Abstract
Tissue homeostasis requires balanced self-renewal and differentiation of stem/progenitor cells, especially in tissues that are constantly replenished like the esophagus. Disruption of this balance is associated with pathological conditions, including eosinophilic esophagitis (EoE), in which basal progenitor cells become hyperplastic upon proinflammatory stimulation. However, how basal cells respond to the inflammatory environment at the molecular level remains undetermined. We previously reported that the bone morphogenetic protein (BMP) signaling pathway is critical for epithelial morphogenesis in the embryonic esophagus. Here, we address how this pathway regulates tissue homeostasis and EoE development in the adult esophagus. BMP signaling was specifically activated in differentiated squamous epithelium, but not in basal progenitor cells, which express the BMP antagonist follistatin. Previous reports indicate that increased BMP activity promotes Barrett’s intestinal differentiation; however, in mice, basal progenitor cell–specific expression of constitutively active BMP promoted squamous differentiation. Moreover, BMP activation increased intracellular ROS levels, initiating an NRF2-mediated oxidative response during basal progenitor cell differentiation. In both a mouse EoE model and human biopsies, reduced squamous differentiation was associated with high levels of follistatin and disrupted BMP/NRF2 pathways. We therefore propose a model in which normal squamous differentiation of basal progenitor cells is mediated by BMP-driven NRF2 activation and basal cell hyperplasia is promoted by disruption of BMP signaling in EoE.
Keywords: Gastroenterology, Stem cells
Introduction
Constant wear-and-tear of squamous epithelium in tissues like the skin and esophagus requires rapid and coordinated self-renewal and differentiation of stem/progenitor cells to replenish the tissue (1, 2). In the esophagus, basal cells serve as progenitor cells to maintain the stratified squamous epithelium (2, 3). Disruption of the balance of basal cell activities underlies multiple pathological conditions, including eosinophilic esophagitis (EoE), in which basal cells become hyperplastic in the setting of high levels of the inflammatory cytokines eotaxin-3, IL-5, and IL-13 (4–6). Studies have shown that these hyperplastic basal cells serve as an important source of cytokines and chemokines that facilitate EoE disease progression (7, 8). However, little is known about how basal progenitor cells respond to the inflammatory environment in the initial stages of the disease. This is partly due to a lack of understanding of the normal mechanisms maintaining epithelial restitution in the esophagus.
Bone morphogenetic proteins (BMPs) belong to the TGF-β superfamily of cytokines and activate downstream signaling through the formation of complexes with type I (BMPR1A or BMPR1B) and type II (BMPR2) receptors. BMP signaling activities can be modulated at multiple levels. For example, BMP antagonists, including noggin, follistatin, and gremlins, bind to BMPs (e.g., BMP4 and BMP7) and block their engagement with the receptors (9, 10). Our previous studies have shown that the BMP pathway is required for morphogenesis of the epithelium in the embryonic esophagus. BMP plays a 2-stage role during the conversion of simple columnar to stratified squamous epithelium in esophageal development. First, noggin suppresses BMP signaling activities to allow epithelial stratification to occur. Second, BMP is activated in the top layers of epithelial cells to drive their differentiation into squamous cells (11). Intriguingly, alterations in BMP signaling activity have been found in multiple esophageal diseases, including gastroesophageal reflux disease (GERD) and Barrett’s esophagus (BE) (12, 13). In addition, microarray analysis of EoE human biopsies has revealed changes in the levels of BMP signaling components (14). However, how changes in the BMP pathway are involved in the disease process remains unknown.
Oxidative stress caused by the intracellular accumulation of ROS contributes to disease and cell death. In contrast to the damaging effects of ROS, there is evidence that ROS at lower, nontoxic levels is important for the regulation of stem cell function in some tissues (15, 16). For example, low levels of endogenous ROS in the hematopoietic system are correlated with stem cell quiescence, whereas high levels of ROS drive stem cells into cell cycling, leading to stem cell exhaustion (17). These findings suggest a more complex role of intracellular ROS levels in cellular biology than was first understood by models of oxidative stress. Oxidative stress responses are mediated by nuclear factor erythroid 2–related factor 2 (NRF2) signaling. Under normal, unstressed circumstances, low cellular levels of NRF2 are maintained through KEAP1-dependent ubiquitination. Upon oxidative stress, ubiquitination processes are blocked, leading to the stabilization and nuclear translocation of NRF2 and subsequent induction of NRF2 target genes including NAD(P)H quinine oxidoreductase-1 (Nqo1), which encodes an enzyme critical for reducing ROS levels (18–20). Interestingly, loss of Keap1 leads to increased NRF2 activation and excessive differentiation of the epithelium in the embryonic esophagus and forestomach, where the lining epithelium is also stratified squamous (21–24). In addition, a recent study showed that NRF2 activation is associated with the pathogenesis of GERD, and its deficiency impairs the maintenance of the barrier function of the epithelium (21), suggesting that the ROS pathway continues to play important roles in the adult esophagus. Here, we used multiple mouse genetic models to reveal what we believe to be a novel finding that BMP activation elicits NRF2-mediated oxidative responses and promotes squamous differentiation of basal progenitor cells. In addition, we found that the BMP pathway is inhibited by increased levels of follistatin in EoE mouse models and human biopsies.
Results
BMP activation in the differentiated suprabasal cells of the adult mouse esophagus.
We first used the BRE-lacZ transgenic reporter line to survey BMP activities in the adult esophagus. In this mouse line, the expression of β-gal is controlled by BMP response elements (BREs) from the human ID1 gene (25). β-gal activity was detected in the suprabasal cells and within the subepithelial compartment, but not in basal progenitor cells labeled with the transcription factor p63 (26) (Figure 1, A and B). Previous studies have shown that several BMP ligands, including BMP4 and BMP7, are expressed in the developing mouse esophagus (11, 27). We asked whether the expression of these ligands is maintained in adults. X-gal staining revealed that BMP4-lacZ was expressed in the mesenchymal cells adjacent to the basal layer, similar to that observed in the developing esophagus (27). Interestingly, we also detected β-gal activity in subpopulations of basal progenitor cells (Figure 1C). By contrast, BMP7 was widely expressed in all of the epithelium, including in the cells at the basal and suprabasal layers in the BMP7-lacZ mouse (Figure 1D). Similarly, we found that BMPR1A (also named ALK3) and BMPR2 were expressed in the epithelium (Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI78850DS1).
Figure 1. BMP signaling activity in the stratified squamous epithelium of the adult mouse esophagus.
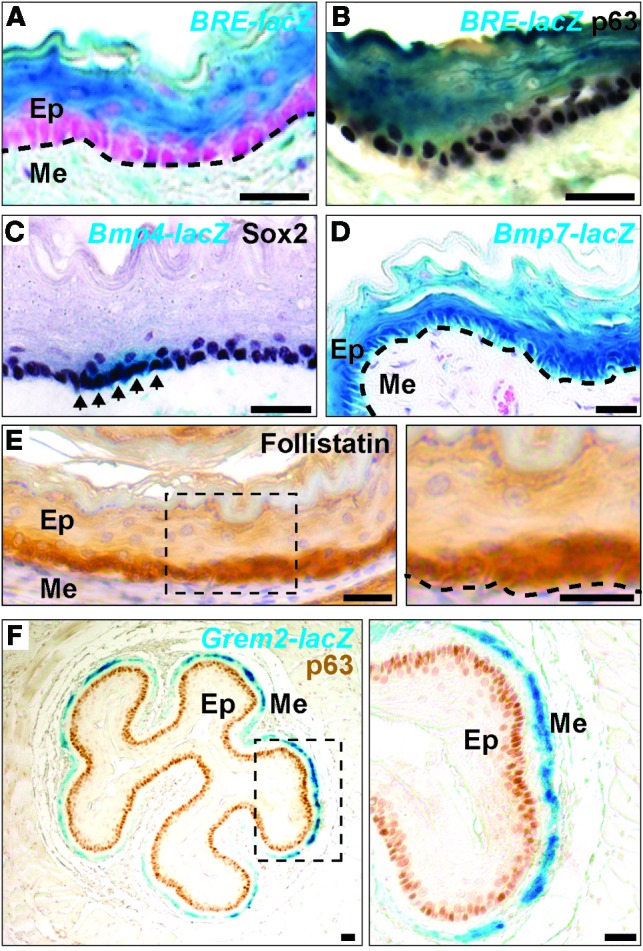
(A and B) BMP signaling is active in differentiated suprabasal cells, but not in basal progenitor cells, as shown by the BMP reporter allele BRE-lacZ. The undifferentiated basal progenitor cells are labeled with p63. Note that some mesenchymal cells in the lamina propria were also positive for X-gal staining. Nuclei were counterstained with nuclear fast red. (C) BMP4 is expressed in subpopulations (arrows) of basal progenitor cells, as shown by the Bmp4-lacZ allele. Basal progenitor cells are labeled with the SOX2 transcription factor. (D) BMP7 was expressed in all of the epithelium, including in basal and suprabasal cells, as demonstrated by the Bmp7-lacZ allele. (E) The BMP inhibitor follistatin was enriched in basal progenitor cells. The boxed region is shown enlarged at right (original magnification, ×40). (F) The BMP inhibitor gremlin 2 was expressed in the muscularis mucosa beneath the lamina propria layer. The boxed region is shown enlarged at right (original magnification, ×40). Dotted line in A, B, D, and E indicates the basement membrane. Ep, epithelium; Me, mesenchyme. Scale bars: 50 μm.
We then asked whether the absence of BMP signaling activity in basal progenitor cells is due to the presence of BMP antagonists. We determined the expression of different BMP antagonists using real-time RT-PCR, immunostaining, and ISH and found that follistatin was enriched in basal progenitor cells (Figure 1E). Low levels of chordin were also detected in the epithelium and adjacent mesenchymal cells (Supplemental Figure 1B). By contrast, the expression of gremlin 2 (Grem2), as revealed by the reporter allele Grem2-lacZ, was restricted to the smooth muscle layer (muscularis mucosa) (Figure 1F), while noggin-lacZ and Grem1-lacZ were weakly expressed in muscle and mesenchymal cells, respectively (Supplemental Figure 1, C and D). Together, these findings suggest that differentiation of the squamous epithelium in the adult mouse esophagus is correlated with BMP activation and that basal cells are protected from BMP activation by the BMP antagonist follistatin.
Activation of BMP signaling inhibits the proliferation and induces the squamous differentiation of basal progenitor cells in vitro.
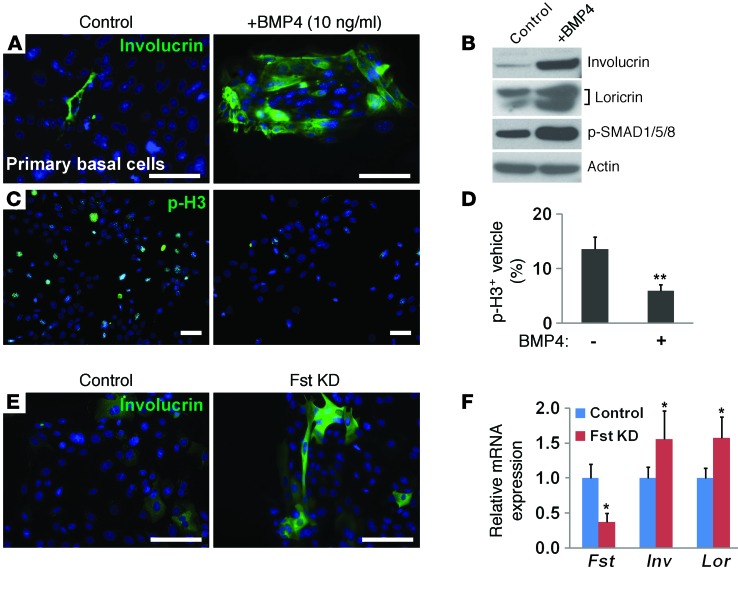
We have previously shown that basal cells are able to proliferate in vitro in serum-free medium supplemented with EGF and FGF (28). After 1 week in culture, more than 95% of the cells maintained p63 expression, suggesting that they were undifferentiated progenitor cells (Supplemental Figure 2, A and B). To directly test whether activated BMP signaling is able to promote the differentiation of basal cells, we cultured mouse basal progenitor cells isolated with the cell surface marker p75 (29) in serum-free medium in the presence of 10 ng/ml BMP4, which is similar to BMP7 in its capability to stimulate BMP activation (30). In addition, we and others have shown that removal of 1 copy of either Bmp4 or Bmp7 is able to rescue foregut defects in noggin-null mutants, suggesting that these 2 ligands have similar potential (27, 31). BMP4 treatment for 5 days led to the expression of the squamous differentiation marker involucrin (Figure 2A). Consistent with the activated BMP signaling and extensive squamous differentiation, high levels of phosphorylated SMAD1/5/8 (p-SMAD1/5/8), involucrin, and loricrin were detected in BMP4-treated cells by Western blot analysis (Figure 2B). Moreover, we found that the differentiation process was accompanied by a 2.3-fold decrease in proliferation, as indicated by phosphorylated histone H3 (p-H3) staining (Figure 2, C and D). BMP4 treatment also reduced the size of esophageospheres formed in a 3D culture system that was initially embedded with single basal progenitor cells (Supplemental Figure 2C).
Figure 2. BMP activation inhibits proliferation and induces squamous differentiation of primary mouse basal progenitor cells in vitro.
(A) BMP4 treatment for 5 days induced the expression of the differentiation marker involucrin in basal progenitor cells. (B) BMP4 treatment led to increased protein levels of involucrin, loricrin, and p-SMAD1/5/8 as shown by Western blot analysis. Actin served as a loading control. (C and D) Proliferation of the epithelium was inhibited by BMP4 treatment. Proliferating cells were labeled with p-H3. Data in D show that the proportion of p-H3 plus vehicle cells in all epithelial cells was significantly reduced after BMP4 treatment (n = 3). (E) Knockdown of follistatin promoted squamous differentiation of basal progenitor cells as indicated by involucrin staining. (F) Knockdown of follistatin led to increased involucrin and loricrin transcript levels (n = 3). Data represent the mean ± SEM. *P < 0.05 and **P < 0.01 by Student’s t test. Scale bars: 50 μm. Fst, follistatin; Inv, involucrin; Lor, loricrin; KD, knockdown.
We then asked whether the presence of the BMP inhibitor follistatin in basal cells is required for maintenance of the undifferentiated state. Significantly, we found that siRNA-mediated downregulation of follistatin promoted the squamous differentiation of basal progenitor cells, as evidenced by increased protein and transcript levels of involucrin and loricrin (Figure 2, E and F). This finding further supports the notion that follistatin is critical for protecting basal progenitor cells from BMP activation and differentiation.
Moreover, activation of BMP signaling also promoted the squamous differentiation of human esophageal progenitor cells. BMP4 treatment promoted differentiation and inhibited the proliferation of primary basal cells isolated from the human esophagus and telomerase-immortalized human esophageal progenitor–like EPC2 cells (EPC2-hTERT) (Supplemental Figure 2, D–F, data not shown, and refs. 28, 32). Notably, we did not detect expression of the columnar differentiation markers KRT8 or CDX2 (Supplemental Figure 2, G–I), suggesting that BMP activation does not promote columnar differentiation of basal progenitor cells. These findings contrast with previous reports that BMP activation is associated with intestinal differentiation of the squamous epithelium (12, 33), one of the hypothetical models for the development of BE.
BMP activation leads to extensive squamous differentiation of esophageal basal progenitors in Krt5-CreER R26caBmpr1a mice.
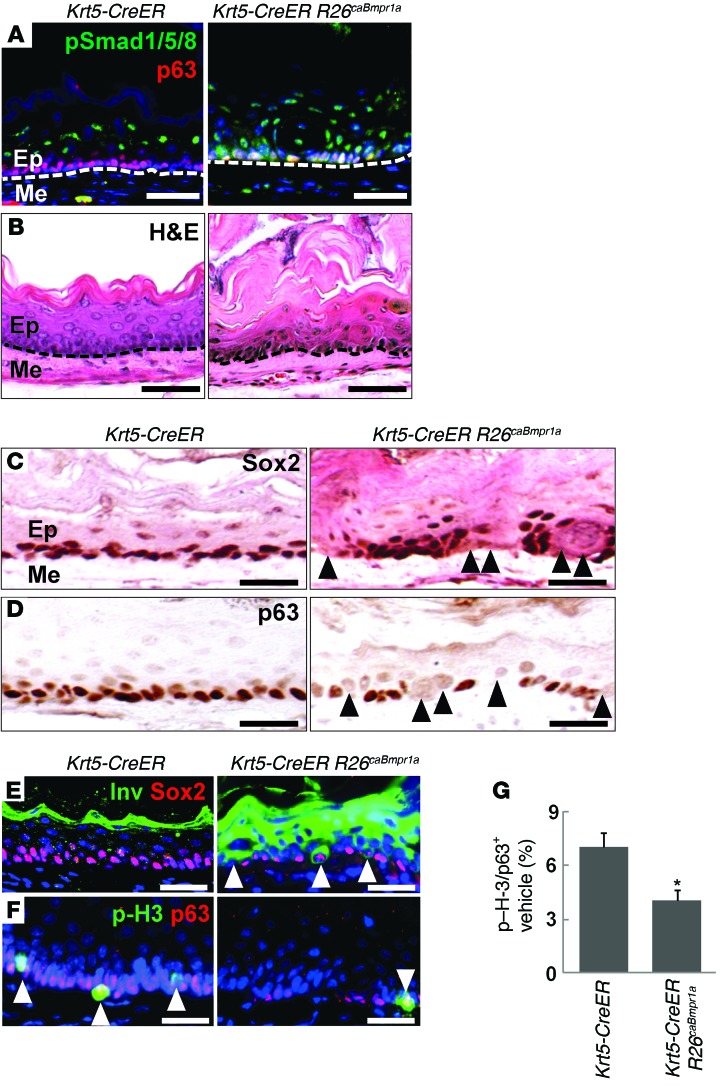
To further test whether activation of the BMP pathway drives squamous, but not intestinal, differentiation of basal progenitor cells in vivo, we generated Krt5-CreER R26caBmpr1a mutant mice in which a constitutively active form of BMPR1A (caBmpr1a) is overexpressed in basal cells and in their derivatives following tamoxifen injection (Supplemental Figure 3, A and B, and refs. 11, 28). Ectopic BMP signaling activity was confirmed by p-SMAD1/5/8 staining in basal cells following 4 injections of tamoxifen (Figure 3A). More important, we found that thickening of keratin layers (hyperkeratosis) developed at the top of the epithelium, indicating excessive squamous differentiation (Figure 3B). In keeping with this finding, segments of basal cells decreased or lost expression of the progenitor markers SOX2 and p63 (Figure 3, C and D), and there was abundant expression of involucrin by immediate suprabasal epithelial cells and by scattered basal epithelial cells (Figure 3E). In addition, ectopic BMP activation also led to a 40% decrease in the proliferation of basal cells as revealed by p-H3 staining (Figure 3, F and G).
Figure 3. Activation of BMP signaling induces extensive squamous differentiation of basal progenitors in the esophagus of Krt5-CreER R26caBmpr1a mutants.
(A) Activation of BMP signaling was detected by p-SMAD1/5/8 immunostaining. Note the ectopic signals in the basal layer. (B) Hyperkeratosis indicative of excessive squamous differentiation developed in response to ectopic BMP activation in basal cells. (C and D) Blocks of basal cells lost SOX2 and p63 expression in the mutant esophagus 1 week after the last tamoxifen injection. (E) Ectopic BMP activation was shown to drive premature squamous differentiation. Arrowheads indicate enlarged cells in the basal and immediate suprabasal layers expressing involucrin. Note that a few differentiating basal cells maintained low levels of SOX2. (F and G) Proliferation of basal cells was reduced in the mutant esophagus, as shown by p-H3 staining (arrowheads). BMP activation significantly reduced cell proliferation (n = 3). Data represent the mean ± SEM. *P < 0.05 by Student’s t test. Scale bars: 50 μm.
Consistent with the in vitro studies, we found that ectopic BMP signaling did not promote expression of the columnar differentiation markers KRT8 or KRT20. We also did not detect the expression of CDX1 or CDX2, both markers for intestinal metaplasia, even in mice that had ectopic BMP activation for up to 14 months (Supplemental Figure 3, C–E, and data not shown). Taken together, these in vivo studies further demonstrate that BMP activation promotes squamous rather than columnar differentiation of basal progenitor cells, accompanied by a reduction in proliferation of these cells.
BMP activation leads to increased intracellular ROS levels and NRF2 function in vitro and in vivo.
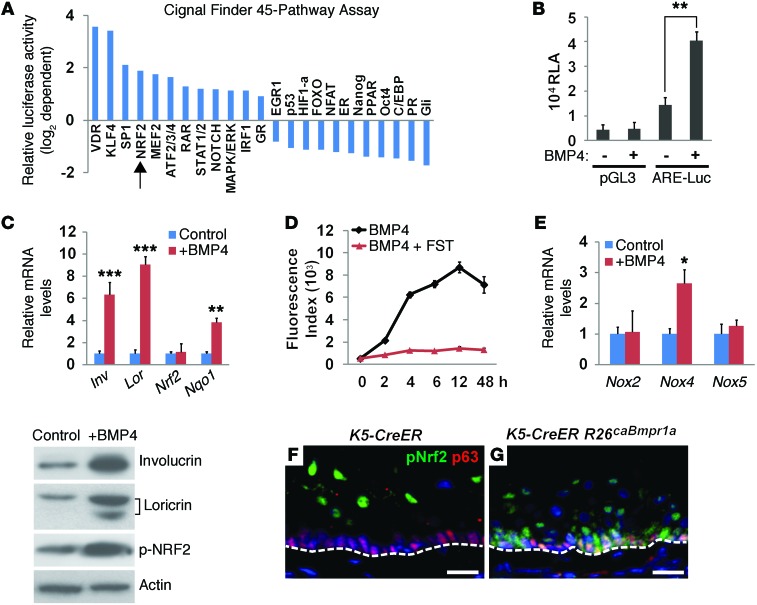
To identify signaling pathways potentially involved in BMP4-induced basal cell differentiation, we used the Cignal Finder 45-Pathway Reporter Arrays, which can survey changes in the activities of 45 pathways via luciferase reporters. BMP4 treatment for 48 hours induced various changes in multiple pathways (Figure 4A and Supplemental Table 1). Notably, the most obvious changes have been found in pathways that are known to be associated with epithelial differentiation including vitamin D receptor (VDR), Kruppel-like factor 4 (KLF4), SP1, and NRF2 (24, 34–36). Intriguingly, excessive squamous differentiation with hyperkeratosis also occurs in the esophagus of Keap1-null mutants, in which NRF2 activity is increased (24). To further confirm that the NRF2 pathway is activated upon BMP4 treatment, we used the antioxidant response element (ARE) (NRF2-binding elements) reporter assay and found that BMP4 induced an approximate 3-fold increase in the luciferase activity of EPC2 cells (Figure 4B). In agreement with this finding, transcript levels of the NRF2 downstream target Nqo1 were increased with BMP4 treatment in EPC2 cells (Figure 4C). Notably, although Nrf2 transcript levels did not change dramatically with BMP4 treatment, p-NRF2 levels were significantly increased (Figure 4C). Increased NFR2 activity is a known reactive response to increased ROS levels (37). Consistently, we also observed increased ROS levels, as indicated by high levels of dichlorodihydrofluorescein (DCF) fluorescence upon BMP4 treatment. This increase was time dependent, and the signals peaked at 12 hours after BMP4 treatment. Moreover, the BMP inhibitor follistatin blocked the ROS production induced by BMP4 (Figure 4D), further confirming the specificity of BMP4 in increasing ROS levels. In addition, we found a 2.5-fold increase in the expression levels of NADPH oxidase 4 (Nox4) mRNA (Figure 4E). NOX proteins facilitated the generation of intracellular ROS, suggesting a functional response to BMP4-induced oxidative stress.
Figure 4. Activation of BMP signaling increases intracellular ROS levels and NRF2 activity.
(A) BMP4 treatment altered the activity of multiple intracellular signaling pathways including the NRF2-mediated oxidative stress pathway (arrow) in EPC2 cells. (B) BMP4 treatment promoted NRF2-mediated transcriptional activation as revealed by the ARE reporter assay. EPC2 cells expressing the carrier vector pGL3 were used as controls (n = 3). Luc, luciferase; RLA, relative luciferase activity. (C) BMP4 treatment increased the transcript levels of Nqo1, a NRF2 downstream target, which was accompanied by increased levels of involucrin and loricrin (n = 3). BMP4 treatment also increased p-NRF2, involucrin, and loricrin protein levels. Note that there were no significant changes in total Nrf2 transcript levels. (D) BMP4 treatment increased ROS levels in a time-dependent manner as visualized by DCF fluorescence intensity, while the presence of the BMP antagonist follistatin inhibited BMP4-induced ROS production. Data represent the average fluorescence intensity from 3 individual experiments. (E) BMP4 treatment increased Nox4 transcript levels, indicating increased intracellular ROS in EPC2 cells as measured by real-time PCR (n = 3). (F and G) BMP activation promoted nuclear accumulation of p-NRF2 in esophageal basal progenitor cells from Krt5-CreER R26caBmpr1a mutants. Note that p-NRF2 was enriched in the differentiated suprabasal cells of the control esophagus. Data represent the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001 by Student’s t test. Scale bars: 50 μm.
In agreement with previous findings, nuclear accumulation of p-NRF2 is limited to differentiated suprabasal cells in the esophagus (Figure 4F and ref. 21). Upon ectopic BMP activation, p-NRF2 was also found in the esophageal basal cells of Krt5-CreER R26caBmpr1a mutants (Figure 4G). Taken together, these in vitro and in vivo findings suggest that BMP activates NRF2 signaling during squamous differentiation of basal progenitor cells.
NRF2 and increased ROS levels are required for squamous differentiation of basal progenitor cells.
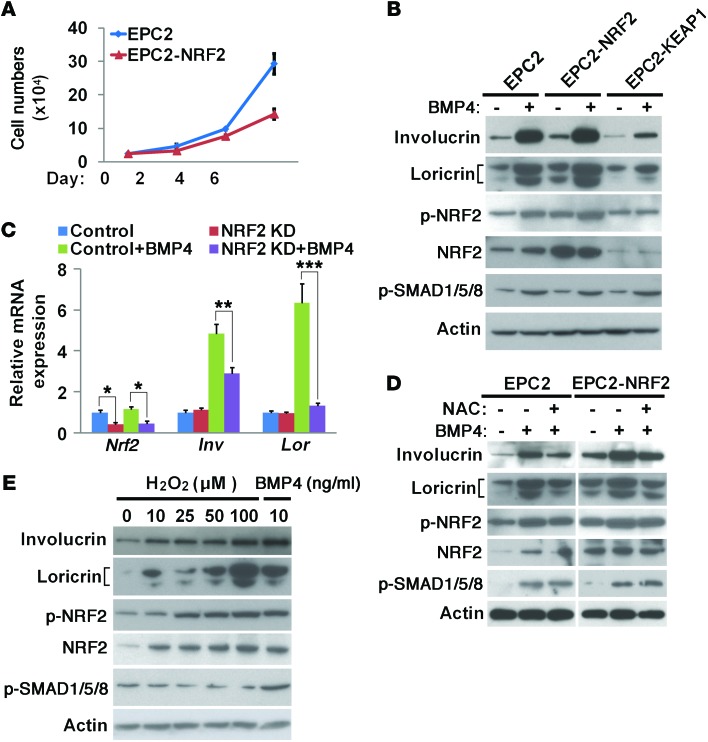
To determine whether NRF2 is directly involved in BMP-mediated squamous differentiation, we generated EPC2 cells overexpressing NRF2 (EPC2-NRF2). NRF2 overexpression reduced the proliferation of EPC2 cells and increased the protein levels of involucrin and loricrin (Figure 5, A and B, and Supplemental Figure 4, A and B). This increase was enhanced by combined treatment with BMP4 (Figure 5B and Supplemental Figure 4, A and B). Conversely, when NRF2 function was suppressed through siRNA-mediated NRF2 knockdown or overexpression of its inhibitor KEAP1, involucrin and loricrin proteins and mRNAs were maintained at low levels, even in the presence of BMP4 (Figure 5, B and C, and Supplemental Figure 4, A and B). In addition, attenuation of intracellular ROS levels with the antioxidant reagent N-acetylcysteine (NAC) also led to low levels of involucrin in the presence of BMP4 (Figure 5D). Conversely, increased intracellular levels of ROS with H2O2 treatment promoted EPC2 cell differentiation in a concentration-dependent manner (Figure 5E). To test whether increased ROS or NRF2 activity can in turn affect BMP signaling in EPC2 cells, we measured the expression of p-SMAD1/5/8 and found that its protein levels were not changed (Figure 5, B, D, and E), suggesting that ROS production and NRF2 activation are downstream events following BMP pathway activation. Collectively, these findings demonstrate that increased ROS and NRF2 activity mediates BMP signaling–induced squamous differentiation of basal progenitor cells.
Figure 5. NRF2 signaling is required for the squamous differentiation of basal progenitor cells.
(A) Overexpression of NRF2 reduced proliferation in EPC2 cells (n = 3). (B) NRF2 overexpression enhanced BMP4-induced squamous differentiation of basal progenitors. By contrast, NRF2 inhibition through KEAP1 overexpression reduced squamous differentiation in BMP4-treated EPC2 cells. NRF2 phosphorylation levels were used as indicators of NRF2 signaling. Total NRF2 levels indicated NRF2 overexpression and inhibition by KEAP1 overexpression. (C) Knockdown of NRF2 inhibited BMP4-induced squamous differentiation of basal progenitor cells as indicated by reduced transcript levels of involucrin and loricrin (n = 3). Cells without BMP4 treatment were used as controls. (D) Attenuation of intracellular ROS levels with the antioxidant reagent NAC inhibited squamous differentiation in the presence of BMP4. (E) H2O2 treatment increased NRF2 signaling and promoted squamous differentiation in a concentration-dependent manner. EPC2 cells treated with BMP4 (10 ng/ml) were used as positive controls. Blot images were derived from samples run on parallel gels. Data represent the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001 by 2-way ANOVA (C).
Expansion of basal progenitor cells is associated with reduced BMP and NRF2 signaling in an IL-13–induced EoE mouse model and in human biopsies.
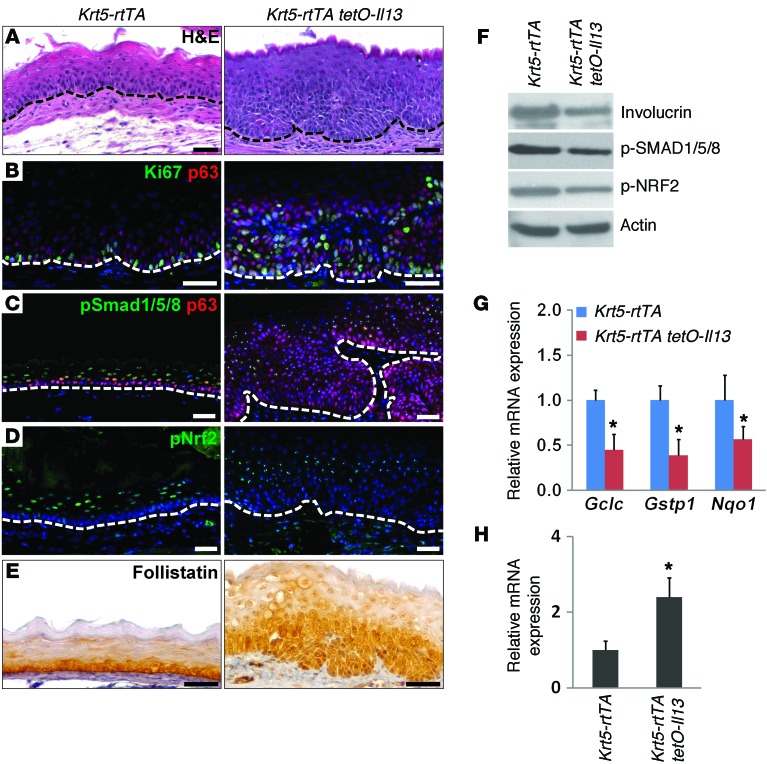
We next tested whether basal cell hyperplasia in EoE is associated with abnormal BMP/NRF2 signaling activity. Previous studies have shown that intratracheal delivery or transgenic overexpression of IL-13 in the lung epithelium leads to EoE-like phenotypes in the esophagus, as evidenced by accumulation of eosinophils and basal cell hyperplasia (38). It was postulated that IL-13 is produced in the lung and swallowed to induce pathological changes in the esophagus (39). To directly test the effects of IL-13 on the esophageal epithelium, we generated Krt5-rtTA tetO-Il13 mice in which IL-13 is specifically overexpressed in basal progenitor cells with the presence of doxycycline (Dox) (Supplemental Figure 5A). As expected, the transcript levels of I13 and its downstream target eotaxin 1 were significantly increased in the esophageal epithelium, and this was accompanied by accumulation of eosinophils upon Dox feeding (Supplemental Figure 5, B and C, and data not shown). More important, expansion of basal progenitor cells became prominent throughout the esophagus 2 weeks after IL-13 induction (Figure 6A). Ki67 immunostaining showed that proliferation of basal cells increased by 1.8-fold (Figure 6B and Supplemental Figure 5D). IL-13 overexpression also blocked the differentiation of basal progenitor cells as measured by involucrin expression (Figure 6F). We next assayed BMP activity in the esophagus. Notably, BMP activity was decreased in the epithelium as revealed by p-SMAD1/5/8 staining and immunoblotting (Figure 6, C and F), concomitant with a reduction in the levels of p-NRF2 (Figure 6, D and F). Consistent with this finding, transcript levels of the NRF2 downstream target Nqo1 were reduced. Additionally, transcript levels of glutamate–cysteine ligase catalytic subunit (Gclc) and glutathione S-transferase P1 (Gstp1), 2 genes critical for ROS production, were also reduced (Figure 6G). Furthermore, follistatin protein levels increased in the epithelium upon IL-13 induction, and its transcript levels also increased by 2.4-fold (Figure 6, E and H). Together, these results indicate that high levels of IL-13 in the mouse esophagus promote prominent basal cell hyperplasia, accompanied by increased levels of follistatin, which in turn inhibits BMP and NRF2 signaling.
Figure 6. Basal cell hyperplasia is associated with decreased BMP signaling in a mouse model of EoE.
(A) The epithelium was hyperplastic 2 weeks after IL-13 overexpression in basal cells of the esophagus in Krt5-rtTA tetO-Il13 compound mice. (B) Basal cell hyperplasia was accompanied by increased proliferation as indicated by Ki67 immunostaining upon IL-13 overexpression. (C) BMP signaling was reduced in the hyperplastic epithelium. (D) Nuclear localization of p-NRF2 was also reduced after IL-13 overexpression. (E) Follistatin accumulated in the hyperplastic basal cells of the mutant esophagus. (F) Protein levels of involucrin, p-SMAD1/5/8, and p-NRF2 in the esophageal epithelium were decreased upon IL-13 overexpression. Actin was used as a loading control. (G) Transcript levels of Gclc, Gstp1, and Nqo1, genes involved in ROS production and NRF2 signaling, were decreased upon IL-13 overexpression as measured by real-time PCR (n = 5). (H) Fst transcript levels were significantly increased after IL-13 overexpression as measured by real-time PCR (n = 5). Data represent the mean ± SEM. *P < 0.05 by Student’s t test. Scale bars: 50 μm.
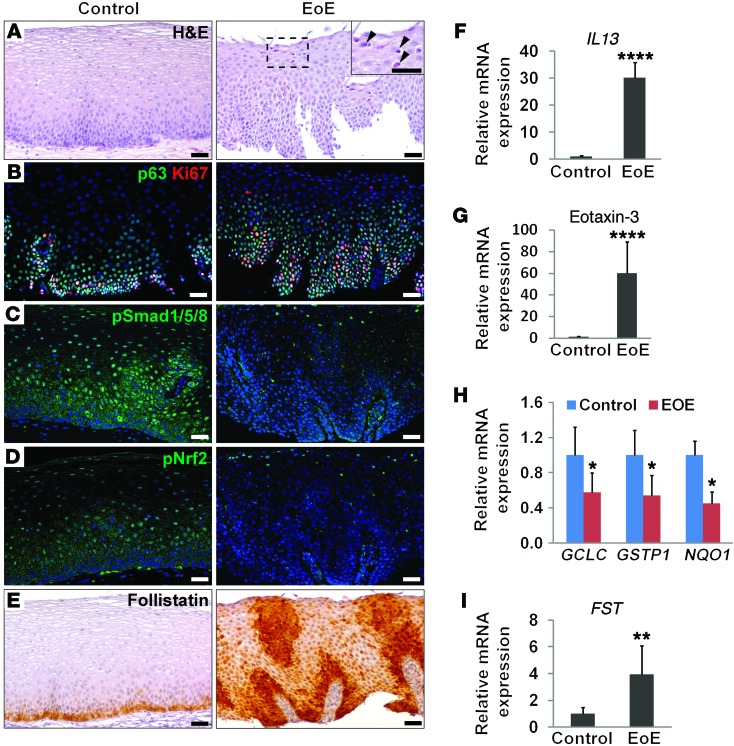
We then asked whether BMP activity is similarly altered in human EoE biopsies. As expected, infiltration of eosinophils and high levels of IL13 and eotaxin-3 were detected in the esophagus (Figure 7, A, F, and G). Basal cell hyperplasia in these biopsies was associated with increased proliferation (Figure 7B). Significantly, we observed decreased expression of p-SMAD1/5/8 and p-NRF2 in these EoE samples (Figure 7, C and D). GCLC and GSTP1 transcript levels were also reduced, suggesting a reduction in ROS production (Figure 7H). Meanwhile, there was an increase in follistatin levels as revealed by immunostaining and real-time PCR (Figure 7, E and I), suggesting that this protein downregulates BMP signaling and allows basal cells to expand during disease progression.
Figure 7. Basal cell hyperplasia in human EoE biopsies is associated with reduced BMP signaling and increased follistatin levels.
(A) H&E staining showed expansion of basal cells in human EoE biopsies. Arrowheads indicate eosinophilic infiltration into the epithelium. The inset is the high-magnification (original magnification, ×40) view of the boxed region. (B) Basal cell hyperplasia was associated with increased proliferation as indicated by p63 and Ki67 staining. (C) BMP activity was absent in the hyperplastic epithelium as shown by p-SMAD1/5/8 staining. (D) Nuclear localization of p-NRF2 was reduced in the EoE epithelium. (E) Basal cell hyperplasia was accompanied by enrichment of follistatin. (F) Transcript levels of IL13 were significantly increased as shown by real-time PCR. (G) Transcript levels of eotaxin-3, a downstream mediator of IL-13, were significantly increased. (H) Transcript levels of GCLC, GSTP1, and NQO1 were decreased in EoE biopsies. (I) Transcript levels of FST were significantly increased in EoE biopsies. Data represent the mean ± SEM. Fourteen biopsies for each group were included. *P < 0.05, **P < 0.01, and ****P < 0.0001 by Student’s t test. Scale bars: 50 μm.
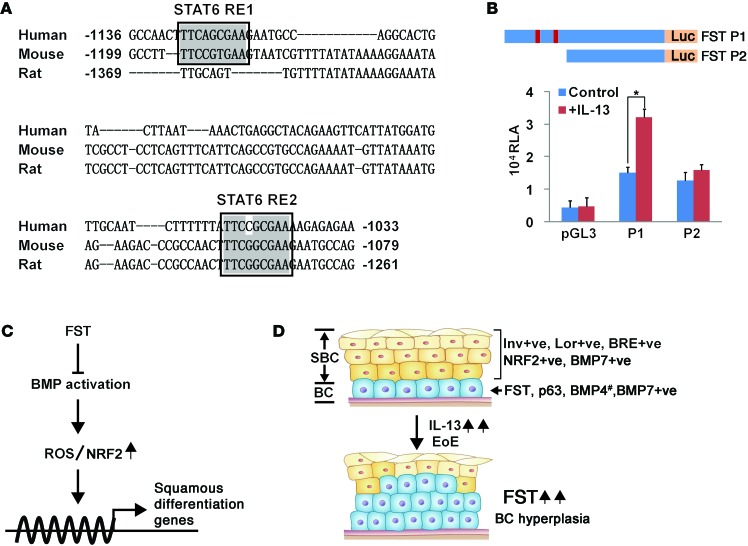
Our data suggest that follistatin is regulated by IL-13. To test whether the IL-13 signaling mediator STAT6 directly modulates transcription of the follistatin gene, we examined the promoter region of follistatin (1.8 kb upstream of the transcription starting site) and found 2 potential STAT6-binding sites that are conserved in humans, mice, and rats (Figure 8A). We then performed promoter analysis and cloned the 1.8-kb promoter region of the mouse follistatin gene into a pGL3 luciferase reporter construct (Figure 8B). We observed a 2.1-fold increase in follistatin promoter–driven luciferase activity upon IL-13 treatment (Figure 8B, P < 0.05). Furthermore, the promoter did not respond to IL-13 treatment when the 2 conserved binding sites were removed, further supporting the notion that follistatin is a direct downstream target of IL-13/STAT6 signaling (Figure 8B).
Figure 8. Follistatin is a direct downstream target of the IL-13/STAT6 signaling pathway.
(A) The follistatin promoter region contains 2 potential STAT6-binding sites within longer, conserved DNA sequences. Sequence comparison revealed the conservation of these 2 STAT6-binding sites in various species. The STAT6-binding sites are highlighted by black boxes. (B) IL-13 treatment significantly increased the promoter activity of the follistatin gene that contains 2 STAT6-binding sites in human EPC2 cells (n = 3). Note that removal of the 2 STAT6-binding sites abrogated the effects induced by IL-13 treatment. The promoter region with or without the 2 STAT6-binding sites was cloned into the pGL3 luciferase reporter and is designated as P1 and P2, respectively. (C) BMP activation promotes squamous differentiation of basal progenitor cells through crosstalk with ROS-activated NRF2 signaling. The BMP inhibitor follistatin protects basal cells from BMP activation initiated by BMP7 and possibly by BMP4 as well. Lack of follistatin in suprabasal cells leads to BMP activation and increases intracellular ROS levels. Subsequent activation of NRF2 signaling promotes squamous differentiation of basal progenitor cells. (D) Basal cell hyperplasia in the IL-13 overexpression–induced mouse EoE model and in human biopsies was associated with increased levels of follistatin and decreased BMP activation. Note that BMP7 was present in all of the epithelium, while BMP4 was expressed by subpopulations of basal progenitor cells (#). Data represent the mean ± SEM. *P < 0.05 by Student’s t test. BC, basal cell; FST, follistatin; RE1, response element 1; RE2, response element 2; SBC, suprabasal cell; ve, vehicle.
Discussion
Balanced self-renewal and differentiation of basal progenitor cells are essential for maintenance of the esophageal epithelium (2, 40). Yet the molecular mechanisms regulating basal progenitor cell homeostasis remain unexplored. In this study, we provide in vitro and in vivo genetic evidence that BMP activation promotes squamous differentiation of basal progenitor cells in the adult esophagus. We demonstrate that basal progenitor cells are protected from BMP activation by the inhibitor follistatin and that ectopic BMP activation drives the premature differentiation of the progenitor cells. Further analysis revealed that BMP activation elicits an NRF2-mediated oxidative response, which is critical for the normal squamous differentiation of basal progenitor cells. Finally, we show that basal cell hyperplasia in both the EoE mouse model and human biopsies is associated with high levels of follistatin, which is regulated through an IL-13/STAT6 mechanism (Figure 8, C and D).
BMP signaling activity is critical for the development of multiple gastrointestinal organs including the intestine and esophagus (11, 41). The BMP inhibitor follistatin is enriched in basal progenitor cells, presumably counteracting BMP activation by the ligand BMP7, which is expressed in both basal progenitors and differentiated suprabasal cells. Intriguingly, we found that BMP4 was also expressed in certain subpopulations of basal progenitor cells. Recent studies present provocative conclusions regarding whether the basal cell population is homogenous and possesses equal potential to self-renew and differentiate (2, 40, 42). Along this line, these BMP4-positive basal cells could be progenitor cells destined to differentiate and detach from the basal layer. Alternatively, the expression of BMP4 in basal cells could be a stochastic phenomenon.
Increased BMP activity has been linked to the development of BE (12, 13). Upregulated BMP4 mRNA levels have been detected in human BE biopsies, with increased nuclear localization of phosphorylated SMAD1/5/8 (12). However, recent studies suggest that multiple cell types can give rise to BE (43). For example, a cell population identified as residual embryonic progenitor cells at the squamous-columnar junction (GEJ) was shown to replace squamous epithelium and give rise to BE under injury-repair circumstances (44). Moreover, gastric cardia stem cells have also been shown to contribute to the pathogenesis of BE in an animal model (45). Interestingly, Mari et al. recently showed that overexpression of BMP4 using a K14 promoter, which also targets the basal progenitor cell population in the esophagus and forestomach, promotes columnar differentiation of the cells located at the GEJ. These cells become BE-like epithelium after combining with bile acid reflux induced by esophageal-jejunostomy surgery (33). These findings suggest that squamous epithelium located at the GEJ has the potential to become intestinalized upon BMP stimulation. Although we did not observe any signs of intestinalization at the GEJ following long-term BMP activation, it will be interesting to determine whether bile acid can similarly facilitate columnar changes in these mice. In addition, recent reports demonstrate that ectopic sonic hedgehog (SHH) signaling activity promotes BE formation via reactivation of the transcription factor FOXA2 and that crosstalk between the SHH and BMP pathways is required for disease progression (13, 46). It will be of interest to test whether ectopic FOXA2 and BMP activation facilitates columnar differentiation of basal progenitor cells.
A previous study showed that deletion of BMP receptor 1a (Bmpr1a) leads to squamous epithelial hyperplasia in the forestomach (47). Nevertheless, the underlying mechanism was unexplored. Here, we found that BMP activation promotes squamous differentiation of basal progenitor cells through the NRF2-mediated oxidative stress pathway. We also found that BMP activation leads to ROS accumulation in esophageal progenitor cells, concomitant with increased levels of NADPH oxidases, including NOX4. Interestingly, recent studies have shown that BMP signaling promotes cell differentiation through the activation of NADPH oxidases and ROS production. For example, BMP2 increases NADPH oxidase activity and oxidant stress to induce differentiation in preosteoblast cells (48, 49). In addition, BMP2 induces a profibrotic phenotype in renal progenitor cells through NOX4 activation (50). Additionally, accumulation of ROS in a high-oxygen environment promotes the differentiation of skin keratinocytes (51). In this study, we were able to block BMP4-induced differentiation by treating progenitor cells with the antioxidant reagent NAC. We further showed that NRF2 is upregulated upon BMP activation and that inhibition of NRF2 activity attenuates BMP4-induced differentiation. Interestingly, constitutive activation of NRF2 in the esophagus of Keap1–/– mutants also causes hyperkeratosis (21, 22, 24), which can be rescued by inhibition of NRF2 in Nrf2–/– Keap1–/– or Maff–/– Mafg–/– Keap1–/– mutants in which MAFs serve as mediators of NRF2 (22, 52), further supporting the idea that functional activation of NRF2 is required for esophageal epithelial differentiation.
We found that follistatin levels were increased in EoE mouse models and human biopsies. EoE is characterized by basal cell hyperplasia and subepithelial fibrosis (53). We propose that follistatin inhibits BMP activities during disease progression, thereby blocking the differentiation of basal progenitor cells. Several EoE mouse models have been established, mimicking the inflammatory environment seen in EoE patients (54). Built on previous findings that IL-13 is a key modulator of EoE, we directly overexpressed IL-13 in basal progenitor cells and observed phenotypic changes, including basal cell hyperplasia, that were similar to those observed in other IL-13–induced EoE models (38). We found that follistatin was dramatically increased after localized IL-13 overexpression. It will be interesting to determine whether this BMP inhibitor is similarly increased in other IL-13 EoE models as well as in those models with activation of other cytokines such as IL-5 (54). Similarly, it will be interesting to examine changes in follistatin levels in other esophageal diseases, e.g., GERD, that are associated with basal cell hyperplasia and proinflammatory responses (55). EoE is an emerging disease for which the underlying mechanism is unknown, with a significant increase in incidence since it was first characterized in 1993 (56). Epithelial expansion featured by basal cell hyperplasia plays a critical role, as multiple cytokines are secreted from the epithelium during disease progression (7, 8). Therefore, identification of the role for the BMP pathway in basal cell hyperplasia could provide important insights into EoE pathogenesis, leading to novel approaches to tackle this disease.
In conclusion, we used in vitro and multiple mouse models to demonstrate that BMP signaling is critical for basal cell–fate determination in the adult esophagus. Specifically, BMP activation promotes squamous differentiation through NRF2-mediated oxidative stress signaling. We found that inhibition of BMP activity by follistatin underlies the expansion of basal progenitor cells during the pathogenesis of EoE. These findings not only provide important information about the normal mechanism by which BMP signaling controls differentiation of basal progenitor cells, but also important insights into the pathogenesis of EoE. Consequently, targeted pharmacological activation of BMP signaling could be a potential therapeutic approach to treat EoE and warrants further testing in EoE animal models in the future.
Methods
Mice.
BRE-lacZ (25), Bmp4lacZ/+ (57), Bmp7lacZ/+ (58), nogginlacZ/+ (59), Grem1lacZ/+ (60), and Grem2lacZ/+ (gift of Richard Harland, University of California, Berkeley, California, USA) reporter mice were maintained on a C57Bl/6 129/SvEv background. The transgenic Krt5-CreER and the conditional R26CAG–loxp–stop–loxp–caBmpr1a (R26caBmpr1a) lines have been described previously and were maintained on a C57BL/6 and 129SvEv mixed background (11, 61). To activate caBmpr1a overexpression in the Krt5-CreER R26caBmpr1a mutants, tamoxifen was injected every other day for a total of 4 times (0.25 mg/g body weight). Mice were then sacrificed 7 days after the final injection, and the esophagi were harvested for further analysis. Krt5-CreER mice injected with tamoxifen were included as controls. To create the EoE mouse model, Krt5-rtTA (62) and tetO-Il13 (63) mice were crossed to generate Krt5-rtTA tetO-Il13 compound mice. To activate IL-13 expression in the esophagus, mice received Dox (2 mg/ml plus 50 mg/ml sucrose) in their drinking water for 7 days. Mice were then sacrificed 2 weeks after Dox treatment, and the esophagi were collected for further analysis. Krt5-rtTA mice treated with Dox were included as controls.
Histology, immunostaining, X-gal staining, and ISH.
The esophagi were dissected in PBS and fixed in 4% paraformaldehyde overnight at 4°C. After dehydration, they were embedded in paraffin for sectioning and IHC. The antibodies used for IHC analysis include: anti-p63 (1:500, MAB4135, EMD Millipore and sc-8343, Santa Cruz Biotechnology Inc.); anti–p-SMAD1/5/8 (1:1,000, 9511S; Cell Signaling Technology); anti-involucrin (1:1,000, MS-126-P0; Thermo Fisher Scientific); anti-follistatin (1:50, MAB669; R&D Systems); anti-p75 (1:100, ab8874; Abcam); anti–p-H3 (1:500, H9908; Sigma-Aldrich); anti-Ki67 (1:500, 550609; BD Biosciences); anti-SOX2 (1:1,000; WRAB-1236, Seven Hills Bioreagents); anti–p-NRF2 (1:200, ab76026; Abcam); anti-KRT8 (1:1,000, SAB2701925; Sigma-Aldrich); and anti-CDX2 (1:200, ab76541; Abcam). The secondary antibodies were either fluorescence or DAB conjugated, and images were taken with a Leica SP1 confocal microscope. For X-gal staining, the esophagi were fixed in 4% paraformaldehyde for 30 minutes, followed by X-gal staining overnight at 37°C. The X-gal–stained samples were then dehydrated with ethanol and embedded in paraffin for sectioning as previously described (11, 22). ISH analysis was performed as previously described (27, 64).
Cell culture, gene expression, and luciferase assay.
Mouse esophageal basal progenitor cells were isolated and plated onto rat type IV collagen–coated (BD Biosciences) tissue culture dishes in MTEC/Plus (DMEM-Ham’s F12 [1:1 v/v; Gibco], insulin, transferrin, and selenium A [1:100; Gibco], bovine pituitary extract [30 ug/ml; Invitrogen], L-glutamine [4 mM; Gibco]) containing 20 ng/ml EGF and FGF2 medium (28). The EPC2 cell line was hTERT immortalized and maintained as previously described (32). Nrf2 and Keap1 were cloned into the pCDH lentiviral vector. EPC2 cells infected with Nrf2- and Keap1-containing virus were selected with 1 μg/ml puromycin. The EPC2 cells with NRF2 overexpression (EPC2-NRF2) and KEAP1 (EPC2-KEAP1) were analyzed by Western blotting and RT-PCR, with EPC2 cells expressing GFP used as controls. For cell growth analysis, EPC2 and EPC2-NRF2 cells were cultured in 6-well plates and cell numbers counted every 2 days. Cignal Finder 45-Pathway Reporters (QIAGEN) or control empty vectors were transfected into EPC2 cells using X-tremeGENE HP DNA Transfection Reagent (Roche) in the presence or absence of 10 ng/ml BMP4. To test whether the follistatin promoter is regulated by IL-13 through STAT6, the 1.7-kb promoter region of the follistatin gene with 2 conserved STAT6-binding sites and a truncated fragment without these 2 binding sites as a control were cloned into the pGL3 luciferase reporter vector and transfected into EPC2 cells with or without IL-13 treatment. Relative luciferase activity was determined 48 hours after transfection using a Dual-Luciferase Reporter kit (Promega).
Cohort of human subjects with EoE and controls.
Control subjects presented with epigastric abdominal pain but had normal endoscopic and microscopic results. Patients were diagnosed with EoE according to current consensus recommendations (65). Specifically, they were required to have symptoms of esophageal dysfunction — at least 15 eosinophils per high-power field on esophageal biopsy — that persisted after a 2-month high-dose proton pump–inhibitor trial and exclusion of other causes of esophageal eosinophilia. Adult patients with EoE were then prospectively enrolled in biorepository/registry studies at each of the contributing sites. Esophageal biopsies were obtained and banked prospectively, and patients provided written informed consent to store the samples for future use. These tissue banks served as the source of the human esophageal biopsies used in this study, and samples were collected and shipped in dry ice to J. Que for either real-time PCR or IHC analysis.
Reverse transcription and real-time PCR.
Total RNA was extracted with TRIzol (Invitrogen) and purified using the RNeasy Mini Kit (QIAGEN). RNA reverse transcription was performed using the SuperScript III First-Strand SuperMix (Invitrogen) according to the manufacturer’s instructions. cDNA was subjected to quantitative real-time PCR using the MyiQ Single-Color Real-Time PCR Detection System (Bio-Rad) and iQ SYBR Green Supermix (Bio-Rad). The transcript levels of genes were normalized to Rhoa expression. All real-time quantitative PCR experiments were performed in triplicate. PCR primers were designed using the Lasergene Core Suite (DNASTAR Inc.), and the sequences are described in Supplemental Table 2.
Western blot analysis.
Protein was extracted from isolated esophageal epithelium or cultured cells in lysis buffer containing protease and phosphatase inhibitors (Roche). Protein (20 μg) for each sample was denatured and separated on a 10% SDS-PAGE gel, followed by immunoblotting. The primary antibodies used were: involucrin (1:1,000, MS-126-P0; Thermo Fisher Scientific); loricrin (1:1,000, ab24722; Abcam); p-SMAD1/5/8 (1:1,000, 9511S; Cell Signaling Technology); p-NRF2 (1:2,000, ab76026; Abcam); NRF2 (1:500, ab62352; Abcam); and β-actin (1:5,000, sc-47778; Santa Cruz Biotechnology Inc.), and the blots were processed as previously described (66).
Measurement of intracellular ROS.
The oxidation-sensitive fluorescent probe 2′,7′-dichlorodihydrofluorescein diacetate (DCF-DA) (Molecular Probes) was used to detect the generation of intracellular ROS as described previously (67). Briefly, EPC2 cells were treated with 10 ng/ml BMP4 for the indicated time points. Cells were washed 3 times with HBSS without phenol red and then incubated with 5 μM DCF-DA for 30 minutes in the dark at 37°C. DCF fluorescence was detected with a fluorescence plate reader. Emitted fluorescence was normalized to the total amount of cells.
Statistics.
Western blot bands were scanned and analyzed for optical density using ImageJ software (NIH). Data are expressed as the mean ± SD. Differences between 2 groups were analyzed by a 2-tailed Student’s t test. A P value of 0.05 or less was considered statistically significant. Differences among 3 or more groups were analyzed using 2-way ANOVA (SAS version 9.2; SAS Institute Inc.).
Study approval.
All mouse experiments were conducted in accordance with procedures approved by the IACUC of the University of Rochester. In the case of human samples, adult participants from a cohort of control subjects or subjects with EoE at the Center for Esophageal Diseases and Swallowing of the University of North Carolina at Chapel Hill had esophageal biopsies prospectively collected and stored. These samples were provided and analyzed under IRB approval. Adult participants from a cohort of control subjects or subjects with EoE being treated at the Division of Gastroenterology of the University of Pennsylvania were also assessed and were provided under IRB approval. All participants in this study or their parents or legal guardians provided written informed consent.
Supplementary Material
Acknowledgments
This work was initiated in Brigid Hogan’s laboratory at Duke University. We are thankful for the stimulating discussions with Mark Noble, Dirk Bohmann, and Douglas Portman of the University of Rochester. We also thank our colleagues, especially Ian Jacobs at the Que laboratory, for critical reading and discussion of the manuscript. We also thank Marc E. Rothenberg for providing the tetO-Il13 transgenic mouse line. This research is supported by grants from the NIH (R01-DK100342, to J. Que and K23-DK090073, to E.S. Dellon); the New York State Stem Cell Research Program (NYSTEM) (C029555, to J. Que); and the National High Technology Research and Development Program of China (863 Program) (2014AA020541, to K. Liu).
Footnotes
Note regarding evaluation of this manuscript: Manuscripts authored by scientists associated with Duke University, The University of North Carolina at Chapel Hill, Duke-NUS, and the Sanford-Burnham Medical Research Institute are handled not by members of the editorial board but rather by the science editors, who consult with selected external editors and reviewers.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2015;125(4):1557–1568. doi:10.1172/JCI78850.
References
- 1.Clayton E, Doupe DP, Klein AM, Winton DJ, Simons BD, Jones PH. A single type of progenitor cell maintains normal epidermis. Nature. 2007;446(7132):185–189. doi: 10.1038/nature05574. [DOI] [PubMed] [Google Scholar]
- 2.Doupe DP, et al. A single progenitor population switches behavior to maintain and repair esophageal epithelium. Science. 2012;337(6098):1091–1093. doi: 10.1126/science.1218835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jovov B, Que J, Tobey NA, Djukic Z, Hogan BL, Orlando RC. Role of E-cadherin in the pathogenesis of gastroesophageal reflux disease. Am J Gastroenterol. 2011;106(6):1039–1047. doi: 10.1038/ajg.2011.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng E, Souza RF, Spechler SJ. Tissue remodeling in eosinophilic esophagitis. Am J Physiol Gastrointest Liver Physiol. 2012;303(11):G1175–G1187. doi: 10.1152/ajpgi.00313.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanchard C, et al. Coordinate interaction between IL-13 and epithelial differentiation cluster genes in eosinophilic esophagitis. J Immunol. 2010;184(7):4033–4041. doi: 10.4049/jimmunol.0903069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aceves S, Hirano I, Furuta GT, Collins MH. Eosinophilic gastrointestinal diseases — clinically diverse and histopathologically confounding. Semin Immunopathol. 2012;34(5):715–731. doi: 10.1007/s00281-012-0324-x. [DOI] [PubMed] [Google Scholar]
- 7.Blanchard C, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116(2):536–547. doi: 10.1172/JCI26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanchard C, et al. Periostin facilitates eosinophil tissue infiltration in allergic lung and esophageal responses. Mucosal Immunol. 2008;1(4):289–296. doi: 10.1038/mi.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hogan BL. Bone morphogenetic proteins in development. Curr Opin Genet Dev. 1996;6(4):432–438. doi: 10.1016/S0959-437X(96)80064-5. [DOI] [PubMed] [Google Scholar]
- 10.Hogan BL. Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev. 1996;10(13):1580–1594. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez P, Da Silva S, Oxburgh L, Wang F, Hogan BL, Que J. BMP signaling in the development of the mouse esophagus and forestomach. Development. 2010;137(24):4171–4176. doi: 10.1242/dev.056077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milano F, et al. Bone morphogenetic protein 4 expressed in esophagitis induces a columnar phenotype in esophageal squamous cells. Gastroenterology. 2007;132(7):2412–2421. doi: 10.1053/j.gastro.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 13.Wang DH, et al. Aberrant epithelial-mesenchymal Hedgehog signaling characterizes Barrett’s metaplasia. Gastroenterology. 2010;138(5):1810–1822. doi: 10.1053/j.gastro.2010.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blanchard C, et al. IL-13 involvement in eosinophilic esophagitis: transcriptome analysis and reversibility with glucocorticoids. J Allergy Clin Immunol. 2007;120(6):1292–1300. doi: 10.1016/j.jaci.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 15.Blanchetot C, Boonstra J. The ROS-NOX connection in cancer and angiogenesis. Crit Rev Eukaryot Gene Expr. 2008;18(1):35–45. doi: 10.1615/CritRevEukarGeneExpr.v18.i1.30. [DOI] [PubMed] [Google Scholar]
- 16.Leslie NR. The redox regulation of PI 3-kinase-dependent signaling. Antioxid Redox Signal. 2006;8(9–10):1765–1774. doi: 10.1089/ars.2006.8.1765. [DOI] [PubMed] [Google Scholar]
- 17.Jang YY, Sharkis SJ. A low level of reactive oxygen species selects for primitive hematopoietic stem cells that may reside in the low-oxygenic niche. Blood. 2007;110(8):3056–3063. doi: 10.1182/blood-2007-05-087759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol Med. 2004;10(11):549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 19.Itoh K, Igarashi K, Hayashi N, Nishizawa M, Yamamoto M. Cloning and characterization of a novel erythroid cell-derived CNC family transcription factor heterodimerizing with the small Maf family proteins. Mol Cell Biol. 1995;15(8):4184–4193. doi: 10.1128/mcb.15.8.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Venugopal R, Jaiswal AK. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc Natl Acad Sci U S A. 1996;93(25):14960–14965. doi: 10.1073/pnas.93.25.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H, et al. Nrf2 deficiency impairs the barrier function of mouse oesophageal epithelium. Gut. 2014;63(5):711–719. doi: 10.1136/gutjnl-2012-303731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen H, et al. Transcript profiling identifies dynamic gene expression patterns and an important role for Nrf2/Keap1 pathway in the developing mouse esophagus. PLoS One. 2012;7(5):e36504. doi: 10.1371/journal.pone.0036504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobs IJ, Ku WY, Que J. Genetic and cellular mechanisms regulating anterior foregut and esophageal development. Dev Biol. 2012;369(1):54–64. doi: 10.1016/j.ydbio.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wakabayashi N, et al. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat Genet. 2003;35(3):238–245. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- 25.Blank U, Seto ML, Adams DC, Wojchowski DM, Karolak MJ, Oxburgh L. An in vivo reporter of BMP signaling in organogenesis reveals targets in the developing kidney. BMC Dev Biol. 2008;8:86. doi: 10.1186/1471-213X-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daniely Y, et al. Critical role of p63 in the development of a normal esophageal and tracheobronchial epithelium. Am J Physiol Cell Physiol. 2004;287(1):C171–C181. doi: 10.1152/ajpcell.00226.2003. [DOI] [PubMed] [Google Scholar]
- 27.Que J, Choi M, Ziel JW, Klingensmith J, Hogan BL. Morphogenesis of the trachea and esophagus: current players and new roles for noggin and Bmps. Differentiation. 2006;74(7):422–437. doi: 10.1111/j.1432-0436.2006.00096.x. [DOI] [PubMed] [Google Scholar]
- 28.Liu K, et al. Sox2 cooperates with inflammation-mediated Stat3 activation in the malignant transformation of foregut basal progenitor cells. Cell Stem Cell. 2013;12(3):304–315. doi: 10.1016/j.stem.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okumura T, Shimada Y, Imamura M, Yasumoto S. Neurotrophin receptor p75(NTR) characterizes human esophageal keratinocyte stem cells in vitro. Oncogene. 2003;22(26):4017–4026. doi: 10.1038/sj.onc.1206525. [DOI] [PubMed] [Google Scholar]
- 30.Oxburgh L, et al. BMP4 substitutes for loss of BMP7 during kidney development. Dev Biol. 2005;286(2):637–646. doi: 10.1016/j.ydbio.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 31.Li Y, Litingtung Y, Ten Dijke P, Chiang C. Aberrant Bmp signaling and notochord delamination in the pathogenesis of esophageal atresia. Dev Dyn. 2007;236(3):746–754. doi: 10.1002/dvdy.21075. [DOI] [PubMed] [Google Scholar]
- 32.Okawa T, et al. The functional interplay between EGFR overexpression, hTERT activation, and p53 mutation in esophageal epithelial cells with activation of stromal fibroblasts induces tumor development, invasion, and differentiation. Genes Dev. 2007;21(21):2788–2803. doi: 10.1101/gad.1544507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mari L, et al. A pSMAD/CDX2 complex is essential for the intestinalization of epithelial metaplasia. Cell Rep. 2014;7(4):1197–1210. doi: 10.1016/j.celrep.2014.03.074. [DOI] [PubMed] [Google Scholar]
- 34.Yang Y, Goldstein BG, Chao HH, Katz JP. KLF4 and KLF5 regulate proliferation, apoptosis and invasion in esophageal cancer cells. Cancer Biol Ther. 2005;4(11):1216–1221. doi: 10.4161/cbt.4.11.2090. [DOI] [PubMed] [Google Scholar]
- 35.Ghaleb AM, Nandan MO, Chanchevalap S, Dalton WB, Hisamuddin IM, Yang VW. Kruppel-like factors 4 and 5: the yin and yang regulators of cellular proliferation. Cell Res. 2005;15(2):92–96. doi: 10.1038/sj.cr.7290271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santini MP, Talora C, Seki T, Bolgan L, Dotto GP. Cross talk among calcineurin, Sp1/Sp3, and NFAT in control of p21(WAF1/CIP1) expression in keratinocyte differentiation. Proc Natl Acad Sci U S A. 2001;98(17):9575–9580. doi: 10.1073/pnas.161299698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chan K, Han XD, Kan YW. An important function of Nrf2 in combating oxidative stress: detoxification of acetaminophen. Proc Natl Acad Sci U S A. 2001;98(8):4611–4616. doi: 10.1073/pnas.081082098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mishra A, Rothenberg ME. Intratracheal IL-13 induces eosinophilic esophagitis by an IL-5, eotaxin-1, and STAT6-dependent mechanism. Gastroenterology. 2003;125(5):1419–1427. doi: 10.1016/j.gastro.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 39.Zuo L, et al. IL-13 induces esophageal remodeling and gene expression by an eosinophil-independent, IL-13R α 2-inhibited pathway. J Immunol. 2010;185(1):660–669. doi: 10.4049/jimmunol.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalabis J, et al. A subpopulation of mouse esophageal basal cells has properties of stem cells with the capacity for self-renewal and lineage specification. J Clin Invest. 2008;118(12):3860–3869. doi: 10.1172/JCI35012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Auclair BA, Benoit YD, Rivard N, Mishina Y, Perreault N. Bone morphogenetic protein signaling is essential for terminal differentiation of the intestinal secretory cell lineage. Gastroenterology. 2007;133(3):887–896. doi: 10.1053/j.gastro.2007.06.066. [DOI] [PubMed] [Google Scholar]
- 42.DeWard AD, Cramer J, Lagasse E. Cellular heterogeneity in the mouse esophagus implicates the presence of a nonquiescent epithelial stem cell population. Cell Rep. 2014;9(2):701–711. doi: 10.1016/j.celrep.2014.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quante M, Abrams JA, Lee Y, Wang TC. Barrett esophagus: what a mouse model can teach us about human disease. Cell Cycle. 2012;11(23):4328–4338. doi: 10.4161/cc.22485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X, et al. Residual embryonic cells as precursors of a Barrett’s-like metaplasia. Cell. 2011;145(7):1023–1035. doi: 10.1016/j.cell.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quante M, et al. Bile acid and inflammation activate gastric cardia stem cells in a mouse model of Barrett-like metaplasia. Cancer Cell. 2012;21(1):36–51. doi: 10.1016/j.ccr.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang DH, et al. Hedgehog signaling regulates FOXA2 in esophageal embryogenesis Barrett’s metaplasia. J Clin Invest. 2014;124(9):3767–3780. doi: 10.1172/JCI66603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maloum F, et al. Epithelial BMP signaling is required for proper specification of epithelial cell lineages and gastric endocrine cells. Am J Physiol Gastrointest Liver Physiol. 2011;300(6):G1065–G1079. doi: 10.1152/ajpgi.00176.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mandal CC, et al. Reactive oxygen species derived from Nox4 mediate BMP2 gene transcription and osteoblast differentiation. Biochem J. 2011;433(2):393–402. doi: 10.1042/BJ20100357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liberman M, Johnson RC, Handy DE, Loscalzo J, Leopold JA. Bone morphogenetic protein-2 activates NADPH oxidase to increase endoplasmic reticulum stress and human coronary artery smooth muscle cell calcification. Biochem Biophys Res Commun. 2011;413(3):436–441. doi: 10.1016/j.bbrc.2011.08.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simone S, et al. BMP-2 induces a profibrotic phenotype in adult renal progenitor cells through Nox4 activation. Am J Physiol Renal Physiol. 2012;303(1):F23–F34. doi: 10.1152/ajprenal.00328.2011. [DOI] [PubMed] [Google Scholar]
- 51.Ngo MA, Sinitsyna NN, Qin Q, Rice RH. Oxygen-dependent differentiation of human keratinocytes. J Invest Dermatol. 2007;127(2):354–361. doi: 10.1038/sj.jid.5700522. [DOI] [PubMed] [Google Scholar]
- 52.Motohashi H, Katsuoka F, Engel JD, Yamamoto M. Small Maf proteins serve as transcriptional cofactors for keratinocyte differentiation in the Keap1-Nrf2 regulatory pathway. Proc Natl Acad Sci U S A. 2004;101(17):6379–6384. doi: 10.1073/pnas.0305902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheng E, Souza RF, Spechler SJ. Tissue remodeling in eosinophilic esophagitis. Am J Physiol Gastrointest Liver Physiol. 2012;303(11):G1175–G1187. doi: 10.1152/ajpgi.00313.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Masterson JC, et al. Local hypersensitivity reaction in transgenic mice with squamous epithelial IL-5 overexpression provides a novel model of eosinophilic oesophagitis. Gut. 2014;63(1):43–53. doi: 10.1136/gutjnl-2012-303631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Souza RF, et al. Gastroesophageal reflux might cause esophagitis through a cytokine-mediated mechanism rather than caustic acid injury. Gastroenterology. 2009;137(5):1776–1784. doi: 10.1053/j.gastro.2009.07.055. [DOI] [PubMed] [Google Scholar]
- 56.Attwood SE, Smyrk TC, Demeester TR, Jones JB. Esophageal eosinophilia with dysphagia. A distinct clinicopathologic syndrome. Dig Dis Sci. 1993;38(1):109–116. doi: 10.1007/BF01296781. [DOI] [PubMed] [Google Scholar]
- 57.Weaver M, Yingling JM, Dunn NR, Bellusci S, Hogan BL. Bmp signaling regulates proximal-distal differentiation of endoderm in mouse lung development. Development. 1999;126(18):4005–4015. doi: 10.1242/dev.126.18.4005. [DOI] [PubMed] [Google Scholar]
- 58.Godin RE, Takaesu NT, Robertson EJ, Dudley AT. Regulation of BMP7 expression during kidney development. Development. 1998;125(17):3473–3482. doi: 10.1242/dev.125.17.3473. [DOI] [PubMed] [Google Scholar]
- 59.Brunet LJ, McMahon JA, McMahon AP, Harland RM. Noggin, cartilage morphogenesis, and joint formation in the mammalian skeleton. Science. 1998;280(5368):1455–1457. doi: 10.1126/science.280.5368.1455. [DOI] [PubMed] [Google Scholar]
- 60.Khokha MK, Hsu D, Brunet LJ, Dionne MS, Harland RM. Gremlin is the BMP antagonist required for maintenance of Shh and Fgf signals during limb patterning. Nat Genet. 2003;34(3):303–307. doi: 10.1038/ng1178. [DOI] [PubMed] [Google Scholar]
- 61.Rock JR, et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci U S A. 2009;106(31):12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Diamond I, Owolabi T, Marco M, Lam C, Glick A. Conditional gene expression in the epidermis of transgenic mice using the tetracycline-regulated transactivators tTA and rTA linked to the keratin 5 promoter. J Invest Dermatol. 2000;115(5):788–794. doi: 10.1046/j.1523-1747.2000.00144.x. [DOI] [PubMed] [Google Scholar]
- 63.Fulkerson PC, Fischetti CA, Hassman LM, Nikolaidis NM, Rothenberg ME. Persistent effects induced by IL-13 in the lung. Am J Respir Cell Mol Biol. 2006;35(3):337–346. doi: 10.1165/rcmb.2005-0474OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fu J, Jiang M, Mirando AJ, Yu HM, Hsu W. Reciprocal regulation of Wnt and Gpr177/mouse Wntless is required for embryonic axis formation. Proc Natl Acad Sci U S A. 2009;106(44):18598–18603. doi: 10.1073/pnas.0904894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dellon ES, et al. ACG clinical guideline: Evidenced based approach to the diagnosis and management of esophageal eosinophilia and eosinophilic esophagitis (EoE). Am J Gastroenterol. 2013;108(5):679–692. doi: 10.1038/ajg.2013.71. [DOI] [PubMed] [Google Scholar]
- 66.Que J, et al. Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm. Development. 2007;134(13):2521–2531. doi: 10.1242/dev.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang H, Joseph JA. Quantifying cellular oxidative stress by dichlorofluorescein assay using microplate reader. Free Radic Biol Med. 1999;27(5–6):612–616. doi: 10.1016/s0891-5849(99)00107-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.