Abstract
MircroRNA functions as tumor suppressor or promoter in hepatocellular carcinoma (HCC). Researchers have found that miR-365 expression was lower in HCC tissues compared with that in adjacent normal tissues. However, its prognostic significance and anti-proliferation effect in HCC remain to be clarified. In this study, we firstly found that miR-365 expression was lower in HCC tissues compared with that in adjacent normal tissues. Then, we analyzed miR-365 expression level and its clinicopathological and prognostic significance. Finally, overexpression of miR-365 inhibits HCC cell proliferation and migration in vitro. Our findings suggest that miR-365 expression was an independent poor prognostic factor for HCC patient overall survival and suppressed tumor cell growth. Therefore, miR-365 may serve as a valuable prognostic marker and promising target for HCC.
Keywords: miR-365, hepatocellular carcinoma, prognosis, proliferation
Introduction
Hepatocellular carcinoma (HCC) is the major pathologic type of liver cancer and the third most common cause of cancer death world-wide, with high incidence and death rates [1-6]. According to the statistics, approximately 750,000 new cases are diagnosed and 700,000 people die from it [7]. HCC is difficult to diagnose in early-stage and characterized by high frequency of recurrence, metastasis after surgical resection, and resistance to common chemotherapy and radiotherapy, resulting in poor survival [5,6]. Despite recent diagnostic and therapeutic advancements, poor prognosis is observed in a large portion of HCC patients. However, overall patient survival remains low [7]. Especially, the five year survival rate for late stage liver cancer is still well below 10% [8].
MicroRNAs (miRNAs) belong to non-coding small RNAs of approximately 22 nucleotides and regulate the translational inhibition of target mRNAs by base-pairing with their 3’-untranslated region (3’-UTR) [9-11]. The abnormal expression of miRNAs has been demonstrated in many types of cancer, whereby miRNAs function as oncogenes or tumor suppressors during tumor development and progression, indicating their potential as biomarkers for diagnosis and treatment [12-15]. Over the past few decades, growing evidence suggests that dysregulation of miRNAs may contribute to various types of cancers, including HCC [16]. In a previous study, researchers found that the expression of miR-365 was significantly downregulated compared with that in matched normal tissue [17]. However, the correlation between the expression of miR-365 and clinicopathological parameters in HCC patients, and the exact mechanisms of miR-365 in HCC development have not yet been previously reported.
In this study, we firstly demonstrated that miR-365 expression was lower in HCC tissues compared with that in adjacent normal tissues. Next, we analyzed miR-365 expression level and its clinicopathological and prognostic significance. Finally, we found that overexpression of miR-365 remarkably suppressed proliferation and migration capacities of HCC cell in vitro experiments. Our findings will help to elucidate the functions of miRNAs and their roles in tumorigenesis.
Materials and methods
Patients
All patients had signed informed consent forms and the study was approved by the Ethics Committee of The Third Xiangya Hospital of Central South University. The selection criteria for patients with HCC were as follows: (1) Pathologically confirmed patients with HCC and two pathologists respectively reviewed all of the cases. (2) The patients haven’t received preoperative anti-tumor therapy. (3) The patients had no other cancer history. All patients were diagnosed and treated between May 2006 and March 2014. The mean follow-up period was 41 months. Clinical staging was performed according to the 6th edition 2002 American Joint Committee on Cancer (AJCC) TNM staging system. Survival time was calculated from the date of the initial surgery to death.
Tissue specimens and cell lines
Human HCC and corresponding normal tissues (located > 2 cm away from the tumor) were obtained from 15 patients who underwent HCC resection at Xiangya Hospital Central South University. The other 32 cases of HCC tissue for test of miRNA-365 expression level were collected from the pathology department of The Third Xiangya Hospital of Central South University. Human HCC cell line HepG2 was purchased from the ATCC (Manassas, VA, USA). HepG2 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, USA), supplemented with 10% fetal bovine serum (FBS) at 37°C with 5% CO2 in a humidified atmosphere.
Quantitative real-time RT-PCR
miRNA from tissues and cells was extracted using TRIzol (Invitrogen) reagent according to the manufacture’s instruction. Real-time qRT-PCR was carried out as described previously [18]. The qRT-PCR primers for miR-365 were forward 5’-CGTAATGCCCCTAAAAAT-3’ and reverse 5’-GTGCAGGGTCCGAGGT-3’. Internal control U6 primers were forward 5’-CTCGCTTCGGCAGCACA-3’ and reverse 5’-AACGCTTCACGAATTTGCGT-3’. The relative expression level of miR-365 was normalized to that of U6 by using the 2−ΔΔCt cycle threshold method [11].
Cell transfection
Transient transfection was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Both miR-365 mimics and miRNA NC mimics were purchased from GenePharma (Shanghai, China). HepG2 cells were seeded and transfected with 50 nM miR-365 mimics or miRNA NC mimics. 48 h after transfection, the fluorescence microscopy and the qRT-PCR were used to check transfection efficiency.
Cell proliferation assay
The cell counting kit-8 (CCK-8, Dojindo Laboratories, Kumamoto, Japan) colorimetric assay was performed to detect cell proliferation. 48 h after transfection, cells were seeded in 96-well plates by 5 × 103 cells per well. Three repeated wells were set in each group. CCK-8 (10 μl) was added to each well at 24, 48, 72 h later, and incubation was performed at 37°C for 2 h. An ELIASA was used to detect absorbency value at 450 nm.
For the colony formation assay, HepG2 cells were seeded in 10 cm dishes (1000/plate) after transfection and maintained in complete culture medium for 21 days. Next, cells were fixed in 4% paraformaldehyde for 15 min and stained with GIMSA dye. Cells were photographed and the number of clones was calculated.
Scratch test
Scratch test was performed to confirm the influence of miR-365 on HCC cell migration. Cells were collected and seeded in 6-well plates at 5 × 105/well and cultured until a complete monolayer was achieved. And then the plate was scratched a straight line using a 1 mL tip. The migration of cells into the scratch-generated gap was observed under microscope cultures at 0, 24 and 48 h.
Statistical analysis
Statistical analyses were carried out using SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism 5 (GraphPad Software Inc., CA, USA). Difference of measurement data was assessed by Student’s t-test or one-way ANOVA. Count data were analyzed by the x2 or Fishers exact tests. Univariate survival analysis was performed using Kaplan-Meier method and the Log-rank test. Multivariate survival analysis was performed using the Cox multivariate analysis model. P < 0.05 was considered statistically significant.
Results
Expression levels of miR-365 in HCC
We used qRT-PCR to examined miR-365 expression in 15 pairs of HCC tissues and the corresponding noncancerous tissues. As shown in Figure 1A, the expression of miR-365 was significantly down-regulated in HCC tissues when compared with para-cancer tissues. The miR-365 level was significantly negatively correlated with clinical stage in HCC tissue specimens (shown in Figure 1B). MiR-365 expression in HCC tissues with various grades of differentiation was also remarkably different (shown in Figure 1C).
Figure 1.
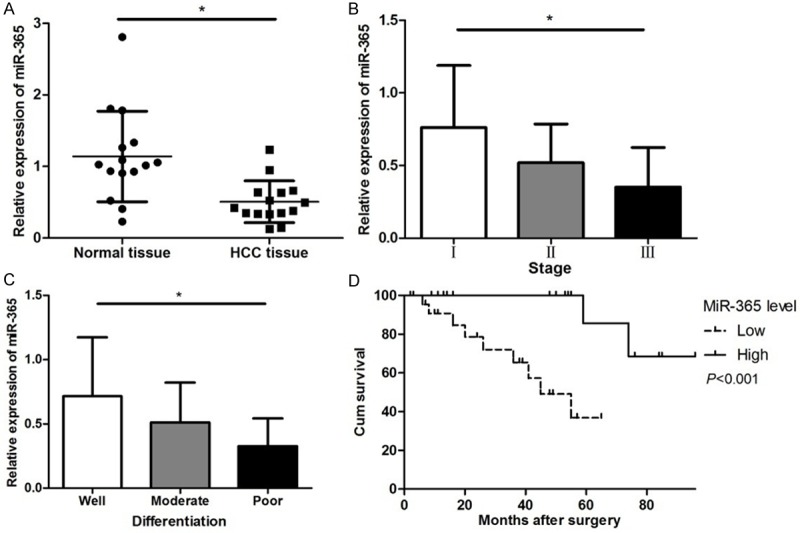
Expression levels of miR-365 and clinicopathological characteristics in HCC. A. Comparison of miR-365 expression levels between HCC tissues and adjacent normal tissues. B. The expression level of miR-365 in HCC patients at different clinical stages. C. The expression level of miR-365 at different differentiation of HCC tissues. D. Overall survival curve of HCC patients with different miR-365 expression. The patients with a lower expression of miR-365 had a lower survival time compared with those with a higher expression of miR-365. (*P < 0.05).
Expression levels of miR-365 and clinicopathological characteristics in HCC
The miR-365 expression levels were classified as high or low in relation to the median value. Low expression of miR-365 was found to significantly correlate with tumor size, clinical stage and tumor differentiation. However, there were no significant difference in miR-365 expression was observed with age, sex distribution, tumor number, cirrhosis and serum AFP value shown in Table 1).
Table 1.
Correlation between the expression of miR-365 and clinicopathological parameters in HCC patients
| Characteristics | Cases | n% | miR-365 expression | P value | ||
|---|---|---|---|---|---|---|
|
| ||||||
| High (n = 24) | Low (n = 23) | |||||
| Age (years) | ||||||
| < 50 | 24 | 65.22 | 11 | 13 | 0.464 | |
| ≥ 50 | 23 | 34.78 | 13 | 10 | ||
| Sex distribution | ||||||
| Male | 30 | 65.22 | 13 | 17 | 0.159 | |
| Female | 17 | 34.78 | 11 | 6 | ||
| Tumor number | 0.477 | |||||
| Solitary | 39 | 82.98 | 19 | 20 | ||
| Multiple | 8 | 17.02 | 5 | 3 | ||
| Tumor size (cm) | 0.029 | |||||
| ≤ 5 | 24 | 39.13 | 16 | 8 | ||
| > 5 | 23 | 45.65 | 8 | 15 | ||
| Vascular invasion | 0.071 | |||||
| Yes | 11 | 23.40 | 3 | 8 | ||
| No | 36 | 76.60 | 21 | 15 | ||
| Clinical stage | 0.018 | |||||
| I | 11 | 23.91 | 8 | 3 | ||
| II | 17 | 36.96 | 11 | 6 | ||
| III | 19 | 39.13 | 5 | 14 | ||
| Differentiation | 0.002 | |||||
| Well | 14 | 30.43 | 11 | 3 | ||
| Moderate | 19 | 39.14 | 11 | 8 | ||
| Poor | 14 | 30.43 | 2 | 12 | ||
| Cirrhosis | 0.900 | |||||
| Negative | 27 | 60.67 | 14 | 13 | ||
| Positive | 20 | 39.13 | 10 | 10 | ||
| AFP (μg/L) | 0.181 | |||||
| < 30 | 16 | 32.61 | 6 | 10 | ||
| ≥ 30 | 31 | 67.39 | 18 | 13 | ||
Low-expression level of miR-365 predicts poor prognosis in HCC patients
Considering that the level of miR-365 expression was remarkably correlated with tumor size, clinical stage and tumor differentiation, we hypothesized that miR-365 might affect the prognosis of HCC patients. Kaplan-Meier analysis with the log-rank test indicated that low miR-365 expression had a significant impact on OS (36.84% vs. 68.57%; P < 0.001; Figure 1D). Univariate and multivariate analyses were utilized to evaluate whether the miR-365 expression level and various clinicopathological features were independent prognostic parameters. Multivariate analysis revealed that miR-365 expression, clinical stage, tumor differentiation and cirrhosis were independently associated with the overall survival (shown in Table 2).
Table 2.
Multivariate analyses for overall survival by Cox regression model
| Wald | P-value | RR | RR 95% CI | ||
|---|---|---|---|---|---|
|
|
|||||
| Lower | Upper | ||||
| Age | 0.559 | 0.455 | 0.398 | 0.036 | 4.45 |
| Gender | 0.785 | 0.384 | 2.192 | 0.374 | 12.831 |
| Tumor size | 2.011 | 0.156 | 0.136 | 0.009 | 2.144 |
| Vascular invasion | 1.566 | 0.211 | 0.224 | 0.022 | 2.332 |
| Clinical stage | 5.842 | 0.016 | 56.803 | 2.146 | 1503.286 |
| Differentiation | 3.256 | 0.071 | 14.643 | 0.793 | 270.258 |
| Cirrhosis | 5.62 | 0.018 | 283.419 | 2.659 | 30203.530 |
| AFP | 2.529 | 0.112 | 0.039 | 0.001 | 2.126 |
| miR-365 | 4.523 | 0.033 | 0.003 | 0.000 | 0.637 |
Overexpression of miR-365 inhibits HepG2 cell proliferation ability
In order to further investigate the role of miR-365 in HCC, miR-365 mimics or NC mimics (Figure 2) was transfected into HepG2 cells. To elucidate whether miR-365 suppressed HepG2 cell proliferation, CCK-8 assays were employed. At 24, 48, 72 and 96 h, the proliferation rates in miR-365 mimic-transfected group (miR-365-HepG2) was significantly slower compared with either the untreated group (HepG2) or control (NC) (Figure 3A). This suggested that the up-regulation of miR-365 expression inhibits HepG2 cell proliferation ability. Colony formation assays were also used to evaluate cell proliferation and plating efficiency after overexpression of miR-365. The results showed that the number of colonies formed of miR-365 group was much lower than that in HepG2 and NC groups (Figure 3B). The results of the CCK-8 and colony formation assays therefore demonstrate that miR-365 overexpression limits HepG2 proliferation suggesting that it may inhibit HCC growth in vivo.
Figure 2.
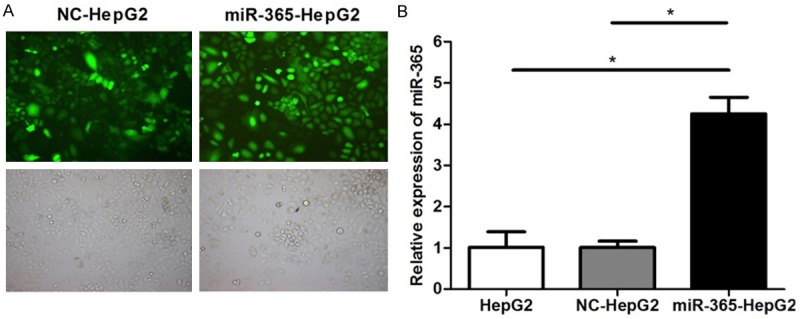
Overexpression of miR-365 in HepG2 cell. HepG2 cells were untransfected (HepG2) or transfected with NC mimics (NC-HepG2) or miR-365 mimics (miR-365-HepG2). A. Representative fluorescence microscopy images (× 100, top) and bright field microscopy images (× 100, bottom) are at left. B. Validation of miR-365 overexpression in HepG2 cells by qRT-PCR analysis. (*P < 0.05).
Figure 3.
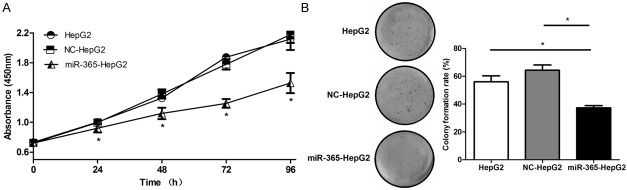
Overexpression of miR-365 inhibits HepG2 cell proliferation. A. CCK-8 assays were performed to examine HepG2 proliferation at the indicated time points. B. Colony formation assays were performed to evaluate HepG2 cell proliferation. (*P < 0.05).
Overexpression of miR-365 inhibits HepG2 cell migration ability
To evaluate the effect of miR-365 interaction on migration of HCC cells, scratch test was used. The results of the scratch test showed that that the closure of the gap area of miR-365 group was significantly less than that of HepG2 and NC-HepG2 groups (Figure 4), which indicated that the migratory ability of HepG2 cells was inhibited by miR-365.
Figure 4.
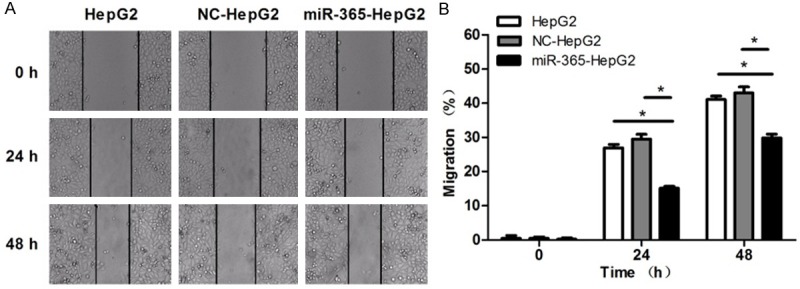
Overexpression of miR-365 inhibits HepG2 cell migration. Representative microscopic images of scratch-induced gap at the indicated time points (× 40) are on the left. Migration rate of each group at 0, 24 and 48 h is showed on the right. (*P < 0.05).
Discussion
A large number of studies suggest that understanding of miRNA function will provide us broad prospects to understand process of tumorigenesis and the behavior of cancer cells. Increasing evidence suggests that the dysregulation of miRNAs participates in HCC progression. For example, Zhang Y [19] et al. demonstrated that the down-regulation of miR-101 in clinical HCC tissues associates with tumor aggressiveness and worse survival of HCC patients. MiR-29b [20] and MiR-122 [21] have been characterized to have anti-metastatic and anti-angiogenic functions in HCC.
Reduced expression of miR-365 has been observed in different types of cancers. MiR-365 has been shown to downregulate expression in lung cancer and inhibit lung cancer cell proliferation through regulating NKX2-1 [22]. MiR-365 induces gemcitabine resistance in pancreatic cancer cells by targeting the proapoptotic molecules SHC1 and BAX [23]. However, miR-365 also has been found to promote cutaneous squamous cell carcinoma (CSCC) through targeting NFIB [24]. All these findings emphasize a fundamental role of miR-365 in development of malignant tumor.
Previously, researchers found that 29 microRNAs exhibited significantly differential expression in the HCC tissues compared to that of normal tissues via noncoding RNA microarray analysis, including miR-365 [17]. However, relevant function and mechanism remain to further explore.
In the present study, our results showed that miR-365 expression was significantly lower in HCC tissues compared with normal adjacent tissues. The aforementioned results were consistent with the study results of Hang Su [17] et al. The relationship of the miR-365 with various clinical features of HCC was analyzed. The results revealed that a low level of miR-365 expression was significantly correlated with tumor size, clinical stage and tumor differentiation, suggesting that miR-365 might be closely related with carcinogenesis and tumor development of HCC. Furthermore, the survival rate of low miR-365 expression group was significantly shorter than that of high miR-365 expression group. In a multivariate Cox model, we found that miR-365 expression was an independent poor prognostic factor for overall survival rate, indicating that low miR-365 level was significantly associated with poor OS in HCC patients. In addition, in order to know more about the effect of miR-365 in vitro, we overexpressed miR-365 in HepG2 cells. The cell proliferation and colony formation assays revealed that miR-365 suppresses proliferation of HepG2 cells. The results of scratch test showed that miR-365 inhibits cell migration ability. In future studies, we will collect more HCC tissues of patents and explore the targeted genes of miR-365 and downstream signaling pathways.
In conclusion, the expression of miR-365 was decreased in HCC. Low expression of miR-365 was significantly associated with tumor progression and decreased survival in patients with HCC. What’s more, miR-365 inhibited HCC cell proliferation and migration. These findings indicate that miR-365 serves as a novel prognostic marker and tumor suppressor in HCC.
Acknowledgements
This work was supported by Project of Natural Science Foundation of Hunan Province of China (No. 13JJ3036).
Disclosure of conflict of interest
None.
References
- 1.Huntzicker EG, Hotzel K, Choy L, Che L, Ross J, Pau G, Sharma N, Siebel CW, Chen X, French DM. Differential effects of targeting Notch receptors in a mouse model of liver cancer. Hepatology. 2014;61:942–952. doi: 10.1002/hep.27566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aravalli RN, Steer CJ, Cressman EN. Molecular mechanisms of hepatocellular carcinoma. Hepatology. 2008;48:2047–2063. doi: 10.1002/hep.22580. [DOI] [PubMed] [Google Scholar]
- 3.Santos NP, Oliveira PA, Arantes-Rodrigues R, Faustino-Rocha AI, Colaco A, Lopes C, Gil da Costa RM. Cytokeratin 7/19 expression in N-diethylnitrosamine-induced mouse hepatocellular lesions: implications for histogenesis. Int J Exp Pathol. 2014;95:191–198. doi: 10.1111/iep.12082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tu ZQ, Li RJ, Mei JZ, Li XH. Down-regulation of long non-coding RNA GAS5 is associated with the prognosis of hepatocellular carcinoma. Int J Clin Exp Pathol. 2014;7:4303–4309. [PMC free article] [PubMed] [Google Scholar]
- 5.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 6.Forner A, Hessheimer AJ, Isabel Real M, Bruix J. Treatment of hepatocellular carcinoma. Crit Rev Oncol Hematol. 2006;60:89–98. doi: 10.1016/j.critrevonc.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 7.Ho C, Wang C, Mattu S, Destefanis G, Ladu S, Delogu S, Armbruster J, Fan L, Lee SA, Jiang L, Dombrowski F, Evert M, Chen X, Calvisi DF. AKT (v-akt murine thymoma viral oncogene homolog 1) and N-Ras (neuroblastoma ras viral oncogene homolog) coactivation in the mouse liver promotes rapid carcinogenesis by way of mTOR (mammalian target of rapamycin complex 1), FOXM1 (forkhead box M1)/SKP2, and c-Myc pathways. Hepatology. 2012;55:833–845. doi: 10.1002/hep.24736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de Leon GP, Chen Y, Finkle D, Venook R, Wu X, Ridgway J, Schahin-Reed D, Dow GJ, Shelton A, Stawicki S, Watts RJ, Zhang J, Choy R, Howard P, Kadyk L, Yan M, Zha J, Callahan CA, Hymowitz SG, Siebel CW. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464:1052–1057. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
- 9.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 10.Zhang K, Zhang C, Liu L, Zhou J. A key role of microRNA-29b in suppression of osteosarcoma cell proliferation and migration via modulation of VEGF. Int J Clin Exp Pathol. 2014;7:5701–5708. [PMC free article] [PubMed] [Google Scholar]
- 11.Lu W, Xu Z, Zhang M, Zuo Y. MiR-19a promotes epithelial-mesenchymal transition through PI3K/AKT pathway in gastric cancer. Int J Clin Exp Pathol. 2014;7:7286–7296. [PMC free article] [PubMed] [Google Scholar]
- 12.Lu Y, Yue X, Cui Y, Zhang J, Wang K. MicroRNA-124 suppresses growth of human hepatocellular carcinoma by targeting STAT3. Biochem Biophys Res Commun. 2013;441:873–879. doi: 10.1016/j.bbrc.2013.10.157. [DOI] [PubMed] [Google Scholar]
- 13.Nelson KM, Weiss GJ. MicroRNAs and cancer: past, present, and potential future. Mol Cancer Ther. 2008;7:3655–3660. doi: 10.1158/1535-7163.MCT-08-0586. [DOI] [PubMed] [Google Scholar]
- 14.Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Braconi C, Henry JC, Kogure T, Schmittgen T, Patel T. The role of microRNAs in human liver cancers. Semin Oncol. 2011;38:752–763. doi: 10.1053/j.seminoncol.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang J, Gusev Y, Aderca I, Mettler TA, Nagorney DM, Brackett DJ, Roberts LR, Schmittgen TD. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res. 2008;14:419–427. doi: 10.1158/1078-0432.CCR-07-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y, Zhuang SM. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 2009;69:1135–1142. doi: 10.1158/0008-5472.CAN-08-2886. [DOI] [PubMed] [Google Scholar]
- 18.Xie P, Tian C, An L, Nie J, Lu K, Xing G, Zhang L, He F. Histone methyltransferase protein SETD2 interacts with p53 and selectively regulates its downstream genes. Cell Signal. 2008;20:1671–1678. doi: 10.1016/j.cellsig.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Guo X, Xiong L, Kong X, Xu Y, Liu C, Zou L, Li Z, Zhao J, Lin N. MicroRNA-101 suppresses SOX9-dependent tumorigenicity and promotes favorable prognosis of human hepatocellular carcinoma. FEBS Lett. 2012;586:4362–4370. doi: 10.1016/j.febslet.2012.10.053. [DOI] [PubMed] [Google Scholar]
- 20.Fang JH, Zhou HC, Zeng C, Yang J, Liu Y, Huang X, Zhang JP, Guan XY, Zhuang SM. MicroRNA-29b suppresses tumor angiogenesis, invasion, and metastasis by regulating matrix metalloproteinase 2 expression. Hepatology. 2011;54:1729–1740. doi: 10.1002/hep.24577. [DOI] [PubMed] [Google Scholar]
- 21.Bai S, Nasser MW, Wang B, Hsu SH, Datta J, Kutay H, Yadav A, Nuovo G, Kumar P, Ghoshal K. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J Biol Chem. 2009;284:32015–32027. doi: 10.1074/jbc.M109.016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang SM, Lee HJ, Cho JY. MicroRNA-365 regulates NKX2-1, a key mediator of lung cancer. Cancer Lett. 2013;335:487–494. doi: 10.1016/j.canlet.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 23.Hamada S, Masamune A, Miura S, Satoh K, Shimosegawa T. MiR-365 induces gemcitabine resistance in pancreatic cancer cells by targeting the adaptor protein SHC1 and pro-apoptotic regulator BAX. Cell Signal. 2014;26:179–185. doi: 10.1016/j.cellsig.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 24.Zhou M, Zhou L, Zheng L, Guo L, Wang Y, Liu H, Ou C, Ding Z. miR-365 promotes cutaneous squamous cell carcinoma (CSCC) through targeting nuclear factor I/B (NFIB) PLoS One. 2014;9:e100620. doi: 10.1371/journal.pone.0100620. [DOI] [PMC free article] [PubMed] [Google Scholar]


