu Z, Yang H, Wang S, Xie S, Liu Q, Liu T, Huang J, Xie W, Li Z, Zhao Y, Wang E, Marincola F, Yao K. Transcriptional patterns, biomarkers and pathways characterizing nasopharyngeal carcinoma of Southern China. J Transl Med. 2008;6:32. doi: 10.1186/1479-5876-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richard C, Saul S, Lawrence W, Noel W. Modern Surgical Pathology. London: W B Saunders; 2002. [Google Scholar]
- 9.Kongruttanachok N, Sukdikul S, Setavarin S, Kerekhjanarong V, Supiyaphun P, Voravud N, Poovorawan Y, Mutirangura A. Cytochrome P450 2E1 polymorphism and nasopharyngeal carcinoma development in Thailand: a correlative study. BMC Cancer. 2001;1:4. doi: 10.1186/1471-2407-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hildesheim A, Anderson L, Chen C, Cheng Y, Brinton L, Daly A, Reed C, Chen I, Caporaso N, Hsu M, Chen J, Idle J, Hoover R, Yang C, Chhabra S. CYP2E1 genetic polymorphisms and risk of nasopharyngeal carcinoma in Taiwan. J Natl Cancer Inst. 1997;89:5. doi: 10.1093/jnci/89.16.1207. [DOI] [PubMed] [Google Scholar]
- 11.Feng B, Huang W, Shugart Y, Lee M, Zhang F, Xia J, Wang H, Huang T, Jian S, Huang P, Feng Q, Huang L, Yu X, Li D, Chen L, Jia W, Fang Y, Huang H, Zhu J, Liu X, Zhao Y, Liu W, Deng M, Hu W, Wu S, Mo H, Hong M, King M, Chen Z, Zeng Y. Genome-wide scan for familial nasopharyngeal carcinoma reveals evidence of linkage to chromosome 4. Nat Genet. 2002;31:4. doi: 10.1038/ng932. [DOI] [PubMed] [Google Scholar]
- 12.Luo Z, Dai Y, Zhang L, Jiang C, Li Z, Yang J, Mccarthy J, She X, Zhang W, Ma J, Xiong W, Wu M, Lu J, Li X, Li X, Xiang J, Li G. miR-18a promotes malignant progression by impairing microRNA biogenesis in nasopharyngeal carcinoma. Carcinogenesis. 2013;34:10. doi: 10.1093/carcin/bgs329. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Li Z, Wu L, Wang Z, Wang X, Yu Y, Zhao Q, Luo F. MiRNA-125a-5p: a regulator and predictor of gefitinib’s effect on nasopharyngeal carcinoma. Cancer Cell Int. 2014;14:24. doi: 10.1186/1475-2867-14-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang G, Zong J, Lin S, Verhoeven R, Tong S, Chen Y, Ji M, Cheng W, Tsao S, Lung M, Pan J, Chen H. Circulating Epstein-Barr virus microRNAs miR-BART7 and miR-BART13 as biomarkers for nasopharyngeal carcinoma diagnosis and treatment. Int J Cancer. 2015;136:E301–12. doi: 10.1002/ijc.29206. [DOI] [PubMed] [Google Scholar]
- 15.Dodd L, Sengupta S, Chen I, Den Boon J, Cheng Y, Westra W, Newton M, Mittl B, Mcshane L, Chen C, Ahlquist P, Hildesheim A. Genes involved in DNA repair and nitrosamine metabolism and those located on chromosome 14q32 are dysregulated in nasopharyngeal carcinoma. Cancer Epidemiol Biomarkers Prev. 2006;15:9. doi: 10.1158/1055-9965.EPI-06-0455. [DOI] [PubMed] [Google Scholar]
- 16.Zhang N, Wang X, Huo Q, Li X, Wang H, Schneider P, Hu G, Yang Q. The oncogene metadherin modulates the apoptotic pathway based on the tumor necrosis factor superfamily member TRAIL (Tumor Necrosis Factor-related Apoptosis-inducing Ligand) in breast cancer. J Biol Chem. 2013;288:11. doi: 10.1074/jbc.M112.395913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu SD, Tseng YT, Shrestha S, Lin YL, Khaleel A, Chou CH, Chu CF, Huang HY, Lin CM, Ho SY, Jian TY, Lin FM, Chang TH, Weng SL, Liao KW, Liao IE, Liu CC, Huang HD. miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2014;42:D78–85. doi: 10.1093/nar/gkt1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, Balakrishnan L, Marimuthu A, Banerjee S, Somanathan DS, Sebastian A, Rani S, Ray S, Harrys Kishore CJ, Kanth S, Ahmed M, Kashyap MK, Mohmood R, Ramachandra YL, Krishna V, Rahiman BA, Mohan S, Ranganathan P, Ramabadran S, Chaerkady R, Pandey A. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009;37:D767–772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Wang D, Li T. LIBGS: a MATLAB software package for gene selection. Int J Data Min Bioinform. 2010;4:7. doi: 10.1504/ijdmb.2010.033525. [DOI] [PubMed] [Google Scholar]
- 20.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 22.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Musahl C, Schulte D, Burkhart R, Knippers R. A human homologue of the yeast replication protein Cdc21. Interactions with other Mcm proteins. Eur J Biochem. 1995;230:5. doi: 10.1111/j.1432-1033.1995.tb20660.x. [DOI] [PubMed] [Google Scholar]
- 24.Watanabe E, Ohara R, Ishimi Y. Effect of an MCM4 mutation that causes tumours in mouse on human MCM4/6/7 complex formation. J Biochem. 2012;152:7. doi: 10.1093/jb/mvs060. [DOI] [PubMed] [Google Scholar]
- 25.Gineau L, Cognet C, Kara N, Lach F, Dunne J, Veturi U, Picard C, Trouillet C, Eidenschenk C, Aoufouchi S, Alcaïs A, Smith O, Geissmann F, Feighery C, Abel L, Smogorzewska A, Stillman B, Vivier E, Casanova J, Jouanguy E. Partial MCM4 deficiency in patients with growth retardation, adrenal insufficiency, and natural killer cell deficiency. J Clin Invest. 2012;122:11. doi: 10.1172/JCI61014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hughes C, Guasti L, Meimaridou E, Chuang C, Schimenti J, King P, Costigan C, Clark A, Metherell L. MCM4 mutation causes adrenal failure, short stature, and natural killer cell deficiency in humans. J Clin Invest. 2012;122:6. doi: 10.1172/JCI60224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shima N, Buske T, Schimenti J. Genetic screen for chromosome instability in mice: Mcm4 and breast cancer. Cell Cycle. 2007;15:5. doi: 10.4161/cc.6.10.4250. [DOI] [PubMed] [Google Scholar]
- 28.Gudas J, Payton M, Thukral S, Chen E, Bass M, Robinson M, Coats S. Cyclin E2, a novel G1 cyclin that binds Cdk2 and is aberrantly expressed in human cancers. Mol Cell Biol. 1999;19:10. doi: 10.1128/mcb.19.1.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caldon C, Sergio C, Burgess A, Deans A, Sutherland R, Musgrove E. Cyclin E2 induces genomic instability by mechanisms distinct from cyclin E1. Cell Cycle. 2013;12:11. doi: 10.4161/cc.23512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai H, Liu L, Peng Y, Wu Y, Li L. Diagnostic assessment by dynamic contrast-enhanced and diffusion-weighted magnetic resonance in differentiation of breast lesions under different imaging protocols. BMC Cancer. 2014;14:366. doi: 10.1186/1471-2407-14-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang C, Peng L, Huang T, Yang G, Chu Q, Liang Y, Cao X, Xie P, Zheng L, Huang H, Cai M, Huang J, Liu R, Zhu Z, Qian C, Huang B. Cancer stem-like cell characteristics induced by EB virus-encoded LMP1 contribute to radioresistance in nasopharyngeal carcinoma by suppressing the p53-mediated apoptosis pathway. Cancer Lett. 2014;344:11. doi: 10.1016/j.canlet.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 32.Li L, Tao Q, Jin H, Van Hasselt A, Poon F, Wang X, Zeng M, Jia W, Zeng Y, Chan A, Cao Y. The tumor suppressor UCHL1 forms a complex with p53/MDM2/ARF to promote p53 signaling and is frequently silenced in nasopharyngeal carcinoma. Clin Cancer Res. 2010;16:9. doi: 10.1158/1078-0432.CCR-09-3178. [DOI] [PubMed] [Google Scholar]
- 33.Mogi Y, Kato J, Horimoto M, Takimoto R, Murakami T, Hirayama A, Kohgo Y, Watanabe N, Niitsu Y. Close correlation between the dephosphorylation of p53 and growth suppression by transforming growth factor-beta 1 in nasopharyngeal carcinoma cells transduced with adenovirus early region genes. Jpn J Cancer Res. 1994;85:4. doi: 10.1111/j.1349-7006.1994.tb02380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steinfeld I, Navon R, Ach R, Yakhini Z. miRNA target enrichment analysis reveals directly active miRNAs in health and disease. Nucleic Acids Res. 2013;41:e45. doi: 10.1093/nar/gks1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szeto C, Lin C, Choi S, Yip T, Ngan R, Tsao G, Li Lung M. Integrated mRNA and microRNA transcriptome sequencing characterizes sequence variants and mRNA-microRNA regulatory network in nasopharyngeal carcinoma model systems. FEBS Open Bio. 2014;4:12. doi: 10.1016/j.fob.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li G, Qiu Y, Su Z, Ren S, Liu C, Tian Y, Liu Y. Genome-wide analyses of radioresistance-associated miRNA expression profile in nasopharyngeal carcinoma using next generation deep sequencing. PLoS One. 2013;8:e84486. doi: 10.1371/journal.pone.0084486. [DOI] [PMC free article] [PubMed] [Google Scholar]


