Abstract
Objective: Previous studies have demonstrated that levels of hypermethylation of paired boxed gene 1 in cervical tissues are associated with the grades of severities of cervical neoplasia in women, which suggests that testing for DNA methylation has a potential role in neoplasma screening. In this study, by testing methylation levels of PAX1 genes in cervical scrapings and cervical tissues of different lesion levels, aims to evaluate the diagnostic value of DNA methylation testing as a biomarker for early detecting cancerous changes in cervical tissues and to compare the efficacy between PAX1 methylation test and HPV test in detecting of cervical cancer. Methods: A total of 121 cervical scrapings were analyzed, including normal (n = 28), cervical intraepithelial neoplasm 1 (CIN1; n = 32), CIN2/3 (n = 34), and invasive cancer (n = 27), which were all diagnosed by pathologic examination. Results: The values of PAX1 methylation reference in invasive cancer (mean [SE], 26.3 [3.5]) was significantly higher than CIN2/3 (13. 2 [2.2]) and the CIN1 (4.5 [0.45]; P < 0.001). The PAX1 promoter was hypermethylated in 100% of invasive cancer tissue compared with 0% of normal tissue, 9% of CIN1, 44% of CIN2/3 (P < 0.01). Methylation levels of cervical scrapings and cervical tissues represent strong consistency within each group. In contrast, the HPV test result was positive in 17% of normal tissue, 81% of CIN1, 91% of CIN2/CIN3, and 92% of invasive cancer. Based on receiver operating characteristic (ROC) analysis, hypermethylation of PAX1 was a significant candidate in segregating cervical cancer from normal/cervical neoplasia cases (P < 0.001). At an optimal cutoff value, sensitivity and specificity between 80% and 93% were obtained. In conclusion, the current results indicated that the methylation density of PAX1 by pyrosequencing in cervical scrapings held a great promise for cervical cancer screening.
Keywords: PAX1, methylation, cervical cancer, HPV, bisulfate pyrosequencing
Introduction
In women, cervical cancer is the second most common malignancy in worldwide, which remains one of the main causes of deaths in women [1,2]. Cervical cancer has a long pre-invasive phase, which can last several years. The Bethesda System is used to define the different grades of dysplasia based on the descriptive terminology [3]. The low-grade squamous intraepithelial lesion (LSIL) comprises mild dysplasia and cervical intraepithelial neoplasia grade 1 (CIN1), whereas high-grade squamous intraepithelial lesion (HSIL) includes moderate to severe dysplasia, (CIN2 and CIN3) [4]. If left to no intervention, it is about 20% CIN2 progress to CIN3, 10-40% progress to invasive cancer [5,6].
Human papilloma virus (HPV) infection is the main risk factor leading to cervical cancer [7,8]. The HPV DNA test is performed as a triage for unequivocal Papanicolaou test results and for primary screening for cervical cancer in cervical scrapings [9]. However, HPV testing may lead to unnecessary referrals for colposcopy and unwarranted concern, which decrease its value in cervical cancer screening [10]. In addition, most infections are subclinical, transient, and uncancerous, which limited HPV test in the diagnosis of cervical cancer. Therefore, the identification of novel biomarkers for cervical cancer screening is important.
It is obviously that many genetic and genes modification occur during tumorigenesis. Among those changes, histone deacetylation and aberrant promoter methylation of tumor-suppressor genes could lead to its silencing functions and the significant carcinogenesis in the development of cancer [11,12]. Since the discovery of aberrant DNA methylation patterns in cancer cells, several studies have emerged, reporting atypical promoter hypermethylation of tumor suppressor genes in cervical cancer [13-15]. The paired boxed gene 1 (PAX1) is a member of highly conserved family of developmentally controlled genes that play a role in pattern formation during embryogenesis in vertebrates at transcriptional level [16]. Several studies show that PAX1 is frequently methylation silenced in cervical and ovarian cancers [17-20]. However, the role of frequent methylation silencing of the tumor suppressor PAX1 in the development of cervical cancer, and the functional role of PAX1 in carcinogenesis remains unknown.
In the present study, we performed quantitative measurement of PAX1 gene methylation in cervical scrapings and tissues, and compared the results between cervical scrapings and tumor tissues from the same patient. We further assess the ability of PAX1 methylation to separate normal, LSIL and HSIL from squamous cell carcinoma (SCC) by using the receiver operating characteristic (ROC) analysis.
Materials and methods
Patients
Approvals for this study were obtained from the Institutional Review Boards of the Central Hospital of Minhang District, Shanghai, China. Samples were obtained from patients with informed consent. Between July 2013 and September 2014, women referred for colposcopic examination or known cervical cancer at the Central Hospital of Minhang District was enrolled in this study. Healthy women were randomly invited into our study as healthy controls from those patients who visited the hospital for routine Papanicolaou test during the same period. Cervical scrapings and cervical tissues were collected (preserved with the liquid nitrogen) and tumor suppressor genes were tested from the samples. Pathologic examination of the cervical tissues were made and set as the golden standard of diagnosis. The exclusion criteria included pregnancy, systemic viral infections, presence of other cancers, absence of the uterine cervix, history of cervical neoplasia and previously diagnosed as immune compromise diseases. A total of 121 women, including 28 controls, 32 with CIN1, 17 with CIN2, 20 with CIN3, and 27 with invasive cancers were engaged in this study.
DNA extraction
Genomic DNA was extracted from the collected cells with a TIANamp Blood DNA Kit (QIAGEN Inc, Valencia, CA) according to the manufacturer’s protocol. The amount of extracted DNA was determined by NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific Inc, Waltham, MA).
Sodium bisulphate treatment and pyrosequencing
Sodium bisulphate treated DNA was prepared using the EpiTect Bisulfate Kit (QIAGEN Inc, Valencia, CA) according to the manufacturer’s instructions. We used bisulfate pyrosequencing (BPS) to detect and quantify the patterns and levels of DNA methylation, which range from 0% to 100% at each CpG site. The BPS primers were described as following, outside forward, 5’-AAGTTTATTTTGGGTTTGGGGT-3’, outside reverse, 5’-ACCCACCTCATCAACCCTCCC-3’; inside forward, 5’-GTGGAGAGTGTTTTGGGAGGG-3’, inside reverse, 5’-AAATAACCRAAACTAAACCC-3’ (5’ cap of the primer is marked with biotin) in a nested PCR master mix reagent Kit (Roche Diagnostic, Mannheim, Germany). In nested PCR, we first used the outside primer (as described above) for amplification. The conditions were set as follows: denaturation at 95°C for 5 minutes, 20 polymerization cycles (95°C for 40 seconds, the appropriate annealing temperature for 40 seconds, and 72°C for 50 seconds), and a final extension of 72°C for 5 minutes. The product of the above PCR were set as templates for the following PCR and the inside primer were used for amplification. The conditions were the same as the above except that the polymerization cycle number were 35. The BPS followed was performed on the PSQ 96 System (Pyrosequencing, Inc, Westborough, Mass), the sequencing primers are described as follows: 5’-GGGTAGGTTTTGGAG-3’.
HPV detection
Briefly, extracted DNA was amplified with biotin labeled primers that hybridized with HPV (PGMY primers) or b2-microglobulin. An aliquot of the amplified product was visualized after agarose gel electrophoresis. The integrity of the extracted DNA and the HPV genotype were confirmed by hybridization with a strip containing probes for 27 HPV types and for the b2-microglobulin control, and visualized with streptavidin and alkaline phosphatase staining (Digene, Silver Spring, Md).
Statistical analysis
Data analysis was carried out using statistical package Statistical Package for the Social Sciences Version 19 (IBM, NY). The Fisher exact and χ2 tests were used to analyze the status of PAX1 methylation in the different groups. Receiver operating characteristic (ROC) curves were generated to confirm the accuracy of diagnosis of PAX1 methylation, and sensitivity and specificity. The statistical significance was set at P < 0.05.
Results
PAX1 methylation levels in cervical scrapings
A total of 121 women enrolled in this study, which was specific enrichment for invasive cervical cancer, CIN2/3, CIN1 and normal categories. Of 37 patients with cytologically proven CIN2/3, 32 had CIN1 and 27 had invasive cancers (Table 1). The status of methylation of the PAX1 gene was determined by pyrosequencing assays in cervical scrapings obtained from the patients with various degrees of cervical neoplasia and from the healthy controls. The levels of PAX1 methylation varied widely, depending on the extent of cervical dysplasia, and displayed a tendency closely related to the progression of cervical carcinogenesis (Figures 1, 2). Specifically, invasive cervical cancers and CIN2/3 had significant higher methylation values than the groups with less severe disease or normal controls (P < 0.001) (Table 1).
Table 1.
The values of PAX1 methylation in the cervical scrapings
| Values of PAX1 methylation | ||||
|---|---|---|---|---|
|
|
||||
| N | Age (Mean ± SE) | Range | (Mean ± SE) | |
| Normal | 28 | 36.1 ± 1.4 | 0-6.1 | 3.3 ± 0.41 |
| CIN1 | 32 | 38.3 ± 1.3 | 0-10.2 | 4.5 ± 0.45 |
| CIN2/3 | 34 | 39.7 ± 1.8 | 0-39.2 | 13.0 ± 2.2Ψ |
| Cancer | 27 | 44.4 ± 1.6 | 0-65.6 | 26.3 ± 3.5* |
P < 0.001, comparing the CIN2/3 with the Normal and CIN1 groups;
P < 0.001, comparing the invasive cancer with CIN2/3 groups.
Figure 1.
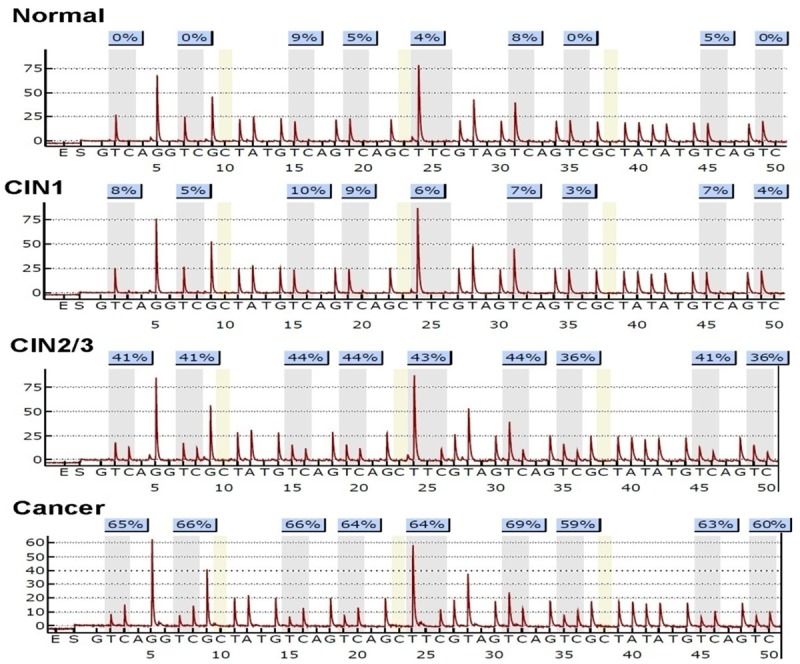
Representation of methylation status in cervical scrapings of healthy controls and cervical neoplasia. The methylation density of pyrosequencing is presented in the top of each tracing as the averaged methylation of the CpG sites analyzed. Normal (n = 28), cervical intraepithelial neoplasia grade 1 (CIN1, n = 32), cervical intraepithelial neoplasia grade 2 and 3 (CIN2/3, n = 34), cancer (n = 27).
Figure 2.
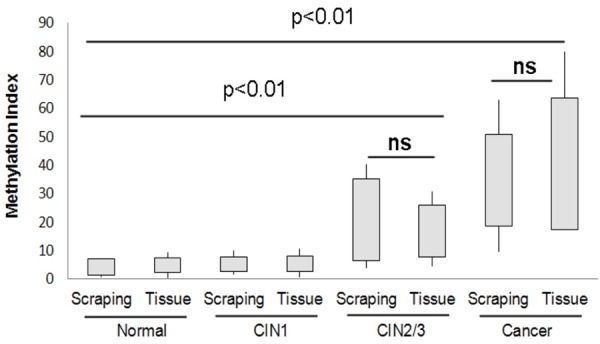
The levels of PAX1 methylation in cervical scrapings and the corresponding tissues was measured by pyrosequencing. Pyrosequencing results for methylation of PAX1 comparing cervical scrapings and tumor tissues from the same patient. Normal (n = 28), cervical intraepithelial neoplasia grade 1 (CIN1, n = 32), cervical intraepithelial neoplasia grade 2 and 3 (CIN2/3, n = 34), cancer (n = 27).
As shown in Table 2, the HPV infection rate in CIN1, CIN2/3 and cancer group was significantly higher than that of the control group, but it was not markedly different between CIN2/3 group and cancer group. Moreover, the high frequency of PAX1 methylation was observed, where 100% were methylated in cervical cancers patients. Interestingly, the percentage of PAX1 methylation in cancer group was significantly higher than that of the control group (P < 0.01). These data confirmed that PAX1 methylation assay pro- ved superior to HPV positive rate testing in the detecting of high grade lesions in cervical neoplasia.
Table 2.
The status of PAX1 methylation and HPV infection in cervical neoplasia and in healthy controls
| N | HPV positive (%) | P | PAX1 methylation | P | |
|---|---|---|---|---|---|
| Normal | 28 | 10 (36%) | 0 (0%) | ||
| CIN1 | 32 | 26 (81%) | < 0.01 | 3 (9%) | < 0.01 |
| CIN2/3 | 34 | 31 (91%) | < 0.05 | 15 (44%) | < 0.01 |
| Cancer | 27 | 25 (92%) | 27 (100%) | < 0.01 |
*Cut-off-value of 4.1.
In this study, we also measured the levels of PAX1 methylation in the corresponding tissues. Similarly, we found that PAX1 methylation correlated with cervical disease grade and was 100% positive in cervical cancers tissues (Figure 2). The PAX1 methylation levels were advanced in CIN2/3 group and cancer group as compared to control group and CIN1 group (Figure 2). There are no significant differences in methylation levels between cervical neoplasia tissues and cervical scrapings within each group, which is not statistically significant. But above all, linear regression analysis indicated that pyrosequencing methylation results were highly correlated between cervical scrapings methylation and tumor tissue methylation of PAX1 in 121 patient samples (Figure 3).
Figure 3.
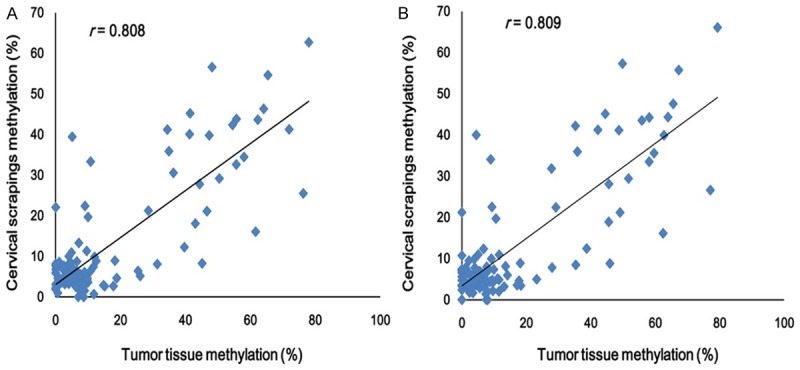
Correlation between cervical scrapings methylation and tumor tissue methylation of PAX1. Linear regression analysis comparing average cervical scrapings methylation and tumor tissue methylation levels in all CpG sites by pyrosequencing (A) and from the first three CpG sites (B) in 121 patients. R ≥ 0.8 means highly correlation between cervical scrapings and tumor tissue PAX1 methylation levels.
Receiver operating characteristic (ROC) curve analysis of PAX1
The cutoff value that distinguishes invasive cancer from noncancerous (CIN1-3 and normal tissue) was further determined on the data of PAX1 methylation rates in the subjects with different severities of cervical neoplasia. As shown in Table 2, a decreasing trend of methylation frequency was shown in invasive cancer followed by CIN2/3, CIN1 and normal tissues. In invasive cancer, the methylation rate was 100%. In CIN2/3, methylation frequency had dropped to 44%. In CIN1/normal, the methylation frequency fell further to 9% and 0% respectively (Figure 2; Table 2). At this cutoff value, specimens positive for invasive cancer were more significant than the noncancerous specimens (P < 0.001). As a comparison, the HPV test result was positive in 92% of the invasive cancer and 69% of noncancerous specimens. Interestingly, HPV was present in 81% and 17% of the CIN1 and the normal tissues, respectively, but PAX1 methylation was present in only 9% and 0%, respectively (Table 2).
In ROC analysis of methylation levels of PAX1 for cervical cancers versus normal and CIN1-3 cases, we found that the area under the curve (AUC) was 0.88, which represents the accuracy of differentiate normal/CIN1-3 and tumor samples in terms of sensitivity (81%) and specificity (93%) (P < 0.001). For segregating cervical cancer from high-grade squamous intraepithelial lesion (CIN2/3), it showed that AUC was 0.51 with a sensitivity of 32% and specificity of 90% (P < 0.01) (Figure 4).
Figure 4.
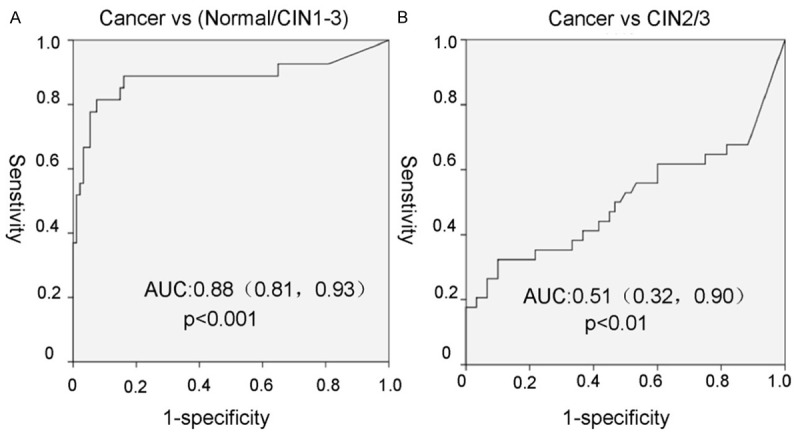
The ROC curve of PAX1 methylation for distinguishes between cervical cancer and cervical intraepithelial neoplasia. The area under the ROC curve (AUC) was calculated for the diagnosis of cancer vs. normal/CIN1-3 (A) and cancer vs. CIN2/3 (B).
Discussion
During the cervical cancer development, normal cervical cells gradually develop pre-cancerous changes that turn into cancer. Cervical cancer evolves from pre-existing noninvasive premalignant lesions referred to as cervical intraepithelial neoplasias (CINs), ranging from CIN1 (mild dysplasia) to CIN2/3 (moderate/severe dysplasia/carcinoma) [21,22]. Infection with high-risk human papillomavirus (hr-HPV) is regarded as the vector that confers susceptibility to neoplastic conversion or that directly incites transmutation to a malignant phenotype in some infected epithelial cells [23]. HPV tests are appealing in cervical cancer. However, the HPV test has low specificity and is not recommended for screening purposes when it was used alone. Thus, we needed more specific and accurate biomarkers for cervical cancer screening. In the development of cancer, hypermethylation of CpG island promoters can inactivate tumor suppressor genes, affecting genes of the cell cycle, DNA repairing, cell-to-cell interactions, apoptosis and angiogenesis [24]. In the scrapings of cervical cancer, the PAX1 gene is silenced by hypermethylation [17]. We here detected the degrees of PAX1 methylation in cervical cancer specimens, CIN2/3, CIN1 and normal specimens. We found a progressive increase in the level of PAX1 methylation in cervical scrapings in the spectrum of cervical neoplasia and a significant increase of methylation levels in CIN2/3 and invasive cancer. It suggests that hypermethylation of the PAX1 gene plays an important role in the development of cervical cancinogenesis. There were two significant findings in this report: (1) we first reported on the levels and status of PAX1 methylation by pyrosequencing assays in cervical scrapings and tumor tissues from the same patient, and the levels of PAX1 methylation was highly correlated in two adjacent tissues; (2) we found that PAX1 methylation testing might significantly improve the efficacy of cervical cancer screening.
In present practice, the Papanicolaou test is useful in screening for cervical cancer, and the HPV testing is helpful in predicting and guiding the management of equivocal or low-grade Papanicolaou test results [25]. However, owing to its high false positive rate, the HPV test cannot be used alone as a screening or diagnostic tool [25,26]. In this study, the methylation levels of the PAX1 gene was determined by pyrosequencing assays, and significantly associated with the cervical carcinogenesis and degrees of the cervix dysplasia. In addition, the methylation test of the PAX1 gene was both sensitive and specific in detecting cervical cancer from cervical scrapings, and thus could be a good candidate for testing of cervical cancer in vitro.
In this study, the method of quantitative detection of PAX1 gene methylation in cervical scrapings by pyrosequencing could well distinguish cervical lesions between low-grade and high-grade, between cancerous and noncancerous, which suggested great values of its clinical application. This method could to some extent distinguish between high-grade lesions and cervical cancer, which has brilliant prospect of development. Methylation Specific PCR (MS-PCR) on cervical tissues is a qualitative study, the result of which showed unsatisfactory sensitivity and positive predictive value [27]. Not only that, the invasive operation of cervical tissues sampling also limits its further clinical application. Our study also compared methylation levels of cervical tissues and cervical scrapings of patients in each group, the results of which represented strong consistency. There were no significant differences in methylation levels between tumor tissues and cervical scrapings within each group, which prompted that the quantitative detection of the levels of methylation of cervical scrapings could well reflect that of the tumor tissues.
In conclusion, our results demonstrated that pyrosequencing PAX1 methylation results were highly correlated between cervical scrapings and tumor tissue. In cervical scrapings, as a biomarker for cervical cancer screening in clinical application, PAX1 methylation test by pyrosequencing assays might significantly improve the efficacy of cervical cancer screening.
Acknowledgements
This work was supported by Scientific Research Funding of Shanghai Health Bureau (2011152) and Wu Jieping Foundation for Clinical Medicine Research (320. 6750. 13152).
Disclosure of conflict of interest
None.
References
- 1.Ferlay J, Forman D, Mathers CD, Bray F. Breast and cervical cancer in 187 countries between 1980 and 2010. Lancet. 2012;379:1390–1391. doi: 10.1016/S0140-6736(12)60595-9. [DOI] [PubMed] [Google Scholar]
- 2.Forouzanfar MH, Foreman KJ, Delossantos AM, Lozano R, Lopez AD, Murray CJ, Naghavi M. Breast and cervical cancer in 187 countries between 1980 and 2010: a systematic analysis. Lancet. 2011;378:1461–1484. doi: 10.1016/S0140-6736(11)61351-2. [DOI] [PubMed] [Google Scholar]
- 3.Solomon D, Davey D, Kurman R, Moriarty A, O’Connor D, Prey M, Raab S, Sherman M, Wilbur D, Wright T Jr, Young N. The 2001 Bethesda System: terminology for reporting results of cervical cytology. JAMA. 2002;287:2114–2119. doi: 10.1001/jama.287.16.2114. [DOI] [PubMed] [Google Scholar]
- 4.Wright TC Jr, Massad LS, Dunton CJ, Spitzer M, Wilkinson EJ, Solomon D. 2006 consensus guidelines for the management of women with cervical intraepithelial neoplasia or adenocarcinoma in situ. Am J Obstet Gynecol. 2007;197:340–345. doi: 10.1016/j.ajog.2007.07.050. [DOI] [PubMed] [Google Scholar]
- 5.McCredie MR, Sharples KJ, Paul C, Baranyai J, Medley G, Jones RW, Skegg DC. Natural history of cervical neoplasia and risk of invasive cancer in women with cervical intraepithelial neoplasia 3: a retrospective cohort study. Lancet Oncol. 2008;9:425–434. doi: 10.1016/S1470-2045(08)70103-7. [DOI] [PubMed] [Google Scholar]
- 6.Castle PE, Schiffman M, Wheeler CM, Solomon D. Evidence for frequent regression of cervical intraepithelial neoplasia-grade 2. Obstet Gynecol. 2009;113:18–25. doi: 10.1097/AOG.0b013e31818f5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nessa A, Rashid MH, E-Ferdous N, Chowdhury A. Screening for and management of high-grade cervical intraepithelial neoplasia in Bangladesh: a cross-sectional study comparing two protocols. J Obstet Gynaecol Res. 2013;39:564–571. doi: 10.1111/j.1447-0756.2012.01998.x. [DOI] [PubMed] [Google Scholar]
- 8.Kaufman RH, Adam E, Icenogle J, Lawson H, Lee N, Reeves KO, Irwin J, Simon T, Press M, Uhler R, Entman C, Reeves WC. Relevance of human papillomavirus screening in management of cervical intraepithelial neoplasia. Am J Obstet Gynecol. 1997;176:87–92. doi: 10.1016/s0002-9378(97)80017-8. [DOI] [PubMed] [Google Scholar]
- 9.Dillner J, Rebolj M, Birembaut P, Petry KU, Szarewski A, Munk C, de Sanjose S, Naucler P, Lloveras B, Kjaer S, Cuzick J, van Ballegooijen M, Clavel C, Iftner T. Long term predictive values of cytology and human papillomavirus testing in cervical cancer screening: joint European cohort study. BMJ. 2008;337:a1754. doi: 10.1136/bmj.a1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eide ML, Debaque H. HPV detection methods and genotyping techniques in screening for cervical cancer. Ann Pathol. 2012;32:e15–23. 401–409. doi: 10.1016/j.annpat.2012.09.231. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Chen FQ, Sun YH, Zhou SY, Li TY, Chen R. Effects of DNMT1 silencing on malignant phenotype and methylated gene expression in cervical cancer cells. J Exp Clin Cancer Res. 2011;30:98. doi: 10.1186/1756-9966-30-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki H, Maruyama R, Yamamoto E, Kai M. Epigenetic alteration and microRNA dysregulation in cancer. Front Genet. 2013;4:258. doi: 10.3389/fgene.2013.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang N, Eijsink JJ, Lendvai A, Volders HH, Klip H, Buikema HJ, van Hemel BM, Schuuring E, van der Zee AG, Wisman GB. Methylation markers for CCNA1 and C13ORF18 are strongly associated with high-grade cervical intraepithelial neoplasia and cervical cancer in cervical scrapings. Cancer Epidemiol Biomarkers Prev. 2009;18:3000–3007. doi: 10.1158/1055-9965.EPI-09-0405. [DOI] [PubMed] [Google Scholar]
- 14.Kim JH, Choi YD, Lee JS, Lee JH, Nam JH, Choi C. Assessment of DNA methylation for the detection of cervical neoplasia in liquid-based cytology specimens. Gynecol Oncol. 2010;116:99–104. doi: 10.1016/j.ygyno.2009.09.032. [DOI] [PubMed] [Google Scholar]
- 15.Chang CC, Huang RL, Wang HC, Liao YP, Yu MH, Lai HC. High methylation rate of LMX1A, NKX6-1, PAX1, PTPRR, SOX1, and ZNF582 genes in cervical adenocarcinoma. Int J Gynecol Cancer. 2014;24:201–209. doi: 10.1097/IGC.0000000000000054. [DOI] [PubMed] [Google Scholar]
- 16.McGaughran JM, Oates A, Donnai D, Read AP, Tassabehji M. Mutations in PAX1 may be associated with Klippel-Feil syndrome. Eur J Hum Genet. 2003;11:468–474. doi: 10.1038/sj.ejhg.5200987. [DOI] [PubMed] [Google Scholar]
- 17.Lai HC, Lin YW, Huang TH, Yan P, Huang RL, Wang HC, Liu J, Chan MW, Chu TY, Sun CA, Chang CC, Yu MH. Identification of novel DNA methylation markers in cervical cancer. Int J Cancer. 2008;123:161–167. doi: 10.1002/ijc.23519. [DOI] [PubMed] [Google Scholar]
- 18.Su HY, Lai HC, Lin YW, Chou YC, Liu CY, Yu MH. An epigenetic marker panel for screening and prognostic prediction of ovarian cancer. Int J Cancer. 2009;124:387–393. doi: 10.1002/ijc.23957. [DOI] [PubMed] [Google Scholar]
- 19.Huang TH, Lai HC, Liu HW, Lin CJ, Wang KH, Ding DC, Chu TY. Quantitative analysis of methylation status of the PAX1 gene for detection of cervical cancer. Int J Gynecol Cancer. 2010;20:513–519. doi: 10.1111/IGC.0b013e3181c7fe6e. [DOI] [PubMed] [Google Scholar]
- 20.Wang ZM. PAX1 methylation analysis by MS-HRM is useful in triage of high-grade squamous intraepithelial lesions. Asian Pac J Cancer Prev. 2014;15:891–894. doi: 10.7314/apjcp.2014.15.2.891. [DOI] [PubMed] [Google Scholar]
- 21.Fabrizii M, Moinfar F, Jelinek HF, Karperien A, Ahammer H. Fractal analysis of cervical intraepithelial neoplasia. PLoS One. 2014;9:e108457. doi: 10.1371/journal.pone.0108457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rakotomahenina H, Garrigue I, Marty M, Brun JL. [Prevention and screening of cervical cancer] . Rev Prat. 2014;64:780–785. [PubMed] [Google Scholar]
- 23.Bosch FX, Lorincz A, Munoz N, Meijer CJ, Shah KV. The causal relation between human papillomavirus and cervical cancer. J Clin Pathol. 2002;55:244–265. doi: 10.1136/jcp.55.4.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 25.Owens CL, Buist DS, Peterson D, Kamineni A, Weinmann S, Ross T, Williams AE, Stark A, Adams KF, Doubeni CA, Field TS. Follow-up and clinical significance of unsatisfactory liquid-based Papanicolaou tests. Cancer Cytopathol. 2014 doi: 10.1002/cncy.21490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tjalma WA, Depuydt CE. Cervical atypical glandular cells and false negative HPV testing: a dramatic reality of the wrong test at the right place. Eur J Gynaecol Oncol. 2014;35:117–120. [PubMed] [Google Scholar]
- 27.Steenbergen RD, Ongenaert M, Snellenberg S, Trooskens G, van der Meide WF, Pandey D, Bloushtain-Qimron N, Polyak K, Meijer CJ, Snijders PJ, Van Criekinge W. Methylation-specific digital karyotyping of HPV16E6E7-expressing human keratinocytes identifies novel methylation events in cervical carcinogenesis. J Pathol. 2013;231:53–62. doi: 10.1002/path.4210. [DOI] [PubMed] [Google Scholar]


