Abstract
Widespread reversion of genetic disease is rare; however, such events are particularly evident in some skin disorders in which normal clones develop on a background of affected skin. We previously demonstrated that mutations in keratin 10 (KRT10) cause ichthyosis with confetti (IWC), a severe dominant disorder that is characterized by progressive development of hundreds of normal skin spots via revertant mosaicism. Here, we report on a clinical and histological IWC subtype in which affected subjects have red, scaly skin at birth, experience worsening palmoplantar keratoderma in childhood, and develop hundreds of normal skin spots, beginning at around 20 years of age, that increase in size and number over time. We identified a causal de novo mutation in keratin 1 (KRT1). Similar to IWC-causing KRT10 mutations, this mutation in KRT1 resulted in a C-terminal frameshift, replacing 22 C-terminal amino acids with an alternate 30-residue peptide. Mutant KRT1 caused partial collapse of the cytoplasmic intermediate filament network and mislocalized to the nucleus. As with KRT10 mutations causing IWC, reversion of KRT1 mutations occurred via mitotic recombination. Because reversion is not observed with other disease-causing keratin mutations, the results of this study implicate KRT1 and KRT10 C-terminal frameshift mutations in the high frequency of revertant mosaicism in IWC.
Keywords: Dermatology, Genetics, Stem cells
Introduction
While somatic loss-of-heterozygosity (LOH) events affecting tumor suppressor alleles are common in carcinogenesis (1), similar events leading to reversion of dominantly inherited disease traits are much less frequent. In ichthyosis with confetti (IWC), which features red, scaly skin, interrupted by thousands of normal skin spots, we have shown that normal spots are genetic revertants resulting from independent mitotic recombination events (2). All IWC mutations reported to date affect the C-terminus of keratin 10 (KRT10), replacing all or a portion of the tail with an arginine-rich frameshift motif, which mislocalizes KRT10 to the nucleolus. Cutaneous somatic reversion events were first described in the blistering disorder epidermolysis bullosa and have been reported to expand to many centimeters overtime, providing evidence of improved fitness of revertant clones (3, 4). While reversion is seen in disorders, including Bloom syndrome and Fanconi anemia, which feature defects in recombination-mediated DNA repair and show a significant proportion of reversion events via mitotic recombination (5, 6), reversion events in most skin disorders appear to be less frequent and more commonly occur via second site mutation, gene conversion, back mutation, and deletion. The high frequency of reversion events via LOH is a notable feature of IWC.
Results and Discussion
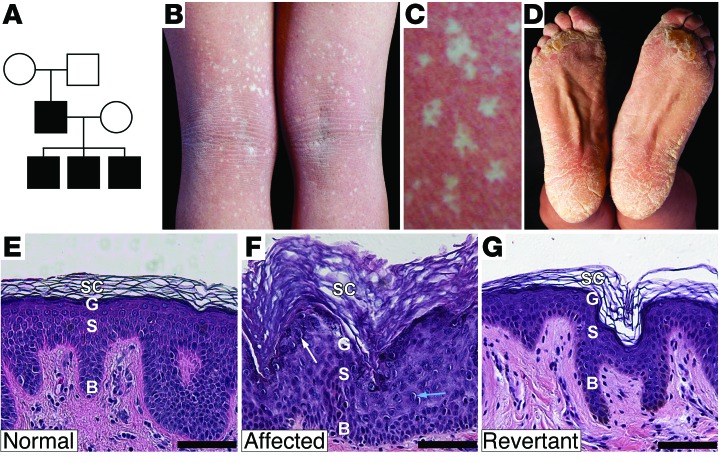
While characterizing our IWC cohort, we identified a kindred with distinct clinical findings, including unique histology and smaller white spots. Recognizing that this kindred represented phenotypic expansion of IWC, we denoted IWC due to KRT10 mutations as IWC-K10 and this new phenotype preliminarily as IWC-II. The index case is a 35-year-old male with unaffected parents, who has 3 affected offspring, ages 3 to 9 years (Figure 1A). As in IWC-K10, affected individuals had bright red skin (erythroderma) at birth and never experienced skin fragility, but palmoplantar hyperkeratosis was disproportionately severe compared with the hyperkeratosis elsewhere. Further, confetti-like spots, which are normal skin on histologic examination (Figure 1G), first appeared in the index case at age 22, are most prominent in the flexures, and reach a maximum size of 4 mm (Figure 1B), while those in IWC-K10 typically appear by age 3, are more widely distributed, and can grow to 1 to 2 cm. IWC-II skin shows thickened stratum corneum without parakeratosis, milder perinuclear vacuolization, and prominent coarse keratohyalin granules (Figure 1C) in contrast to IWC-K10–affected skin, which features parakeratosis, pronounced perinuclear vacuolization, and absent keratohyalin granules (2). These distinct clinical features suggested a novel genetic determinant for IWC-II, and sequencing of KRT10 in the index case identified no mutations.
Figure 1. IWC pedigree, clinical features, and histology of affected and revertant skin.
(A) Pedigree. (B) Index case popliteal fossa shows numerous revertant spots. (C) Revertant clones have a phylloid configuration, with intervening focal affected skin islands. (D) Thick scale on the feet. Histology of (E) normal, (F) affected, and (G) revertant skin, showing basal layer (B), spinous layer (S), granular layer (G), and stratum corneum (SC). Affected skin shows hypercellularity, increased epidermal thickness, prominent keratohyalin granules (white arrow), and perinuclear vacuolization with rare binucleate cells (blue arrow). Revertant skin shows normal histology. Scale bar: 50 mm.
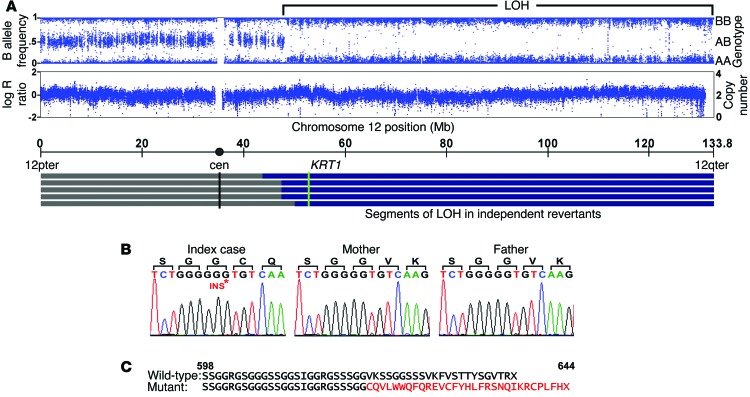
Presuming that revertant keratinocytes had lost a disease-causing de novo dominant mutation, we pursued a LOH mapping strategy to identify the IWC-II locus. We used SNP genotyping analysis to compare genotypes of revertant cell DNA to those of peripheral blood DNA. Of 8 revertant keratinocyte cultures, 2 showed fractional loss of heterozygous genotype calls (B allele deviation), beginning near the centromere on chromosome 12q and extending to the telomere, without change in copy number (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI64415DS1). We suspected admixture between affected and revertant cells, consistent with the clinical finding of affected skin islands within white spots (Figure 1C). Therefore, we used laser capture microdissection (LCM) on 4 additional revertant spots, finding long segments of copy-neutral LOH. Altogether, LOH events began between 44.3 and 49.1 Mb and extended to the telomere of chromosome 12 (Figure 2A). These findings are consistent with mitotic recombination as the mechanism of LOH, with the disease-causing mutation lying distal to 49.1 Mb. This segment contains keratin 1 (KRT1) at 53.1 Mb, which encodes KRT1, the binding partner for KRT10. Sequencing of KRT1 in affected subjects, unaffected family members, and controls revealed a de novo single-base insertion in KRT1 exon 9 (c.1866insG) of the index case that is transmitted to all 3 affected offspring (Figure 2B). This mutation introduces a frameshift, replacing the last 22 amino acids of KRT1 with a novel 30–amino acid peptide (p.622V>CfsX30) (Figure 2C). Revertant spots showed loss of the mutant allele (Supplemental Figure 2). The finding of a de novo mutation in KRT1 that is concordant with IWC-II and is transmitted to 3 affected offspring and which is lost in genetic revertants provides unequivocal evidence that this mutation causes IWC-II, which we subsequently denote as IWC-K1. To exclude the possibility that a distinct de novo mutation causes IWC-II, we performed whole-exome sequencing of the affected case and both unaffected parents and found no additional de novo mutations, including missense, nonsense, or splice site mutations, after filtering against dbSNP, 1000 genomes, and 2,577 in-house controls.
Figure 2. LOH mapping of the IWC-II locus and identification of a de novo mutation in KRT1.
(A) Genotyping of LCM revertant tissue DNA shows heterozygous genotypes from 12pter to 48-Mb base pairs, while from 48-Mb base pairs to 12qter, genotypes are homozygous, indicating LOH. Genotype intensity remains stable over this interval of LOH (logR ratio), and copy number remains at 2, supporting a copy-neutral mechanism. LOH intervals from 5 revertant spots are shown, with breakpoints occurring between 44.3- and 49.1-Mb base pairs. KRT1 lies at 53.1-Mb base pairs. (B) A de novo single-base (G) insertion in KRT1 exon 9 is present in the index case but neither parent; this mutation is transmitted to affected offspring (data not shown). TA cloned sequence is shown for clarity. (C) This mutation results in a C-terminal frameshift replacing the last 22 amino acids of the C-terminus of KRT1 with a novel 30–amino acid peptide (p.622V>CfsX30) (shown in red below the corresponding normal KRT1 tail).
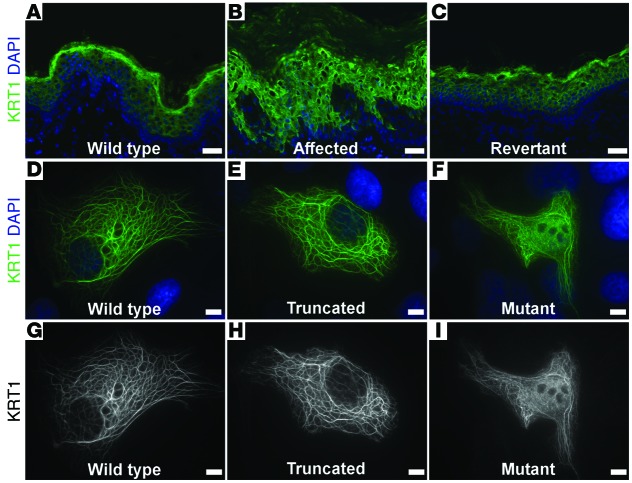
KRT1 is a cytoplasmic intermediate filament protein in suprabasal keratinocytes. Via immunolocalization, we studied KRT1 in affected and revertant skin from our index case and a normal control. Consistent with a pathogenic role of the KRT1 mutation, partial collapse of the intermediate filament network was evident as bright perinuclear rings within affected tissue (Figure 3, A–C). Transmission electron microscopy confirmed this finding, showing perinuclear filament retraction in affected, but not normal or revertant, skin (Supplemental Figure 3).
Figure 3. Mutant KRT1 filaments are mislocalized in vivo and in vitro.
(A–C) Immunolocalization of KRT1 reveals collapsed filament networks in affected skin (B) visible as bright perinuclear rings, which are absent in (A) normal and (C) revertant skin. (D–I) Immunolocalization of KRT1 in primary liver hepatoma cells transfected with (D) wild-type KRT1, (E) KRT1 truncated at the site of the frameshift identified in the index case, and (F) mutant KRT1IWC. Wild-type and truncated KRT1 integrate into the cytoplasmic filament network, while cells expressing KRT1IWC show filament network collapse and nuclear accumulation. (G–I) KRT1 staining alone reveals that (I) the mutant protein accumulates in the nucleus. Scale bars: 50 mm (A–C); 5 mm (D–I).
To further investigate KRT1 mutation pathobiology in vitro, we generated expression constructs for wild-type and mutant KRT1 and KRT1 truncated at the mutation position of the index case (KRT1IWC). These were transfected into primary liver hepatoma cells, which have a well-developed keratin intermediate filament network, and KRT1 immunolocalization was performed. Wild-type KRT1 or KRT1 truncated at the frameshift site was incorporated into normal, well-arborized cytoplasmic filament networks, while KRT1IWC expression produced a collapsed intermediate filament network and led to nuclear accumulation of KRT1 (Figure 3, D–I). These findings implicate the novel peptide resulting from the KRT1 frameshift mutation in disruption of the intermediate filament network and in disease pathogenesis.
KRT1 and KRT10 are coordinately expressed during epidermal differentiation and form heterodimers. Like other keratins, KRT1 has glycine/serine-rich amino-terminal and carboxy terminal nonhelical domains and 2 highly conserved rod domains. The rod domains are critical for intermediate filament assembly, while the nonhelical end domains permit filament stabilization and elongation (7). Keratin end domains have further been shown to be critical for modulation of cell cycle and proliferation (8), and their glycine/serine repeats are thought to assume an omega-loop configuration, which facilitates interactions with loricrin and other nonkeratin molecules (9).
IWC-K1 is distinct from other diseases caused by mutations in KRT1, which have distinct clinical and histologic findings, without evidence of revertant mosaicism. These include missense mutations in the rod domains of KRT1, resulting in epidermolytic palmoplantar keratoderma and epidermolytic ichthyosis characterized by scaling, skin fragility, and keratoderma (10). Skin fragility evident on clinical or histopathologic examination distinguishes both epidermolytic palmoplantar keratoderma and epidermolytic ichthyosis from IWC-K1. Frameshift mutations of KRT1, which replace the C-terminal tail with an alanine-rich peptide, cause ichthyosis hystrix Curth-Macklin with severe palmoplantar keratoderma and distinctive spiny scale (11) or a mild striate palmoplantar keratoderma (12).
Frameshift mutations into the same reading frame of KRT1, as seen in our IWC-II cases, have been reported in two individuals as atypical variants of other disorders. One, a 17-year-old individual, was diagnosed with mild epidermolytic ichthyosis without skin fragility [c.1751insG, p.584S>RfsX69]; the other, a 5-year-old individual, was diagnosed with mild ichthyosis hystrix Curth-Macklin [c.1861insT, p.621S>WfsX32]. Since initial publication, the 17-year-old subject has been lost to follow-up (G. Yosipovitch, unpublished observation), and the 5-year-old case has not developed white spots (Y. Kubo, unpublished observations).
For revertant keratinocytes to persist in the skin and to become clinically evident, reversion events must occur in a renewing cell population and confer improved fitness over mutant skin. This implies that IWC mutations adversely affect the fitness of keratinocyte progenitors that express KRT1 and KRT10 or their progeny (13, 14). The later age at first appearance and smaller size of revertant clones in IWC-K1 versus those in IWC-K10 suggest that somatic reversion events may be less frequent in IWC-K1, that they may require a specific stimulus, or that revertant clones have a lesser selective advantage. Evidence for this includes our inability to successfully culture pure revertant cells from IWC-K1 white spots, their smaller size, and the clinically apparent affected skin islands within revertant spots (Figure 1C).
While somatic revertant mosaicism has been described in other diseases, including Wiskott-Aldrich syndrome, severe combined immune deficiency, Bloom syndrome, Fanconi anemia, dyskeratosis congenita, and epidermolysis bullosa (4, 15, 16), the number of independent clonal revertants in IWC-K10 and IWC-K1 is remarkable. Moreover, while in these other disorders multiple mechanisms for reversion have been identified in the same individual, all revertants studied in this IWC-K1 kindred, as well those previously identified in IWC-K10 kindreds, result from mitotic recombination (2). DNA double-strand breaks are a common antecedent to mitotic recombination (17), and their induction may play a role in the high rate of reversion in IWC. This could result from impaired cell cycle progression or DNA repair defects; however, the mechanism is presently unknown. Notably, none of the other 74 reported mutations in KRT1 or KRT10 that cause other disorders show reversion. The frameshift mutations, but not C-terminal–truncating mutations, in IWC-K10 and IWC-K1 disrupt the filament network and lead to mislocalization of KRT1 and KRT10 to the nucleus. These findings implicate these frameshift peptides in disruption of the normal interaction of KRT1/KRT10 tails, resulting in dysfunctional cytokeratin filament networks and possibly contributing to cellular events that result in genetic reversion.
Methods
For additional details, see the Supplemental Methods.
Keratinocyte culture.
Skin biopsies were transported in DMEM (Invitrogen) with 2X penicillin-streptomycin (Invitrogen) and incubated overnight in 1X dispase (BD Biosciences), and epidermal sheets were harvested. Keratinocytes were isolated via 0.05% trypsin–2% EDTA incubation and were plated upon a feeder layer of mitomycin-C–treated 3T3 cells in DMEM/F12 medium with supplements as described previously (18). Cells were subsequently propagated in EpiLife (Invitrogen).
LCM.
5-micron frozen tissue sections were cut onto polyethylene naphthalate membrane slides (Leica), and laser capture of histologically normal tissue was performed using a LS-AMD microscope (Leica). Tissue was gravity collected into lysis buffer and DNA prepared using the QIAamp DNA Micro Kit (Qiagen).
SNP analysis.
SNP genotyping (370CNV and 1M arrays, Illumina) was performed at the Yale Center for Genome Analysis. DNA from LCM samples underwent whole-genome amplification (Qiagen) and purification (Gen Elute PCR Clean-Up, Sigma-Aldrich) prior to genotyping. Illumina Genome Studio software (version 2011.1) was used for genotyping analysis, which uses cluster files representing 100 samples from the HapMap populations for genotype calling. To identify LOH breakpoints, the B allele frequency of 20 SNP bins was plotted along the length of chromosome 12, identifying both the position of initial B allele deviation and its total extent. Manual inspection of B allele frequency of individual SNPs was used to confirm breakpoints, and the logR ratio of intensity was examined for deviation from baseline to assess for copy number alteration at sites of observed B allele deviation (19). Data from arrays have been deposited in the database of Genotypes and Phenotypes (dbGAP) (study accession phs000744.v1.p1).
Whole-exome sequencing.
DNA from blood was sheared, bar-coded libraries were prepared (TruSeq DNA v2 Sample Preparation Kit, Illumina), and exome capture was performed (EZ Exome 2.0, Roche), followed by sequencing using the Illumina HiSeq platform with samples pooled 6 per lane. After alignment to the hg19 reference using ELAND (Illumina), we used a Perl script, which trimmed sequence to the targeted intervals, removed PCR duplicates, and identified single nucleotide variations (SNVs), deletions, and insertions using SAMtools software (20). Perl scripts were used to exclude variants in dbSNP (build 132) and 1000 Genomes (release 05/2011) from further analysis and to annotate the remaining variants for functional impact (21). Heterozygous SNVs (missense, nonsense, and splice site variants) and indels in the proband not present in 2,577 control exomes with a SAMtools quality score (QS) ≥ 60 and 200, respectively, were selected for further analysis.
De novo variant analysis.
For SNVs and indels/insertions, coverages for corresponding major and minor alleles were extracted from both parents with minor read frequencies of less than 10%. A Bayesian algorithm with the following elements was used to assist de novo mutation calls: probability of the proband being heterozygous at the test position; probability that parents are homozygous for the reference allele, given frequency of reference and nonreference reads and probability of heterozygosity in offspring; and probability that a variant is de novo, given a region of segmental duplication and its population frequency. The resulting Bayesian QSs (BQSs) were scaled from 0 to 100. Previously, we used this approach in which Sanger validation of 181 putative mutations distributed across the BQS spectrum showed strong correlation (R2 = 0.89) and calls with a BQS ≥ 50 had a positive predictive value of virtually 100% (22). Aligned reads at these positions were examined with the Broad Institute Integrative Genome Viewer in the index case and both of his parents to eliminate false positives (23). Sequencing data have been deposited into dbGAP (study accession phs000744.v1.p1).
Study approval.
All study participants provided consent to a protocol approved by the Yale Human Investigation Committee, which included permission for deposition of genetic data into public databases. All participants provided a blood sample, and affected and revertant skin biopsies were collected from the index case.
Supplementary Material
Acknowledgments
We thank our study subjects and their families for contributing to and supporting this study. We thank M. Choi for assistance with LOH analyses, L. Boyden for helpful discussions, and S. Mane and the Yale Center for Genome Analysis for technical support. K.A. Choate was supported by a grant from the National Institute of Arthritis and Musculoskeletal and Skin Diseases/NIH (5K08AR056305) and a Clinical Scientist Development Award from the Doris Duke Charitable Foundation. R.P. Lifton is an investigator of the Howard Hughes Medical Institute. This work was partly supported by a National Center for Research Resources High-End Instrumentation grant, the Yale Center for Mendelian Genomics (NIH U54 HG006504); and the Yale Center for Clinical Investigation (NIH UL1 TR000142).
Footnotes
Conflict of interest: Richard P. Lifton has served on the scientific advisory board of Merck.
Reference information:J Clin Invest. 2015;125(4):1703–1707. doi:10.1172/JCI64415.
References
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Choate KA, et al. Mitotic recombination in patients with ichthyosis causes reversion of dominant mutations in KRT10. Science. 2010;330(6000):94–97. doi: 10.1126/science.1192280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jonkman MF, et al. Revertant mosaicism in epidermolysis bullosa caused by mitotic gene conversion. Cell. 1997;88(4):543–551. doi: 10.1016/S0092-8674(00)81894-2. [DOI] [PubMed] [Google Scholar]
- 4.Jonkman M, Pasmooj AM. Revertant mosaicism — patchwork in the skin. N Engl J Med. 2009;360(16):1680–1682. doi: 10.1056/NEJMc0809896. [DOI] [PubMed] [Google Scholar]
- 5.Lo Ten Foe JR, et al. Somatic mosaicism in Fanconi anemia: molecular basis and clinical significance. Eur J Hum Genet. 1997;5(3):137–148. [PubMed] [Google Scholar]
- 6.Ellis NA, Lennon DJ, Proytcheva M, Alhadeff B, Henderson EE, German J. Somatic intragenic recombination within the mutated locus BLM can correct the high sister-chromatid exchange phenotype of Bloom syndrome cells. Am J Hum Genet. 1995;57(5):1019–1027. [PMC free article] [PubMed] [Google Scholar]
- 7.Lee CH, Coulombe PA. Self-organization of keratin intermediate filaments into cross-linked networks. J Cell Biol. 2009;186(3):409–421. doi: 10.1083/jcb.200810196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Magin TM, Vijayaraj P, Leube RE. Structural and regulatory functions of keratins. Exp Cell Res. 2007;313(10):2021–2032. doi: 10.1016/j.yexcr.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Zhou XM, Idler WW, Steven AC, Roop DR, Steinert PM. The complete sequence of the human intermediate filament chain keratin 10. Subdomainal divisions and model for folding of end domain sequences. J Biol Chem. 1988;263(30):15584–15589. [PubMed] [Google Scholar]
- 10.Rothnagel JA, et al. Mutations in the rod domains of keratins 1 and 10 in epidermolytic hyperkeratosis. Science. 1992;257(5073):1128–1130. doi: 10.1126/science.257.5073.1128. [DOI] [PubMed] [Google Scholar]
- 11.Richardson ES, Lee JB, Hyde PH, Richard G. A novel mutation and large size polymorphism affecting the V2 domain of keratin 1 in an African-American family with severe, diffuse palmoplantar keratoderma of the ichthyosis hystrix Curth-Macklin type. J Invest Dermatol. 2006;126(1):79–84. doi: 10.1038/sj.jid.5700025. [DOI] [PubMed] [Google Scholar]
- 12.Whittock NV, et al. Frameshift mutation in the V2 domain of human keratin 1 results in striate palmoplantar keratoderma. J Invest Dermatol. 2002;118(5):838–844. doi: 10.1046/j.1523-1747.2002.01750.x. [DOI] [PubMed] [Google Scholar]
- 13.Ezhkova E, et al. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136(6):1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koçer SS, Djurić PM, Bugallo MF, Simon SR, Matic M. Transcriptional profiling of putative human epithelial stem cells. BMC Genomics. 2008;9:359. doi: 10.1186/1471-2164-9-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis BR, Candotti F. Genetics. Mosaicism — switch or spectrum? Science. 2010;330(6000):46–47. doi: 10.1126/science.1195991. [DOI] [PubMed] [Google Scholar]
- 16.Jongmans MC, et al. Revertant somatic mosaicism by mitotic recombination in dyskeratosis congenita. Am J Hum Genet. 2012;90(3):426–433. doi: 10.1016/j.ajhg.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 18.Schwartz PM, Barnett SK, Atillasoy ES, Milstone LM. Methotrexate induces differentiation of human keratinocytes. Proc Natl Acad Sci U S A. 1992;89(2):594–598. doi: 10.1073/pnas.89.2.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peiffer DA, et al. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res. 2006;16(9):1136–1148. doi: 10.1101/gr.5402306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scholl UI, et al. Seizures, sensorineural deafness, ataxia, mental retardation, and electrolyte imbalance (SeSAME syndrome) caused by mutations in KCNJ10. Proc Natl Acad Sci U S A. 2009;106(14):5842–5847. doi: 10.1073/pnas.0901749106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zaidi S, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498(7453):220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.