Abstract
Tumor cell migration is a key process for cancer cell dissemination and metastasis that is controlled by signal-mediated cytoskeletal and cell matrix adhesion remodeling. Using a phagokinetic track assay with migratory H1299 cells, we performed an siRNA screen of almost 1,500 genes encoding kinases/phosphatases and adhesome- and migration-related proteins to identify genes that affect tumor cell migration speed and persistence. Thirty candidate genes that altered cell migration were validated in live tumor cell migration assays. Eight were associated with metastasis-free survival in breast cancer patients, with integrin β3–binding protein (ITGB3BP), MAP3K8, NIMA-related kinase (NEK2), and SHC-transforming protein 1 (SHC1) being the most predictive. Examination of genes that modulate migration indicated that SRPK1, encoding the splicing factor kinase SRSF protein kinase 1, is relevant to breast cancer outcomes, as it was highly expressed in basal breast cancer. Furthermore, high SRPK1 expression correlated with poor breast cancer disease outcome and preferential metastasis to the lungs and brain. In 2 independent murine models of breast tumor metastasis, stable shRNA-based SRPK1 knockdown suppressed metastasis to distant organs, including lung, liver, and spleen, and inhibited focal adhesion reorganization. Our study provides comprehensive information on the molecular determinants of tumor cell migration and suggests that SRPK1 has potential as a drug target for limiting breast cancer metastasis.
Keywords: Cell Biology, Oncology
Introduction
Cancer cell invasion into neighboring tissue and dissemination from the primary tumor, causing metastasis to distant organs, remains the major cause of cancer deaths. Individual or collective tumor cell migration constitutes critical steps in this complex process, involving extracellular matrix (ECM) remodeling, and dynamic reorganization of cell adhesions with neighboring cells and with the underlying connective tissue (e.g., focal adhesions). Focal adhesions (FAs) are dynamic multiprotein complexes consisting of over 200 different molecules, including signaling proteins as well as cytoskeleton-associated scaffolding components, that mediate cell adhesion to the ECM in a coordinated manner (1–3). Upstream signaling by proto-oncogenic growth factor receptors was shown to affect the dynamics of FA formation and turnover, through the integrated phosphorylation and activation of FA-associated kinases and adapter proteins, as well as the dynamic reorganization of the actin cytoskeletal network. These locally and globally coordinated processes determine the directionality, speed, and persistence of cancer cell migration, which ultimately affect the metastatic phenotype in vivo.
Various genes involved in cell migration are associated with tumor cell motility and cancer metastasis. Increased signaling through EGF or HGF receptor family members promotes actin reorganization and cancer cell invasiveness and metastasis, and is associated with poor prognosis (4–6). Alternatively, activation of TGF-β receptor induces the transition of otherwise poorly migratory epithelial cancer cells into migratory mesenchymal cells (7). Downstream effectors, such as the Rho-GTPase family member RhoC, further stimulate invasiveness and cancer cell dissemination (8). Similarly, integrin adhesome molecules such as integrin α5β1, focal adhesion kinase (FAK), Src, and p130Cas were shown to be associated with metastatic activity (9–14).
Although the roles of some individual FA-associated proteins in tumor cell migration and invasion are well documented, a systematic functional survey of the involvement of individual signaling and cell adhesion components in tumor cell migration and metastatic disease is still lacking. A limited number of RNA-interference and cDNA gain-of-function screens were performed to identify kinases that regulate collective and single cell migration (15, 16). Since nonmotile and noninvasive cells were used, the identification of genes that drive the highly motile behavior of aggressive cancer cells was impossible. Moreover, these screens focused on kinases and several genes associated with breast cancer (BC), precluding the identification of the individual roles of cell adhesion and migration machinery components.
Here, we used a quantitative, multiparametric, high-content imaging-based RNA-interference screen to identify genes involved in the regulation of different cell migratory features, including migration speed, directionality, and persistence. Using the highly migratory H1299 cells as a model, we screened 1,429 genes in total, of which 136 genes either stimulated or inhibited cell migration behavior. Of 64 carefully selected genes, 30 were validated by transfection with more than 3 individual siRNA duplexes. We further selected 8 genes for in-depth characterization: all showed a strong, significant association with cancer metastasis-free survival (MFS) in BC patients. In addition, we focused in greater depth on SRSF protein kinase 1 (SRPK1), which is highly expressed in aggressive cell lines of the basal (A and B) BC subtype. We show that depletion of SRPK1 from basal BC cells inhibited their migration, and inhibited FA dynamics. Furthermore, SRPK1 is required for metastasis to occur in a spontaneous orthotopic tumor metastasis mouse model.
Results
RNA-interference screening identifies novel regulators of tumor cell migration.
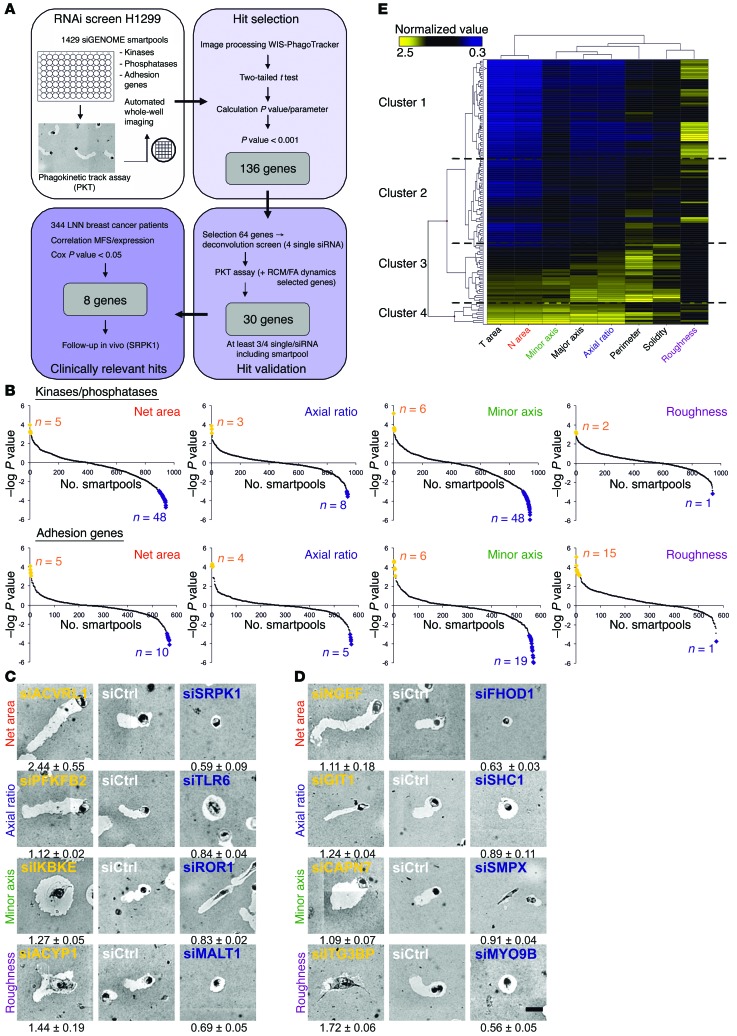
To discover novel regulators of tumor cell migration, we used the highly migratory and well-characterized H1299 human non–small cell lung carcinoma cell line, in a high-content, imaging-based phagokinetic track (PKT) assay in which cells migrate on a fibronectin-coated surface covered by a monolayer of beads (15). The bead-free tracks formed upon cell locomotion correspond to the cell’s protrusion activity and migratory path. Multiparametric image analysis of individual cell tracks enables the quantitative assessment of the migratory strategy of individual cells, by determining 8 different “migratory phenotypes” (“total area,” “net area,” “axial ratio,” “major axis,” “minor axis,” “roughness,” “perimeter,” and “solidity”) (15).
Cell migration involves spatially and temporally regulated lamellipodial protrusion and cell adhesion, regulated by upstream signaling cues mostly through specific phosphorylation and dephosphorylation events. We used siGENOME SMARTpool libraries (Dharmacon) targeting all known human kinases (n = 779) and phosphatases (n = 198), as well as a “custom adhesome library” targeting integrin adhesion components and cytoskeletal regulators (n = 576). In total, 1,429 unique genes (after removal of duplicates) were screened in 2 independent experiments on duplicate plates (see schematic overview in Figure 1A). Candidate genes affecting H1299 cell migration were identified, on the basis of statistical comparison with nontargeting siCtrl. Four main parameters (net area, axial ratio, minor axis, and roughness) were selected for identifying “hits.” P values for each parameter and each gene were calculated using a 2-tailed t test: the cutoff value for hit identification was set at P < 0.001 in at least 1 of the 4 mentioned parameters, and visually represented in scatter plots (Figure 1B and Supplemental Table 1; supplemental material available online with this article; doi:10.1172/JCI74440DS1). This method enabled the selection of strong candidate genes that upon knockdown either enhance (e.g., activin A receptor type II–like 1 [ACVRL1], neuronal guanine nucleotide exchange factor [NGEF]) or inhibit (e.g., SRPK1, formin homology 2 domain containing 1 [FHOD1]) migration; or diminish directionality (e.g., inhibitor of kappaB kinase epsilon [IKBKE], calpain 7 [CAPN7]) or migration strategy (e.g., receptor tyrosine kinase-like orphan receptor 1 [ROR1], GPCR kinase interacting ArfGAP 1 [GIT1]) (Figure 1, C and D). In total, we identified 136 genes that significantly affected H1299 cell migration. The total data set and all images can be browsed at the Cellular Phenotype Database created at the European Bioinformatics Institute of the European Molecular Biology Laboratory (EMBL-EBI) (http://www.ebi.ac.uk/fg/sym; accession number P2_SyM). Unsupervised hierarchical clustering of all 136 genes and all 8 migration parameters defined 4 large clusters of genes with common roles in a particular migration strategy. For example, genes that strongly increased the roughness of tracks (cluster 1) also decreased the cells’ overall migratory capacity. Genes that increased the overall length of the migratory track (cluster 4) typically have an increased axial ratio, but not an increased roughness, indicating that these cells have an increased cell migration speed and directionality (Figure 1E and Supplemental Figure 1).
Figure 1. RNA-interference screen identifies novel tumor cell migration regulators.
(A) Schematic representation of screen setup. Starting upper left, clockwise: Screening of 1,429 individual SMARTpools (kinases, phosphatases, and adhesion genes), making use of the PKT assay in H1299 to examine the effect of knockdown of a gene on cell migration. In a deconvolution screen, 30 of 64 tested genes were validated with at least 3 of 4 single siRNAs. Expression of validated genes was correlated with MFS in a BC patient cohort, which resulted in 8 clinically relevant genes. (B) For each tested SMARTpool in the primary screen, the P value is calculated for each parameter using a 2-tailed t test. The extremes of the graph indicate the significant SMARTpools (P < 0.001), visualized as yellow and blue data points. A negative score indicates a decrease compared with siGFP control. (C and D) Examples of identified genes (representative image chosen from 4 replicates) in the kinase and phosphatase library are shown in C, and results for the adhesion-related genes in D. Normalized values (± SD) are written underneath each image (full table Supplemental Table 1). Scale bar: 50 μm. (E) Unsupervised hierarchical clustering on the 136 identified genes revealed signatures for specific migratory behavior such as increased net area combined with increased axial ratio, or decreased net area combined with increased roughness (for details see Supplemental Figure 1).
For our deconvolution screen, we narrowed our list of candidates by selecting genes that had an inhibitory effect on cell migration upon knockdown in the primary screen, which may then act as targets for therapeutic inhibition in relation to cancer metastasis. We also included hits of other migratory features, to ensure the reproducibility and power of our methods. A final filtering step was based on available knowledge in the cancer literature, eventually leading to the selection of 64 genes of interest, out of the 136 candidate genes from our primary screen for detailed validation.
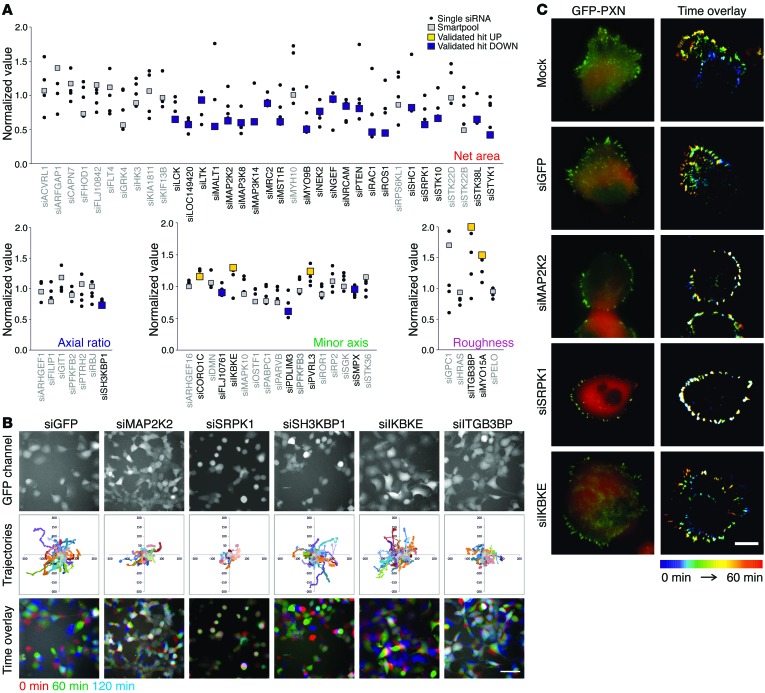
In a secondary deconvolution PKT screen, we tested the effect of the 4 individual siRNA duplexes that constitute each SMARTpool. In total, 30 high-confidence genes were defined, on the basis of the observation that their effect in the primary screen was confirmed in at least 3 of 4 single sequences, and in the SMARTpool, in the secondary screen (Figure 2A). The strongest validated hits included overall inhibitors of cell migration (net area): lymphocyte-specific tyrosine kinase (LCK), mucosa-associated lymphoid tissue lymphoma translocation protein 1 (MALT1), myosin IXB (MYO9B), RAC1, ROS protooncogene 1 (ROS1), and SRPK1; and genes involved in cell migration mode (axial ratio, minor axis, and roughness): SH3-domain kinase binding protein 1 (SH3KBP1), IKBKE, poliovirus receptor-related 3 (PVRL3), ITGB3BP, and myosin XVA (MYO15A).
Figure 2. Validated high-confidence hits affect cell migration in association with modulated FA dynamics.
(A) Sixty-four selected genes were tested in a deconvolution PKT screen with 4 single siRNAs per gene. Validated high-confidence genes (confirmed effect in primary screen in at least 3 of 4 single siRNAs and SMARTpool) are highlighted in yellow (increased compared with control) or blue (decreased). (B) Hits for each parameter were selected and tested in a live cell imaging–based migration assay. Stills from a representative movie chosen from 2 independent experiments with each 3 replicates per siRNA are shown. Scale bar: 100 μm. (C) FA dynamics was studied using TIRF imaging of H1299 cells expressing GFP-paxillin (PXN). In each experiment (mock n = 12, siGFP n = 10, siMEK2 n = 3, siSRPK1 n = 12, and siIKBKE n = 2) at least 10 cells were imaged, and representative images are shown. Decreased cell migration after knockdown of SRPK1 and MAP2K2 was related to increased stabilization of FA as indicated by the presence of “white” FAs in the time color overlay. Knockdown of IKBKE, affecting minor axis, does not affect cell migration per se, but resulted in increased cell area, reflecting the increase in minor axis in the PKT screen. Scale bar: 20 μm.
Inhibition of cell migration diminishes FA dynamics.
To confirm the migratory phenotype detected in the static PKT assay in a tertiary screen, we also tested the effects caused by knockdown of our 30 high-confidence genes in a live cell imaging–based cell migration assay using the same H1299 cells, but this time ectopically expressing GFP, which enabled visualization of the cells with real-time epifluorescence microscopy (Figure 2B, Supplemental Figure 2, and Supplemental Videos 1–6; and the Cellular Phenotype database created at EMBL-EBI for all 30 movies). Interestingly, while depletion of SRPK1 or ITGB3BP inhibited cell migration, cells without ITGB3BP, in particular, still displayed dynamic membrane ruffling, in agreement with an increase in the roughness parameter in the PKT assay (Figure 1D).
FA reorganization is essential for cell migration; therefore, we monitored FA organization for several genes. Their role in FA dynamics was evaluated in H1299 cells expressing GFP-tagged paxillin. In control and siGFP conditions, continuous, polarized remodeling of FA occurred at the leading edge of the cell (Supplemental Videos 7 and 8). In contrast, depletion of SRPK1 or MAP2K2, both of which inhibited cell migration, stabilized FA structures throughout the cell periphery, and hardly any reorganization occurred (Supplemental Videos 9 and 10). Knockdown of IKBKE, however, which caused a depolarization of lamellipodia formation (Figure 2B), was associated with enhanced FA remodeling along the entire cell border (Figure 2C and Supplemental Video 11). Taken together, these data indicate that changes in cell migratory behavior caused by the different candidate migration genes are largely dominated by changes in cell matrix adhesion dynamics.
Genes affecting cell migration are associated with BC metastasis-free survival.
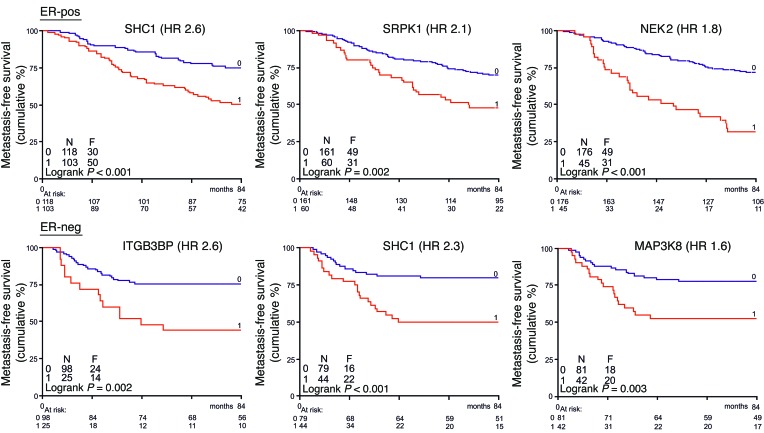
Excessive invasive cell migration associated with enhanced cell survival is a hallmark of metastatic cancer (17). Consequently, we checked which of our 30 high-confidence genes showed an association with BC progression. We used an expression data set from a cohort of 344 BC patients. All patients were lymph node–negative at the time of presentation in the clinic, and none had received (neo) adjuvant systemic therapy. Some of our candidate genes were previously described as being associated with cancer progression (e.g., SHC1, ref. 18; macrophage-stimulating protein receptor (MST1R), refs. 19, 20; and ROR1, ref. 21). We further determined the correlation between expression levels of the identified genes: MFS, hazard ratio, and Cox P values were calculated. This analysis indicated that of our 30 high-confidence genes, ITGB3BP, LCK, MAP2K2, MAP3K8, NEK2, ROS1, SHC1, and SRPK1 showed significant clinical association with tumor aggressiveness. The expression level of those genes (low versus high, split by optimal cutoff point as well as median split point) significantly correlates with the MFS of either estrogen receptor–negative (ER-negative) (n = 123), ER-positive (n = 221), or all profiled BC patients (Cox P value < 0.05; Supplemental Table 2). Kaplan-Meier MFS curves for these genes further highlighted the relationship with respect to BC clinical outcome (Figure 3 and Supplemental Figures 3 and 4). Increased expression of NEK2 and SRPK1 in ER-positive BC patients is related to poor MFS and, similarly, increased expression of ITGB3BP and MAP3K8 in ER-negative BC patients. High expression levels of SHC1 are correlated with poor prognosis. The association of SRPK1 and NEK2 was validated in a combined set of 3 independent BC patient cohorts (Supplemental Figure 5).
Figure 3. Clinical relevance of candidate tumor cell migration hits in BC metastasis-free survival.
Gene expression data of a lymph node–negative BC patient cohort (n = 344) without prior treatment were used. Expression of identified tumor cell migration genes was correlated with MFS, and hazard ratio (HR) and Cox P values were calculated for ER-negative (n = 123) and -positive (n = 221) BC patients as well as all patients combined. Eight of 30 validated genes showed clinical relevance (2-sided P < 0.05, boldface in Supplemental Table 2). Kaplan-Meier curves are shown for genes with highest HR for MFS, drawn after determination of the optimal cutoff point (low log-rank P value). Low expression is represented by blue lines, and high expression by red lines.
As these 30 selected genes were the highest-confidence hits that control tumor cell migration, we examined their expression levels in a panel of 22 luminal-like and 13 basal A/B human BC cell lines, and calculated the ratio of basal over luminal cell lines (Supplemental Figure 6 and ref. 22). Interestingly, most genes showed enhanced expression in basal BC cells, which are ER-negative and more aggressive than the luminal, mostly ER-positive, cells. This confirmed the strength of our phenotypic screen to discover new genes that might be good candidates to inhibit the migratory phenotype of highly metastatic cells such as those in the basal human BC cell lines.
Increased SRPK1 protein levels are correlated with poor prognosis in BC patients.
SRPK1, a splicing factor kinase that has not been previously studied in connection with cancer metastasis, was one of the genes most strongly associated with BC. Moreover, RNA splicing is becoming increasingly relevant to cancer progression (23). Therefore, we decided to focus in particular on SRPK1. Its expression levels were increased in more aggressive basal BC cell lines (Supplemental Figure 6), and our gene expression profiling revealed a correlation between SRPK1 mRNA expression level and BC disease outcome.
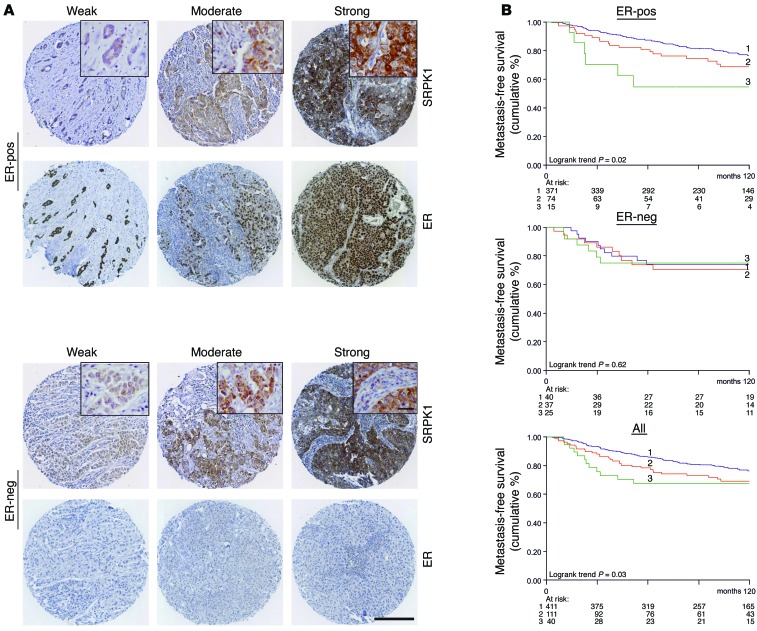
To test the latter correlation at the protein level, we performed immunostaining for SRPK1 on tissue microarrays (TMAs) containing 562 lymph node–negative BC patient samples. Intensity levels were scored as weak, moderate, or strong (Figure 4A), and correlated with the rate of metastatic tumor formation. While only 19% of the ER-positive patients (n = 460) scored moderately or strongly positive for SRPK1 expression, the increased SRPK1 protein levels in these BC patients correlated significantly with poor MFS. In the smaller subset of ER-negative BC patients (n = 102), such a correlation was not observed, but here, the majority of patients (61%) showed already enhanced SRPK1 protein levels, making it difficult to divide this group on the basis of different expression levels with enough statistical power.
Figure 4. SRPK1 protein levels correlate with poor prognosis in BC patients.
(A) TMAs of lymph node–negative BC patients (n = 562) without prior treatment were stained for SRPK1 and scored. Examples of weak (score 1), moderate (score 2), and strong (score 3) SRPK1 intensity in the TMA are shown for ER-positive and ER-negative samples. Scale bar: 200 μm, inset 20 μm. (B) Increased expression of SRPK1 correlated with decreased MFS in ER-positive patients (n = 460, log-rank P < 0.05, top graph), but not ER-negative patients (n = 102, middle graph). For all patients combined, SRPK1 expression was associated with worse prognosis (log-rank P < 0.05, bottom graph).
When all patients were analyzed, regardless of their ER status, increased SRPK1 protein expression was significantly associated with poor disease outcome (Figure 4B). Since the highest significance was observed in luminal BC cell lines, we looked further into luminal A and the more aggressive luminal B subtype. On the basis of our array data, SRPK1 expression was in particular significantly correlated with the luminal B subtype (Supplemental Figure 7). Moreover, TMA staining indicated a significant correlation between high SRPK1 and high Ki67 staining in ER-positive BC (Pearson’s χ2 statistics; n = 438, P = 0.0012), while Ki67 itself correlated with the luminal B subtype (Pearson’s χ2 statistics; overlap TMA and gene expression array BC patients n = 40, P = 0.011). Taken together, these findings support an association between SRPK1 and luminal B BC.
SRPK1 knockdown in ER-negative BC cells reduces their migratory capacity and stabilizes FAs.
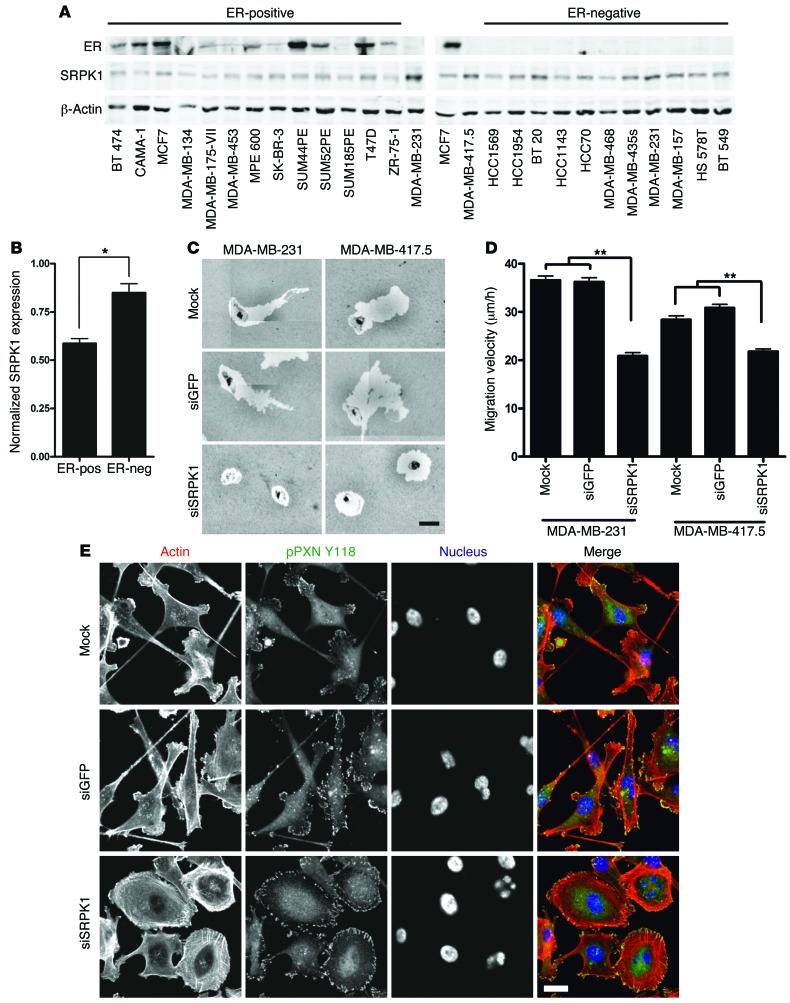
Since SRPK1 expression significantly correlated with poor MFS outcome in BC patients, we set out to define the role of SRPK1 in BC metastasis. First, we determined SRPK1 protein levels in a panel of human BC cell lines. Although the SRPK1 protein was detected in all cell lines tested, its expression was significantly higher in the more aggressive, ER-negative basal BC cells (Figure 5, A and B).
Figure 5. Knockdown of SRPK1 in human ER-negative BC cells blocks cell migration and results in a more epithelial-like morphology.
(A) Western blot analysis showed the increased expression of SRPK1 in ER-negative human BC cells compared with ER-positive cells. (B) Quantification of Western blot (2-tailed t test, *P < 0.05). (C) Transient knockdown of SRPK1 in MDA-MB-231 and MDA-MB-417.5 blocked cell migration in a PKT assay. Scale bar: 50 μm. (D) Transient knockdown inhibited random cell migration of MDA-MB-231 and -417.5 cells (2-tailed t test, **P < 0.01). (E) Knockdown MDA-MB-417.5 cells were stained for actin with rhodamine phalloidin (red) and phospho Tyr(118)-paxillin (pPXN) to visualize FAs (green). After depletion of SRPK1, cells had a more epithelial-like morphology. Scale bar: 20 μm.
Next, we determined the role of SRPK1 in cell migration in the ER-negative and highly migratory MDA-MB-231 cell line and its lung metastatic variant, MDA-MB-417.5 (24). As shown, depletion of SRPK1 significantly reduced the velocity of both cell lines, as measured by both the PKT assay (Figure 5C) and live cell imaging (Figure 5D and Supplemental Videos 12–17). Similar effects were observed in 2 other basal BC cell lines, HCC70 and Hs578T (Supplemental Figure 8). This reduced migratory phenotype was accompanied by a radical morphological switch: from a mesenchymal-like phenotype with extensive protrusions and multiple small FAs, to a more sessile shape with nearly no membrane protrusions, and large FAs at the cell periphery associated with F-actin stress fibers, similar to those seen in H1299 following SRPK1 knockdown (compare Figure 5E and Figure 2C).
SRPK1 is essential for BC metastasis to different target organs.
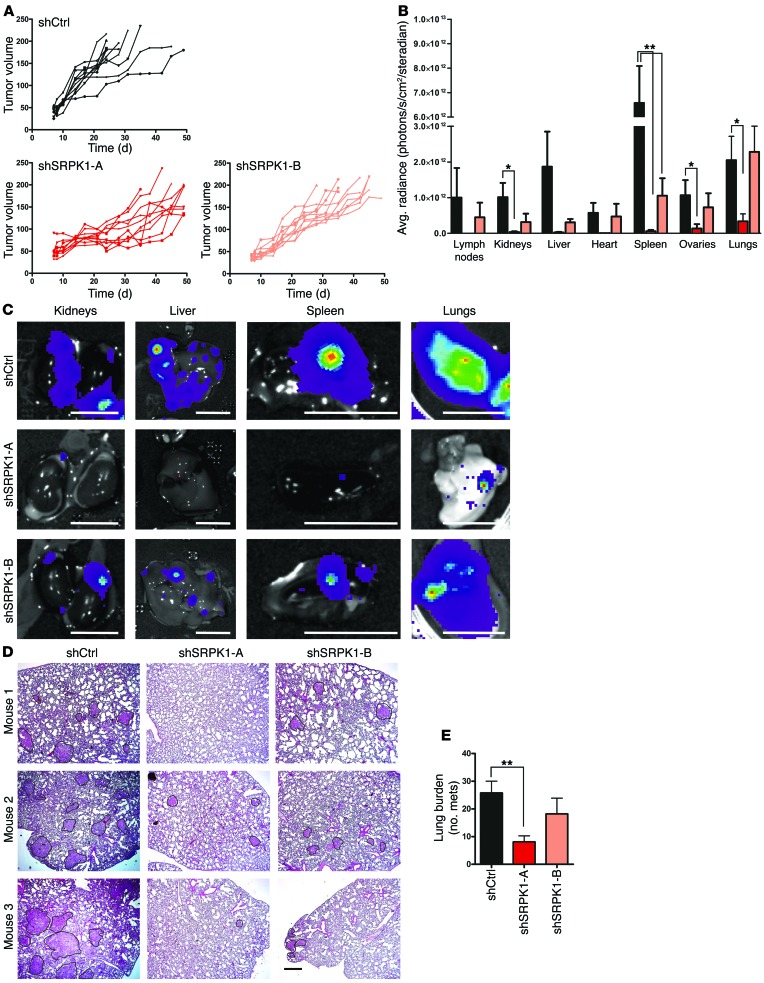
Finally, we determined the role of SRPK1 expression in BC metastasis in vivo. We used the well-characterized GFP and luciferase-expressing MDA-MB-417.5 cells that metastasize to the lung in an orthotopic mouse tumor metastasis model (24, 25). Toward that end, we generated independent, stable SRPK1 lentiviral shRNA knockdown MDA-MB-417.5 cell lines using 5 different shRNA sequences, and selected 2 cell lines that showed nearly complete depletion of SRPK1 protein levels (Supplemental Figure 9, A and B). Knockdown of SRPK1 slightly affected proliferation of these cells in culture (Supplemental Figure 10A). In vivo, SRPK1 knockdown slightly affected primary tumor growth in the mammary fat pad for both individual shRNA constructs, compared with shCtrl (Figure 6A). Despite the fact that expression of the proliferation marker Ki67 was unaffected (Supplemental Figure 10B), we removed every individual tumor, once they reached a size of 7 × 7 mm (Supplemental Figure 11, A and B), to enable equal numbers of tumor cell dissemination from the primary tumor, and ensure that the total duration of the experiment was the same for all groups (Supplemental Figure 11C). We confirmed reduced SRPK1 levels in primary tumors by Western blotting (Supplemental Figure 11, D and E).
Figure 6. SRPK1 is required for BC metastasis in vivo.
MDA-MB-417.5 cells were transduced with lentiviral shRNA specific for 2 independent sequences of SRPK1 (shSRPK1-A and shSRPK1-B) and a nontargeting shRNA (shCtrl). (A) MDA-MB-417.5 cells were injected in 7- to 9-week-old Rag2–/– Il2rg–/– mice (n = 10), and tumor growth was followed over time. (B) One to five weeks after primary tumor removal, mice were sacrificed, and average radiance was quantified by bioluminescence imaging of the organs after isolation for each animal (2-tailed t test, *P < 0.05, **P < 0.01). (C) Representative bioluminescence images for the main organs and each group are shown. Scale bars: 1 cm. (D) The small lung lobes were processed for immunohistological analysis and stained with H&E (scale bar: 200 μm). (E) The number of metastases (mets) was counted (2-tailed t test, **P < 0.01). See examples of metastases delineated by a black line in D. Knockdown of SRPK1 significantly reduced lung burden.
Metastatic spread was monitored by bioluminescence imaging over time, which revealed that the overall metastatic potential of SRPK1-depleted cells was significantly reduced (Figure 6, B and C, and Supplemental Figure 11F). In particular, metastatic spread to lungs was examined in greater detail by H&E staining, and showed decreased tumor lung burden in SRPK1 knockdown groups compared with control (Figure 6, D and E). Lung metastatic lesions of shSRPK1-A and -B still displayed reduced SRPK1 levels compared with shCtrl, although levels were somewhat higher than in the primary tumor (Supplemental Figure 12).
To evaluate further the mode of action of SRPK1 knockdown on metastasis, we additionally evaluated the metastatic potential of shSRPK1 cells using an experimental metastasis model in which MDA-MB-417.5 cells were injected directly in the circulation through tail vein injection. Under these circumstances shSRPK1 cells metastasized to the different target organs in a manner similar to that of shCtrl cells: the lung metastasis burden of shSRPK1 cells was similar to that of shCtrl, and the metastatic lesions were comparable in size (Supplemental Figure 13, A–D). These data indicate that shSRPK1 does not affect proliferation to the extent that it will block metastatic outgrowth. Moreover, these findings support a role for SRPK1 in the regulation of tumor dissemination from the primary tumor.
In addition, we repeated this experiment in the 4T1 cell line, another ER-negative basal mammary carcinoma from mouse. Interestingly, depletion of SRPK1 in this instance resulted in an almost complete eradication of the metastatic potential of 4T1 cells to colonize the lungs (Supplemental Figure 14, A–G).
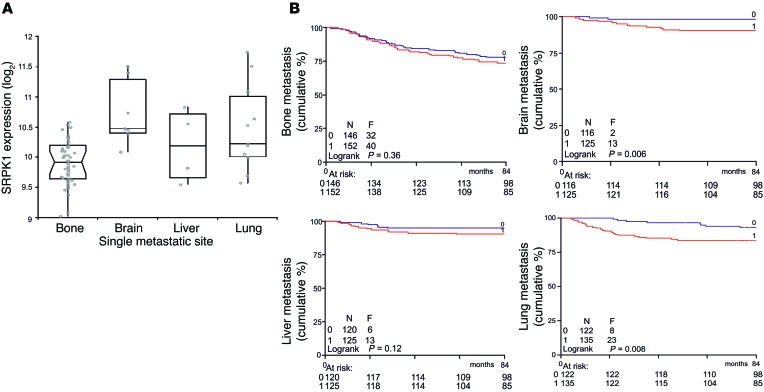
Given this overall strong effect of SRPK1 on lung colonization, we next selected from our BC patient gene expression database those patients in whom cancer had metastasized to a single site, in order to determine whether homing of the cells to the lungs was related to SRPK1 expression. Primary breast tumors with increased SRPK1 levels showed preference for homing to the lung and brain (Figure 7A). Comparison of Kaplan-Meier curves between patients without relapse and patients with lung relapse (optimal cutoff point) demonstrated that high SRPK1 expression is associated with lung and brain metastasis (Figure 7B), but not with bone and liver metastasis, indicating that SRPK1 exerts an organ-specific rather than an overall effect on metastasis.
Figure 7. High SRPK1 expression is associated with preferred metastasis to lung and brain.
(A) Within the BC patient gene expression data set, only patients with metastasis to a single site were selected, and this was related to SRPK1 expression. In the box plot, median SRPK1 expression is depicted by the horizontal line, and the box starts at the first quartile and ends at the third quartile. Whiskers are extending to furthest observations within ± 1.5 interquartile range. (B) Kaplan-Meier curves between patients without relapse and patients with lung relapse (optimal cutoff point, low log-rank P value) were compared. Low expression is represented by blue lines, and high expression by red lines.
SRPK1 is involved in controlling the NF-κB signaling network.
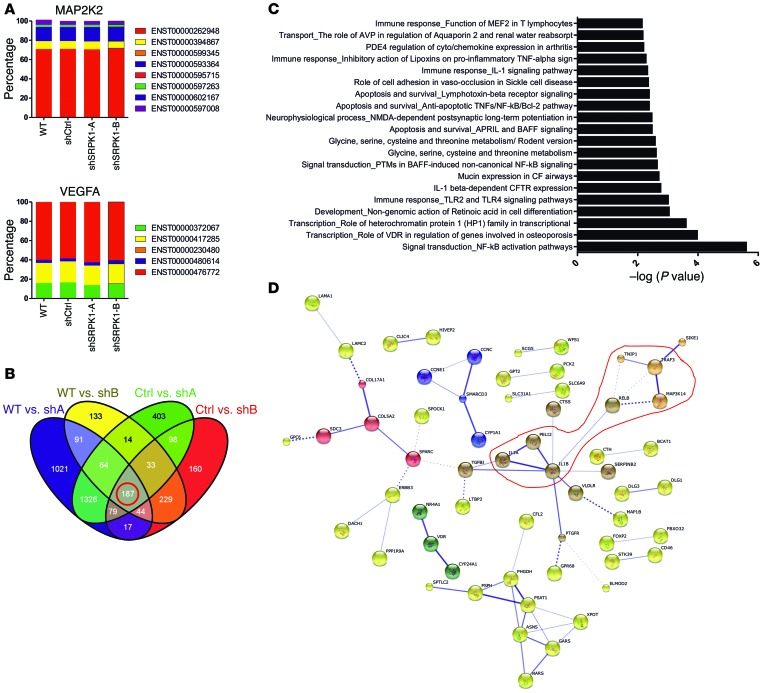
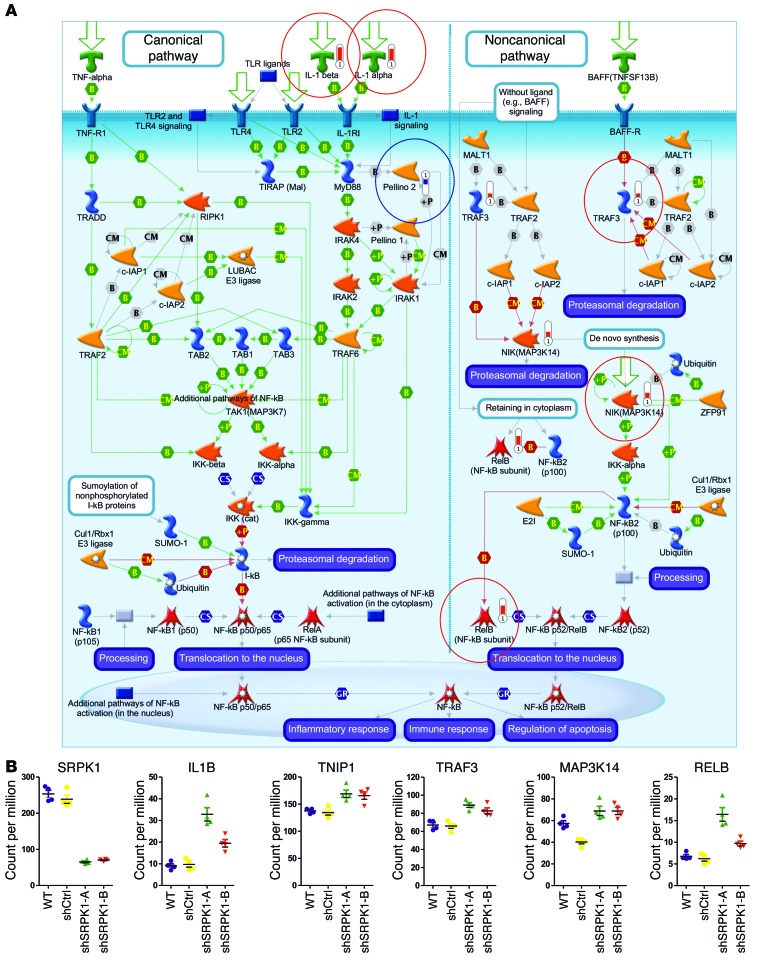
SRPK1 belongs to the serine-arginine protein kinase family, a relatively new subfamily within the serine-threonine kinases. Given the role of SRPK1 in both constitutive and alternative splicing (26), we anticipated that SRPK1 might affect splicing patterns of specific genes and/or the expression of particular genes or gene networks. To investigate this we performed RNA deep sequencing of MDA-MB-417.5 WT, shCtrl, and shSRPK1-A and -B cell lines (Supplemental Figure 15) followed by bioinformatics analysis. We analyzed the output data of deep sequencing with 2 different RNA-seq analysis tools and could not identify significant altered splicing events. We specifically checked MAP2K2 and VEGFA, which were both previously shown to undergo differential splicing upon SRPK1 knockdown in other cell types (27, 28), but no differential splicing was detected in our sequencing data (Figure 8A). This supports our finding that upon SRPK1 knockdown angiogenesis was not perturbed, as we did not observe a difference in CD31-positive blood vessel density within the primary tumors (Supplemental Figure 14H). The differential expression analysis performed in a pairwise comparison resulted in a list of 187 overlapping confident differential regulated genes after SRPK1 knockdown (Figure 8B); SRPK1 was the most downregulated gene in both shSRPK1-A and -B cell lines (Figure 9B). Pathway analysis revealed that the NF-κB pathway was most significantly affected, involving increased gene expression of the canonical pathway ligands IL1A and IL1B and of the noncanonical pathway components TNF receptor-associated factor 3 (TRAF3), MAP3K14, and V-rel avian reticuloendotheliosis viral oncogene homolog B (RELB) (Figure 8, C and D, and Figure 9, A and B). These data suggest an involvement of the NF-κB pathway in the control of breast tumor cell migration and metastasis formation.
Figure 8. Knockdown of SRPK1 does not affect splicing but results in the differential expression of almost 200 genes when analyzed with deep sequencing.
(A) No difference in splicing of MAP2K2 and VEGFA was detected. (B) Pairwise comparison to identify common genes between the 4 groups that are significantly differentially expressed (log cpm >1.4; FDR <0.1). (C) A pathway analysis performed with MetaCore shows that the NF-κB signaling is the most significantly altered. (D) Similarly, the 187 genes were used in the STRING 9.1 database to visualize potential protein networks (highlighted in red are proteins of the NF-κB signaling). The confidence view is shown: stronger association between proteins is represented by thicker lines (disconnected nodes are hidden).
Figure 9. Knockdown of SRPK1 affects expression of components of NF-κB signaling.
(A) Components of the NF-κB signaling that are significantly upregulated (circled in red) and downregulated (circled in blue) in both SRPK1 knockdown cell lines. The network was generated through the use of MetaCore. (B) Expression levels (expressed in count per million) of SRPK1 and the NF-κB signaling–related genes in WT, shCtrl, and shSRPK1-A and -B cell lines.
Discussion
In this study, we used a multiparametric, quantitative, high-content, imaging-based RNA-interference screen to identify novel regulators of tumor cell migration. Since such migration is likely to be controlled by enhanced oncogenic (receptor-mediated) signaling that drives the dynamic rearrangement of the actin cytoskeletal network and cell matrix adhesions, we focused on kinases, phosphatases, and multiple adhesion-related genes. Our work led to the identification of 30 high-confidence genes that control tumor cell migration; of those, 8 were clinically relevant candidate metastasis genes, of which SRPK1 was shown to regulate BC metastasis.
The PKT assay enabled us to quantitatively assess different modes of cell migration, as the track morphology reveals the effects on overall migration of effective speed, persistence, and lateral membrane activity. We focused in particular on hits that affected the parameters net area, minor axis, axial ratio, and roughness. These turned out to be the most independent parameters associated with different migratory phenotypes in relation to either enhanced or decreased migration, which is essential for translation to cancer metastasis. Most of the high-confidence hits decreased the overall net area, indicating inhibition of cell migration, as validated by the live cell imaging cell migration assay. Interestingly, several receptor tyrosine kinases (leukocyte receptor tyrosine kinase [LTK], MST1R/RON, ROS1, and serine/threonine/tyrosine kinase 1 [STYK1]), and likely downstream effectors, including adapters (SHC1), nonreceptor tyrosine kinases or MAPK family members (LCK, MAP2K2, MAP3K8, and MAP3K14), Rho-GTPases, and upstream regulators (RAC1, NGEF, and MYO9B), were identified as well. While none of these genes affected cell survival, they are important regulators of H1299 cancer cell motility. While only 2 of the 5 genes that we re-evaluated for roughness were validated, other genes in the primary screen, including NGEF, DnaJ homolog, subfamily C, member 27 (RBJ), Rho GEF 12 (ARHGEF12), peptidyl-tRNA hydrolase 2 (PTRH2), gap junction protein α5 (GJA5), glypican 1 (GPC1), glutathione S-transferase Mμ 1 (GSTM1), HRAS, integrin αD (ITGAD), ITGB3BP, galectin 3 (LGALS3), myristoylated alanine-rich protein kinase C substrate (MARCKS), MYO15A, protein pelota homolog (PELO), protein phosphatase 1, regulatory subunit 13B (PPP1R13B), and TATA box–binding protein–associated factor 1 RNA polymerase II, 210kDa-like (TAF1L), resulted in increased roughness (Figure 1E, Supplemental Table 1, and Cellular Phenotype Database, http://www.ebi.ac.uk/fg/sym). Since enhanced roughness involves enhanced lamellar activity, it is remarkable that several of the identified roughness hits have previously been linked to Rac1, an important driver of lamellipodia formation. These genes include MARCKS (29), HRAS (30, 31), ARHGEF12 (32), and NGEF (33). Importantly, ITGB3BP (also known as TAP20) had the strongest effect on roughness: it blocked cell migration upon knockdown (Figure 2), and its expression levels in BC were associated with poor MFS outcome in ER-negative BC patients (Figure 3).
Furthermore, our screen provides a detailed, comprehensive source of information on the role of individual genes in cell migration. Dynamic remodeling of cell matrix adhesion complexes/FA is essential for cell migration (34). Indeed, we found that almost all genes that stimulate cell migration, including SRPK1 and MAP2K2, downregulated FA organization in H1299 cells. Previously, 44 kinases/phosphatases/adhesion-related genes that affected the structural organization of FAs had already been identified in an RNA-interference screen involving HeLa cells (35). Hardly any overlap was observed between the different genes that affect cell migration in H1299 cells, and FA organization in HeLa cells. This difference can be attributed to the broad differences in migratory behavior between our highly motile H1299 cells and the rather sessile HeLa cells, as well as to the different oncogenic programs found in these cell lines. Regardless of cell differences, IKBKE (inhibitor of NF-κB kinase subunit e) was identified in both screens as a strong hit. Interestingly, it was also identified in a separate screen using MCF10-A cells, where it impaired the collective migration of these cells (16). IKBKE has been identified as an oncogene, and is amplified and overexpressed in over 30% of breast carcinomas and BC cell lines (36). Detailed analysis of FA dynamics in H1299 cells demonstrated that knockdown of IKBKE strongly affected the axial ratio, causing a circular cell track that indicated a loss of persistent cell migration, and disturbing the polarized distribution of FAs, but not their dynamics. This finding is in contrast to SRPK1 and MAP2K2 (also known as MEK2), both of which showed upon their knockdown almost complete inhibition of FA dynamics in association with loss of cell motility. Focal adhesion kinase (FAK) is essential to the dynamics of FAs, as well as to BC metastasis (11, 37, 38). The inhibition of cell migration by SRPK1 knockdown was not affected by altered expression of FAK; however, FAK phosphorylation was reduced in SRPK1 knockdown cells (Supplemental Figure 16A). The phosphorylation status of other FA signaling and scaffolding proteins, including paxillin and p130Cas, was also affected by SRPK1 knockdown (Supplemental Figure 16B). Altogether, these findings suggest that modulation of tumor cell migration is associated with altered FA dynamics and signaling.
An important strength of our current work is that we systematically determined the relationship between genes that affect tumor cell migration ex vivo, and metastatic disease in vivo. We used expression profiling of a BC patient cohort to determine the possible clinical relevance of the hits found in the migration screen. We established that ITGB3BP, LCK, MAP2K2, MAP3K8, NEK2, ROS1, SHC1, and SRPK1 are strongly associated with MFS. Some of these genes are already established modulators of BC progression, confirming our approach to target discovery. In particular, activation of SH2 signaling adapter domain–containing transforming protein 1 (SHC1), which acts directly downstream of extracellular signals, is associated with poor BC patient prognosis (39). Lymphocyte-specific protein tyrosine kinase (LCK) was highly expressed in ER-negative tumors compared with ER-positive tumors. High membrane LCK expression was significantly associated with improved survival (40). This is in agreement with our data, which also show that patients with high LCK levels have favorable MFS compared with patients with low levels of this kinase (Figure 3A). Given the fact that LCK inhibited migration in H1299 cells, this contradictory association with MFS is possibly related to a different cancer cell type (as knockdown of LCK in MDA-MB-231 increases cell migration; data not shown) or differential regulation of LCK activity in vitro versus the in vivo condition. Since our siRNA screen was performed in H1299 cells, we cannot exclude the possibility that we missed other critical BC cell migration modulators.
SRPK1 strongly associates with MFS in BC patients, which we confirmed in 2 independent orthotopic BC metastasis models.
SRPK1 overexpression has been associated with increasing tumor grade in several other cancers (27). SRPK1 belongs to the serine-arginine protein kinase family, a relatively new subfamily within the serine-threonine kinases. Although SRPK1 is known to play a role in both constitutive and alternative splicing (26), we could not identify significant altered splicing using our current culture conditions and RNA-seq analysis pipeline. In addition, MAP2K2 and VEGFA, which previously have been associated as targets for RNA splicing under the control of SRPK1, were not altered upon SRPK1 knockdown. This could possibly be related to some remaining SRPK1 in our stable shSRPK1-A and -B cell lines, as was also witnessed by our RNA-seq data (Figure 9B). Despite the lack of clear splicing modulation, we did observe strong inhibitory effects of SRPK1 knockdown on tumor cell migration and metastasis formation. At this point we cannot fully rule out that SRPK1 does contribute to modulation of RNA splicing events in cancer progression in the in vivo situation in a particular subset of cells that is actively disseminating from the primary tumor. On the other hand, interestingly, we did observe a significant modulation of several gene targets that are components of the canonical and noncanonical NF-κB pathway. NF-κB signaling is critically linked with BC progression (41); however, a possible role for SRPK1 in the regulation of this pathway in the context of BC metastasis formation needs further experimental confirmation.
In summary, a high-content imaging-based RNA-interference migration screen enabled us to identify 30 high-confidence tumor cell migration regulators. In particular, we focused on the splicing-related gene SRPK1 as an important, clinically relevant metastasis-modifying factor. We showed that high SRPK1 levels in BC patients were associated with poor disease prognosis, and increased tumor aggressiveness. Importantly, despite clear differences between cell culture conditions and the 3D microenvironment in intact tumor tissue, a systematic analysis of in vitro screening data, with a clear clinical cancer progression end point and in vivo testing, constitutes a powerful experimental pipeline for identifying novel, likely drug-target candidates.
Methods
Cell culture.
H1299 cells (ATCC-CRL-5803), MDA-MB-231 (ATCC-HTB-26), MDA-MB-417.5 (kindly provided by Joan Massagué, Cancer Biology and Genetics Program, Memorial Sloan Kettering Cancer Center, New York, New York, USA and Howard Hughes Medical Institute, Chevy Chase, Maryland, USA), and all other human BC cell lines (originally purchased from ATCC or provided by others; see Hollestelle et al., ref. 22) were cultured in RPMI medium (GIBCO, Life Technologies) supplemented with 10% FBS (PAA Laboratories), 100 IU/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). For live cell imaging, phenol red–free culture medium was used. Cells were maintained in a 5% CO2 humidified chamber at 37°C.
Antibodies.
Mouse anti-SRPK1 (no. 611072) was purchased from BD Biosciences. Rabbit anti-ER (sc-543) and mouse anti–β-actin (sc-47778) were from Santa Cruz Biotechnology. Mouse anti-tubulin (T-9026) was from Sigma-Aldrich.
Transient siRNA-mediated gene knockdown.
Human protein kinases and phosphatase libraries and a custom-designed adhesome siRNA library were purchased in siGENOME format from Dharmacon. Plates were diluted to 1 μM working concentration in complementary 1× siRNA buffer in a 96-well plate format. A 50-nM reverse transfection was performed according to the manufacturer’s guidelines. Complex time was 20 minutes, and 2,400 cells were added. The plate was placed overnight in the incubator, and the next morning, medium was refreshed. After 48–72 hours, cells were used for various assays.
Stable shRNA-mediated gene knockdown.
MDA-MB-417.5 cells were transduced with Sigma-Aldrich MISSION lentiviral shRNA constructs coding a nontargeting control sequence (shCtrl) and 2 independent sequences for SRPK1 (shSRPK1-A: CCG GGT GGC AAT GAA AGT AGT TAA ACT CGA GTT TAA CTA CTT TCA TTG CCA CTT TTT; shSRPK1-B: CCG GCC ATA ACT AAA GGA TCA GGA TCT CGA GAT CCT GAT CCT TTA GTT ATG GTT TTT) and selected by puromycin (in collaboration with Rob Hoeben, Leiden University Medical Center).
Phagokinetic track assay (PKT).
Glass-bottom 96-well imaging plates (Whatman) were coated with 10 μg/μl fibronectin (Sigma-Aldrich) and stored overnight at 4°C. The next day, plates were washed twice with PBS using a Hydroflex platewasher (Tecan). White 0.40-μm surfactant-free carboxylated beads (coefficient of variation = 6.8%, solid = 4.1%; Molecular Probes, Invitrogen) were prepared by washing of 1.84 ml per plate twice in PBS. Volume was then adjusted to 4 ml with PBS, and beads were stored at 4°C. The PBS on the fibronectin-coated plates was removed, and 40 μl bead solution per well was added. The plate was then incubated for 1 hour at 37°C, then washed 7 times with PBS. Before cell seeding, PBS was replaced with culture medium. Knockdown cell suspensions were prepared 65 hours after transfection by washing twice in PBS and addition of 50 μl trypsin/EDTA (Invitrogen). One part of the cell suspension was diluted in culture medium, plated on the bead-coated plates, and placed in the incubator. After 7 hours’ migration time, cells were fixed using a 4% PFA solution and washed twice in PBS. Plates were then stored at 4°C for later imaging. For each transfection, duplicate bead plates were generated; the screening of the siRNA libraries was also performed in duplicate.
PKT imaging and analysis.
To image the bead-free tracks, an automatic screening microscope fitted with a ×10/0.20 NA objective was used. The transmitted light signal was acquired for 25 images per well, and an autofocus was set, prior to each image. Montage images per well were exported and analyzed using WIS-PhagoTracker software (15). After that, the data were log2-transformed. Next, normalization was performed, using a ratio of raw measurement to the mean of the negative control (nontargeting siCtrl). After normalization, for each of the 4 replicates, its distance to the mean of the other 3 replicates was calculated and compared with the standard deviation of the other 3 replicates. Outliers were identified when the distance was 3 times greater than the standard deviation; these were removed before hit identification. For hit identification, a 2-tailed t test to compare each condition (4 replicates) with the negative control was used, and genes were ranked according to P value. Normalized values are average values of 4 replicates, with removal of outliers.
PKT image database.
All screening data associated with this publication have been submitted to the Cellular Phenotype Database at EMBL-EBI (http://www.ebi.ac.uk/fg/sym), under accession number P2_SyM. This prototype repository was developed as part of the Systems Microscopy Network of Excellence and stores data derived from high-throughput phenotypic studies. It aims at providing easy access to phenotypic data, and at facilitating the integration of independent phenotypic studies. The data can be browsed by gene ID, RNAi reagent ID, or phenotypic descriptions. All RNAi reagent sequences are mapped to the latest Ensembl release of the genome before data loading, in order to update their target information and ensure correct annotation.
Live cell imaging of cell migration.
Glass-bottom 96-well plates (PAA Laboratories) were coated with 10 μg/μl fibronectin (Sigma-Aldrich) for H1299, or 20 μg/μl collagen type I (isolated from rat tails) for MDA-MB-231, for 1 hour at 37°C. Cells were plated directly upon transfection. After 72 hours, medium was refreshed, and cells were placed on a Nikon Eclipse TE2000-E microscope fitted with a 37°C incubation chamber, ×20 objective (0.75 NA, 1.00 WD), and perfect focus system. Automatically, 3 positions per well were defined, and a GFP signal was acquired at 3 positions every 390 seconds, for a total imaging period of 12 hours. Plates were then fixed using 4% formaldehyde and stored for later immunostaining. All data were converted and analyzed using custom-made ImagePro Plus (MediaCybernetics) macros (42). Cell migration was quantified by tracking of the GFP signal over time using a custom ImageJ-based macro.
Total internal reflection fluorescence microscopy of FA dynamics.
CELLview glass-bottom dishes with 4 compartments were coated with 10 μg/μl fibronectin (Sigma-Aldrich) for 1 hour at 37°C. H1299 cells stably expressing GFP-paxillin were used for this assay. Knockdown cells were seeded 24 hours after transfection, and 48 hours later, cells were placed on a Nikon Eclipse TE2000-E microscope fitted with a 37°C incubation chamber, ×60 oil objective (1.49 NA, 0.12 WD), and perfect focus system. FAs were visualized using total internal reflection fluorescence (TIRF) with the 488 laser for 1 hour, at 1-minute intervals. The cytoplasmic GFP signal was acquired every 5 minutes, using wide-field fluorescence. Data were exported as TIFF images, and both channels were merged while creating AVI files. FA dynamics was analyzed by production of time color overlays, using a custom-made plug-in for ImagePro Plus (MediaCybernetics).
Gene expression profiling of human BC patients and cell lines.
A total of 344 lymph node–negative BC patients who had not received any adjuvant systemic treatment (chemotherapy and/or endocrine therapy) were included. The data set is available in the National Center for Biotechnology Information/Gene Expression Omnibus (GEO), entries GSE2034 and GSE5327. Clinical characteristics and treatment details were previously described (43–45). The cohort consists of 221 estrogen receptor–positive (ER-positive) patients and 123 ER-negative patients. ER status was determined according to ER expression measured on microarrays (43). In total, 118 patients developed a distant metastasis, which was counted as an event in the survival analysis. Patients who died without evidence of metastatic disease were censored at the time of last follow-up. Gene expression data were obtained using Affymetrix HG-U133A chips. Preprocessing was performed as previously described (44). Stata (release 11; StataCorp) was used to calculate the Cox proportional hazard ratio. Log2-transformed gene expression data were evaluated as continuous variables, with MFS times as end point. For the Kaplan-Meier survival curves, log-rank tests were used to assess the equality of survivor functions across the 2 groups. For each gene, either a median split was used in the expression data or an optimal cut point was established (low log-rank P value) to visualize survival differences in the groups. The optimal cut point was determined using the srd algorithm in Stata. A 2-sided P value of 0.05 was considered statistically significant.
For the BC cell lines, microarray data from Affymetrix HG-U133A chips were used. The expression data of 39 BC cell lines are available in GEO (series GSE16795). Additional details are described by Hollestelle et al. (22).
Associations between SRPK1 and KI67, as well as these 2 genes and luminal subtypes, were analyzed using Pearson’s χ2 test. Luminal subtypes were assigned using a subtype clustering model present in the genefu R package, as described by Desmedt et al. (46).
Tissue microarray analysis of SRPK1.
In this study, a tissue microarray (TMA) was used that included formalin-fixed paraffin-embedded primary tumors of 1,350 consecutive BC patients who entered the Erasmus University Medical Center Rotterdam (Netherlands) during the years 1985–2000 for local treatment of their primary disease, and from whom complete clinical follow-up information was available. To enable analysis of pure prognosis without the confounding effects of adjuvant therapy, only the 562 lymph node–negative patients who did not receive adjuvant systemic therapy were included in our statistical analyses. From all specimens, tumor grading according to Scarff, Bloom, and Richardson and histology were available. The Medical Ethical Committee of the Erasmus Medical Center Rotterdam (Netherlands) approved our study design (MEC 02.953). This retrospective study was conducted in accordance with the Code of Conduct of the Federation of Medical Scientific Societies in the Netherlands (http:/www.federa.org/). The median age of patients included in the analysis at the time of surgery (258 pre- and 304 postmenopausal patients) was 54 years (range, 26–92 years). Two hundred ninety-one patients (52%) underwent mastectomy, while the remaining were treated with breast-conserving therapy. MFS was defined as the time from diagnosis to the first distant metastasis or, for patients still surviving, as the time from diagnosis to the last contact. Of the 562 patients included in the analysis of MFS, 143 were counted as failures for the entire cohort, 115 of the 460 ER-positive and 28 of the 102 ER-negative subgroups analyzed. Patients diagnosed with secondary contralateral BC during follow-up were censored for MFS, at the date of diagnosis of the secondary BC. Patients who died without evidence of disease were censored at last follow-up in the analysis of MFS. Slides from the TMA were cut (4 μm) and stained for SRPK1 (BD Biosciences) using the EnVision method (Dako). After staining, slides were converted to digital images with a Virtual Slide Scanner (NanoZoomer 2.0-HT) and uploaded into our TMA database (Distiller; SlidePath). All images were scored manually for staining intensity (0 = negative; 1 = weak; 2 = moderate; 3 = strong). Only staining on invasive breast tumor cells was recorded. The mean of the positive stained breast tumor cells was calculated from the total cell count yielded by the 3 separate images of the cores.
Western blotting.
Cells were scraped in ice-cold TSE (10 nM Tris-HCl, 250 mM sucrose, 1 mM EGTA, pH 7.4) supplemented with inhibitors. Tumor samples were lysed in 20 mM Tris-HCl, 137 mM NaCl, 2 mM EDTA, 1% Triton, and 10% glycerol, pH 7.4, supplemented with inhibitors. After sonication of cell or tumor lysates, protein concentration was determined by a Bio-Rad protein assay, using IgG as the internal standard. Total cellular protein (30 μg) was separated on 7.5% or 10% SDS-PAGE and transferred to PVDF membranes (Millipore). Blots were blocked in 5% wt/vol BSA in TBST (0.5 M NaCl, 20 mM Tris-HCl, 0.05% vol/vol Tween-20, pH 7.4) and probed with primary antibody (overnight, 4°C), followed by incubation with either Dylight-649 or HRP-coupled secondary antibody (Jackson ImmunoResearch Laboratories Inc.). Dylight-649 was directly visualized by scanning on a Typhoon imager 9400 (Amersham Biosciences). HRP probes were first activated by incubation with Enhanced Chemiluminescence Plus reagent (Amersham Biosciences). Full uncut gels are shown in the Supplemental Material.
Immunofluorescence staining.
Cells were fixed in 4% PFA in PBS for 10 minutes at room temperature. Coverslips were blocked in TBP (0.1% vol/vol Triton-X and 0.5% wt/vol BSA in PBS) for 1 hour at room temperature and incubated overnight with primary antibody, followed by 1 hour of incubation with fluorescently labeled secondary antibody. Next, cells were incubated with 2 μg/ml Hoechst 33258 (Sigma-Aldrich) to visualize nuclei. Cells were imaged using a Nikon Eclipse TE2000-E confocal microscope fitted with a ×20 objective (0.75 NA, 1.00 WD) and 4× zoom.
Orthotopic mouse BC model.
Female 7- to 9-week-old Rag2–/– Il2rg–/– mice were obtained from in-house breeding. Animals were housed in individually ventilated cages under sterile conditions. Housing and experiments were performed according to the Dutch guidelines for the care and use of laboratory animals (UL-DEC-11244). Sterilized food and water were provided ad libitum. In total, 2 × 106 MDA-MB-417.5 cells in 0.1 ml PBS were injected into the fourth mammary fat pad (n = 10). Size of the primary tumors was measured with a caliper. Animals were anesthetized when tumor volume reached 7 × 7 mm; the primary tumor was then removed. Lung metastasis formation was followed over time using bioluminescence imaging. At 7–35 days after primary tumor removal, mice were sacrificed, and lung metastasis was determined using bioluminescence imaging (total lung burden), and imaging of GFP (individual lung metastases) using confocal laser scanning microscopy. Tumors and lungs were embedded in paraffin and sectioned (4 μm thickness) onto 3-aminopropyltriethoxysilane–coated slides. After deparaffinization, the tumor and lung sections were stained for H&E, and lungs were examined for metastasis formation. The number of lung metastases per paraffin section was counted.
Generation of RNA samples.
To perform a deep sequencing experiment, we cultured the cells following strict planning to generate 4 biological replicates of each cell line (WT, shCtrl, shSRPK1-A, shSRPK1-B). First, we made several vials containing each exactly the same number of cells of the 4 lines with the same passage. Once a vial was taken in culture, cells were allowed to grow for 3 days in a flask, and then 2 million cells were plated in P150 dishes containing 30 ml of complete medium. After 3 days of culture, cells were lysed and RNA was isolated with the RNeasy Kit (QIAGEN). RNA samples were checked for their purity and further processed for the sequencing.
Next-generation sequencing of RNA samples.
RNA was sequenced on Illumina HiSeq2000 using the protocol for strand-specific RNA sequencing (47). Paired-end reads (100 nt each end) with the average insert size of 220 nt were generated.
Bioinformatic analysis of RNA sequencing data.
Sequencing data obtained from 4 biological replicates of 4 sample groups (WT, control, shRNA-A, and shRNA-B; 16 samples in total) underwent quality check with FastQC toolkit (Babraham Bioinformatics, http://www.bioinformatics.babraham.ac.uk/projects/fastqc) and were clipped (48) and trimmed to remove adapter sequences and low-quality bases (Phred score below 20). The remaining reads (between 20 × 106 and 50 × 106) were aligned to the human reference genome hg19 (http://hgdownload.cse.ucsc.edu/goldenpath/hg19/chromosomes) with Tophat version 2.0.9 (49) with default options.
Gene expression values were calculated with HTSeq (50) following the protocol recommended by developers using the Ensembl (http://www.ensembl.org/index.html) gene annotation GRCh37.p12. Differential gene expression analysis was performed with edgeR version 2.13 (51). A generalized linear model glmLRT with default function parameters was used as described in the developer’s manual. Differential expression analysis was performed twice. In the first analysis, the group of WT and control samples was compared with the group of all knockdown samples (shSRPK1-A and shSRPK1-B). The list of differentially expressed genes in pooled comparison resulted in 1,809 genes (data not shown). In the second analysis (shown in Figure 8B) we made all pairwise comparisons between the 4 experimental groups. The network as shown in Figure 9A was generated through the use of MetaCore (Thomson Reuters).
Differential splicing analysis was performed at the exon level using DEXSeq version 1.8.0 (52) and at the transcript level using Cuffdiff version 2.1.1 (53). Cuffdiff was used with the default parameters in the annotation-restricted mode (using the Ensembl transcript annotation). The genes tagged as low-quality/low-coverage ones (tag “LOWDATA” and/or “NOTEST” in the “*.splicing.diff”) were excluded from further analysis. Genes with differences in transcript levels were retrieved from the splicing.diff output file. DEXSeq was used following the protocol recommended by the developers using the default settings. All exons with Benjamini-Hochberg adjusted P value below 0.05 were considered as the indicators of genes with potential differential splicing.
Statistics.
A 2-tailed Student’s t test was used to determine significant differences between 2 means. A P value less than 0.05 was considered significant. Values are represented as mean ± SEM. Significant differences are marked in the graphs.
Study approval.
The study involving human BC patients was approved by the Medical Ethical Committee of the Erasmus Medical Center Rotterdam (Netherlands) (MEC 02.953). This retrospective study was conducted in accordance with the Code of Conduct of the Federation of Medical Scientific Societies in the Netherlands (http:/www.federa.org/). Mouse experiments and housing were performed according to the Dutch guidelines for the care and use of laboratory animals (UL-DEC-11244).
Supplemental Methods.
For all methods not listed here, please refer to the online Supplemental Methods.
Supplementary Material
Acknowledgments
We thank Zvi Kam for help and advice during image acquisition and analysis. We thank Vasiliki-Maria Rogkoti for help with the Western blot samples of the human BC cell lines; Bart Jacobse for help with acquisition of the TIRF movies; Reshma Lalai and Lizette Haazen for technical assistance during in vivo experiments; and Michiel Fokkelman and Lynn Wester for technical support. The authors are grateful to Barbara Morgenstern for her expert help in the style editing of the manuscript. This work was financially supported by grants from the Dutch Cancer Society (UL2007-3860) and the EU FP7 Health Programs MetaFight project (grant agreement 201862) to B. van de Water; Systems Microscopy NoE project (grant agreement 258068) to B. van de Water and B. Geiger; and an EMBO Short-Term Fellowship (ASTF 109-2009) to W. van Roosmalen. The authors are grateful for financial support from the European Research Council (ERC): Syn-Ad Advanced Grant, ERC Grant Agreement 294852 to B. Geiger, and ERC Grant Agreement 322737 to J.A. Foekens, J.W.M. Martens, and B. van de Water. B. Geiger holds the Erwin Neter Professorial Chair in Cell and Tumor Biology.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2015;125(4):1648–1664. doi:10.1172/JCI74440.
References
- 1.Zaidel-Bar R, Geiger B. The switchable integrin adhesome. J Cell Sci. 2010;123(pt 9):1385–1388. doi: 10.1242/jcs.066183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zaidel-Bar R, Itzkovitz S, Ma’ayan A, Iyengar R, Geiger B. Functional atlas of the integrin adhesome. Nat Cell Biol. 2007;9(8):858–867. doi: 10.1038/ncb0807-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geiger B, Yamada KM. Molecular architecture and function of matrix adhesions. Cold Spring Harb Perspect Biol. 2011;3(5):201–221. doi: 10.1101/cshperspect.a005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5(5):341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 5.Rijken PJ, Hage WJ, van Bergen en Henegouwen PM, Verkleij AJ, Boonstra J. Epidermal growth factor induces rapid reorganization of the actin microfilament system in human A431 cells. J Cell Sci. 1991;100(pt 3):491–499. doi: 10.1242/jcs.100.3.491. [DOI] [PubMed] [Google Scholar]
- 6.Xue C, et al. Epidermal growth factor receptor overexpression results in increased tumor cell motility in vivo coordinately with enhanced intravasation and metastasis. Cancer Res. 2006;66(1):192–197. doi: 10.1158/0008-5472.CAN-05-1242. [DOI] [PubMed] [Google Scholar]
- 7.Zhang HJ, et al. Transforming growth factor-beta1 promotes lung adenocarcinoma invasion and metastasis by epithelial-to-mesenchymal transition. Mol Cell Biochem. 2011;355(1):309–314. doi: 10.1007/s11010-011-0869-3. [DOI] [PubMed] [Google Scholar]
- 8.Rosenthal DT, et al. p38gamma promotes breast cancer cell motility and metastasis through regulation of RhoC GTPase, cytoskeletal architecture, and a novel leading edge behavior. Cancer Res. 2011;71(20):6338–6349. doi: 10.1158/0008-5472.CAN-11-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danen EH. Integrins: regulators of tissue function and cancer progression. Curr Pharm Des. 2005;11(7):881–891. doi: 10.2174/1381612053381756. [DOI] [PubMed] [Google Scholar]
- 10.Di Stefano P, et al. The adaptor proteins p140CAP and p130CAS as molecular hubs in cell migration and invasion of cancer cells. Am J Cancer Res. 2011;1(5):663–673. [PMC free article] [PubMed] [Google Scholar]
- 11.van Nimwegen MJ, Verkoeijen S, van Buren L, Burg D, van de Water B. Requirement for focal adhesion kinase in the early phase of mammary adenocarcinoma lung metastasis formation. Cancer Res. 2005;65(11):4698–4706. doi: 10.1158/0008-5472.CAN-04-4126. [DOI] [PubMed] [Google Scholar]
- 12.Zhang XH, et al. Latent bone metastasis in breast cancer tied to Src-dependent survival signals. Cancer Cell. 2009;16(1):67–78. doi: 10.1016/j.ccr.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Flier S, et al. Bcar1/p130Cas protein and primary breast cancer: prognosis and response to tamoxifen treatment. J Natl Cancer Inst. 2000;92(2):120–127. doi: 10.1093/jnci/92.2.120. [DOI] [PubMed] [Google Scholar]
- 14.Winograd-Katz SE, Fassler R, Geiger B, Legate KR. The integrin adhesome: from genes and proteins to human disease. Nat Rev Mol Cell Biol. 2014;15(4):273–288. doi: 10.1038/nrm3769. [DOI] [PubMed] [Google Scholar]
- 15.Naffar-Abu-Amara S, et al. Identification of novel pro-migratory, cancer-associated genes using quantitative, microscopy-based screening. PLoS One. 2008;3(1):e1457. doi: 10.1371/journal.pone.0001457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simpson KJ, et al. Identification of genes that regulate epithelial cell migration using an siRNA screening approach. Nat Cell Biol. 2008;10(9):1027–1038. doi: 10.1038/ncb1762. [DOI] [PubMed] [Google Scholar]
- 17.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 18.Rajendran M, Thomes P, Zhang L, Veeramani S, Lin MF. p66Shc — a longevity redox protein in human prostate cancer progression and metastasis: p66Shc in cancer progression and metastasis. Cancer Metastasis Rev. 2010;29(1):207–222. doi: 10.1007/s10555-010-9213-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu HS, et al. An unusual function of RON receptor tyrosine kinase as a transcriptional regulator in cooperation with EGFR in human cancer cells. Carcinogenesis. 2010;31(8):1456–1464. doi: 10.1093/carcin/bgq100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wagh PK, Zinser GM, Gray JK, Shrestha A, Waltz SE. Conditional deletion of β-catenin in mammary epithelial cells of Ron receptor, Mst1r, overexpressing mice alters mammary tumorigenesis. Endocrinology. 2012;153(6):2735–2746. doi: 10.1210/en.2011-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rebagay G, Yan S, Liu C, Cheung NK. ROR1 and ROR2 in human malignancies: potentials for targeted therapy. Front Oncol. 2012;2:34. doi: 10.3389/fonc.2012.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hollestelle A, et al. Distinct gene mutation profiles among luminal-type and basal-type breast cancer cell lines. Breast Cancer Res Treat. 2010;121(1):53–64. doi: 10.1007/s10549-009-0460-8. [DOI] [PubMed] [Google Scholar]
- 23.Padgett RA. New connections between splicing and human disease. Trends Genet. 2012;28(4):147–154. doi: 10.1016/j.tig.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minn AJ, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436(7050):518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Graauw M, et al. Annexin A2 depletion delays EGFR endocytic trafficking via cofilin activation and enhances EGFR signaling and metastasis formation. Oncogene. 2013;33(20):2610–2619. doi: 10.1038/onc.2013.219. [DOI] [PubMed] [Google Scholar]
- 26.Ghosh G, Adams JA. Phosphorylation mechanism and structure of serine-arginine protein kinases. FEBS J. 2011;278(4):587–597. doi: 10.1111/j.1742-4658.2010.07992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayes GM, Carrigan PE, Miller LJ. Serine-arginine protein kinase 1 overexpression is associated with tumorigenic imbalance in mitogen-activated protein kinase pathways in breast, colonic, and pancreatic carcinomas. Cancer Res. 2007;67(5):2072–2080. doi: 10.1158/0008-5472.CAN-06-2969. [DOI] [PubMed] [Google Scholar]
- 28.Amin EM, et al. WT1 mutants reveal SRPK1 to be a downstream angiogenesis target by altering VEGF splicing. Cancer Cell. 2011;20(6):768–780. doi: 10.1016/j.ccr.2011.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalwa H, Sartoretto JL, Sartoretto SM, Michel T. Angiotensin-II and MARCKS: a hydrogen peroxide- and Rac1-dependent signaling pathway in vascular endothelium. J Biol Chem. 2012;287(34):29147–29158. doi: 10.1074/jbc.M112.381517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Appledorn DM, Dao KH, O’Reilly S, Maher VM, McCormick JJ. Rac1 and Cdc42 are regulators of HRasV12-transformation and angiogenic factors in human fibroblasts. BMC Cancer. 2010;10:13. doi: 10.1186/1471-2407-10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim IK, Hong KW, Kwak IH, Yoon G, Park SC. Translocational inefficiency of intracellular proteins in senescence of human diploid fibroblasts. Ann N Y Acad Sci. 2001;928:176–181. doi: 10.1111/j.1749-6632.2001.tb05647.x. [DOI] [PubMed] [Google Scholar]
- 32.Jaiswal M, et al. Mechanistic insights into specificity, activity, and regulatory elements of the regulator of G-protein signaling (RGS)-containing Rho-specific guanine nucleotide exchange factors (GEFs) p115, PDZ-RhoGEF (PRG), and leukemia-associated RhoGEF (LARG). J Biol Chem. 2011;286(20):18202–18212. doi: 10.1074/jbc.M111.226431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shamah SM, et al. EphA receptors regulate growth cone dynamics through the novel guanine nucleotide exchange factor ephexin. Cell. 2001;105(2):233–244. doi: 10.1016/S0092-8674(01)00314-2. [DOI] [PubMed] [Google Scholar]
- 34.Huttenlocher A, Horwitz AR. Integrins in cell migration. Cold Spring Harb Perspect Biol. 2011;3(9):a005074. doi: 10.1101/cshperspect.a005074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winograd-Katz SE, Itzkovitz S, Kam Z, Geiger B. Multiparametric analysis of focal adhesion formation by RNAi-mediated gene knockdown. J Cell Biol. 2009;186(3):423–436. doi: 10.1083/jcb.200901105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hutti JE, et al. Phosphorylation of the tumor suppressor CYLD by the breast cancer oncogene IKKepsilon promotes cell transformation. Mol Cell. 2009;34(4):461–472. doi: 10.1016/j.molcel.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berrier AL, Yamada KM. Cell-matrix adhesion. J Cell Physiol. 2007;213(3):565–573. doi: 10.1002/jcp.21237. [DOI] [PubMed] [Google Scholar]
- 38.Schlaepfer DD, Hauck CR, Sieg DJ. Signaling through focal adhesion kinase. Prog Biophys Mol Biol. 1999;71(3):435–478. doi: 10.1016/s0079-6107(98)00052-2. [DOI] [PubMed] [Google Scholar]
- 39.Ursini-Siegel J, Muller WJ. The ShcA adaptor protein is a critical regulator of breast cancer progression. Cell Cycle. 2008;7(13):1936–1943. doi: 10.4161/cc.7.13.6205. [DOI] [PubMed] [Google Scholar]
- 40.Elsberger B, et al. Breast cancer patients’ clinical outcome measures are associated with Src kinase family member expression. Br J Cancer. 2010;103(6):899–909. doi: 10.1038/sj.bjc.6605829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karin M, Greten FR. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5(10):749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 42.van Roosmalen W, Le Devedec SE, Zovko S, de Bont H, van de Water B. Functional screening with a live cell imaging-based random cell migration assay. Methods Mol Biol. 2011;769:435–448. doi: 10.1007/978-1-61779-207-6_29. [DOI] [PubMed] [Google Scholar]
- 43.Foekens JA, et al. Multicenter validation of a gene expression-based prognostic signature in lymph node-negative primary breast cancer. J Clin Oncol. 2006;24(11):1665–1671. doi: 10.1200/JCO.2005.03.9115. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 45.Yu JX, et al. Pathway analysis of gene signatures predicting metastasis of node-negative primary breast cancer. BMC Cancer. 2007;7:182. doi: 10.1186/1471-2407-7-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Desmedt C, et al. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin Cancer Res. 2008;14(16):5158–5165. doi: 10.1158/1078-0432.CCR-07-4756. [DOI] [PubMed] [Google Scholar]
- 47.Parkhomchuk D, et al. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009;37(18):e123. doi: 10.1093/nar/gkp596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. 2011;17(1):10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 49.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012;22(10):2008–2017. doi: 10.1101/gr.133744.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.