Abstract
The mechanistic target of rapamycin (mTOR) is hyperactivated in many types of cancer, rendering it a compelling drug target; however, the impact of mTOR inhibition on metabolic reprogramming in cancer is incompletely understood. Here, by integrating metabolic and functional studies in glioblastoma multiforme (GBM) cell lines, preclinical models, and clinical samples, we demonstrate that the compensatory upregulation of glutamine metabolism promotes resistance to mTOR kinase inhibitors. Metabolomic studies in GBM cells revealed that glutaminase (GLS) and glutamate levels are elevated following mTOR kinase inhibitor treatment. Moreover, these mTOR inhibitor–dependent metabolic alterations were confirmed in a GBM xenograft model. Expression of GLS following mTOR inhibitor treatment promoted GBM survival in an α-ketoglutarate–dependent (αKG-dependent) manner. Combined genetic and/or pharmacological inhibition of mTOR kinase and GLS resulted in massive synergistic tumor cell death and growth inhibition in tumor-bearing mice. These results highlight a critical role for compensatory glutamine metabolism in promoting mTOR inhibitor resistance and suggest that rational combination therapy has the potential to suppress resistance.
Keywords: Metabolism, Oncology
Introduction
Most cancer cells consume glucose at a surprisingly high rate compared with normal cells and secrete most of the glucose-derived carbon as lactate rather than oxidizing it completely, a phenomenon known as the Warburg effect (1). In addition to glucose, amino acids can also funnel into the tricarboxylic acid (TCA) cycle, as their catabolism results in the production of TCA cycle intermediates. Glutamine has an important role in cell growth and energy metabolism, with the TCA cycle as well as glucose as the carbon source (2, 3). The first 2 reactions of glutamine catabolism are the conversion of glutamine into glutamate regulated by glutaminase (GLS) and the conversion of glutamate into α-ketoglutarate (αKG), catalyzed by glutamate dehydrogenase (GDH). These are followed by subsequent enzymatic reactions generating aspartate, malate, pyruvate, citrate, alanine, and lactate to support mitochondrial respiration and adenosine triphosphate (ATP) production (4). Glutamate can also participate in glutathione synthesis to help regulate the antioxidant status of the cell. It has been widely accepted that cancer cells favor glutamine as a source of energy, and this phenomenon has been observed in many cancers. Many studies have shown that inhibiting glutaminolysis by preventing the activity of these key enzymes significantly suppresses cancer cell growth and proliferation (5–7). Consequently, GLS has become an attractive target for the therapeutic intervention of malignant gliomas.
The mechanistic target of rapamycin (mTOR) is a protein kinase that integrates oncogenic signaling from growth factor receptors through the phosphoinositide 3-kinase (PI3K) pathway with cellular energy and nutrient status to activate downstream signaling pathways that promote tumor growth and survival (8). Thus, mTOR has emerged as an important molecular target in PI3K-driven cancers. mTOR acts through the canonical PI3K pathway via 2 distinct complexes, mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2), to mediate cell growth and proliferation and potentially tumor cell survival (9). In particular, mTORC1 uniquely integrates growth factor and metabolic signaling through PI3K with downstream signaling through S6K1, underscoring its value as a cancer target (10). We have shown previously that mTOR is a critical effector of downstream signaling in EGFR-mutated, PTEN-deficient glioblastoma multiforme (GBM), mediating resistance to EGFR tyrosine kinase inhibitors (11). However, the clinical failure of the allosteric mTOR inhibitor rapamycin has been caused by AKT activation, resulting in the loss of negative feedback, consistent with the homeostatic regulatory role of mTORC1 as a negative regulator of PI3K/AKT signaling (12). Furthermore, we demonstrated that mTORC2 mediated rapamycin resistance through AKT- and mTORC1-independent signaling pathways (13). These results have highlighted the role for mTOR kinase inhibitors, which block mTORC1 and mTORC2, in the treatment of GBM (14). Moreover, uncovering molecular and metabolic responses to mTOR kinase inhibition provides precious insight into identifying new drug targets and the resistance mechanism for mTOR-targeted therapies.
Here, we performed integrated analyses of GBM cell lines, in vivo xenograft models, and clinical samples to examine the importance of glutamine metabolism in response to mTOR-targeted treatments. We demonstrated that GLS is highly expressed in a large number of clinical GBM samples compared with normal brain tissues, suggesting an attractive target for therapeutic intervention. Interestingly, mTOR-targeted treatments affected glutamine utilization and elicited a switch in the pathways used to deliver glutamine carbon to the TCA cycle, with increasing expression of GLS. Suppression of GLS expression with RNA interference or an inhibitor with compound 968, which has been shown to block GLS activity in cancer cells (7, 15–17), sensitized EGFRvIII-expressing GBM cells to mTOR-targeted therapies. Combined mTOR and GLS inhibition showed a synergic suppression of tumor growth in vivo. GBM cells required GLS to survive mTOR inhibition in an αKG-dependent manner. These results demonstrate that the inhibition of mTOR signaling is sufficient to change the metabolic characteristics of GBM cells and point to what we believe is a previously unrecognized function of GLS in promoting acquired resistance to mTOR-targeted therapy, indicating the need for glutaminolysis inhibition in combination with mTOR-targeted therapies.
Results
Glutamine metabolism and GLS expression in GBM patients in situ.
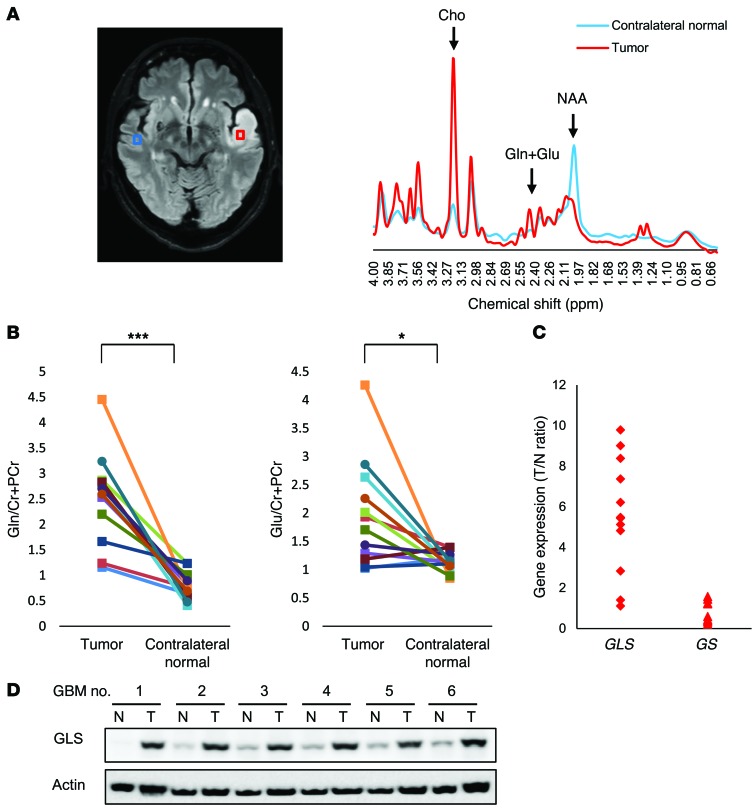
For most mammalian cells in culture, glucose and glutamine are catabolized in appreciable quantities, supplying carbon, nitrogen, and free energy and reducing equivalents necessary to support cell growth and division (1, 18). Aerobic glycolysis, also known as the Warburg effect, and glutaminolysis are core hallmarks of cancer and are required in cancer cells. To explore the clinical implications of intracellular metabolism, we examined the glucose and glutamine metabolism in tumor tissues and adjacent normal brain tissue from several GBM patients. Magnetic resonance spectroscopy (MRS) of a 68-year-old man presenting with GBM in the left temporal lobe showed significantly higher choline and lower N-acetyl-l-aspartate (NAA) peaks in tumors than those in the contralateral normal brain (Figure 1A). A decrease in the NAA/choline ratio is a common marker in predicting increased malignancy in gliomas (19). However, what is most important and is an essential change in this study is the discovery of increasing glutamine and glutamate levels in tumors compared with those in the contralateral normal brain shown in the MRS (Figure 1A). As detected by subsequent pairwise comparisons in 12 GBM patients, glutamine and glutamate levels significantly increased in tumors relative to contralateral normal brain tissue, revealing that glutamine metabolism is strongly involved in metabolic reprogramming in GBM cells (Figure 1B). Glucose and lactate levels were also statistically higher in tumor than in contralateral normal brain tissue, showing the well-known phenomenon of aerobic glycolysis in GBM cells (Supplemental Figure 1, A and B; supplemental material available online with this article; doi:10.1172/JCI78239DS1). Next, to determine how glutamine metabolism was influenced in GBM cells, we examined the expression of GLS, a glutamine-to-glutamate converting enzyme, and the level of glutamine synthetase (GS), a glutamate-to-glutamine converting enzyme, in tumor samples and normal brain tissues in 12 GBM patients. Gene-expression analysis demonstrated that the level of GLS was frequently elevated in tumor samples compared with the GS level (Figure 1C), suggesting a potential metabolic flux from glutamine to glutamate for the high rates of glutamine catabolism. Immunoblot analysis of lysates obtained from surgical samples of 6 GBM patients confirmed increases of GLS expression in tumor tissue relative to normal brain tissue (Figure 1D). Taken together, these findings suggest that glutamine is also the major nutrient for GBM cells and that GLS could be a good target of metabolic genes for GBM treatments.
Figure 1. Glutamine and glutamate levels and GLS expression are elevated in the tumors of GBM patients.
(A) MRS studies targeting glutamine and glutamate for tumors (red) and contralateral normal brain (blue) regions in a 68-year-old patient with GBM. The peaks of choline, glutamine and glutamate complex, and NAA are around 3.22, 2.4, and 2.0 ppm of chemical shift, respectively. Cho, choline; Cr, Creatine; Gln, glutamine; Glu, glutamate. (B) Glutamine and glutamate levels in MRS studies for tumors and contralateral normal brain regions in 12 GBM patients. The relative level of glutamine and glutamate was calculated with respect to creatine and phosphocreatine for VOIs of tumor and contralateral normal brain. *P < 0.05; ***P < 0.001, according to 2-tailed Student’s t test. (C) mRNA levels of key enzymes including GLS and GS between glutamine and glutamate in 12 GBM patients. The relative levels of GLS and GS are presented as the tumor/normal brain (T/N) expression ratio. (D) Immunoblot analysis of GLS staining in tumor and normal brain tissue obtained at tumor resection from 6 patients with GBM. See also Supplemental Figure 1.
GLS and intracellular glutamate levels rise in GBMs in vitro and in vivo in response to mTOR inhibitors.
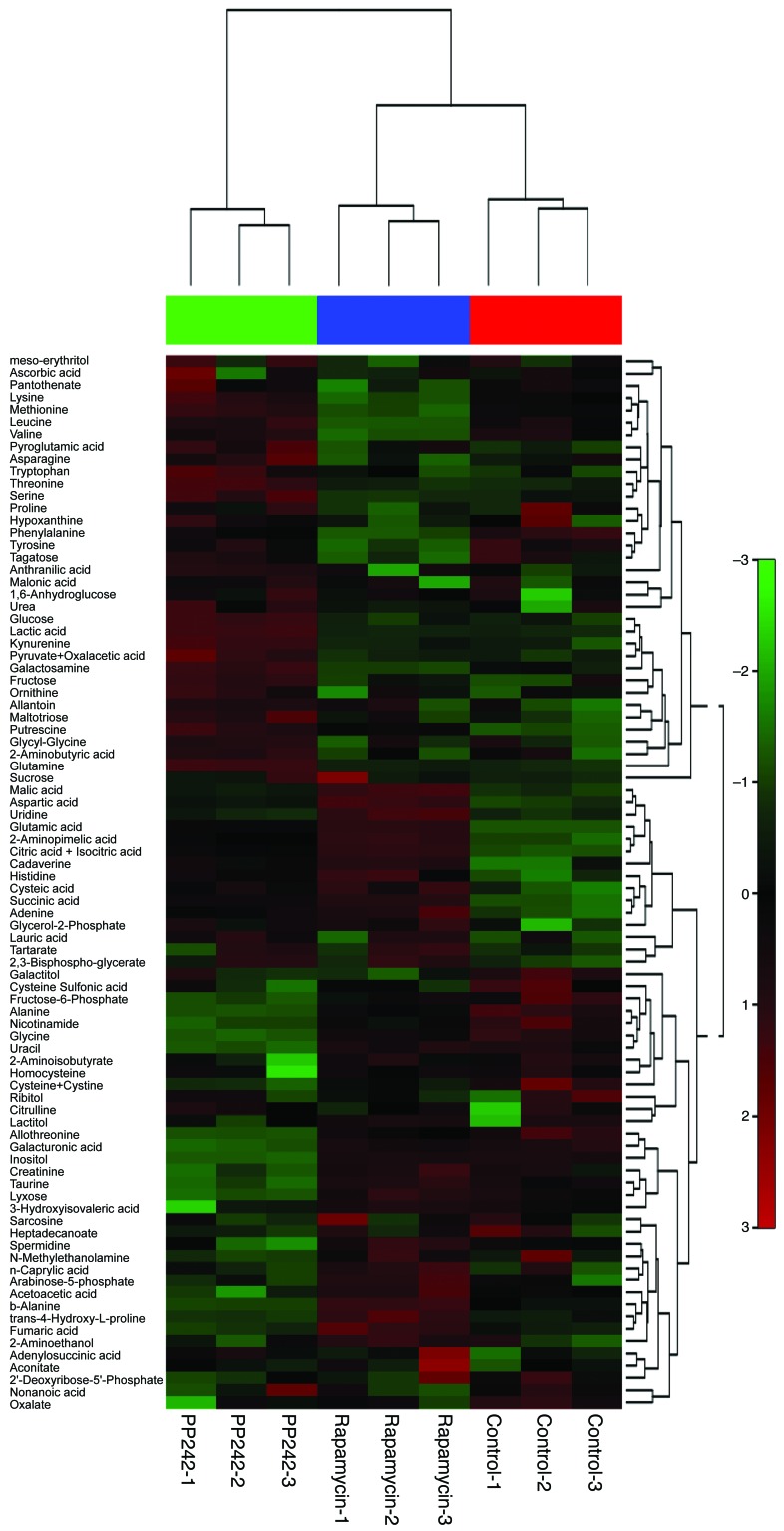
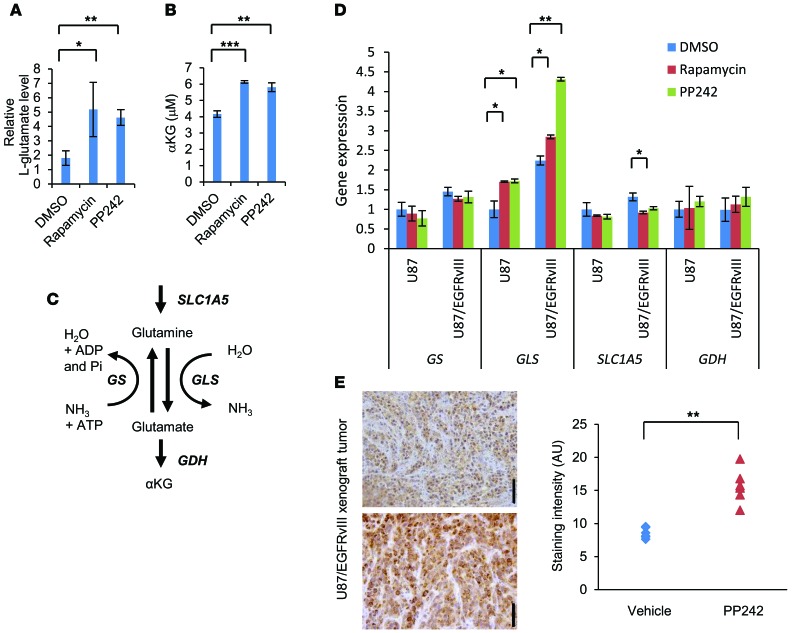
In the EGFR/PI3K pathway stimulating glucose uptake and utilization, mTOR has a well-described role in directing available amino acids into protein synthesis. Glutamine uptake also appears to be critical for lipid synthesis and carbon supply to operate the TCA cycle. We overexpressed the EGFR-activating mutation (EGFRvIII) in the U87 glioma cell line, which has been demonstrated to increase both mTORC1 and mTORC2 signaling (20, 21). Using gas chromatography–mass spectroscopy (GC/MS) of U87 and U87/EGFRvIII cells treated with mTOR inhibitors (rapamycin or PP242) for 48 hours, we identified 91 metabolites whose levels significantly changed in response to the allosteric mTOR inhibitor rapamycin or the ATP-competitive mTOR inhibitor PP242 (Figure 2 and Supplemental Table 1). We have previously shown that rapamycin has minimal activity against mTORC2 signaling in GBM cell lines in in vivo models and patients treated with the drug, whereas PP242 blocks both mTORC1 and mTORC2 signaling in GBM cells (12, 13, 22). The principal component analysis (PCA) of variation in the metabolites for each treatment group demonstrated distinct clustering or a clear separation of the 3 groups (Supplemental Figure 2, A and B). The key differentiating metabolites were glutamic acid (glutamate), aspartic acid, citric (or isocitric) acid, and succinic acid (Supplemental Figure 2B). Particularly, some intermediates of glutaminolysis and the TCA cycle showed an increase with mTOR inhibitor treatment, raising the possibility of efficient metabolism of glutamine (Supplemental Figure 2C). In fact, both mTOR inhibitors significantly suppressed glucose consumption, lactate production, and cell proliferation, but did not increase cell death in U87/EGFRvIII models (Supplemental Figure 3, A and B). Consistent with the findings of Csibi et al. (23), intracellular l-glutamate was elevated in U87 and U87/EGFRvIII cells, allowing them to survive mTOR-targeted treatments (Figure 3A). Also, we found that intracellular αKG, ATP, and ammonia levels were elevated or at least preserved after mTOR inhibition treatments, demonstrating compensatory increase of glutamine metabolism (Figure 3B and Supplemental Figure 4, A and B). These results suggested the potential aspects of GBM cells that were resistant to mTOR inhibitors. Next, to identify how inhibition of mTOR signaling affected the metabolic pathway, we treated U87 and U87/EGFRvIII cells with mTOR inhibitors to test the gene expression of key enzymes in the glycolysis and glutaminolysis pathways (Figure 3C). Notably, mTOR inhibitor treatments of U87/EGFRvIII and, to a lesser extent, U87 cells resulted in the upregulation of GLS (Figure 3D). In particular, of the 2 GLS variants (7, 15), expression of the longer kidney type glutaminase (KGA) mRNA increased markedly, while expression of the shorter glutaminase C (GAC) mRNA decreased after mTOR inhibitor treatments (Supplemental Figure 4C). Glucose transporter 1 (Glut1), pyruvate dehydrogenase kinase 1 (PDK1), and lactate dehydrogenase A (LDHA) levels were decreased upon rapamycin or PP242 treatment, consistent with the effect of mTOR inhibition on glycolysis (Supplemental Figure 4D). Finally, to determine whether GLS expression could be detected in vivo in response to mTOR-targeted treatment, we analyzed EGFRvIII-expressing tumor tissues from a xenograft model after 5 days of treatment. After PP242 treatment, GLS expression was significantly elevated relative to that of controls (Figure 3E). Similar results were found after treatment with CC214, which is the other mTOR kinase inhibitor (22) (Supplemental Figure 5).
Figure 2. Comparative metabolomics identifies glutamate as a potential metabolite that promotes resistance to mTOR inhibitor treatment.
Heat map representation of a 2D hierarchical clustering of metabolites identified as differentially expressed among U87/EGFRvIII GBM cells treated with 1 nM rapamycin, 1 μM PP242, and control DMSO for 48 hours. Each column represents a treatment group, and each row represents a metabolite.
Figure 3. Compensatory elevation of GLS protein and glutamate levels enables GBM cells to survive mTOR inhibitor treatment.
(A and B) Intracellular levels of l-glutamate (A) and αKG (B) in U87/EGFRvIII GBM cells treated with 1 nM rapamycin, 1 μM PP242, or control DMSO for 48 hours. Data represent the mean ± SEM of 3 independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001, according to 2-tailed Student’s t test. (C) Schematic showing the enzymes involved in glutaminolysis that were targeted in this study. ADP, adenosine diphosphate; Pi, phosphate. (D) mRNA levels of GS, GLS, solute-linked carrier family A1 (SLC1A5), and GDH in U87 and 87/EGFRvIII cells. Cells were treated with 1 nM rapamycin, 1 μM PP242, or control DMSO for 48 hours. Data represent the mean ± SEM of 3 independent experiments. *P < 0.05; **P < 0.01, according to 2-tailed Student’s t test. (E) Immunohistochemical images of GLS obtained from EGFRvIII-expressing U87 xenografts that received treatment with PP242 (n = 2) or control DMSO (n = 2) for 5 days. Tissue was counterstained with hematoxylin. Scale bars: 50 μm. Relative staining intensity was measured from 6 independent images for each group. **P < 0.01, according to 2-tailed Student’s t test. See also Supplemental Figures 2-5 and Supplemental Table 1.
GLS inhibition sensitizes GBM cells to mTOR-targeted treatments.
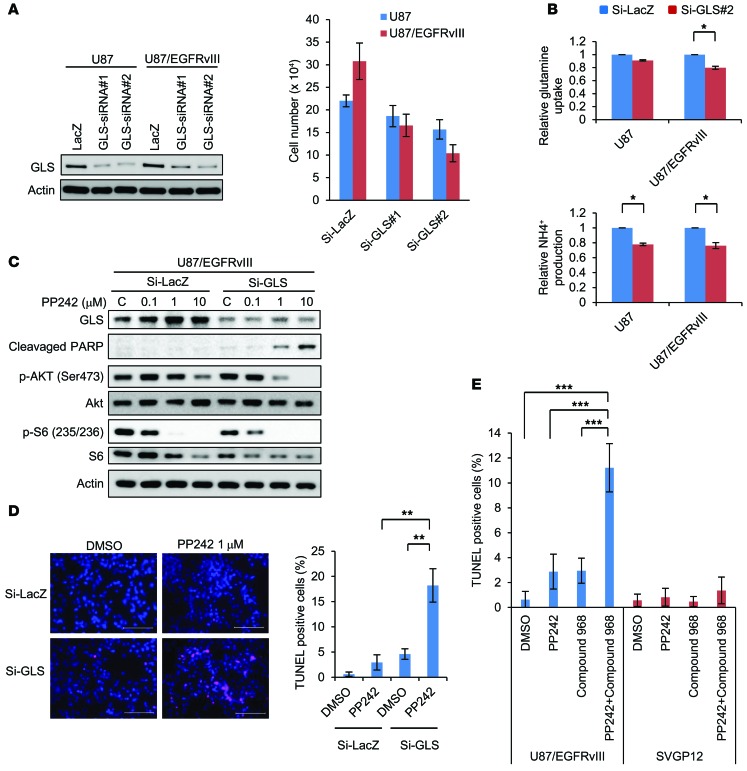
Because of the enhanced activity of PP242 relative to rapamycin on both mTOR complexes in GBM cells, we mainly used PP242 for further functional analysis (12, 13, 24, 25). Before confirming a specific role for GLS in mTOR-targeted therapies for GBMs, we tested to determine whether glutamine availability influenced PP242-mediated cell death. TUNEL analysis demonstrated that glutamine deprivation dramatically sensitized U87/EGFRvIII GBM cells to PP242-mediated cell death, which was confirmed by analysis of polyADP ribose polymerase (PARP) cleavage and TUNEL staining (Supplemental Figure 6, A and B). Next, we induced siRNA-mediated GLS knockdown in U87/EGFRvIII GBM cells and assessed its impact on the response to PP242. Genetic depletion of GLS by siRNA transfection inhibited the proliferation of all GBM cells tested (U87MG and U87/EGFRvIII), with enhanced antiproliferative effects in EGFRvIII-expressing tumor cells (Figure 4A). GLS knockdown was confirmed to inhibit glutamine uptake and NH4+ production from glutamine via the enzymatic activity of GLSs in U87 and U87/EGFRvIII GBM cells (Figure 3C and Figure 4B). Of note, knockdown of GLS significantly reversed mTOR-targeted treatment resistance, effectively sensitizing U87-EGFRvIII cells to PP242-mediated cell death, as indicated by cleaved PARP–positive and TUNEL-positive cells (Figure 4, C and D). Next, to assess the possibility that pharmacological inhibition of GLS could be used to sensitize GBMs to mTOR-targeted treatment, we tested the effect of the GLS inhibitor compound 968 on mediation of the cellular response to PP242. Compound 968 decreased the uptake of glutamine and secretion of by-products of glutamine catabolism such as ammonia in a dose-dependent manner, inhibiting GLS activity (Supplemental Figure 7A). Following this inhibition of GLS activity, compound 968 significantly suppressed GBM cell proliferation in a dose-dependent manner (Supplemental Figure 7B). In the TUNEL-staining assay, compound 968 significantly enhanced PP242-mediated cell death of EGFRvIII-expressing GBM cells, although neither PP242 nor compound 968 alone promoted extensive tumor cell death (Figure 4E and Supplemental Figure 8B). Compound 968 also increased PARP cleavage of EGFRvIII-expressing GBM cells treated with PP242 (Supplemental Figure 8A). Importantly, combined treatment with compound 968 and PP242 did not induce significant cell death of normal human astroglia cells (SVGP12) (Figure 4E and Supplemental Figure 8C), suggesting that normal astroglia cells have characteristics different from those of GBM cells and that there is a potential clinical use of GLS inhibition in selectively sensitizing GBM cells to mTOR-targeted therapies. To test the effect of compound 968 and PP242 on GBM patient–derived tumor cells, we established primary cultured cells (KMG02) from a GBM patient, showing positive staining for the EGFR/PI3K axis (p-EGFR [Y1068], p-AKT [S473], and p-S6 [S235/236]) as well as GLS (Supplemental Figure 9A). TUNEL assays showed that compound 968 significantly enhanced PP242-mediated cell death of KMG02 cells (Supplemental Figure 9B). Compound 968 also increased PARP cleavage of KMG02 cells treated with PP242 (Supplemental Figure 9C). Taken together, these results demonstrate a previously unknown role for GLS in mediating mTOR kinase inhibitor resistance through glutamine metabolism in GBM cells.
Figure 4. GLS inhibition sensitized GBM cells to mTOR-targeted treatment.
(A) U87 and U87/EGFRvIII cells were transfected with 2 types of GLS siRNA and scrambled control siRNA constructs for 24 hours and changed to 10% FBS medium for 2 days. Cell number represents the mean ± SEM of 3 independent experiments. (B) Relative glutamine uptake and NH4+ production in control versus GLS knockdown U87 and U87/EGFRvIII cells. Data represent the mean ± SEM of 3 independent experiments. *P < 0.05, according to 2-tailed Student’s t test. (C) Immunoblot analysis using indicated antibodies of U87/EGFRvIII cells with the siRNA constructs against GLS or control LacZ treated with PP242 or control DMSO for 2 days. Images of GLS, p-AKT (Ser473), and AKT were obtained from another gel using the same cell lysate. (D) Representative images of U87/EGFRvIII GBM cells with TUNEL staining. Cells were transfected with siRNA against GLS and control LacZ treated with 1 μM PP242 or DMSO for 2 days. Quantification of TUNEL-positive cells was performed with ImageJ software. Data represent the mean ± SEM of 3 independent images for each group. **P < 0.01, according to 2-tailed Student’s t test. Scale bars: 100 μm. (E) TUNEL staining in U87/EGFRvIII and SVGP12 cells treated with 1 μM PP242 and/or 1 μM compound 968 for 2 days. Quantification of TUNEL-positive cells was performed with ImageJ software. Data represent the mean ± SEM of 3 independent images for each group. Tukey-Kramer honest significance testing was performed for multiple comparisons testing. ***P < 0.001. See also Supplemental Figures 6–9.
GBM cells required GLS to survive mTOR inhibition in an αKG-dependent manner.
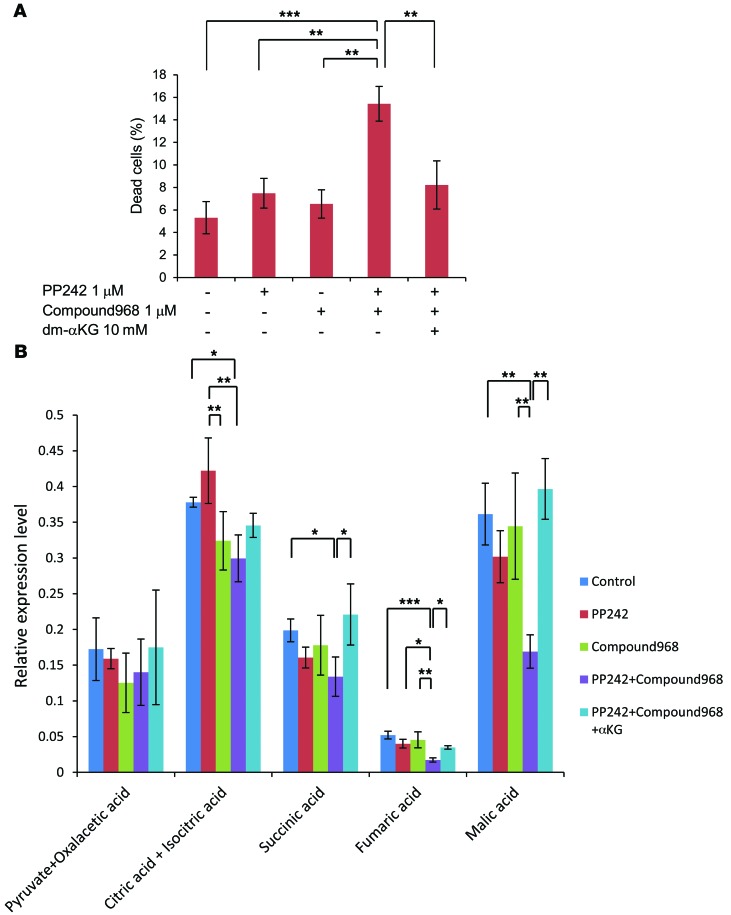
To determine the downstream mechanism by which GLS mediates mTOR-targeted treatment resistance, we examined the involvement of αKG as a downstream metabolite. Interestingly, dimethyl αKG (dm-αKG), an analogue of αKG, obviously rescued the viability of the U87/EGFRvIII GBM cells treated with mTOR and GLS inhibition (Figure 5A), suggesting that apoptotic resistance is mediated through αKG, which is required for the TCA cycle. To determine the metabolic change in the TCA cycle, we analyzed intracellular metabolites by GC/MS, using U87/EGFRvIII GBM cells treated with PP242 and compound 968 in the culture medium containing dm-αKG. Comprehensive metabolome analysis indicated marked reduction in TCA cycle metabolites, including citric acid and isocitric acid, succinic acid, fumaric acid, and malic acid in the cells treated with combined PP242 and compound 968. Importantly, dm-αKG rescued the decrease in the levels of immediately downstream metabolites of αKG in the TCA cycle, succinic acid, fumaric acid, and malic acid, which occurred in response to combined mTOR and GLS inhibition, but the levels of metabolites upstream of αKG, citric and isocitric acid, were not significantly rescued (Figure 5B). These results raise the possibility that glucose and glutamine metabolism coordinately regulate cancer metabolic reprogramming and that GLS may potentially be involved in driving the oxidative TCA cycle through αKG-dependent anaplerosis to enable GBM cells to survive mTOR inhibition.
Figure 5. In response to mTOR inhibition, GLS promotes GBM cell survival in an αKG-dependent manner.
(A) The metabolite dm-αKG (10 mM) was tested for the ability to rescue the viability in U87/EGFRvIII cells treated with both 1 μM PP242 and 1 μM compound 968 for 3 days. Cell death was calculated by trypan blue exclusion. Data represent the mean ± SEM of 3 independent experiments. Tukey-Kramer honest significance testing was performed for multiple comparisons testing. **P < 0.01; ***P < 0.001. (B) GC/MS analysis targeting TCA cycle–related metabolites, pyruvate and oxaloacetic acid, citric and isocitric acid, succinic acid, fumaric acid, and malic acid in U87/EGFRvIII cells treated with 1 μM PP242 and/or 1 μM compound 968 with the normal DMEM or DMEM with dm-αKG (10 mM) for 24 hours. Data represent the mean ± SEM of 3 independent experiments. Tukey-Kramer honest significance testing was performed for multiple comparisons testing. *P < 0.05; **P < 0.01; ***P < 0.001.
Combined GLS and mTOR inhibition causes GBM cell death and blocks tumor growth in xenograft models.
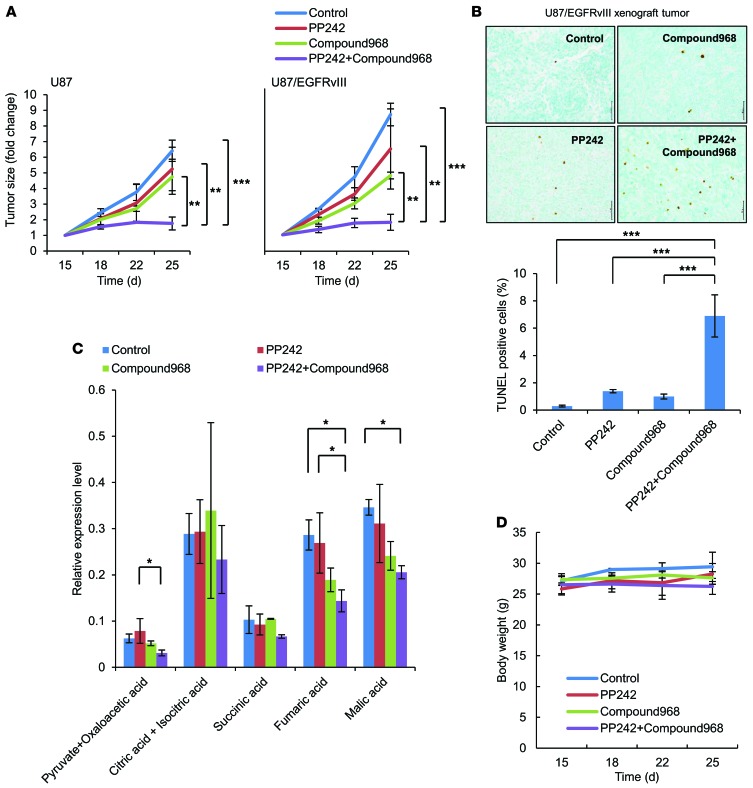
To determine whether GLS inhibition sensitizes GBM cells to mTOR kinase inhibitors in vivo, we analyzed the effect of combining compound 968 with PP242 on tumor cell metabolites, cell death, and tumor size in U87 and U87/EGFRvIII GBM xenografts. Mice were divided into 4 treatment arms and treated for 15 days with intraperitoneal injections of PP242 (5 mg/kg), compound 968 (5 mg/kg), or a combination of PP242 (5 mg/kg) and compound 968 (5 mg/kg). Compound 968 or PP242 alone decreased tumor size by approximately 20%–50% (Figure 6A) without significant induction of apoptosis. Importantly, combined treatment with compound 968 and PP242 resulted in approximately 80% tumor shrinkage for U87/EGFRvIII and, to a lesser extent, U87 xenograft models (Figure 6A). In the immunohistochemical analysis of the U87/EGFRvIII xenograft model, compound 968 led to a 6-fold increase in apoptotic cells in treatment with PP242 (Figure 6B). Importantly, in vivo global metabolic analysis indicated a significant reduction in some TCA cycle metabolites in the EGFRvIII-expressing GBM tumors treated with combined PP242 and compound 968 (Figure 6C), suggesting that both mTOR signaling and glutamine metabolism play an important role in fueling the TCA cycle in GBM cells. Most importantly, such treatment had no apparent effect on body weight throughout the course of the experiment in mice (Figure 6D). Finally, to determine the adverse effects of drugs on normal organs and motor function, non–tumor-bearing mice were treated similarly for 15 days and tested on a rota-rod apparatus (26, 27). No apparent effect on body weight was observed throughout the treatment, and no deterioration of motor function was found after the whole treatment (Supplemental Figure 10A and B). There was no significant difference of cell morphology and no induction of dead cells in brain (cortex and hippocampus), liver, and kidney among mice treated with PP242, compound 968 alone, or a combination (Supplemental Figures 11–13). Taken together, these results demonstrate that GLS inhibition can reverse mTOR-targeted therapy resistance in vivo and that it acts synergistically with PP242 to induce tumor cell death by regulating tumor bioenergetic and molecular synthesis (Figure 7). These results also suggest that the cytotoxic effect of the drug combination was enhanced in tumor tissue, rather than normal tissue.
Figure 6. Both mTOR and GLS inhibition demonstrated synergic tumor growth suppression in vivo.
(A) Each mouse was subcutaneously injected with 1 × 106 U87 (on the left flank) and U87/EGFRvIII (on the right flank) cells. Mice bearing tumors were treated with intraperitoneal injections of PP242 (5 mg/kg) and/or compound 968 (5 mg/kg). The control group received an equal volume of DMSO. Treatment started 15 days after implantation. Tumor volume (in fold change) was measured at the indicated time points from each xenograft of 5 mice. Data represent the mean ± SEM of 3 independent experiments. **P < 0.01; ***P < 0.001. (B) Representative images of U87/EGFRvIII xenograft samples with TUNEL staining (brown cells) to assess apoptotic cells. Quantification of TUNEL staining was performed with ImageJ software. Data represent the mean ± SEM of 3 independent images for each group. ***P < 0.001. Scale bars: 50 μm. (C) GC/MS analysis targeting TCA cycle–related metabolites, pyruvate and oxaloacetic acid, citric and isocitric acid, succinic acid, fumaric acid, and malic acid in U87/EGFRvIII xenograft samples. Data represent the mean ± SEM of 3 independent xenografts for each group. *P < 0.05. (D) Effect of daily intraperitoneal injection of PP242 and/or compound 968 on changes in mouse body weight per group. Tukey-Kramer honest significance testing was performed for multiple comparisons testing. See also Supplemental Figures 10–13.
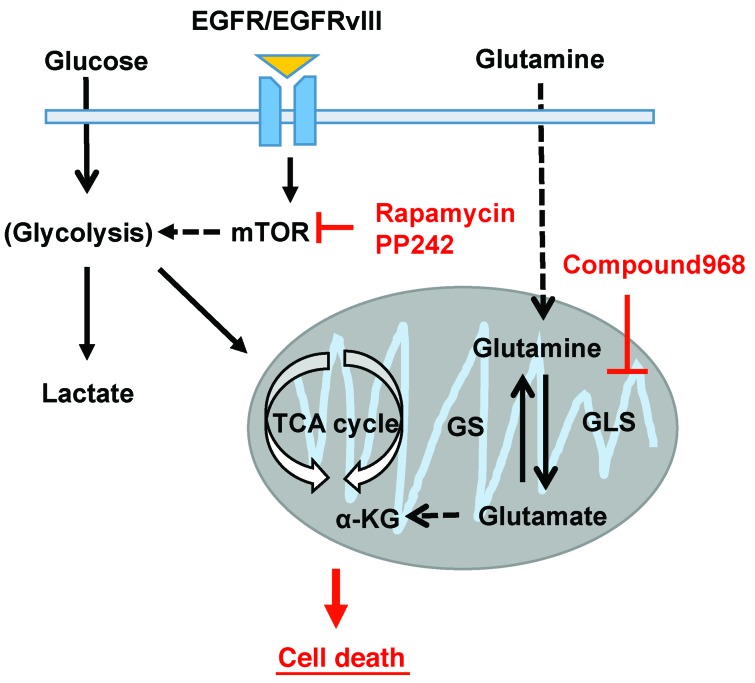
Figure 7. Both mTOR and GLS inhibition attenuate energy production from the TCA cycle, leading to cell death in GBM.
GLS and glutamate levels are elevated following mTOR kinase inhibitor treatment of GBM cells. Combining mTOR kinase inhibitors with GLS inhibition provides a rational therapeutic strategy for patients with GBM.
Discussion
Cancer cell proliferation and oncogenesis are coupled to metabolic reprogramming (18). Commonly occurring oncogenic signal transduction pathways initiated by receptor tyrosine kinases or Ras engage PI3K/AKT signaling to directly stimulate glycolytic metabolism (28). In addition to this aerobic glycolysis, called the Warburg effect, the involvement of glutamine metabolism in cancers has received increasing attention as a potential avenue for the development of new therapeutic agents for cancer treatment. Our study demonstrated that pharmacological impairment of PI3K signaling by mTOR-targeted therapy enhanced glutamine metabolism with increasing GLS activity, leading to glutamine-dependent GBM cells. Altered metabolism is considered to be a key aspect of cancer progression, survival, and drug resistance in most cancers and can help with the design of effective molecular inhibitors and strategies for the treatment of GBM patients.
Cancer cells take up and metabolize glucose and glutamine to a degree that far exceeds their needs for these molecules in anabolic macromolecular synthesis (4, 29). We have demonstrated that glutamine and glutamate and GLS levels are frequently elevated in GBM samples, including in patients imaged in vivo. With the PI3K signaling pathway closely linked to both growth control and metabolic reprogramming, mTOR signaling stimulates specific metabolic pathways, including glycolysis, the oxidative arm of the pentose phosphate pathway, and de novo lipid biosynthesis (10), and mTOR signaling may play an important role in regulating glutamine metabolism. Glutamine can regulate the mTORC1 pathway by facilitating the uptake of leucine (30) and by promoting mTORC1 assembly and lysosomal localization (31, 32), suggesting that glutamine serves both as a signal to mTORC1 and as a source to promote protein translation. Further highlighting the functional link between mTORC1 and glutamine metabolism, a recent study demonstrated that mTORC1 inhibition decreases glutamine metabolism through upregulation of SIRT4 expression to suppress GDH activity (23). We found that GBM cells increase glutamine metabolism with elevated GLS expression in response to mTOR-targeted treatments, and our data may suggest a different pathway because the GBM cells used in the present study are able to resist mTOR inhibition while those used for the previous paper did not (23). Although GDH levels were not changed after mTOR inhibition treatment in our study, we found that ammonia, intracellular glutamate, αKG, and ATP levels were elevated or at least preserved, consistent with increased glutamine metabolism. These results also suggest a potential mechanism underlying the resistance to mTOR kinase inhibition, at least in some GBM cells.
We have previously shown that EGFRvIII, the most common EGFR mutation in GBM, reprograms cellular metabolism to drive tumor growth by activating the Warburg effect through the upregulation of c-MYC, and we have identified complementary mTORC1- and mTORC2-dependent pathways. mTORC1 regulates c-MYC activity through splicing of the MYC-interacting protein MAX, and mTORC2 regulates MYC activity by controlling its levels through a FOXO-acetylation–dependent mechanism (20, 21). The results presented here underscore the importance of mTOR-mediated regulation of glutamine metabolism by showing that GBM cells adapt to mTOR inhibitors by increasing GLS expression to elevate glutamate levels. This could be a necessary compensation for the loss of TCA intermediates that occurs when mTOR inhibition causes a deficit in glycolysis. However, further studies will be needed to test this model and to explore the potential role of other pathways as mediators of GLS expression and activity.
These results also delineate metabolic reprogramming against molecular targeted treatments and/or some metabolic stress. Under glucose limitation, the acquisition of KRAS pathway mutations has been driven with highly expressed levels of GLUT1 in human cancers (33) and the TCA cycle could also be reprogrammed and driven solely by glutamine, generating citrate that consists only of glutamine carbons in a certain type of cancer cell (34). AKT inhibition has stimulated glutamine metabolism with increasing GDH activity (6). Inhibition of mitochondrial pyruvate metabolism may increase glutamate-dependent anaplerosis and promote reductive metabolism of αKG to supply citrate for lipid synthesis (35). It is not surprising that cancer cells are dependent on glutamine metabolism (4). Importantly, our data suggest that maintaining the TCA cycle through αKG-dependent anaplerosis could be involved in promoting GBM survival and resistance to mTOR-targeted treatments. These findings may have important implications for combining mTOR kinase inhibitors with GLS inhibition for patients with GBM and possibly other mTOR-activated cancers.
Methods
Detailed protocols are found in the Supplemental Experimental Procedures.
Cell lines.
U87, U87-EGFRvIII, and U87-EGFR isogenic GBM cell lines were obtained as described previously (36). U251, LN229, T98, and A172 human GBM cell lines and SVGP12 human astroglia cells (ATCC) were cultured in DMEM (Nacalai Tesque) supplemented with 10% FBS (Biological Industries) and 100 U/ml penicillin and streptomycin (Nacalai Tesque) in a humidified 5% CO2 incubator at 37°C.
Antibodies and reagents.
Antibodies obtained were directed against the following: EGFR (Cell Signaling, catalog 2232), p-EGFR Tyr1068 (Cell Signaling, catalog 2236), p-AKT Ser473 (Cell Signaling, catalog 4060), AKT (Cell Signaling, catalog 9272), p-S6 Ser235/236 (Cell Signaling, catalog 4858), S6 (Cell Signaling, catalog 2217), PARP (Cell Signaling, catalog 9542), cleaved PARP (Cell Signaling, catalog 5625), GLS (Abcam, catalog ab60709), GLS1 (Sigma-Aldrich, catalog HPA036223), GLS2 (Sigma-Aldrich, catalog HPA038608), β-actin (Ambion), and EGFR/EGFRvIII cocktail antibody (Novocastra). Reagents used were rapamycin (LC Laboratories), PP242 (Chemdea), GLS inhibitor (compound 968, Calbiochem), and dm-αKG (Sigma-Aldrich).
RNA extraction and real-time PCR.
Total RNA from cell lines, tumor samples, and normal brain tissues was extracted using a mirVana miRNA Isolation Kit (Applied Biosystems). First-strand cDNA was synthesized from 20 ng of total RNA using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Real-time PCR was performed with 3 μl of diluted cDNA using TaqMan gene-expression assays (Applied Biosystems) following the manufacturer’s instructions. All reactions were performed in triplicate. 18S ribosomal RNA was used as the endogenous control. Quantitative mRNA expression data were acquired and analyzed by the ΔΔCt method using an Applied Biosystems 7500 real-time PCR system (Applied Biosystems). The following TaqMan gene-expression assays with FAM-MGB dye were used for this study: SLC2A1 (SMID: Hs00892681_m1), LDHA (SMID: Hs00855332_g1), PDK1 (SMID: Hs01561850_m1), SLC1A5 (SMID: Hs01056542_m1), GLS (SMID: Hs00248163_m1), KGA (SMID: Hs01014019_m1), GAC (SMID: Hs01022166_m1), GLUL (SMID: Hs00365928_g1), GLUD1 (SMID: Hs03989560_s1), 18S (SMID: Hs99999901_s1).
Plasmid and siRNA transfection.
Transfection of siRNA into GBM cell lines was carried out using Lipofectamine RNAiMAX (Invitrogen) in full serum, with a medium change after 24 hours. On-TARGETplus SMARTpool siRNAs (Thermo Scientific, Dharmacon Division) specifically targeting GLS (catalog L-004548-01-0005) and nontargeting control pools of siRNAs were used at 10 nM.
Immunohistochemical staining and image analysis–based scoring.
Paraffin-embedded tissue slides were obtained from the Pathology Histology and Tissue Core Facility at Kobe University Hospital. Slides were counterstained with hematoxylin to visualize nuclei. Staining intensities were scored independently by 2 pathologist and/or 2 neurooncologists who were unaware of the findings of the molecular analyses. Quantitative image analysis was performed with Soft Imaging System software, the utility of which has been previously demonstrated for measuring drug-specific effects in GBM samples in clinical trials with targeted agents (11, 12).
Metabolite measurements.
Glucose, lactate, glutamine, glutamate, and ammonium in the media of cultured cells were measured using the BioProfile 400 analyzer (Nova Biomedical). Cells were cultured in fresh media with various conditions, and metabolite concentrations in the media were measured 48 hours later and normalized to the number of cells in each sample.
l-glutamate levels.
Cultured cells or snap-frozen tissue samples were lysed and homogenized with phosphate buffer. The intracellular l-glutamate level was measured following the manufacturer’s instructions (Yamasa l-glutamate Assay Kit).
GC/MS analysis.
Metabolites from biological samples were extracted and derivatized as described previously, with some modifications (37, 38). The GC/MS analysis was performed using a GC/MSQP2010 Ultra (Shimadzu Co.) with a fused silica capillary column (CP-SIL 8 CB low bleed/MS; 30 m × 0.25 mm inner diameter, 0.25 μm film thickness; Agilent Co.) according to the method described in previous reports (39, 40). The resulting data were exported in the CSV-format file and analyzed using in-house analytical software (AI output). This software enables peak identification and quantification using the in-house metabolites library. All data were normalized to the peak height of the internal standard 2-isopropylmalic acid. To assess the technical variation in the metabolomics experiments, each of the samples was extracted, derivatized, and measured in 3 replicates.
MRS studies.
GBM patients with newly diagnosed or recurrent gliomas underwent preoperative MRI and MRS. A 3T MRI/MRS scanner (Achieva, Philips Medical Systems) was used to acquire the MR spectral data. An 8–channel head MRI coil was used to receive the signal, and the quadrature body MRI coil was used to transmit the radio-frequency (RF) pulses. Single-voxel localized MR spectra were acquired using the double-echo PRESS sequence. The MRS acquisition parameters were as follows: 1.5 × 1.5 × 1.5 cm3, TR/TE = 2000/35 ms, 128 averages, and 1024 complex points for the spectral data. Volumes of interest (VOIs) were localized to representative areas of solid tumor, as determined by a board-certified neuroradiologist or technician. As a control, voxels were also placed in the same anatomical location on the contralateral (nontumor) side of the brain to obtain control spectra. Concentration estimates in absolute units of mM/l VOI were obtained with a user-independent fitting routine (LCModel, developed by Steven Provencher [http://s-provencher.com/pages/lcmodel.shtml]), which is based on a library of model spectra of individual metabolites (41, 42). Quantification was obtained for levels of choline (3.22 ppm), NAA (2.0 ppm), glucose (3.44 ppm), lactate (1.33 ppm), glutamine (2.45 ppm), and glutamate (2.35 ppm). Metabolite ratios were also calculated with respect to the total creatine (creatine plus phospho-creatine, 3.0 ppm), a level that is explicitly assumed to be stable in normal as well as in many pathologic states (43).
Xenograft model.
U87 and U87-EGFRvIII cells were implanted into immunodeficient BALB/cAJcl-nu/nu mice (Clea Japan) for subcutaneous xenograft studies. For subcutaneous implantation, exponentially growing tumor cells in culture were trypsinized, enumerated by trypan blue exclusion, and resuspended at 1 × 106 cells/100 μl in a solution of Dulbecco’s PBS and Matrigel (BD Biosciences). Tumor growth was monitored with calipers by measuring the perpendicular diameter of each subcutaneous tumor. Tumors were treated with intraperitoneal injections of PP242 (5 mg/kg) and/or GLS inhibitor (5 mg/kg) and normal saline every day. Appropriate measures were taken to minimize animal discomfort, and appropriate sterile surgical techniques were utilized in tumor implantation and drug administration. Animals that became moribund or had necrotic tumors were compassionately euthanized.
Statistics.
Results are shown as mean ± SEM. PCA was performed using commercially available SIMCA-P Plus Software, version 12.0.1 (Umetrics). Tukey-Kramer honest significance testing was performed for multiple comparison testing. Other comparisons were performed with 2-tailed Student t test, unless otherwise noted. Statistical significance was defined as P < 0.05.
Study approval.
Glioma tissues were obtained from therapeutic procedures performed as routine clinical management at the Department of Neurosurgery, Kobe University. Tissue samples and peripheral brain tissues were resected during surgery and immediately frozen in liquid nitrogen for subsequent investigation. Each patient or his or her legal guardian provided written, informed consent to use all clinical data and resected tissue specimens for research purposes. This study was approved by the Ethics Committee at Kobe University (approval numbers: 1497 for GC/MS and MRS studies of glioma patients; 1579 for use of glioma samples). All mice were bred and kept under defined-flora pathogen-free conditions at Kobe University in accordance with Laboratory Animal Resources Commission standards. All experiments were approved by the Institutional Animal Care and Use Committees of Kobe University.
Supplementary Material
Acknowledgments
We would like to thank all members of the Brain Tumor Translational Resources staff for biospecimen and biorepository support at Kobe University. We would also like to express our gratitude to Izumi Takayama (Department of Radiation Oncology, Kobe University Graduate School of Medicine) for helping with the TUNEL assay, Yukiko Takeuchi (Division of Evidenced-Based laboratory medicine, Kobe University Graduate School of Medicine) for helping with the GC/MS analysis, and Ryosuke Doi and Mitsuharu Endo (Department of Physiology and Cell Biology, Kobe University Graduate School of Medicine) for helping with the ATP assay. K. Tanaka is supported in part by a Grant-in-Aid for Scientific Research (KAKENHI) (24791501) and the Takeda Science Foundation. T. Sasayama, K. Hosoda, and E. Kohmura are also supported in part by a Grant-in-Aid for Scientific Research (KAKENHI) (25462258, 24592126, and 25293309, respectively). P.S. Mischel is supported by the Ludwig Institute for Cancer Research and grants from the National Institute for Neurological Diseases and Stroke (NS73831), the National Cancer Institute (CA151819), the Ben and Catherine Ivy Foundation, and the Defeat GBM Research Collaborative, a subsidiary of the National Brain Tumor Society.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2015;125(4):1591–1602. doi:10.1172/JCI78239.
References
- 1.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchakjian MR, Kornbluth S. The engine driving the ship: metabolic steering of cell proliferation and death. Nat Rev Mol Cell Biol. 2010;11(10):715–727. doi: 10.1038/nrm2972. [DOI] [PubMed] [Google Scholar]
- 3.Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Ann Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 4.Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010;35(8):427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seltzer MJ, et al. Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res. 2010;70(22):8981–8987. doi: 10.1158/0008-5472.CAN-10-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang C, Sudderth J, Dang T, Bachoo RM, McDonald JG, DeBerardinis RJ. Glioblastoma cells require glutamate dehydrogenase to survive impairments of glucose metabolism or Akt signaling. Cancer Res. 2009;69(20):7986–7993. doi: 10.1158/0008-5472.CAN-09-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang JB, et al. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell. 2010;18(3):207–219. doi: 10.1016/j.ccr.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12(1):9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 10.Duvel K, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39(2):171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mellinghoff IK, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353(19):2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 12.Cloughesy TF, et al. Antitumor activity of rapamycin in a Phase I trial for patients with recurrent PTEN-deficient glioblastoma. PLoS Med. 2008;5(1):e8. doi: 10.1371/journal.pmed.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka K, et al. Oncogenic EGFR signaling activates an mTORC2-NF-κB pathway that promotes chemotherapy resistance. Cancer Discov. 2011;1(6):524–538. doi: 10.1158/2159-8290.CD-11-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang H, Rudge DG, Koos JD, Vaidialingam B, Yang HJ, Pavletich NP. mTOR kinase structure, mechanism and regulation. Nature. 2013;497(7448):217–223. doi: 10.1038/nature12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katt WP, Ramachandran S, Erickson JW, Cerione RA. Dibenzophenanthridines as inhibitors of glutaminase C and cancer cell proliferation. Mol Cancer Ther. 2012;11(6):1269–1278. doi: 10.1158/1535-7163.MCT-11-0942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang W, et al. A proposed role for glutamine in cancer cell growth through acid resistance. Cell Res. 2013;23(5):724–727. doi: 10.1038/cr.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simpson NE, Tryndyak VP, Pogribna M, Beland FA, Pogribny IP. Modifying metabolically sensitive histone marks by inhibiting glutamine metabolism affects gene expression and alters cancer cell phenotype. Epigenetics. 2012;7(12):1413–1420. doi: 10.4161/epi.22713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell. 2012;21(3):297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poptani H, Gupta RK, Roy R, Pandey R, Jain VK, Chhabra DK. Characterization of intracranial mass lesions with in vivo proton MR spectroscopy. AJNR Am J Neuroradiol. 1995;16(8):1593–1603. [PMC free article] [PubMed] [Google Scholar]
- 20.Babic I, et al. EGFR mutation-induced alternative splicing of Max contributes to growth of glycolytic tumors in brain cancer. Cell Metab. 2013;17(6):1000–1008. doi: 10.1016/j.cmet.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masui K, et al. mTOR complex 2 controls glycolytic metabolism in glioblastoma through FoxO acetylation and upregulation of c-Myc. Cell Metab. 2013;18(5):726–739. doi: 10.1016/j.cmet.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gini B, et al. The mTOR kinase inhibitors, CC214-1 and CC214-2, preferentially block the growth of EGFRvIII-activated glioblastomas. Clin Cancer Res. 2013;19(20):5722–5732. doi: 10.1158/1078-0432.CCR-13-0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Csibi A, et al. The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell. 2013;153(4):840–854. doi: 10.1016/j.cell.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarbassov DD, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22(2):159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 25.Efeyan A, Sabatini DM. mTOR and cancer: many loops in one pathway. Curr Opin Cell Biol. 2010;22(2):169–176. doi: 10.1016/j.ceb.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodrigues TB, Granado N, Ortiz O, Cerdan S, Moratalla R. Metabolic interactions between glutamatergic and dopaminergic neurotransmitter systems are mediated through D(1) dopamine receptors. J Neurosci Res. 2007;85(15):3284–3293. doi: 10.1002/jnr.21302. [DOI] [PubMed] [Google Scholar]
- 27.Hamm RJ, Pike BR, O’Dell DM, Lyeth BG, Jenkins LW. The rotarod test: an evaluation of its effectiveness in assessing motor deficits following traumatic brain injury. J Neurotrauma. 1994;11(2):187–196. doi: 10.1089/neu.1994.11.187. [DOI] [PubMed] [Google Scholar]
- 28.Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell. 2008;13(6):472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 29.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7(1):11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 30.Nicklin P, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136(3):521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duran RV, et al. Glutaminolysis activates Rag-mTORC1 signaling. Mol Cell. 2012;47(3):349–358. doi: 10.1016/j.molcel.2012.05.043. [DOI] [PubMed] [Google Scholar]
- 32.Kim SG, et al. Metabolic stress controls mTORC1 lysosomal localization and dimerization by regulating the TTT-RUVBL1/2 complex. Mol Cell. 2013;49(1):172–185. doi: 10.1016/j.molcel.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yun J, et al. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science. 2009;325(5947):1555–1559. doi: 10.1126/science.1174229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le A, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15(1):110–121. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hitosugi T, et al. Tyrosine phosphorylation of mitochondrial pyruvate dehydrogenase kinase 1 is important for cancer metabolism. Mol Cell. 2011;44(6):864–877. doi: 10.1016/j.molcel.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang MY, et al. Mammalian target of rapamycin inhibition promotes response to epidermal growth factor receptor kinase inhibitors in PTEN-deficient and PTEN-intact glioblastoma cells. Cancer Res. 2006;66(16):7864–7869. doi: 10.1158/0008-5472.CAN-04-4392. [DOI] [PubMed] [Google Scholar]
- 37.Yoshie T, et al. Regulation of the metabolite profile by an APC gene mutation in colorectal cancer. Cancer Sci. 2012;103(6):1010–1021. doi: 10.1111/j.1349-7006.2012.02262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wise DR, et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci U S A. 2011;108(49):19611–19616. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshida M, et al. Diagnosis of gastroenterological diseases by metabolome analysis using gas chromatography-mass spectrometry. J Gastroenterol. 2012;47(1):9–20. doi: 10.1007/s00535-011-0493-8. [DOI] [PubMed] [Google Scholar]
- 40.Nakamizo S, et al. GC/MS-based metabolomic analysis of cerebrospinal fluid (CSF) from glioma patients. J Neurooncol. 2013;113(1):65–74. doi: 10.1007/s11060-013-1090-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Provencher SW. Automatic quantitation of localized in vivo 1H spectra with LCModel. NMR Biomed. 2001;14(4):260–264. doi: 10.1002/nbm.698. [DOI] [PubMed] [Google Scholar]
- 42.Andronesi OC, et al. Detection of 2-hydroxyglutarate in IDH-mutated glioma patients by in vivo spectral-editing and 2D correlation magnetic resonance spectroscopy. Sci Transl Med. 2012;4(116):116ra4. doi: 10.1126/scitranslmed.3002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tartaglia MC, et al. Choline is increased in pre-lesional normal appearing white matter in multiple sclerosis. J Neurol. 2002;249(10):1382–1390. doi: 10.1007/s00415-002-0846-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.