Abstract
Macrophages and CD4+ T-cells are the major reservoirs for HIV-1 infection. CD63 is a tetraspanin transmembrane protein, which has been shown to play an essential role during HIV-1 replication in macrophages. In this study, we further confirm the requirement of CD63 in HIV-1 replication events in primary human CD4+ T-cells, dendritic cells, and a CD4+ cell line. Most interestingly, we also show the evidences for the co-localization and internalization of CD63 and HIV-1 major receptor CD4 in primary human macrophages and CD4+ cell line by confocal microscopy and Co-Immunoprecipitation assay. Analysis revealed that CD63-depleted CD4+ T-cells, dendritic cells, and a cell line showed significant decrease in HIV-1 production. Further analysis showed that CD63 down regulation reduced production of the early HIV protein Tat, and affected HIV protein Gag by CD63-Gag interaction. In agreement, CD63 silencing also inhibited production of the late protein p24. Furthermore, we revealed that CD63 silencing has no effect on HIV-1 replication with extensive viral challenge (MOI > 0.2). These findings suggest that CD63 plays a dual-role both in early and late HIV-1 life cycle with a range of HIV-1 infection (MOI < 0.2).
Keywords: HIV-1, CD63, siRNA, CD4+ T-cells, dendritic cells, life cycle, protein-protein-interactions (PPIs)
Introduction
Macrophages and CD4+ T lymphocytes are major target cells for HIV-1 infection and replication, and play critical roles in multiple aspects of viral pathogenesis. Numerous host determinants have been identified that facilitate HIV-1 replication by performing essential steps of the viral life cycle from viral entry to egress [1,2]. A recent functional genomics screen using siRNA technology revealed over 250 host factors critical for HIV-1 function at every step of the virus life cycle [3]. HIV encodes 15 proteins: Gag and Env structural proteins MA (matrix), CA (capsid), NC (nucleocapsid), p6, SU (surface), and TM (transmembrane); the Pol enzymes PR (protease), RT (reverse transcriptase), and IN (integrase); the gene regulatory proteins Tat and Rev; and the accessory proteins Nef, Vif, Vpr, and Vpu. Thus, HIV must take advantage of multiple host proteins for productive infection [3,4]. HIV uses a CD4 receptor [5], as well as CCR5 and CXCR4 co-receptors [6,7] for entry into the host cells. Upon membrane fusion, the viral core containing the viral nucleocapsid along with HIV RNA, reverse transcriptase, integrase, protease, and viral accessory proteins is released into the cytoplasm. Uncoating involves viral proteins as well as a variety of host factors that promote dissociation of the matrix protein from the plasma membrane [8]. After uncoating, reverse transcriptase converts viral RNA into DNA which is then transported across the nucleus and integrates into host chromosome. HIV genes are transcribed using host enzymes, and viral mRNA is transported outside the nucleus, and is used as a blueprint for producing new HIV proteins and enzymes. Some studies have reported reverse transcription to be largely associated with cellular enzymes or co-factors in vitro [9-12]. HIV integration and proviral transcription also require the use of host cell factors [13,14]. Following viral gene expression, the cellular GTPase Rab9 facilitates trafficking of viral proteins from the late endosome to the trans-Golgi [15]. HIV assembly and release utilizes TSG101 and the Endosomal Sorting Complexes Required for Transport (ESCRTs), a network of cellular factors that sort proteins and facilitate HIV budding [16,17].
CD63 is a type II cellular membrane protein and belongs to the tetraspanin superfamily [18,19]. It has been reported that CD63 incorporates into HIV-1 virions [20-22] and colocalizes with HIV-1 Env and Gag proteins in HIV-1 producing cells [21,23-26]. In addition, CD63 has been shown to be associated with tetraspanin-enriched mircrodomains (TEMs) that act as the gateway for HIV-1 budding at the plasma membrane [27]. A recent study showed CD63 suppresses chemokine receptor CXCR4 trafficking of the cell surface, which affects HIV-1 entry into T-lymphocytes [28]. Another recent paper showed CD63 is not required for HIV-1 infection of human macrophages, which is controversial with our studies [29].
We have previously demonstrated a role for CD63 in HIV-1 infection events in macrophages and a CD4+ cell line. Our earlier studies showed that anti-CD63 monoclonal antibody treatment 30 min prior to and during infection markedly reduced HIV replication in macrophages [30]. Inhibition was shown to occur during early infection, suggestive of involvement in virus entry or reverse transcription. Our more recent studies confirm the requirement of CD63 in HIV-1 replication by pretreatment with CD63-specific siRNA in both macrophages and a CD4+ cell line [12,31]. In this study, we further investigated the role of CD63 in HIV-1 replication cycle in primary CD4+ lymphocytes, dendritic cells (DCs), and a CD4+ cell line. These studies revealed that CD63-depleted CD4+ T cells showed a significant decrease in HIV-1 production by affecting subsequent events of the HIV-1 life cycle in both early and late events. These findings support a role for CD63 in HIV-1 replication cycle in human primary lymphocytes and cells lines.
Materials and methods
Cells and culture conditions
U373-MAGI-CCR5 cells (obtained from NIH AIDS Research and Reference Reagent Program, and contributed by Drs. Michael Emerman and Adam Geballe) are a cell line derived from a glioblastoma that has been modified by stable transfection of LTR-galactosidase which is transactivated by HIV Tat protein during virus replication [32,33]. U373-MAGI-CCR5 cells express CD4 and human chemokine receptor CCR5 on the cell surface, which allow infection by HIV R5 strains [33]. U373-MAGI-CCR5 cells were propagated in Dulbecco’s modified Eagle’s (DMEM) (Gibco, CA) supplemented with 10% fetal bovine serum (Hyclone, IL), 0.2 mg/ml G418, 0.1 mg/ml hygromycin B, and 1.0 mg/ml puromycin. For infection experiments, U373-MAGI-CCR5 cells were maintained in 90% DMEM, 10% fetal bovine serum, and 1% penicillin/streptomycin (Sigma, MO).
Monocytes and peripheral blood lymphocytes (PBLs) were separated using adherence on plastic after Ficoll gradient. PBLs were activated for 3 days in stimulation media (RPMI 1640 media with 20% Fetal calf serum (FCS); 1% Penicillin/Streptomycin; 5 μg/ml phytohemagglutinin) and incubated at 37°C with 5% CO2 for 72 h. PBLs were then collected by centrifugation and resuspended in growth media (RPMI 1640 with 1% L-glutamine; 1% Penicillin/Streptomycin; 20% FCS; 20 units/ml IL-2). Monocytes were differentiated to immature DCs by incubation for 7 days with 50 ng/mL of recombinant human granulocyte-macrophage colony-stimulating factor and 20 ng/mL of interleukin-4 (R&D Systems, Minneapolis, MN). Alternatively, monocytes were differentiated to macrophages (MDMs) by incubation for 7 days with 50 ng/mL of human recombinant granulocyte-macrophage colony-stimulating factor alone in plastic tissue culture dishes as described previously [34]. Briefly, PBMCs were purified by Ficoll-Hypaque centrifugation from buffy coats of healthy HIV-negative blood donors prepared by the UTMB Blood Bank (Galveston, TX). MDMs were obtained by adherence for 6 days to plastic Petri dishes initially coated with human AB serum [35]. During differentiation, macrophages were cultured in Iscove’s modified Dulbecco’s medium (IMDM) supplemented with 20% FCS; 1% L-glutamine and 1% Penicillin/streptomycin.
U1/HIV-1 cells (1 × 106) obtained from the NIH AIDS Research and Reference Reagent Program [36] were plated in 24-well microtiter plates on day 1. Cells were transfected with siRNA (200 nM final concentration) using Oligofectamine (Invitrogen) following the manufacturer’s instructions. Complete cell media (RPMI 1640 containing 2.0 mM L-glutamine; 10% heat-inactivated fetal bovine serum) was changed after 24 h. Forty-eight hours post-transfection (day 3), complete RPMI media plus phorbol 12-myristate 13-acetate (PMA) was added to cells. Intracellular and extracellular virus was measured in the cells lysates and culture supernatant on day 5 by p24 ELISA.
Viruses
HIV-1 SX, which is a chimeric R5-tropic virus encoding the majority of the HIV-1 JRFL envelope protein in an HIV-1NL4-3 backbone, and dual-tropic (R5/X4) HIV-189.6, which is a HIV-1 laboratory adapted strain originally isolated from infected individuals, were purchased from the Virology Core Facility, Center for AIDS Research at Baylor College of Medicine, Houston, TX. HIV-1SX stock containing 69.681 ng/ml of HIV p24 with 65,325 TCID50/ml was used to infect macrophages and U373 cells. HIV-189.6 stock containing 49.977 ng/ml of HIV p24 with 261,300 TCID50/ml was used to infect PBLs. HIV-1 stocks were titrated, and for experiments, the inoculum was 42 ng of p24 per 1.5 × 105 cells (MOI 0.6). In most experiments, MDMs, PBLs, DCs and U373-MAGI-CCR5 cells were infected with HIV-1 SX or HIV-1 89.6 (MOI 0.02).
siRNA transfections and infectivity assay
The CD63- and CD4-specific target siRNAs were designed and synthesized by Dharmacon (Lafayette, CO), ERBB2IP non-HIV specific siRNA served as a negative control in each experiment. The target sequences for siGENOME SMARTpool siRNA of CD63 includes D-017256 (01-GAGAUAAGGUGAUGUCAGA; 02-AAGGAGAACUAUUGUCUUA; 03-GGAUUAAUUUCAACGAGAA; 04-GAUGGAGAAUUACCCGAAA). The target sequences for siGENOME SMARTpool siRNA of CD4 includes D-005234 (01-GAACUGACCUGUACAGCUU; 02-AAUCAGGGCUCCUUCUUAA; 03-GAAGAAGAGCAUACAAUUC; 04-AUUACCAAGUGCCGGACUA); The target sequences for siGENOME SMARTpool siRNA of ERBB2IP includes D-031861 (01-UGAAACAGCUCACAUAUUU; 02-UGUGAAAUCUCAUAGCAUA; 03-CGAAGAGCCAAAUAUAAUA; 04-CCAAACGACCGACUUAUUC). siRNAs transfections were performed using oligofectamine (Invitrogen, CA) following the manufacturer’s instructions or The Nucleofector™ Technology by Amaxa (Lonza, Switzerland) following the Nucleofector Kits and devices. In brief, MDMs and DCs adherent for 5 days were plated in 24-well plates (5 × 105 cells/well), and transfected with 50 nM siRNAs using oligofectamine in serum-free Opti-MEM I medium (Gibco) 24 h after plating. Cells were maintained in Iscove’s medium and 20% FCS after 4 h to terminate transfection. In order to measure down regulation of target genes, cells were lysed by cell lysis buffer (Promega, WI) and lysates were collected for Western blot analysis, or RNA was extracted for real time quantitative PCR analysis at 2 days post-transfection. U373-MAGI-CCR5 cells were transfected in 24-well plates (1 × 105 cells/well), using oligofectamine and 50 nM siRNAs (final concentration in supernatants). Subsequently, cells were fed with DMEM with 10% FCS after 4 h to terminate transfection, followed by detection of proteins or target genes down regulation as described above. PBLs were transfected in 6-well plates (5 × 105 cells/well), using The Nucleofector®, working with single cuvettes for standard applications following instruction of the device of Amaxa.
Forty eight hours post-transfection, MDMs, DCs or U373-MAGI-CCR5 cells were infected with HIV-1 SX, and PBLs were infected with HIV-1 89.6 (m.o.i. = 0.02). Culture supernatants or cell lysates were harvested for p24 detection on day 5 post-infection in PBLs and U373-MAGI-CCR5 cells, and on day 7 post-infection in MDMs and DCs using p24 Capture ELISA kit (ImmunoDiagnostics, Woburn, MA).
In order to assess Tat protein expression, U373-MAGI-CCR5 cells were lysed on day 3 post-infection, and analyzed for β-galactosidase activity using the Beta-Glo Assay System (Promega) and a Dynex MLX Luminometer.
HIV-1 Env pseudovirus production and titration
Stocks of single-round-infection HIV-1 Env pseudovirus were produced by cotransfecting 293T cells (1.7 × 107 cells per T75 flask) with 2 μg of an HIV-1 rev/env expression plasmid and 12 μg of an env-deficient HIV-1 backbone plasmid (PNL4.3 ΔEnv) using Lipofectamine transfection reagent (Invitrogen). Pseudovirus-containing supernatant was harvested 24 h following transfection and clarified by centrifugation and 0.45-μm filtration. Single-use aliquots (1.0 ml) were stored at -80°C. The 50% tissue culture infectious dose (TCID50) for each pseudovirus preparation was determined by infection of TZM.bl cells.
In situ proximity ligation assay (in situ PLA)
Cultures, treated as indicated, were immediately fixed in 4% PFA on ice for 30 min and thereafter subjected to in situ PLA using Duolink Detection kit (Olink Bioscience, Uppsala, Sweden) according to the manufacturer’s instructions for Duolink Blocking solution and Detection protocol. Briefly, slides were blocked, incubated with antibodies directed against CD63 and HIV-1 Gag, and thereafter incubated with PLA probes, which are secondary antibodies (anti-mouse and anti-rabbit) conjugated to unique oligonucleotides. Briefly, 50 μg monoclonal antibody anti-CD63 (Santa Cruz) or anti-HIV-Gag (R&D) was added the cells. Circularization and ligation of the oligonucleotides was followed by an amplification step. The products were detected by a complementary fluorescently labelled probe. Slides were mounted using Vectashield (Vector Laboratories Inc, Burlingame, CA) and evaluated using an LSM 510 META confocal microscope (Carl Zeiss). Z-stack micrographs taken with the 40 ×/63 × objectives were obtained. The number of heterodimers, visualized as bright fluorescent signals, was counted in 10-15 fields/well. n (number of wells) = 6 for HSaVEC analyses, and n (number of individual EBs) = 8 for EB analyses. Representative results are shown from experiments repeated at least three times. Cell images obtained were exported using the AxioVision software (Carl Zeiss) in TIF format for further analysis and determination of heterodimers/cell in Blob-Finder image analysis software (Version 2.5).
Real time quantitative PCR
Primers and probes for real time PCR were custom ordered from Sigma-Aldrich. Full genomic DNA was isolated from MDMs or cell lines using the Qiagen Blood and Tissue Kit (Qiagen, Inc., CA). Small non-genomic DNA, such as reverse transcribed viral cDNA and 2-LTR circles were isolated using a Qiagen Miniprep kit. To detect strong stop of reverse transcription, primers M667 and AA55, along with probe MH603 [37] were used; to detect full length HIV DNA, primers M667 and M661 [38], along with probe MH603 were used; to identify 2-LTR circle formation, as described in our previous study [39], primers MH535 and MH536 were used with the MH603 probe [37]. Total mRNA was isolated from siRNA-transfected cells and MDMs using RNeasy Mini Kits (Qiagen). For integration, a two-step PCR reaction was performed. For the initial PCR amplification, Alu forward and HIV-1 gag reverse primers were used [39,40]. The second step real-time PCR used the primers LTR forward, LTR reverse, and LTR probe [39,40]. CD63 specific primers and probe were purchased from Applied Biosytems (Carlsbad, CA). All reactions were performed using Applied Biosystems TaqMan Universal Master Mix and run using an Applied Biosystems 7500 Fast Real Time PCR system and 7500 Fast System Software. Silencing of target genes was determined by normalizing target gene expression to GAPDH expression (n = 3).
Statistical analysis
Study results were summarized for each group and results were expressed as the mean ± standard deviation (SD) of at least four wells. Comparison between two groups was performed by multiple comparisons testing/least significant difference or Tukey’s procedure depending on the ANOVA F test Prism software (version 4; GraphPad Software) by a nonparametric Mann-Whitney U test. A pairwise t test was used to compare the significance of changes in CD63 siRNA silencing experiments. Values of P < 0.05 (*), and P < 0.01 (**) were considered significant or very significant, respectively.
Results
CD63 mRNA down regulation by siRNA in human MDMs which are challenged with different concentration of HIV-1
To confirm the down regulation of CD63 by siRNA in MDMs, mRNA was extracted and analyzed by quantitative reverse transcriptase PCR (qRT-PCR). CD63 mRNA expression was silenced by > 90% following CD63 siRNA transfections of MDM (Figure 1A). CD63 mRNA was not significantly reduced by transfections of other siRNAs (CD4, ERBB2IP) or FDA approved HIV-1 inhibitors (AZT, RT inhibitor; Raltegravir, integrase inhibitor) used throughout this study indicating specificity for CD63 siRNA down regulation in MDMs, T lymphocytes, DCs and U373-MAGI-CCR5 cells. ERBB2IP siRNA was used as a cellular target negative control, as our previous studies showed that silencing ERBB2IP does not inhibit HIV-1 [15]. Western blot analysis revealed that CD63 protein expression was significantly reduced in CD63-siRNA transfectants compared to the cells transfected with control siRNAs or non-transfected cells (data not shown), consistent with our previous findings [12,31].
Figure 1.
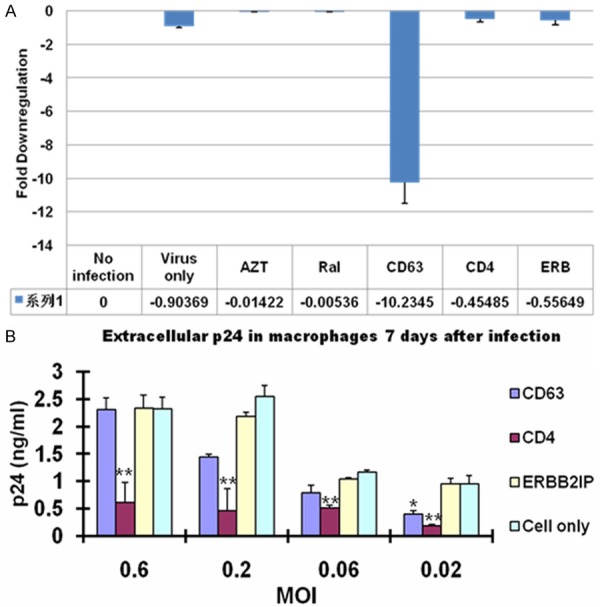
CD63 mRNA down regulation by siRNA in human MDMs which are challenged with different concentration of HIV-1. A. MDMs (5 × 105 cells/well) were plated in 24-well plates and transfected with 50 nM siRNAs (CD63, CD4, and ERBB2IP). For controls, cells were treated with AZT (1 mM) or raltegravir (20 mM). Controls also included untreated cells or cells infected with HIV-1 SX (m.o.i. = 0.02) only. Total mRNA was isolated from each well using the Qiagen RNeasy kit 48 h after transfection. Quantitative RT-PCR was used to determine relative CD63 expression levels, normalizing to GAPDH expression as an internal control. B. MDMs were challenged with different concentration of HIV-1 SX (m.o.i. = 0.6, 0.2. 0.06 and 0.02, respectively). Supernatants were harvested for p24 detection on day 7 post-infection for MDMs using p24 Capture ELISA kit (ImmunoDiagnostics, Woburn, MA).
To gauge the effect of CD63 down regulation on viral production in MDMs, the experiment with different concentration of viral infection was performed. MDMs were transfected with various specific siRNAs or ERBB2IP control siRNA, followed by different concentration of HIV-1 SX infection (MOI = 0.6, 0.2, 0.06 and 0.02, respectively) 48 h post-transfection. HIV-1 SX virus production was assessed in culture supernatants by p24 ELISA at 7 days post-infection of MDMs (Figure 1B). As shown in Figure 1B, with higher concentration of viral infection (MOI = 0.6, 0.2 and 0.06), virus production has no effect following CD63 silencing compared to ERBB2IP siRNA transfected cells. However, when challenging cells with lower concentration (MOI = 0.02), virus production was significantly reduced following CD63 or CD4 silencing compared to ERBB2IP control (P < 0.05). Dose-response experiment determined that CD63 silencing affects HIV-1 production in MDMs with dependence of concentration of viral challenge as low as MOI 0.02.
CD63 down regulation affects HIV-1 production in human PBLs and DCs
Our previous studies showed CD63 silencing affects HIV-1 production in MDMs and CD4+ U373-MAGI-CCR5 cells. In order to further elucidate the effect of CD63 down regulation on viral production in primary human PBLs and DCs, PBLs and DCs were transfected with various specific siRNAs or ERBB2IP control siRNA, followed by HIV-1 SX or HIV-1 89.6 infection 48 h post-transfection. To confirm the down regulation of CD63 by siRNA in PBLs or DCs, mRNA was extracted and analyzed by quantitative reverse transcriptase PCR (qRT-PCR). CD63 mRNA expression was silenced by > 92% following CD63 siRNA transfections of PBLs (Figure 2A). HIV-1 production was assessed in culture supernatants by p24 ELISA at 5 days post-infection of PBLs (Figure 2B) or 7 days post-infection of DCs (Figure 2C). As shown in Figure 2B and 2C, virus production in both cell types was significantly reduced following CD63 silencing compared to ERBB2IP siRNA transfected cells (P < 0.05), especially, HIV-1 production in DCs completely disappeared by CD63 silencing. Our data proved that CD63 functions for HIV-1replication in both PBLs and DCs.
Figure 2.
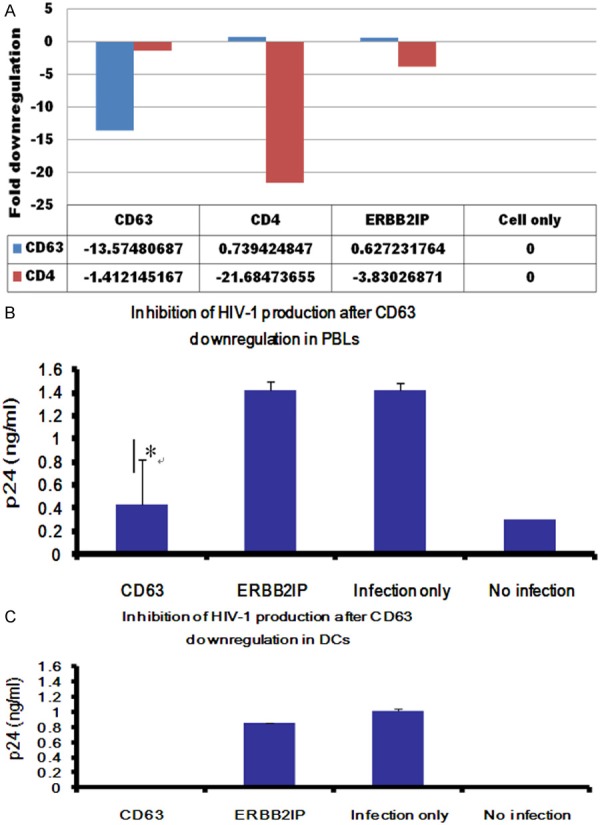
Effects of CD63 silencing on HIV-1 replication in human PBLs and DCs. A. PBLs (1 × 105 cells/well) were plated in triplicate in 24-well plates and transfected with 50 nM siRNAs (CD63, CD4, and ERBB2IP). Controls included untreated cells or cells treated with virus infection only. Total mRNA was isolated from each well using the Qiagen RNeasy kit 48 h after transfection. Quantitative RT-PCR was used to determine relative CD63 expression levels, normalizing to GAPDH expression as an internal control. B. PBLs cells (1 × 105 cells/well) or (C) DCs cells (5 × 105 cells/well) were plated in triplicate in 24-well plates and transfected with 50 nM siRNAs (CD63, CD4, and ERBB2IP). Controls included untreated cells or cells treated with virus infection only. Forty eight hours post-transfection, cells were infected with HIV-1 89.6 (m.o.i. = 0.02). Supernatants were harvested for p24 detection on day 5 post-infection for PBLs cells, and on day 7 post-infection for DCs using p24 Capture ELISA kit (ImmunoDiagnostics, Woburn, MA). C. Cell lysates from MDMs on day 7 post-infection were also harvested for detection of intracellular p24. *P < 0.05, **P < 0.01, compared with ERBB2IP control group, respectively.
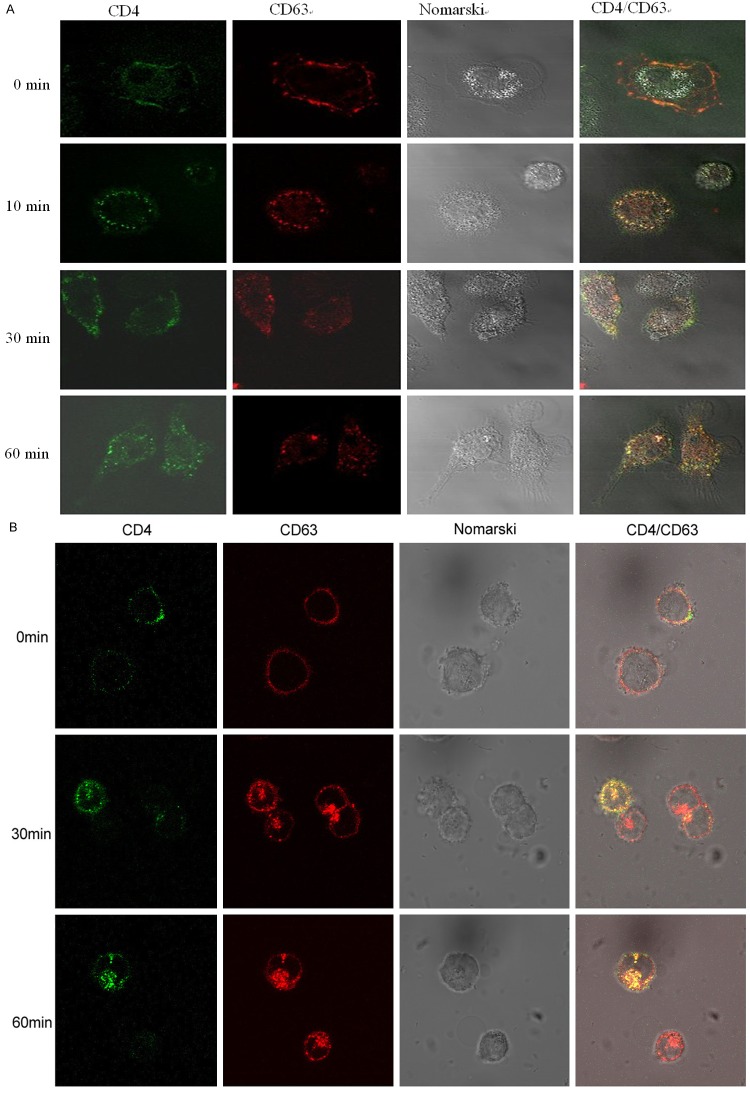
Colocalization and internalization of CD63 and CD4 in human MDMs and U373-MAGI-CCR5 cells
We investigated the effect of CD63 on HIV-1 attachment by virus-cell binding assay using anti-CD63 in our previous study [12]. Anti-CD63 showed no inhibition of HIV-1 attachment to human MDMs and U373-MAGI-CCR5. However, we still suppose that CD63 like HIV-1 co-receptors CCR5/CXCR4 facilitates HIV-1 infection by interaction with CD4, so here we investigated the colocalization and internalization of CD63 and CD4 in human MDMs and U373-MAGI-CCR5 cells. After blocking with non-specific antibodies, MDMs and U373 cells were incubated with anti-CD63 antibody (Caltag, labeled by Alexa 568) and/or anti-CD4 (Becton-Dickinson, labeled by Alexa 488). As shown in Figure 3, immunofluorescent microscopy showed that CD4 and CD63 were located and co-localized at the plasma membrane at beginning time point. They were internalized via clathrin-mediated endocytosis at different rates in MDMs (Figure 3A) and U373 cells (Figure 3B). After 30 min at 37°C, 20% of labeled proteins had been internalized, and when 60 min at 37°C, 50% of labeled proteins had been internalized. We also saw co-ip of CD4 and CD63 in both cell types (Data not shown). Our findings proved that CD63 and CD4 are co-localized and internalized together, suggesting that CD63 may facilitate HIV-1 infection via CD4 signaling.
Figure 3.
Colocalization and internalization of CD63 and CD4 in human MDMs and U373-MAGI-CCR5 cells. After blocking with non-specific antibodies, A. MDMs (5 × 105 cells/well) and (B) U373 cells (5 × 105 cells/well) were incubated with anti-CD63 antibody (Caltag, labeled by Alexa 568) and/or anti-CD4 (Becton-Dickinson, labeled by Alexa 488). For time 0 min, cells were fixed with 4% paraformaldehyde and then stained with Alexa-labeled antibodies for 30 min at RT, and then fixed for an additional 20 min. For all other samples, cells were incubated with antibody for the indicated times at 37°C, and then washed twice in cold PBS and fixed with 4% paraformaldehyde.
Virus Env-mediated entry in CD63-silenced human MDMs
In order to determine the role of CD63 in early events of HIV-1 infection, we first examined the effects of CD63 on virus Env-mediated entry, which is one of the initial events that occur during HIV-1 replication. We used Env-specific pseudovirus to infect CD63-silenced MØ. Our preliminary data (Figure 4) shows that when viral entry is redirected through the endocytic pathway by pseudotyping virions with MLV or VSV, infectivity is not affected in CD63-silenced MØ. However, when viral entry is mediated through CD4 and coreceptor CCR5 using R5 and dual-tropic pseudotyped envelopes, infectivity is significantly reduced. MLV and VSV enter the cell via endocytosis, independent of CD4 and chemokine receptor usage. This indicates a role for CD63 on virus-Env mediated entry or fusion events facilitated through CD4 and CCR5, and that the entry events are specific for HIV-1 and not other retroviruses.
Figure 4.
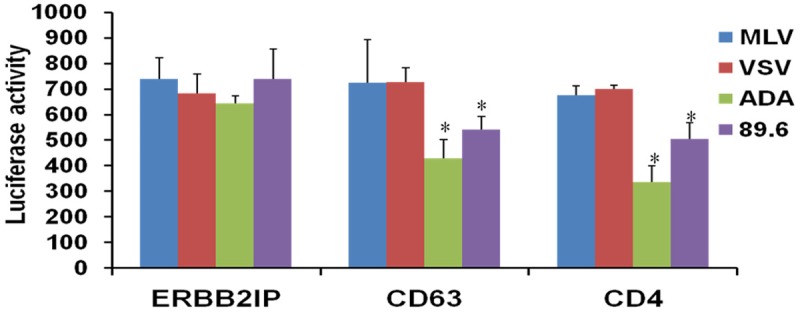
Virus Env-mediated entry in CD63-silenced human MDMs. MDMs (5 × 105 cells/well) were transfected with 50 nM siRNAs (CD63, CD4, and ERBB2IP). Forty eight hours post-transfection, cells were infected with various Env-specific pseudoviruses (m.o.i. = 0.02). Cell lysates were collected on day 3 post infection, and luciferase activity was analyzed to monitor pseudovirus integration. MLV and VSV were used as controls because they enter the cell via endocytosis. ERBB2IP siRNA was used as a cellular target negative control because ERBB2IP does not inhibit HIV-1 {Murray 2005}. CD4 siRNA was used as a control because it is the main receptor for entry of HIV. R5 (ADA) & dual-tropic (89.6) viruses were used because MDMs are R5-tropic and uses the main receptor CD4 and CCR5 co-receptor to enter the cell. All experiments were performed in quadruplicate. *P < 0.05, **P < 0.01, compared with ERBB2IP group.
CD63 down regulation also affects HIV-1 replication in late events of life cycle
To further assess the role of CD63 in the late stage of HIV replication, CD63-specific siRNA was transfected into U1/HIV-1 cells, which are chronically infected monocytoid cells harboring 2 integrated copies of provirus per cell. Early steps of HIV replication are not required for virus production in these cells, and virus production can be induced with 3 phorbol 12-myristate 13-acetate (PMA). HIV-1 p24 protein in cell lysates with CD63 silencing was reduced slightly compared to ERBB2IP control, but no significance between them. However, HIV viral production in supernatants was reduced significantly in CD63-depleted U1 cells compared to ERBB2IP control (P < 0.05), which shows the same level as in cells treated with TSG 101-specific siRNA (Figure 5B). TSG 101 is involved in post-integration trafficking of HIV proteins from the late endosome to the trans-Golgi [15]. These findings further support the role of CD63 in later stages of HIV replication, specifically occurring after proviral integration.
Figure 5.
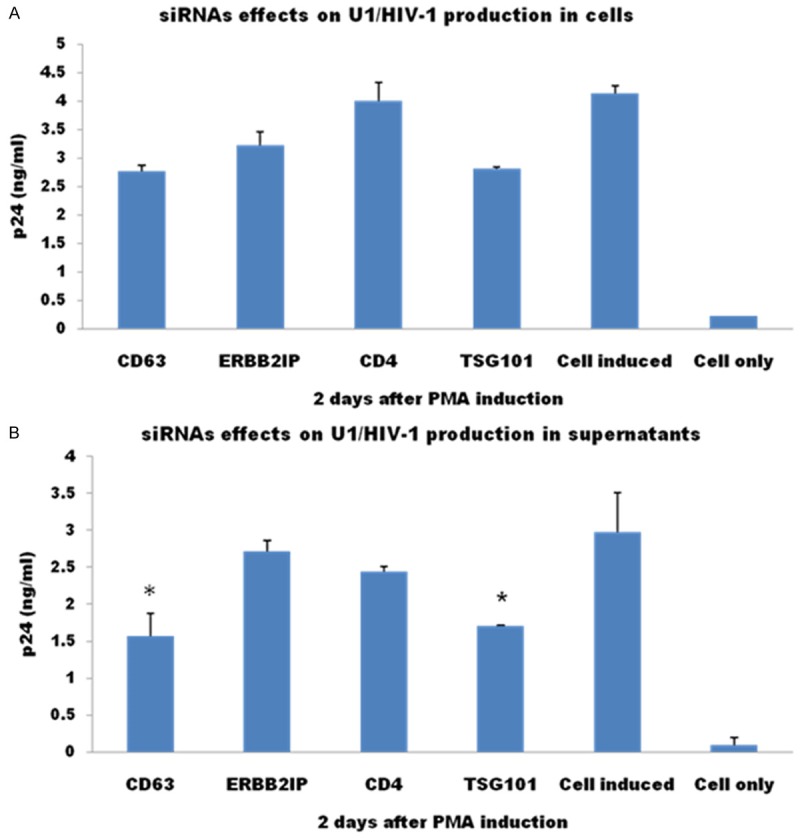
CD63 down regulation also affects HIV-1 replication in late events of life cycle. U1/HIV-1 cells (5 × 105 cells/well) were transfected with CD63-specific siRNA. Two days after transfection, cells were induced with 3 phorbol 12-myristate 13-acetate (PMA, 5 × 10-6 M). Three days later, HIV-1 p24 protein in cell lysates (A) and culture supernatants (B) were detected with DCs using p24 Capture ELISA kit (ImmunoDiagnostics, Woburn, MA).
Co-localization and in situ PLA reveals interaction of CD63 and HIV-1 gag
We hypothesized that CD63 functions same way like TSG 101 and involves in the trafficking of HIV proteins. To demonstrate the interaction of CD63 and HIV-1 proteins, we used confocal and in situ PLA to investigate the relationship between CD63 and HIV-1 Gag protein in U1 cells. U1/HIV-1 cells were transfected with 50 nM siRNAs (CD63, ERBB2IP and TSG 101). Controls included cells induced. Forty eight hours post-transfection, cells were treated with 5 × 10-6 M PMA. Cells were fixed on day 2 post PMA inductions, followed by double staining of CD63 and HIV-1 Gag with PE-labelled anti- CD63 and FITC labelled anti-HIV Gag. Confocal images showed co-localization of CD63 with HIV-1 Gag in U1 cells (Figure 6A). CD63 silencing cells showed little interaction between CD63 and HIV-1 Gag (Yellow signal, Figure 6Ab). However, U1 cell only, CERBB2IP transfected cells, and TSG101 silencing cells showed more positive signals of co-localization between CD63 and HIV-1 Gag (Figure 6Aa, 6Ac, 6Ad respectively). To further confirm the interaction between CD63 and HIV-Gag protein, we used an experimental design where cells were transfected with 50 nM siRNAs (CD63, ERBB2IP and TSG 101). Controls included cells induced. Cells were fixed on day 2 post PMA inductions and then probed with primary antibodies raised in different species and directed towards the proteins of CD63 and Gag 17, or integrin α4 as positive control. This was followed by incubation with two sets of secondary antibodies conjugated with oligonucleotide, unique for each type of secondary antibody. Ligation of the oligonucleotides by a bridging probe in a proximity-dependent manner, allows a rolling-circle amplification. Finally, this product is detected by complementary fluorescent probes. As shown in Figure 6B, cells of transfection with CD63 and TSG 101 siRNAs showed decreased formation of heterodimers in situ, distributed on the plasma membrane as well as in the cytoplasm (Figure 6Ba, 6Bc). The number of heterodimers/cell with ERBB2IP siRNAs transfection increased 100-fold from basal (Figure 6Bb). Our results indicate that CD63 also involves in the HIV-1 proteins trafficking in late events of HIV-1 replication (Figure 7).
Figure 6.
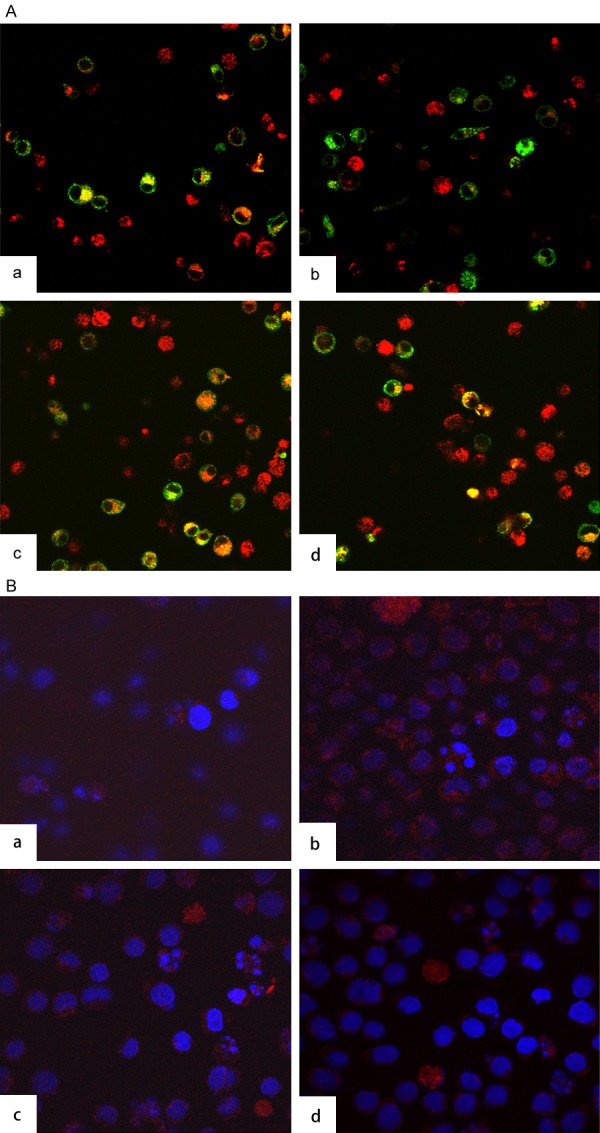
Co-localization and In situ PLA reveals interaction of CD63 and HIV-1 Gag. A. Co-localization of CD63 and HIV-1 Gag in U1 cells. U1/HIV-1 cells (2 × 106 cells/well) were plated in triplicate at 6-well plates and transfected with 50 nM siRNAs (CD63, ERBB2IP and TSG 101). Controls included cells induced. Forty eight hours post-transfection, cells were treated with 5 × 10-6 M PMA. Cells were fixed on day 2 post PMA induction, followed by double staining of CD63 and HIV-1 Gag with PE-labelled anti-CD63 and FITC labelled anti-HIV Gag. Confocal images showed co-localization of CD63 with HIV-1 Gag in (a) U1 cells, (b) CD63 siRNA transfected cells, (c) ERBB2IP siRNA transfected cells, and (d) TSG101 siRNA transfected cells, respectively. B. Detection of CD63 and Gag 17 in U1 cells using Duolink with two primary antibodies. U1/HIV-1 cells were treated with (a) CD63 specific siRNA; (b). ERBB2IP siRNA; (c) Tsg 101 siRNA; (d) No treatment, two primary antibodies were anti-CD63 and anti-integrin α4 as positive control. Two primary antibodies for a, b, and c were anti-CD63 and anti Gag 17.
Figure 7.
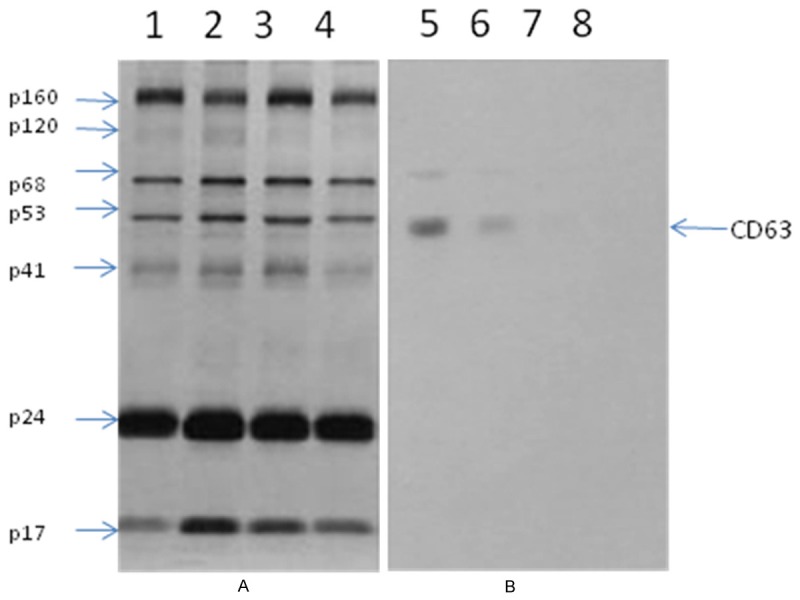
CD63 presents in macrophages produced HIV-1 virions. HIV-1 virions produced by macrophages and PBLs were concentrated. HIV-1 viral proteins were separated by electrophoresis, then transferred to a membrane which was incubated with (A) HIV-1 IgG, and (B) anti-CD63, followed by incubating with an enzyme- labelled antibody. A substrate was added for visualization. Lane 1, 2, 5, 6 were loaded with macrophage produced virions, and lane 3, 4, 7, 8 were PBLs produced virions.
Discussion
Extensive studies have revealed that HIV-1 exploits multiple host proteins during infection [1,3,41-45]. These proteins participate in a broad array of cellular functions and implicate different pathways in the viral life cycle. However, the mechanisms by which these proteins play relevant roles in HIV-1 pathogenesis are subjects of continued exploration. CD63 has also been implicated in HIV infection. CD63 is incorporated into virus particles [20,21] in culture cells or in T-cell lines, and is present at sites where particles assemble [27,46,47]. Indeed, our previous studies have suggested CD63 has important functions in HIV-1 replication [12,16,31]. However, how CD63 is targeted to the virus infection or virus assembly compartment and incorporated into budding HIV-1 particles is still unclear. In addition, there are no specific interactions between CD63 and viral components or relevant receptors have been demonstrated to date. In this study, we revealed the role of CD63 during early/late viral protein formation, and virion release from infected human MDMs and U373-MAGI-CCR5 cells. We found that CD63 was required for efficient HIV-1 entry and subsequent steps from viral protein translation to egress. Viral entry mechanisms including cell membrane docking and cell-cell fusion life cycle steps that precede HIV-1 un-coating were related with CD63. Our data suggests that CD63 may affect HIV-1 entry by interacting with HIV-1 major receptor CD4 pathway. In all cases, the down regulation of CD63 via specific siRNA significantly reduced HIV-1 replication in MDMs, U1/HIV-1 cells, U373 cells, T lymphocytes and DCs cell types, suggesting that CD63 is associated with a common mechanism in early events of the HIV-1 life cycle. It is possible that HIV-1 may be linked to an intracellular signaling event involving CD63. CD63 has been shown to induce signaling through tyrosine-dependent interaction of its carboxyterminal cytoplasmic tail with the AP-3 adaptor subunit [48]. In addition, when HIV-1 gp120 binds to CD4 and CCR5 on the surface of macrophages, it has been shown to induce phosphorylation of receptor tyrosine kinases such as p56Lck, which in turn activates the Raf/MEK/ERK and PIP3 kinase pathways and intracellular calcium channels [49], implying that an intracellular signaling route is likely to be important for the HIV-1 life cycle. Our findings show CD63 may link to CD4 pathway to affect HIV-1 infection in early entry event, which still remains to be further explored.
Although recent studies [29,50] showed that CD63 expression is not required for either the production or the infectivity of HIV-1, numerous studies have reported that CD63 incorporates into HIV-1 virions and have the potential to interfere with viral infection [20-27]. Our previous studies [12,30,31] demonstrated that CD63 plays an important supportive role in HIV-1 replication in macrophages and cell lines. Although controversial data have been published in the last few years concerning effects of CD63 on HIV-1 replication in macrophages and other cell types, it is possible that this effect may be dependent on the cell types, viral strains, and virus concentration. Even though MDMs display a great heterogeneity in their capacities to replicate HIV-1, depending on the donors (up to a 3 log difference in viral production) [51-53], we performed experiments in macrophages from several different donors as well U373 cells, U1/HIV-1 cells, T lymphocytes and DCs, infecting with HIV-1 R5 virus strain or dual tropic strain, and our results consistently present similar outcomes. Specially, in current study, we did dose-dependent experiments to show that the effect of CD63 depends on challenged viral concentration, which means CD63 affects HIV-1 infection within a range of viral concentration (moi <0.02).
Understanding the virus-host interactions that lead to limited HIV replication in these cell types can be a powerful tool to identify molecular mechanisms of innate/intrinsic restriction of HIV replication. We have found CD63 silencing also affects HIV-1 late protein trafficking. Co-localization and interaction between CD63 and HIV-1 Gag indicated that effect of CD63 on HIV-1 late event also is associated with HIV-1 Gag protein trafficking. This is first findings as we know to show the interaction between CD63 and HIV-1 components.
A key pharmacological strategy for treating individuals living with HIV has been to simultaneously target multiple virus-encoded enzymes required for replication to overcome emergence of drug resistance. Our laboratory has pursued an alternate strategy by identifying host factors, such as CD63, that are required for HIV-1 replication, as HIV-1 should be less likely to evolve resistance to drugs targeting cellular proteins. In conclusion, we have revealed that CD63 plays an essential role both in early entry event of the HIV-1 replication cycle and late viral proteins assembly. CD63 interacts directly with HIV-1 Gag, and major receptor CD4, altering host dependent factors potentially required for HIV-1 infection and replication. Further research is needed to gain insight into the molecular signaling mechanisms, including CD63, CD4, PTK and viral proteins that possibly facilitate early or late events in the HIV replication cycle.
Acknowledgements
We acknowledge the NIH AIDS Research and Reference Reagent Program for providing the U373-MAGI-CCR5 cells. We thank Edward Siwak, Ph.D., Associate Director of Virology Core Facility, Center for AIDS Research at Baylor College of Medicine, Houston, TX for providing HIV-1SX. Also, we greatly appreciate Merck & CO., Inc. for generously providing raltegravir used in our studies.
Disclosure of conflict of interest
None.
References
- 1.Bergamaschi A, Pancino G. Host hindrance to HIV-1 replication in monocytes and macrophages. Retrovirology. 2010;7:31. doi: 10.1186/1742-4690-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedrich BM, Dziuba N, Li G, Endsley MA, Murray JL, Ferguson MR. Host factors mediating HIV-1 replication. Virus Res. 2011;161:101–114. doi: 10.1016/j.virusres.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 4.Kammula EC, Mötter J, Gorgels A, Jonas E, Hoffmann S, Willbold D. Brain transcriptome-wide screen for HIV-1 Nef protein interaction partners reveals various membrane-associated proteins. PLoS One. 2012;7:e51578. doi: 10.1371/journal.pone.0051578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman JC, Montagnier L. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature. 1984;312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 6.Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 7.Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA, Cayanan C, Maddon PJ, Koup RA, Moore JP, Paxton WA. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 8.Aiken C. Viral and cellular factors that regulate HIV-1 uncoating. Curr Opin HIV AIDS. 2006;1:194–199. doi: 10.1097/01.COH.0000221591.11294.c1. [DOI] [PubMed] [Google Scholar]
- 9.Hooker CW, Harrich D. The first strand transfer reaction of HIV-1 reverse transcription is more efficient in infected cells than in cell-free natural endogenous reverse transcription reactions. J Clin Virol. 2003;26:229–238. doi: 10.1016/s1386-6532(02)00121-x. [DOI] [PubMed] [Google Scholar]
- 10.Warrilow D, Meredith L, Davis A, Burrell C, Li P, Harrich D. Cell factors stimulate human immunodeficiency virus type 1 reverse transcription in vitro. J Virol. 2008;82:1425–1437. doi: 10.1128/JVI.01808-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warrilow D, Warren K, Harrich D. Strand transfer and elongation of HIV-1 reverse transcription is facilitated by cell factors in vitro. PLoS One. 2010;5:e13229. doi: 10.1371/journal.pone.0013229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li G, Dziuba N, Friedrich B, Murray JL, Ferguson MR. A post-entry role for CD63 in early HIV-1 replication. Virology. 2011;412:315–324. doi: 10.1016/j.virol.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodarzi G, Pursley M, Felock P, Witmer M, Hazuda D, Brackmann K, Grandgenett D. Efficiency and fidelity of full-site integration reactions using recombinant simian immunodeficiency virus integrase. J Virol. 1999;73:8104–8111. doi: 10.1128/jvi.73.10.8104-8111.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greene WC. The molecular biology of human immunodeficiency virus type 1 infection. N Engl J Med. 1991;324:308–317. doi: 10.1056/NEJM199101313240506. [DOI] [PubMed] [Google Scholar]
- 15.Murray JL, Mavrakis M, McDonald NJ, Yilla M, Sheng J, Bellini WJ, Zhao L, Le Doux JM, Shaw MW, Luo CC, Lippincott-Schwartz J, Sanchez A, Rubin DH, Hodge TW. Rab9 GTPase is required for replication of human immunodeficiency virus type 1, filoviruses, and measles virus. J Virol. 2005;79:11742–11751. doi: 10.1128/JVI.79.18.11742-11751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von Schwedler UK, Stuchell M, Müller B, Ward DM, Chung HY, Morita E, Wang HE, Davis T, He GP, Cimbora DM, Scott A, Kräusslich HG, Kaplan J, Morham SG, Sundquist WI. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 17.Morita E, Sundquist WI. Retrovirus budding. Annu Rev Cell Dev Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- 18.Stipp CS, Kolesnikova TV, Hemler ME. Functional domains in tetraspanin proteins. Trends Biochem Sci. 2003;28:106–112. doi: 10.1016/S0968-0004(02)00014-2. [DOI] [PubMed] [Google Scholar]
- 19.Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol. 2005;6:801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 20.Chertova E, Chertov O, Coren LV, Roser JD, Trubey CM, Bess JW Jr, Sowder RC 2nd, Barsov E, Hood BL, Fisher RJ, Nagashima K, Conrads TP, Veenstra TD, Lifson JD, Ott DE. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J Virol. 2006;80:9039–9052. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pelchen-Matthews A, Kramer B, Marsh M. Infectious HIV-1 assembles in late endosomes in primary macrophages. J Cell Biol. 2003;162:443–455. doi: 10.1083/jcb.200304008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sato K, Aoki J, Misawa N, Daikoku E, Sano K, Tanaka Y, Koyanagi Y. Modulation of human immunodeficiency virus type 1 infectivity through incorporation of tetraspanin proteins. J Virol. 2008;82:1021–1033. doi: 10.1128/JVI.01044-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raposo G, Moore M, Innes D, Leijendekker R, Leigh-Brown A, Benaroch P, Geuze H. Human macrophages accumulate HIV-1 particles in MHC II compartments. Traffic. 2002;3:718–729. doi: 10.1034/j.1600-0854.2002.31004.x. [DOI] [PubMed] [Google Scholar]
- 24.Grigorov B, Arcanger F, Roingeard P, Darlix JL, Muriaux D. Assembly of infectious HIV-1 in human epithelial and T-lymphoblastic cell lines. J Mol Biol. 2006;359:848–862. doi: 10.1016/j.jmb.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 25.Jolly C, Sattentau QJ. Human immunodeficiency virus type 1 assembly, budding, and cell-cell spread in T cells take place in tetraspanin-enriched plasma membrane domains. J Virol. 2007;81:7873–7884. doi: 10.1128/JVI.01845-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perlman M, Resh MD. Identification of an intracellular trafficking and assembly pathway for HIV-1 gag. Traffic. 2006;7:731–745. doi: 10.1111/j.1398-9219.2006.00428.x. [DOI] [PubMed] [Google Scholar]
- 27.Nydegger S, Khurana S, Krementsov DN, Foti M, Thali M. Mapping of tetraspanin-enriched microdomains that can function as gateways for HIV-1. J Cell Biol. 2006;173:795–807. doi: 10.1083/jcb.200508165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshida T, Kawano Y, Sato K, Ando Y, Aoki J, Miura Y, Komano J, Tanaka Y, Koyanagi Y. A CD63 mutant inhibits T-cell tropic human immunodeficiency virus type 1 entry by disrupting CXCR4 trafficking to the plasma membrane. Traffic. 2008;9:540–58. doi: 10.1111/j.1600-0854.2007.00700.x. [DOI] [PubMed] [Google Scholar]
- 29.Ruiz-Mateos E, Pelchen-Matthews A, Deneka M, Marsh M. CD63 is not required for production of infectious human immunodeficiency virus type 1 in human macrophages. J Virol. 2008;82:4751–4761. doi: 10.1128/JVI.02320-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.von Lindern JJ, Rojo D, Grovit-Ferbas K, Yeramian C, Deng C, Herbein G, Ferguson MR, Pappas TC, Decker JM, Singh A, Collman RG, O’Brien WA. Potential role for CD63 in CCR5-mediated human immuno-deficiency virus type 1 infection of macrophages. J Virol. 2003;77:3624–3633. doi: 10.1128/JVI.77.6.3624-3633.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen H, Dziuba N, Friedrich B, von Lindern J, Murray JL, Rojo DR, Hodge TW, O’Brien WA, Ferguson MR. A critical role for CD63 in HIV replication and infection of macrophages and cell lines. Virology. 2008;379:191–196. doi: 10.1016/j.virol.2008.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harrington RD, Geballe AP. Cofactor requirement for human immunodeficiency virus type 1 entry into a CD4-expressing human cell line. J Virol. 1993;67:5939–5947. doi: 10.1128/jvi.67.10.5939-5947.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vodicka MA, Goh WC, Wu LI, Rogel ME, Bartz SR, Schweickart VL, Raport CJ, Emerman M. Indicator cell lines for detection of primary strains of human and simian immunodeficiency viruses. Virology. 1997;233:193–198. doi: 10.1006/viro.1997.8606. [DOI] [PubMed] [Google Scholar]
- 34.O’Brien WA, Koyanagi Y, Namazie A, Zhao JQ, Diagne A, Idler K, Zack JA, Chen IS. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature. 1990;348:69–73. doi: 10.1038/348069a0. [DOI] [PubMed] [Google Scholar]
- 35.Rich EA, Chen IS, Zack JA, Leonard ML, O’Brien WA. Increased susceptibility of differentiated mononuclear phagocytes to productive infection with human immunodeficiency virus-1 (HIV-1) J Clin Invest. 1992;89:176–183. doi: 10.1172/JCI115559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Folks TM, Kessler SW, Orenstein JM, Justement JS, Jaffe ES, Fauci AS. Infection and replication of HIV-1 in purified progenitor cells of normal human bone marrow. Science. 1988;242:919–22. doi: 10.1126/science.2460922. [DOI] [PubMed] [Google Scholar]
- 37.Butler SL, Hansen MS, Bushman FD. A quantitative assay for HIV DNA integration in vivo. Nat Med. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- 38.Zack JA, Arrigo SJ, Weitsman SR, Go AS, Haislip A, Chen IS. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 39.Friedrich B, Li G, Dziuba N, Ferguson MR. Quantitative PCR used to assess HIV-1 integration and 2-LTR circle formation in human macrophages, peripheral blood lymphocytes and a CD4+ cell line. Virol J. 2010;7:354. doi: 10.1186/1743-422X-7-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Doherty U, Swiggard WJ, Jeyakumar D, McGain D, Malim MH. A sensitive, quantitative assay for human immunodeficiency virus type 1 integration. J Virol. 2002;76:10942–10950. doi: 10.1128/JVI.76.21.10942-10950.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bouazzaoui A, Kreutz M, Eisert V, Dinauer N, Heinzelmann A, Hallenberger S, Strayle J, Walker R, Rübsamen-Waigmann H, Andreesen R, von Briesen H. Stimulated trans-acting factor of 50 kDa (Staf50) inhibits HIV-1 replication in human monocyte-derived macrophages. Virology. 2006;356:79–94. doi: 10.1016/j.virol.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 42.Hamamoto S, Nishitsuji H, Amagasa T, Kannagi M, Masuda T. Identification of a novel human immunodeficiency virus type 1 integrase interactor, Gemin2, that facilitates efficient viral cDNA synthesis in vivo. J Virol. 2006;80:5670–5677. doi: 10.1128/JVI.02471-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krishnan V, Zeichner SL. Alterations in the expression of DEAD-box and other RNA binding proteins during HIV-1 replication. Retrovirology. 2004;1:42. doi: 10.1186/1742-4690-1-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar A, Zloza A, Moon RT, Watts J, Tenorio AR, Al-Harthi L. Active beta-catenin signaling is an inhibitory pathway for human immunodeficiency virus replication in peripheral blood mononuclear cells. J Virol. 2008;82:2813–2820. doi: 10.1128/JVI.02498-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 46.Booth AM, Fang Y, Fallon JK, Yang JM, Hildreth JE, Gould SJ. Exosomes and HIV Gag bud from endosome-like domains of the T cell plasma membrane. J Cell Biol. 2006;172:923–935. doi: 10.1083/jcb.200508014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nguyen DG, Booth A, Gould SJ, Hildreth JE. Evidence that HIV budding in primary macrophages occurs through the exosome release pathway. J Biol Chem. 2003;278:52347–52354. doi: 10.1074/jbc.M309009200. [DOI] [PubMed] [Google Scholar]
- 48.Rous BA, Reaves BJ, Ihrke G, Briggs JA, Gray SR, Stephens DJ, Banting G, Luzio JP. Role of adaptor complex AP-3 in targeting wild-type and mutated CD63 to lysosomes. Mol Biol Cell. 2002;13:1071–1082. doi: 10.1091/mbc.01-08-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Popik W, Pitha PM. Binding of human immunodeficiency virus type 1 to CD4 induces association of Lck and Raf-1 and activates Raf-1 by a Ras-independent pathway. Mol Cell Biol. 1996;16:6532–6541. doi: 10.1128/mcb.16.11.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krementsov DN, Weng J, Labele M, Roy NH, Thali M. Tetraspanins regulate cell-to-cell transmission of HIV-1. Retrovirol. 2009;6:64. doi: 10.1186/1742-4690-6-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bol SM, van Remmerden Y, Sietzema JG, Kootstra NA, Schuitemaker H, van't Wout AB. Donor variation in vitro HIV-1 susceptibility of monocyte-derived macrophages. Virology. 2009;390:205–211. doi: 10.1016/j.virol.2009.05.027. [DOI] [PubMed] [Google Scholar]
- 52.Chang J, Naif HM, Li S, Sullivan JS, Randle CM, Cunningham AL. Twin studies demonstrate a host cell genetic effect on productive human immunodeficiency virus infection of human monocytes and macrophages in vitro. J Virol. 1996;70:7792–7803. doi: 10.1128/jvi.70.11.7792-7803.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eisert V, Kreutz M, Becker K, Königs C, Alex U, Rübsamen-Waigmann H, Andreesen R, von Briesen H. Analysis of cellular factors influencing the replication of human immunodeficiency virus type I in human macrophages derived from blood of different healthy donors. Virology. 2001;286:31–44. doi: 10.1006/viro.2001.0940. [DOI] [PubMed] [Google Scholar]