Abstract
Aims and background: Colorectal cancer (CRC) is common, especially in developed countries. CRC is a multifactorial disease influenced by both environmental and genetic factors. In this study, we investigated the role of genetic polymorphisms in the dual specificity protein phosphatase 10 (DUSP10) gene especially in sex-specific. Methods: We selected nine DUSP10 tag single nucleotide polymorphisms (tSNPs) previously reported to be associated with colorectal cancer risk of in a case-control study from Xi’an city of China. Results: In females, three SNPs were associated with decreased CRC risk: rs11118838, rs12724393, and rs908858. However, in males, only one SNP, rs908858, was associated with decreased CRC risk. Using a log-additive model, the rs11118838 “C” allele and the rs12724393 “G” allele were associated with decreased CRC risk in females, while the rs908858 “G” allele was associated with decreased CRC risk in both females and males. In addition, haplotype analysis also found “CG” and “CCT” were associated with the decreased CRC risk in females. Conclusions: Our findings suggest that DUSP10 polymorphisms influence the risk of developing CRC in Han Chinese and emphasize that sex should be considered in the design and analysis of health studies and biomedical research.
Keywords: Colorectal cancer, case-control study, DUSP10, tag single nucleotide polymorphism (tSNP)
Introduction
Colorectal cancer (CRC) is the third most common cancer and the fourth leading cause of cancer death worldwide [1]. The incidence of CRC has been increasing, especially in developed countries [2], and China is no exception [3]. Patients with advanced-stage CRC have a poor prognosis [1]. It has been shown that environment and diet affect CRC risk [4]. However, the etiology of CRC is complex and much remains unknown. The disease aggregates in families, being two-to-three times more common among first degree-relatives of cases than in those of population-based controls [5]. This suggests genetics may play an important role in determining CRC risk. Previous analyses have indicated that dual specificity protein phosphatase 10 (DUSP10/MKP-5) is frequently dysregulated in CRC [6]. DUSP10 inactivates p38 and JNK and plays a significant role in mediating innate and adaptive immunity [7]. In this study, we investigated whether DUSP10 polymorphisms were associated with CRC risk in the Han Chinese population.
Materials and methods
Study participants
We recruited 203 individuals (80 females, 123 males; median age 57 years) diagnosed with CRC at Shaanxi Province Hospital in Xi’an City, China. All subjects were newly diagnosed and at least 18 years old at the time of diagnosis, and CRC had been confirmed histologically. None of the subjects had been diagnosed with other cancers or undergone chemotherapy or radiotherapy. We selected 296 unrelated healthy individuals (112 females, 184 males; median age 53 years) as a control group, according to standard recruitment and exclusion criteria. In general, all control subjects were healthy, without chronic diseases or disorders related to vital organs. To ensure that controls were cancer-free, they were tested for the presence of plasma carcinoembryonic antigen and alpha-fetoprotein. All participants were Han Chinese, living in Xi’an City and its environs.
Demographic and clinical data
Demographic and personal data were collected through an in-person interview using a standardized epidemiological questionnaire, which collected data on age, sex, ethnicity, residential region, diet, education status, family history of cancer, etc. For patients, related information was also collected through consultation with treating physicians or medical chart reviews. All participants signed informed consent forms, and then 5 ml peripheral blood was drawn from each subject. The study protocol was approved by the Clinical Research Ethics Committees of Shaanxi Province Hospital and Northwest University.
Single nucleotide polymorphism selection and genotyping
Based on previous research on DUSP10 and CRC, we selected nine tag single nucleotide polymorphisms (tSNPs) in DUSP10 with minor allele frequencies (MAFs) > 5% in the HapMap Han Chinese population from Beijing. Genomic DNA was extracted from whole blood using the GoldMag-Mini Whole Blood Genomic DNA Purification Kit (GoldMag Co. Ltd., Xi’an City, China). DNA concentration was measured using the NanoDrop 2000 (Thermo Scientific, Waltham, Massachusetts, USA). Sequenom MassARRAY Assay Design 3.0 Software (Sequenom, San Diego, California, USA) was used to design a Multiplexed SNP MassEXTEND assay (Sequenom) [8]. A Sequenom Typer 4.0 Software was used for data management and analysis [8,9]. Laboratory personnel were blinded to genotyping results.
Statistical analysis
Microsoft Excel and the SPSS 16.0 statistical package (SPSS, Chicago, IL) were used to perform statistical analyses. All P values in this study were two-sided, and P ≤ 0.05 was considered the threshold for statistical significance. Each SNP’s frequency in controls was tested for departure from Hardy-Weinberg Equilibrium (HWE) using Fisher’s exact test. We tested for differences in tSNP genotype distribution between cases and controls using the χ2 test [10]. Odds ratios (ORs) and 95% confidence intervals (CIs), adjusted for age, were calculated using unconditional logistic-regression models [11]. We also investigated the association between genotype and CRC risk under different genetic models (co-dominant, dominant, recessive, over-dominant, and log-additive) using software available at the SNPStats website: http://bioinfo.iconcologia.net/snpstats/start.htm [12]. We then stratified by sex and analyzed the association between genotype and CRC risk using each of these models. The Haploview software package (version 4.2) and SHEsis software platform (http://www.nhgg.org/analysis/) were used to assess linkage disequilibrium, haplotype construction, and the genetic association between polymorphisms, with a D’ > 0.8 indicating that related tSNPs formed a single block [13,14].
Results
Table 1 shows the MAF of all nine SNPs genotyped; the rs12041033 SNP deviated from HWE in male controls. Using the χ2 test, we identified three susceptibility tSNPs in females and one in males that were significantly associated with CRC risk. The SNPs rs11118838 (OR = 0.58; 95% CI: 0.36-0.93; P = 0.022), rs12724393 (OR = 0.44; 95% CI: 0.23-0.85; P = 0.013), and rs908858 (OR = 0.45; 95% CI: 0.29-0.68; P = 0.0001) were all associated with decreased CRC risk in females. In males, rs908858 (OR = 0.69; 95% CI: 0.49-0.95; P = 0.025) was associated with decreased CRC risk.
Table 1.
Association between nine DUSP10 SNPs and colorectal cancer
| SNP ID | Chromosome position | Minor allele | MAFb (Cases) | MAF (Controls) | HWE P-value | OR (95% CI)a | P-value | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Females | Males | Females | Males | Females | Males | Females | Males | Females | Males | |||
| rs11118838 | 221897346 | C | 0.233 | 0.229 | 0.344 | 0.266 | 0.349 | 0.985 | 0.58 (0.36-0.93) | 0.82 (0.56-1.20) | 0.022* | 0.302 |
| rs12036163 | 221897105 | G | 0.056 | 0.049 | 0.054 | 0.071 | 0.549 | 0.228 | 1.05 (0.43-2.56) | 0.67 (0.33-1.36) | 0.909 | 0.263 |
| rs12044821 | 221898307 | T | 0.058 | 0.049 | 0.054 | 0.074 | 0.549 | 0.277 | 1.10 (0.45-2.70) | 0.65 (0.32-1.31) | 0.839 | 0.224 |
| rs12724393 | 221896525 | G | 0.084 | 0.136 | 0.174 | 0.120 | 0.359 | 0.254 | 0.44 (0.23-0.85) | 1.16 (0.72-1.89) | 0.013* | 0.541 |
| rs17010629 | 221895393 | C | 0.260 | 0.254 | 0.341 | 0.264 | 0.449 | 0.935 | 0.69 (0.43-1.07) | 0.95 (0.66-1.38) | 0.094 | 0.793 |
| rs6689705 | 221904615 | T | 0.162 | 0.240 | 0.241 | 0.231 | 0.800 | 0.451 | 0.61 (0.36-1.03) | 1.05 (0.72-1.54) | 0.064 | 0.804 |
| rs12041033 | 221894890 | G | 0.099 | 0.074 | 0.067 | 0.077 | 0.448 | 0.044 | 1.53 (0.72-3.22) | 0.97 (0.52-1.80) | 0.265 | 0.923 |
| rs6604668 | 221901624 | T | 0.384 | 0.363 | 0.428 | 0.377 | 0.607 | 0.996 | 0.83 (0.54-1.27) | 0.94 (0.67-1.32) | 0.397 | 0.733 |
| rs908858 | 221883225 | G | 0.353 | 0.397 | 0.550 | 0.489 | 0.333 | 0.138 | 0.45 (0.29-0.68) | 0.69 (0.49-0.95) | 0.0001* | 0.025* |
P ≤ 0.05;
OR: odds ratio; 95% CI: 95% confidence interval; HWE P ≤ 0.01 is excluded;
MAF: minor allele frequency.
We hypothesized that harboring the minor allele of each SNP was a risk factor, as compared to possessing the wild-type allele. The results of the various genetic models are displayed in Tables 2, 3 and 4. Using a co-dominant model, we established that the “CA” genotype (OR = 0.53; 95% CI: 0.28-0.99; P = 0.036) for rs11118838 (Table 2) and the “CG” genotype (OR = 0.42; 95% CI: 0.20-0.87; P = 0.018) for rs12724393 (Table 3) were associated with decreased CRC risk in females. In females, the “AG” (OR = 0.41; 95% CI: 0.21-0.83; P = 4e-04) and “GG” (OR = 0.17; 95% CI: 0.06-0.44; P = 4e-04) genotypes for rs908858 were also associated with decreased CRC risk in a co-dominant model, while in males, the “GG” genotype (OR = 0.45; 95% CI: 0.21-0.96; P = 0.11) was associated with decreased risk. Using a dominant model, the “CA” and “CC” genotypes for rs11118838 (OR = 0.49; 95% CI: 0.27-0.89; P = 0.018), the “CG” and “GG” genotypes for rs12724393 (OR = 0.40; 95% CI: 0.19-0.82; P = 0.0091), and the “AG” and “GG” genotypes for rs908858 (OR = 0.17; 95% CI: 0.06-0.44; P = 9e-04) were associated with decreased CRC risk in females (Tables 2, 3 and 4). In the recessive model, the “GG” genotype for rs908858 was associated with decreased CRC risk (OR = 0.30; 95% CI: 0.13-0.69; P = 0.0025; Table 4); in the log-additive model, the “C” allele of rs11118838 (OR = 0.52; 95% CI: 0.32-0.87; P = 0.01) and the “G” allele of rs12724393 (OR = 0.43; 95% CI: 0.21-0.89; P = 0.019) were associated with decreased CRC risk in females (Tables 2 and 3). The “G” allele of rs908858 (Table 4) was associated with decreased CRC risk in both females (OR = 0.41; 95% CI: 0.26-0.65; P = 1e-04) and males (OR = 0.68; 95% CI: 0.47-0.98; P = 0.036).
Table 2.
Association between rs111188381 genotypes and colorectal cancer
| Model | Genotype | Females | Males | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Controls | Cases | OR (95% CI) | P-value | Controls | Cases | OR (95% CI) | P-value | ||
| Co-dominant | A/A | 46 (41.1%) | 43 (57.3%) | 1.00 | 0.036* | 99 (53.8%) | 72 (60.0%) | 1.00 | 0.64 |
| C/A | 55 (49.1%) | 29 (38.7%) | 0.53 (0.28-0.99) | 72 (39.1%) | 41 (34.2%) | 0.78 (0.47-1.30) | |||
| C/C | 11 (9.8%) | 3 (4.0%) | 0.27 (0.07-1.04) | 13 (7.1%) | 7 (5.8%) | 0.91 (0.32-2.61) | |||
| Dominant | A/A | 46 (41.1%) | 43 (57.3%) | 1.00 | 0.018* | 99 (53.8%) | 72 (60.0%) | 1.00 | 0.36 |
| C/A-C/C | 66 (58.9%) | 32 (42.7%) | 0.49 (0.27-0.89) | 85 (46.2%) | 48 (40.0%) | 0.80 (0.49-1.30) | |||
| Recessive | A/A-C/A | 101 (90.2%) | 72 (96%) | 1.00 | 0.11 | 171 (92.9%) | 113 (94.2%) | 1.00 | 1,00 |
| C/C | 11 (9.8%) | 3 (4.0%) | 0.37 (0.10-1.36) | 13 (7.1%) | 7 (5.8%) | 1.00 (0.36-2.83) | |||
| Over-dominant | A/A-C/C | 57 (50.9%) | 46 (61.3%) | 1.00 | 0.13 | 112 (60.9%) | 79 (65.8%) | 1.00 | 0.35 |
| C/A | 55 (49.1%) | 29 (38.7%) | 0.63 (0.34-1.14) | 72 (39.1%) | 41 (34.2%) | 0.79 (0.47-1.30) | |||
| Log-additive | - | - | - | 0.52 (0.32-0.87) | 0.01* | - | - | 0.86 (0.57-1.29) | 0.46 |
P ≤ 0.05.
Table 3.
Association between rs12724393 genotypes and colorectal cancer
| Model | Genotype | Females | Males | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Controls | Cases | OR (95% CI) | P-value | Controls | Cases | OR (95% CI) | P-value | ||
| Co-dominant | C/C | 75 (67.0%) | 64 (83.1%) | 1.00 | 0.018* | 141 (76.6%) | 91 (75.2%) | 1.00 | 0.64 |
| C/G | 35 (31.2%) | 13 (16.9%) | 0.42 (0.20-0.87) | 42 (22.8%) | 27 (22.3%) | 0.95 (0.53-1.70) | |||
| G/G | 2 (1.8%) | 0 (0.0%) | 0.00 (0.00-NA) | 1 (0.5%) | 3 (2.5%) | 5.64 (0.47-67.99) | |||
| Dominant | C/C | 75 (67.0%) | 64 (83.1%) | 1.00 | 0.0091* | 141 (76.6%) | 91 (75.2%) | 1.00 | 0.36 |
| C/G-G/G | 37 (33.0%) | 13 (16.9%) | 0.40 (0.19-0.82) | 43 (23.4%) | 30 (24.8%) | 1.03 (0.59-1.82) | |||
| Recessive | C/C-C/G | 110 (98.2%) | 77 (100%) | 1.00 | 0.14 | 183 (99.5%) | 118 (97.5%) | 1.00 | 1.00 |
| G/G | 2 (1.8%) | 0 (0.0%) | 0.00 (0.00-NA) | 1 (0.5%) | 3 (2.5%) | 5.71 (0.47-68.55) | |||
| Over-dominant | C/C-G/G | 77 (68.8%) | 64 (83.1%) | 1.00 | 0.019* | 142 (77.2%) | 94 (77.7%) | 1.00 | 0.35 |
| C/G | 35 (31.2%) | 13 (16.9%) | 0.43 (0.21-0.89) | 42 (22.8%) | 27 (22.3%) | 0.93 (0.52-1.65) | |||
| Log-additive | - | - | - | 0.40 (0.20-0.80) | 0.0062* | - | - | 1.13 (0.67–1.90) | 0.66 |
P ≤ 0.05.
Table 4.
Association between rs908858 genotypes and colorectal cancer
| Model | Genotype | Females | Males | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Controls | Cases | OR (95% CI) | P-value | Controls | Cases | OR (95% CI) | P-value | ||
| Co-dominant | A/A | 20 (18.0%) | 31 (39.7%) | 1.00 | 4e-04* | 43(23.4%) | 42 (34.7%) | 1.00 | 0.11* |
| A/G | 60 (54.0%) | 39 (50.0%) | 0.41 (0.21-0.83) | 102(55.4%) | 62 (51.2%) | 0.72 (0.41-1.26) | |||
| G/G | 31 (27.9%) | 8 (10.3%) | 0.17 (0.06-0.44) | 39 (21.2%) | 17 (14.1%) | 0.45 (0.21-0.96) | |||
| Dominant | A/A | 20 (18.0%) | 31 (39.7%) | 1.00 | 9e-04* | 43 (23.4%) | 42 (34.7%) | 1.00 | 0.1 |
| A/G-G/G | 91 (82.0%) | 47 (60.3%) | 0.33 (0.17-0.64) | 141 (76.6%) | 79 (65.3%) | 0.64 (0.37-1.09) | |||
| Recessive | A/A-A/G | 80 (72.1%) | 70 (89.7%) | 1.00 | 0.0025* | 145 (78.8%) | 104 (86%) | 1.00 | 0.077 |
| G/G | 31 (27.9%) | 8 (10.3%) | 0.30 (0.13-0.69) | 39 (21.2%) | 17 (14.1%) | 0.56 (0.29-1.08) | |||
| Over-dominant | A/A-G/G | 51 (46.0%) | 39 (50.0%) | 1.00 | 0.55 | 82 (44.6%) | 59 (48.8%) | 1.00 | 0.91 |
| A/G | 60 (54.0%) | 39 (50.0%) | 0.84 (0.47-1.50) | 102 (55.4%) | 62 (51.2%) | 0.97 (0.60-1.58) | |||
| Log-additive | - | - | - | 0.41 (0.26-0.65) | 1e-04* | - | - | 0.68 (0.47-0.98) | 0.036* |
P ≤ 0.05.
Using haplotype analysis, two blocks were detected among the DUSP10 SNPs (Figure 1). Block 1 contains rs17010629 and rs12724393, and Block 2 contains rs12044821, rs6604668, and rs6689705. The associations between the DUSP10 block haplotypes and CRC risk are listed in Table 5. In females, the ‘‘CG’’ haplotype for Block 1 was associated with a decreased risk of CRC (OR = 0.39; 95% CI: 0.19-0.80; P = 0.01). In Block 2, the ‘‘CCT’’ haplotype was also associated with decreased CRC risk (OR = 0.51; 95% CI: 0.26-0.99; P = 0.047) in females.
Figure 1.
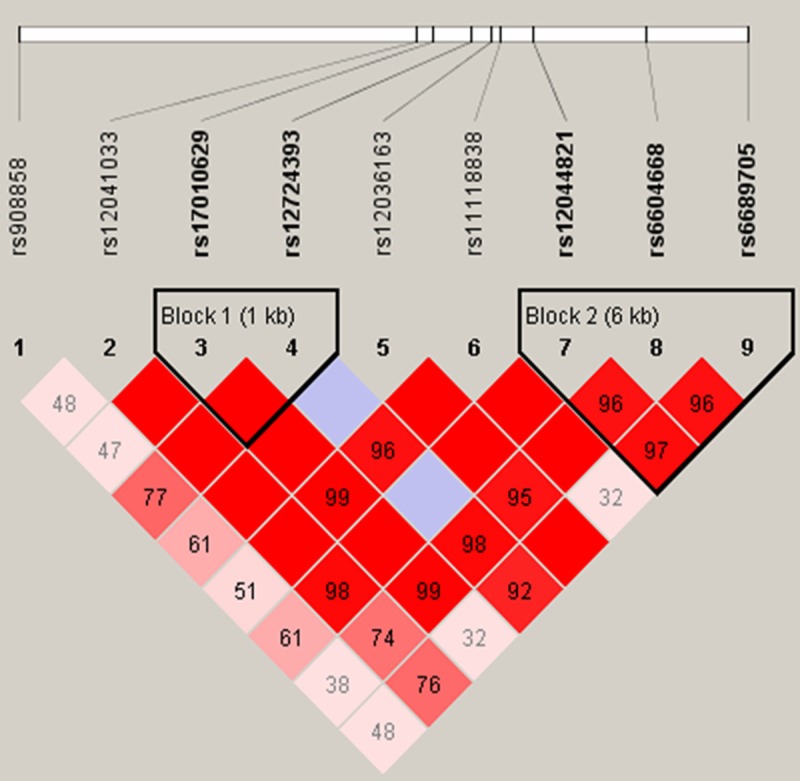
Haplotype block map for the nine DUSP10 tSNPs genotyped in this study. Block 1 includes rs17010629 and rs12724393; Block 2 includes rs12044821, rs6604668, and rs6689705.
Table 5.
DUSP10 haplotype frequencies and their association with colorectal cancer risk
| Block | Haplotype | Frequency | OR (95% CI)a | p-value | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Females | Males | Females | Males | Females | Males | ||
| 1 | TC | 0.691 | 0.739 | 1.00 | 1.00 | --- | --- |
| CC | 0.172 | 0.135 | 0.89 (0.51-1.55) | 0.98 (0.58-1.64) | 0.67 | 0.93 | |
| CG | 0.137 | 0.125 | 0.39 (0.19-0.80) | 1.10 (0.65-1.88) | 0.01* | 0.72 | |
| 2 | CTA | 0.590 | 0.620 | 1.00 | 1.00 | --- | --- |
| CCA | 0.199 | 0.165 | 0.99 (0.57-1.73) | 1.17 (0.73-1.88) | 0.98 | 0.52 | |
| CCT | 0.154 | 0.143 | 0.51 (0.26-0.99) | 1.08 (0.64-1.85) | 0.047* | 0.77 | |
| TCT | 0.057 | 0.062 | 1.08 (0.42-2.74) | 0.82 (0.40-1.68) | 0.87 | 0.6 | |
P ≤ 0.05;
OR: odd ratio; 95% CI: 95% confidence interval; Block 1 includes rs17010629 and rs12724393; Block 2 includes rs12044821, rs6604668, and rs6689705.
Discussion
In this study, we found that polymorphisms at rs11118838 and rs12724393 are associated with CRC risk in females, and rs908858 polymorphisms are associated with CRC risk in both sexes. Several previous studies have demonstrated that rs6691170 and rs66867758 polymorphisms in DUSP10 are associated with CRC risk. To date, a number of studies have also suggested that the incidence of colorectal cancer varies considerably according to age, gender, and race [15-17].
DUSP10 is a member of the dual-specificity protein phosphatase (DUSP) family. More specifically, it is a member of the dual kinase phosphatase family, which is associated with cellular proliferation and differentiation; its members have been shown to act as tumor suppressors [18]. DUSPs inactivate their target kinases by dephosphorylating both phosphoserine/threonine and phosphotyrosine residues [18]. They regulate at multiple levels, playing a role in fine-tuning signaling cascades. They negatively regulate members of the mitogen-activated protein kinase (MAPK) superfamily [19]. MAPKs are implicated in cell proliferation, survival, and migration, activities that are often dysregulated in cancer. MAPKs are also involved in tumor cell response to anti-cancer treatments, and many studies have demonstrated that abnormal MAPK signaling has important consequences for the development and progression of human cancer [19]. DUSPs have already have been implicated as major modulators of critical signaling pathways dysregulated in various diseases [18]. For example, DUSP1/MKP-1 is overexpressed in the early phases of cancer and diminishes during tumor progression [6]. Similarly, altered DUSP2/PAC-1 and DUSP7/MKP-X expression is associated with acute leukemia [20-22]. Finally, studies on lung cancer suggest that DUSP6/MKP-3 may be involved in human cancer pathogenesis [23].
There is abundant evidence that DUSP10, in particular, may play an important role in tumorigenesis. It inactivates p38 and JNK in vitro [24], and it is frequently upregulated in CRC [25]. The JNK protein is activated by the protein kinase G (PKG)/MEKK1/SEK1/JNK cascade, and this pathway is important for cell proliferation and inducing apoptosis [26]. Recent work has also implicated p38 in the promotion of cellular senescence as a means of evading oncogene-induced transformation; it participates in cell cycle regulation, functioning as a suppressor of cell proliferation and tumorigenesis [27]. Thus, the evidence suggests that DUSP10 may have an important role in cancer induction and progression. For this reason, we predicted that changes in the DUSP10 gene could lead to altered CRC risk. Just as MAPKs are regulated by MKPs, MAPK activity impinges on many of the processes involved in the initiation and genesis of cancer. Abnormal MAPK signaling plays an important role in processes critical to the development and progression of CRC. MAPK signaling also plays a key role in determining the response of tumor cells to conventional cancer therapies, and MAPK signaling pathways have been implicated in a wide range of human malignancies.
In summary, in this study of a Han Chinese sample, we identified three novel DUSP10 tSNPs associated with CRC susceptibility in females and one in males. The loci we identified are likely to provide new insights into the etiology of CRC, especially differences in risk according to sex. Further studies are required to determine the functional consequences of these polymorphisms in tumorigenesis and the biological mechanism underlying the sex differences in CRC risk that we identified.
Acknowledgements
This work was supported by the National 863 High-Technology Research and Development Program (No. 2012AA02A519) and Natural Science Basic Research Plan in Shaanxi Province of China (Program No. 2013JM4031).
Disclosure of conflict of interest
None.
References
- 1.Cui R, Okada Y, Jang SG, Ku JL, Park JG, Kamatani Y, Hosono N, Tsunoda T, Kumar V, Tanikawa C, Kamatani N, Yamada R, Kubo M, Nakamura Y, Matsuda K. Common variant in 6q26-q27 is associated with distal colon cancer in an Asian population. Gut. 2011;60:799–805. doi: 10.1136/gut.2010.215947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Center MM, Jemal A, Ward E. International Trends in Colorectal Cancer Incidence Rates. Cancer Epidemiology Biomarkers Prevention. 2009;18:1688–1694. doi: 10.1158/1055-9965.EPI-09-0090. [DOI] [PubMed] [Google Scholar]
- 3.Fengju S, Guanglin W, Kexin C. Incidence of colon cancer in Tianjin, China, 1981-2000. Asia Pac J Public Health. 2005;17:22–25. doi: 10.1177/101053950501700106. [DOI] [PubMed] [Google Scholar]
- 4.Wei EK, Giovannucci E, Wu K, Rosner B, Fuchs CS, Willett WC, Colditz GA. Comparison of risk factors for colon and rectal cancer. Int J Cancer. 2004;108:433–442. doi: 10.1002/ijc.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Marchand L. Genome-Wide Association Studies and Colorectal Cancer. Surg Oncol Clin N Am. 2009;18:663–668. doi: 10.1016/j.soc.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bermudez O, Pagès G, Gimond C. The dual-specificity MAP kinase phosphatases: critical roles in development and cancer. Am J Physiol Cell Physiol. 2010;299:189–202. doi: 10.1152/ajpcell.00347.2009. [DOI] [PubMed] [Google Scholar]
- 7.Popescu G, Robert A, Howe JR, Auerbach A. Reaction mechanism determines NMDA receptor response to repetitive stimulation. Nature. 2004;430:790–793. doi: 10.1038/nature02775. [DOI] [PubMed] [Google Scholar]
- 8.Gabriel S, Ziaugra L, Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc Hum Genet. 2009 doi: 10.1002/0471142905.hg0212s60. Chapter 2: Unit 212. [DOI] [PubMed] [Google Scholar]
- 9.Thomas RK, Baker AC, Debiasi RM, Winckler W, Laframboise T, Lin WM, Wang M, Feng W, Zander T, MacConaill L, Lee JC, Nicoletti R, Hatton C, Goyette M, Girard L, Majmudar K, Ziaugra L, Wong KK, Gabriel S, Beroukhim R, Peyton M, Barretina J, Dutt A, Emery C, Greulich H, Shah K, Sasaki H, Gazdar A, Minna J, Armstrong SA, Mellinghoff IK, Hodi FS, Dranoff G, Mischel PS, Cloughesy TF, Nelson SF, Liau LM, Mertz K, Rubin MA, Moch H, Loda M, Catalona W, Fletcher J, Signoretti S, Kaye F, Anderson KC, Demetri GD, Dummer R, Wagner S, Herlyn M, Sellers WR, Meyerson M, Garraway LA. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 10.Adamec C. [Example of the Use of the Nonparametric Test. Test X2 for Comparison of 2 Independent Examples] . Cesk Zdrav. 1964;12:613–619. [PubMed] [Google Scholar]
- 11.Bland JM, Altman DG. Statistics notes. The odds ratio. BMJ. 2000;320:1468. doi: 10.1136/bmj.320.7247.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sole X, Guino E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22:1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- 13.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 14.Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97–98. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- 15.Gao RN, Neutel CI, Wai E. Gender differences in colorectal cancer incidence, mortality, hospitalizations and surgical procedures in Canada. J Pub Health. 2008;30:194–201. doi: 10.1093/pubmed/fdn019. [DOI] [PubMed] [Google Scholar]
- 16.Abotchie PN, Vernon SW, Du XL. Gender Differences in Colorectal Cancer Incidence in the United States, 1975-2006. Journal of Women’s Health. 2012;21:393–400. doi: 10.1089/jwh.2011.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woods SE, Narayanan K, Engel A. The Influence of Gender on Colon Cancer Stage. JJ Womens Health (Larchmt) 2005;14:502–6. doi: 10.1089/jwh.2005.14.502. [DOI] [PubMed] [Google Scholar]
- 18.Patterson KI, Brummer T, O’Brien PM, Daly RJ. Dual-specificity phosphatases: critical regulators with diverse cellular targets. Biochem J. 2009;418:475–489. doi: 10.1042/bj20082234. [DOI] [PubMed] [Google Scholar]
- 19.Keyse SM. Dual-specificity MAP kinase phosphatases (MKPs) and cancer. Cancer Metastasis Rev. 2008;27:253–261. doi: 10.1007/s10555-008-9123-1. [DOI] [PubMed] [Google Scholar]
- 20.Kim SC, Hahn JS, Min YH, Yoo NC, Ko YW, Lee WJ. Constitutive activation of extracellular signal-regulated kinase in human acute leukemias: combined role of activation of MEK, hyperexpression of extracellular signal-regulated kinase, and downregulation of a phosphatase, PAC1. Blood. 1999;93:3893–3899. [PubMed] [Google Scholar]
- 21.Levy-Nissenbaum O, Sagi-Assif O, Raanani P, Avigdor A, Ben-Bassat I, Witz IP. cDNA microarray analysis reveals an overexpression of the dual-specificity MAPK phosphatase PYST2 in acute leukemia. Methods Enzymol. 2003;366:103–113. doi: 10.1016/s0076-6879(03)66009-x. [DOI] [PubMed] [Google Scholar]
- 22.Levy-Nissenbaum O, Sagi-Assif O, Raanani P, Avigdor A, Ben-Bassat I, Witz IP. Overexpression of the dual-specificity MAPK phosphatase PYST2 in acute leukemia. Cancer Lett. 2003;199:185–192. doi: 10.1016/s0304-3835(03)00352-5. [DOI] [PubMed] [Google Scholar]
- 23.Okudela K, Yazawa T, Woo T, Sakaeda M, Ishii J, Mitsui H, Shimoyamada H, Sato H, Tajiri M, Ogawa N, Masuda M, Takahashi T, Sugimura H, Kitamura H. Down-regulation of DUSP6 expression in lung cancer: its mechanism and potential role in carcinogenesis. Am J Pathol. 2009;175:867–881. doi: 10.2353/ajpath.2009.080489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanoue T, Moriguchi T, Nishida E. Molecular cloning and characterization of a novel dual specificity phosphatase, MKP-5. J Biol Chem. 1999;274:19949–19956. doi: 10.1074/jbc.274.28.19949. [DOI] [PubMed] [Google Scholar]
- 25.Nomura M, Shiiba K, Katagiri C, Kasugai I, Masuda K, Sato I, Sato M, Kakugawa Y, Nomura E, Hayashi K, Nakamura Y, Nagata T, Otsuka T, Katakura R, Yamashita Y, Sato M, Tanuma N, Shima H. Novel function of MKP-5/DUSP10, a phosphatase of stress-activated kinases, on ERK-dependent gene expression, and upregulation of its gene expression in colon carcinomas. Oncol Rep. 2012;28:931–936. doi: 10.3892/or.2012.1862. [DOI] [PubMed] [Google Scholar]
- 26.Fang JY, Richardson BC. The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 2005;6:322–327. doi: 10.1016/S1470-2045(05)70168-6. [DOI] [PubMed] [Google Scholar]
- 27.Han J, Sun P. The pathways to tumor suppression via route p38. Trends Biochem Sci. 2007;32:364–371. doi: 10.1016/j.tibs.2007.06.007. [DOI] [PubMed] [Google Scholar]


