Abstract
Nonmuscle myosin-2 is the primary enzyme complex powering contractility of the F-actin cytoskeleton in the model organism Drosophila. Despite myosin’s essential function in fly development and homeostasis, its kinetic features remain elusive. The purpose of this in vitro study is a detailed steady-state and presteady-state kinetic characterization of the Drosophila nonmuscle myosin-2 motor domain. Kinetic features are a slow steady-state ATPase activity, high affinities for F-actin and ADP, and a low duty ratio. Comparative analysis of the overall enzymatic signatures across the nonmuscle myosin-2 complement from model organisms indicates that the Drosophila protein resembles nonmuscle myosin-2s from metazoa rather than protozoa, though modulatory aspects of myosin motor function are distinct. Drosophila nonmuscle myosin-2 is uniquely insensitive toward blebbistatin, a commonly used myosin-2 inhibitor. An in silico modeling approach together with kinetic studies indicate that the nonconsensus amino acid Met466 in the Drosophila nonmuscle myosin-2 active-site loop switch-2 acts as blebbistatin desensitizer. Introduction of the M466I mutation sensitized the protein for blebbistatin, resulting in a half-maximal inhibitory concentration of 36.3 ± 4.1 µM. Together, these data show that Drosophila nonmuscle myosin-2 is a bona fide molecular motor and establish an important link between switch-2 and blebbistatin sensitivity.—Heissler, S. M., Chinthalapudi, K., Sellers, J. R. Kinetic characterization of the sole nonmuscle myosin-2 from the model organism Drosophila melanogaster.
Keywords: actin, blebbistatin, cytoskeleton, transient kinetics, rotamer
The sole nonmuscle myosin-2 in the fruit fly Drosophila is encoded by the zipper (zip) gene, named after an extensive defect in cytokinesis that leads to failure in dorsal closure and embryonic lethality (1). Structurally, the nonmuscle myosin-2 heavy chain follows a prototypic architecture, including an N-terminal catalytic motor domain, a light chain binding neck region, followed by a coiled-coil-forming domain that terminates in a short nonhelical tailpiece (2). The quaternary structure of the myosin holoenzyme is hexameric, composed of 2 heavy chains, which associate with a set of essential [cytoplasmic myosin light chain (mlc-c)] and regulatory [spaghetti squash (sqh)] light chains. Under physiologic conditions, multiple myosins oligomerize into defined structures of bipolar minifilaments, which interact with F-actin in an ATP-dependent manner and power cytoplasmic contractility (2).
Nonmuscle myosin-2 plays pivotal roles in cell shape changes and tissue dynamics in morphogenic processes such as organogenesis, myogenesis, and the establishment of left-right asymmetry during Drosophila development (3–5). Furthermore, essential roles of nonmuscle myosin-2 in tension transmission and contraction in cytokinesis, cell proliferation, adhesion, tissue patterning, border cell migration in the egg chamber, and nurse cell dumping are established (1, 6–11). The functional spectrum of Drosophila nonmuscle myosin-2 is similar to the mechanisms underlying developmental and physiologic functions of mammalian nonmuscle myosin-2 paralogs (12). Therefore, it is not surprising that Drosophila has been successfully used as a model system to study human nonmuscle myosin-2 (MYH9)-related diseases (13). This raises the general question if human disease phenotypes that are manifested in mutations within the catalytically active myosin motor domain could be predicted from Drosophila as a model system. To address this question, we performed a detailed kinetic characterization of a Drosophila nonmuscle myosin-2 subfragment 1 (S1)–like construct. Comparative analysis of kinetic signatures with selected nonmuscle myosin-2 paralogs from model organisms suggests a similar overall kinetic mechanism between Drosophila and mammalian nonmuscle myosin-2s, underlining the evolutionary closer relationship among metazoa compared with protozoa.
Blebbistatin is a widely used myosin-2-specific inhibitor whose potency has some variation among different myosin-2 isoforms (14, 15). Drosophila nonmuscle myosin-2 is insensitive toward blebbistatin, as shown by the failure of blebbistatin to inhibit cytokinesis in Drosophila S2 cells (15). We show that blebbistatin in the concentration range up to 200 µM does not inhibit the actin-activated ATPase activity of Drosophila nonmuscle myosin-2. In contrast, a switch-2 mutant in which Met466 is replaced with isoleucine, hereafter referred to as M466I, is partially inhibited by blebbistatin with a half-maximal inhibitory concentration (IC50) of 36.3 ± 4.1 µM. This finding indicates that switch-2 plays a pivotal role in the establishment of blebbistatin sensitivity and the allosteric communication between nucleotide and inhibitor binding site.
MATERIALS AND METHODS
Chemicals and reagents
Standard chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA). Mant-fluorophores were from BIOLOG Life Science Institute (Bremen, Germany). Racemic blebbistatin was purchased from Calbiochem (San Diego, CA, USA). Rabbit skeletal muscle acetone powder was from Pel-Freez Biologicals (Rogers, AR, USA).
Cloning, recombinant overproduction, and protein purification
The cDNA of a myosin S1-like motor domain construct corresponding to amino acids 1–841 of Drosophila nonmuscle myosin-2 (isoform C, accession number NP_001014553.1) was amplified and inserted into a modified pFastBac1 vector, encoding a C-terminal Flag-tag. The switch-2 mutation M466I was introduced with standard cloning techniques. A polycistronic vector comprising the cDNAs for Sqh (19.1 kDa, accession number NP_511057.1) and Mlc-c (16.6 kDa, accession number NP_511049.1) under the control of the polyhedrin and P10 promoter, respectively, was a generous gift from Adam C. Martin (Massachusetts Institute of Technology, Cambridge, MA, USA). Transposition, production of recombinant baculoviruses, and Spodoptera frugiperda (Sf9) insect cell infection were performed as recommended by the manufacturer (Invitrogen, Carlsbad, CA, USA). The myosin heavy chain constructs were overproduced along with the respective light chains and purified to electrophoretic homogeneity as described earlier (16). F-actin was prepared according to Lehrer and Kerwar (17) and labeled with N-(1-pyrene)iodoacetamide (pyrene) as described in Criddle et al. (18).
Kinetic assays
Steady-state ATPase assays were performed as described previously at 25°C in buffer containing 10 mM 3-(N-morpholino) propanesulfonic acid (MOPS) (pH 7.0), 2 mM MgCl2, 0.15 mM EGTA, 40 U/ml l-lactic dehydrogenase, 200 U/ml pyruvate kinase, 200 µM β-NAD, 1 mM phosphoenolpyruvate, 50 mM NaCl, and 2 mM ATP (16). The myosin concentration was between 0.2 and 0.68 µM. To assay the effect of blebbistatin on myosin’s steady-state ATPase activity, dilutions of the compound were made in 90% DMSO. The reaction mixture was incubated with blebbistatin for 10 min at room temperature prior to the assay. To minimize the impact of the solvent on the assay, the DMSO concentration was kept constant at 1.8%.
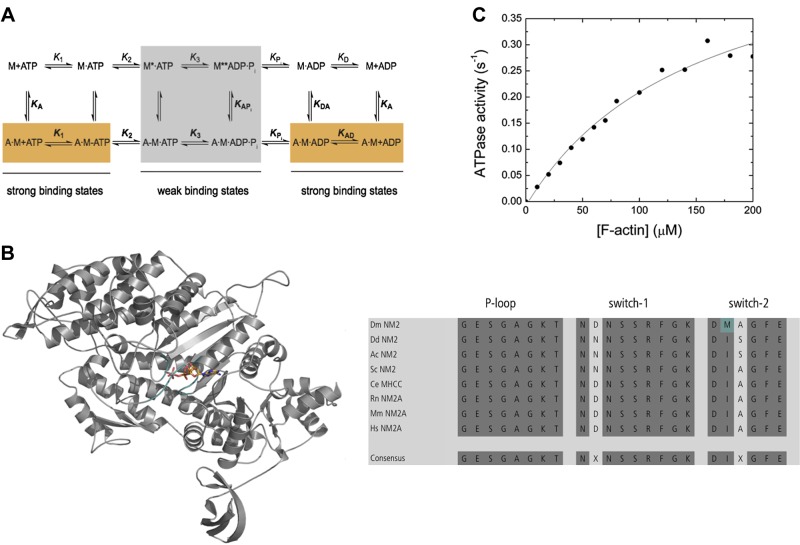
Transient kinetic assays were carried out in an SF-61DX2 stopped-flow system (Hi-Tech Scientific, Bradford-on-Avon, United Kingdom) at 20°C in a buffer containing 25 mM MOPS (pH 7.0), 100 mM KCl, 5 mM MgCl2, and 0.1 mM EGTA, unless stated otherwise. Fluorescence excitation/emission setups were as follows: tryptophan, 297/320 nm cutoff filter; pyrene and mant-nucleotides, 365/390 nm long-pass filter; and Förster transfer from tryptophan to the mant moiety, 297/390 nm long-pass filter. Light-scattering assays were performed at a wavelength of 320 nm, and the signal was detected in a 90° angle to the incident beam. For F-actin binding and release assays, the KCl concentration was reduced to 2.5 mM. In F-actin binding assays, the [myosin]/[F-actin] ratio was kept between 1:5 and 1:7.5 and the reaction mixture treated with 0.1 U/ml apyrase prior to the assays for apo conditions, when required. Concentrations stated throughout the text are postmixing (1:1). Data storage and initial data analysis were accomplished with Kinetic Studio 2.20 (Hi-Tech Scientific). Secondary plots were made with Origin 7 (OriginLab, Northampton, MA, USA). Kinetic rate and equilibrium constants were interpreted as described earlier based on the proposed kinetic features established for skeletal muscle myosin-2 (Fig. 1A; where bold indicates rate and equilibrium constants in the lower pathway and in pathways perpendicular to the lower that are involved in actin-bound states) (19).
Figure 1.
A) Minimum kinetic scheme of the myosin and actomyosin ATPase cycle. KA represents the affinity of myosin for F-actin, KD is the affinity of ADP for myosin, KAD is the affinity of ADP for the actomyosin complex, and KDA is the affinity of F-actin for myosin in the presence of saturating [ADP]. M refers to myosin. The asterisk represents different conformational states of the myosin motor domain detected by the intensity of the intrinsic tryptophan fluorescence. Kinetic parameters in the presence and absence of F-actin are in boldface (k+1 and K1) vs. normal (k+1 and K1) notation. Subscripts A and D refer to actin (KA) and ADP (KD), respectively. Dissociation equilibrium constants are calculated according to Kx = k-x/k+x. B) The overview figure shows the homology model of the Drosophila nonmuscle myosin-2 motor domain in cartoon representation. The nucleotide is in stick representation; switch-1, switch-2, and P-loop are in teal color. Sequence analysis is shown for the conserved structural elements P-loop, switch-1, and switch-2 in the nucleotide binding pocket. Dark gray highlights conserved amino acids. The nonconsensus amino acid Met466 in Drosophila nonmuscle myosin-2 is shown in teal. Dm, Drosophila melanogaster; Dd, Dictyostelium discoideum; Ac, Acanthamoeba castellanii; Sc, Saccharomyces cerevisiae; Ce, Caenorhabditis elegans; Rn, Rattus norvegicus; Mm, Mus musculus; Hs, Homo sapiens. C) Actin-activated steady-state ATPase activity. The dependence of the steady-state ATPase activity on [F-actin] was measured in the concentration range up to 200 µM. The data are corrected for the kbasal. Fitting of the data with the Michaelis-Menten equation yields the parameters kcat = 0.54 ± 0.06 s−1 and Kapp = 152 ± 39 µM.
Homology modeling
The structure of the Drosophila nonmuscle myosin-2 motor domain (amino acids 1–780) was modeled using the structure of the human nonmuscle myosin-2C motor domain [Protein Data Bank identification (PDB ID) 2YCU] in the prepower stroke state as a template. Both proteins share 72% sequence identity and 82% homology. The model was built using Modeler 9.14 (20). For the blebbistatin binding site analysis, the Drosophila nonmuscle myosin-2 homology model was superimposed onto the Dictyostelium nonmuscle myosin-2 motor domain in complex with blebbistatin (PDB ID 1YV3). In silico mutagenesis was performed to mutate Met466 to Ile466, and the residues in the binding site were minimized for structural analysis. Side-chain conformations in the blebbistatin binding sites were optimized using backbone-dependent rotamer libraries (21) included in the University of California (San Francisco, CA, USA) Chimera molecular visualization package (22).
RESULTS
Sequence analysis and comparison with other nonmuscle myosin-2s
The Drosophila nonmuscle myosin-2 motor domain core shares around 71 and 86% sequence identity and similarity, respectively, with vertebrate nonmuscle myosin-2 isoforms from human, rat, zebrafish, and Xenopus at the protein level. Those numbers/similarities reduce to 45 and 71% for unicellular nonmuscle myosin-2s from Dictyostelium, Acanthamoeba, and Saccharomyces, underlining the phylogenetic closer relationship to higher eukaryotes (Supplemental Fig. 1S). Sequence analysis further indicates that the nucleotide binding pocket comprises the conserved structural elements P-loop (GESGAGKT) and switch-1 (NXSSRFGK). Switch-2, the third active-site loop involved in nucleotide coordination, deviates from the consensus sequence (DIXGFE) and comprises the amino acids DMAGFE (Fig. 1B).
Protein production and steady-state ATPase activity
An S1-like fragment of Drosophila nonmuscle myosin-2 was overproduced along with the respective light chains in Sf9 insect cells. Approximately 1–2 mg protein was obtained per liter of cells and purified to electrophoretic homogeneity.
The steady-state ATP turnover of the recombinant protein is activated ∼33-fold by [F-actin] from a basal activity of 0.016 s−1 to a maximum calculated catalytic activity (kcat) of 0.54 ± 0.06 s−1 (Fig. 1C). The Kapp of 152 ± 39 µM is very high, resulting in a low coupling efficiency kcat /Kapp of ∼0.004 µM−1 s−1. A low kcat /Kapp is a characteristic of all kinetically described nonmuscle myosin-2 paralogs (Table 1).
TABLE 1.
Steady-state ATPase parameters obtained in the present study for Drosophila nonmuscle myosin S1
| Parameter | Signal | Drosophila nonmuscle myosin-2 | Acanthamoeba nonmuscle myosin-2 (23) | Dictyostelium nonmuscle myosin-2 (24) | Human nonmuscle myosin-2A (25) |
|---|---|---|---|---|---|
| kbasal (s−1) | NADH assay | 0.016 | 0.05 | 0.08 ± 0.02 | 0.013 ± 0.004 |
| kcat (s−1)a | NADH assay | 0.54 ± 0.06 | 4.68 ± 0.73 | 2.6 ± 1 | 0.17 ± 0.005 |
| Kapp (µM)a | NADH assay | 152 ± 39 | 428 ± 97 | 102 ± 20 | 72 ± 4 |
| kcat /Kapp (µM−1 s−1) | NADH assay | 0.004 | 0.01b | 0.025c | 0.0025 |
For comparison, selected kinetic parameters from nonmuscle myosin-2s are listed. aMichaelis-Menten parameters obtained from a hyperbolic fit to the data set. bMaximum steady-state ATPase at 270 µM F-actin. cFrom the initial slope of the steady-state ATPase vs. [F-actin] plot.
ATP interaction
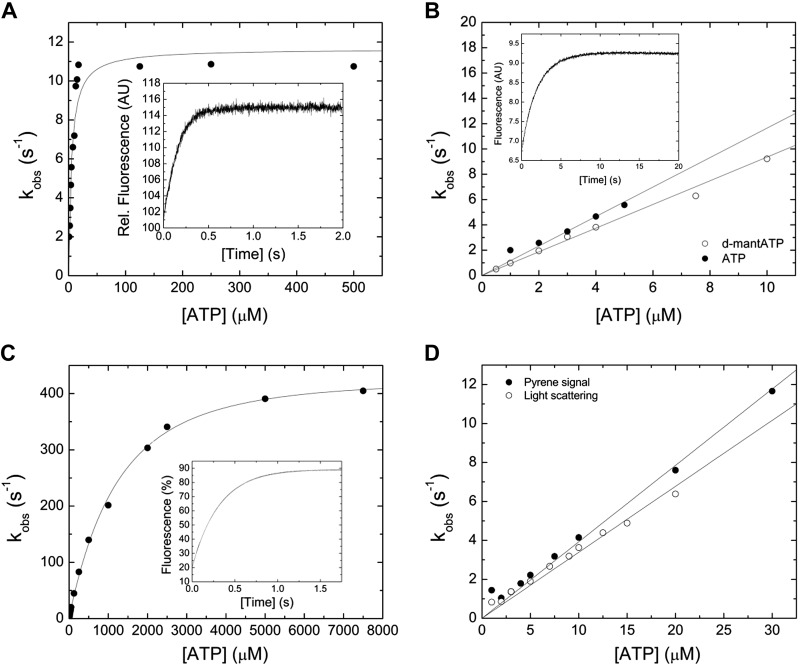
ATP binding to the myosin motor fragment results in a single-exponential transient increase in tryptophan fluorescence (Fig. 2A, inset). The concentration dependence of the observed rate constant (kobs) on [ATP] can be fitted with a hyperbolic function, yielding a maximum value k+3 + k−3 of 11.65 ± 0.56 s−1 (Fig. 2A). Half-saturation (1/K1) is observed at [ATP] = 3.73 ± 1.32 µM (Fig. 2A). A linear fit of the data at low [ATP] yields a second-order rate constant for ATP binding, K1k+2, of 1.16 ± 0.06 µM−1 s−1(Fig. 2B). A single-exponential time evolution of the fluorescence signal is also observed upon mixing 2′-deoxy-3′-O-(N′-methylanthraniloyl) (d-mant) ATP with myosin under pseudo first-order conditions. A representative transient is shown in the inset of Fig. 2B. A secondary plot of the kobs versus [d-mantATP] results in a linear dependence describing the second-order rate constant K1k+2 to 0.94 ± 0.02 µM−1 s−1 (Fig. 2B).
Figure 2.
Interaction between myosin and actomyosin and ATP. A) Increasing [ATP] was mixed under pseudo first-order conditions with 0.25 µM myosin in a stopped-flow spectrophotometer. The dependence of kobs on [ATP] is hyperbolic, yielding the parameter k+3 + k-3 of 11.65 ± 0.56 s−1 at saturation. 1/K1 is observed at [ATP] = 3.73 ± 1.32 µM. The inset shows a representative trace of the transient increase in tryptophan fluorescence after mixing 7.5 µM ATP with 0.25 µM myosin. Single-exponential fit to the data yields an amplitude of 14.2% and a kobs of 6.6 s−1. B) Comparison of the second-order rate constants for ATP and dmantATP binding to myosin at low [nucleotide]. The dependence of kobs on [nucleotide] is linear and results in a K1k+2 of 1.16 ± 0.06 µM−1 s−1 (ATP) and K1k+2 to 0.94 ± 0.02 µM−1 s−1 (d-mantATP). A fluorescence transient of the bimolecular interaction between 0.15 µM myosin and 3 µM d-mantATP is shown in the inset of (B). Single-exponential fit to the data set yields an amplitude of 2.8% and a kobs of 3.06 s−1. C) ATP-induced dissociation of the actomyosin complex. Fluorescence signals were obtained after mixing pyrene-actomyosin under pseudo first-order conditions with increasing [ATP] in a stopped-flow spectrophotometer. Hyperbolic fit to the data yields a k+2 of 482.8 ± 7.85 s−1 at saturating [ATP]. 1/K1 is observed at 1224.37 ± 88.39 µM [ATP]. Inset shows a representative transient of the interaction between 0.5 µM pyrene-actomyosin and 7.5 µM ATP. Single-exponential fit to the data set yields an amplitude of 69% and a kobs of 3.18 s−1. D) Comparison of the kobs upon the ATP-induced dissociation of the actomyosin complex. Monitored is either the postmixing pyrene or the light-scattering signal. Linear fits to the data sets up to 30 µM [ATP] define the second-order rate constants K1k+2 of 0.39 ± 0.01 µM−1 s−1 (pyrene), and K1k+2 of 0.34 ± 0.01 µM−1 s−1 (light-scattering).
ATP binding to pyrene-actomyosin dissociates the complex and results in a transient increase in pyrene fluorescence, as shown in the inset of Fig. 2C. A kobs versus [ATP] plot is hyperbolic (Fig. 2C). A maximum value of 482.8 ± 7.85 s−1, representing the isomerization constant k+2, can be extrapolated from the data set. Half-saturation is observed at 1222.3 ± 60.29 μM ATP. The data are linear at low [ATP] up to 30 µM, yielding the second-order rate constant K1k+2 of 0.39 ± 0.01 µM−1 s−1 (Fig. 2D). In agreement, the light-scattering signal upon the ATP-induced dissociation of the actomyosin complex results in the second-order rate constant K1k+2 of 0.34 ± 0.01 µM−1 s−1 (Fig. 2D). Table 2 lists all kinetic constants for the interaction between myosin and actomyosin with ATP.
TABLE 2.
Transient kinetic parameters obtained in the present study for Drosophila nonmuscle myosin-2 S1
| Parameter | Signal or calculation | Drosophila nonmuscle myosin-2 | Acanthamoeba nonmuscle myosin-2 (23) | Dictyostelium nonmuscle myosin-2 (24, 26–28) | Human nonmuscle myosin-2A (25) |
|---|---|---|---|---|---|
| ATP interaction | |||||
| K1k+2 (µM−1 s−1) | Tryptophan | 1.16 ± 0.06 | 1.33 ± 0.04 | 0.86 | 0.56 ± 0.01 |
| K1k+2 (µM−1 s−1) | d-mantATP | 0.94 ± 0.02 | 1.93 ± 0.02 | 1.13 | 1.03 ± 0.14 |
| 1/K1 (µM) | Tryptophan | 3.73 ± 1.32 | 5.19 ± 0.58 | 25.8 | n.d. |
| k3 + k-3 (s−1) | Tryptophan | 11.65 ± 0.56 | 19.0 ± 0.5 | 30 | 14.1 ± 0.5 |
| K1k+2 (µM−1 s−1) | Pyrene-actin | 0.39 ± 0.01 | 2.11 ± 0.03 | 0.25 | 0.21 ± 0.04 |
| K1k+2 (µM−1 s−1) | Light scattering | 0.34 ± 0.01 | 0.73 ± 0.02 | n.d. | 0.14 ± 0.003 |
| 1/K1 (µM) | Pyrene-actin | 1222.3 ± 60.29 | 651.3 ± 68.4 | 509 | ∼900 |
| k+2 (s−1) | Pyrene-actin | 482.80 ± 7.85 | 779.6 ± 24.9 | 380 | ∼190 |
| ADP interaction | |||||
| k+D (µM−1 s−1) | d-mantADP | 0.52 ± 0.04 | 0.50 ± 0.03 | 1.02 | 0.55 ± 0.06 |
| k−D (s−1) | d-mantADPa | 2.42 ± 0.13 | 2.29 ± 0.20 | n.d. | 0.54 ± 0.23 |
| k−D (s−1) | d-mantADPb | 2.92 ± 0.03 | 1.53 ± 0.01 | 1.6 | 1.12 ± 0.13 |
| k−D (s−1) | Tryptophanb | 2.35 ± 0.01 | 1.68 ± 0.02 | n.d. | n.d. |
| KD (µM) | k−D/k+D | 4.65 ± 0.44 | 4.58 ± 0.49 | 1.6 | 1.5 ± 0.4 |
| k+AD (µM−1 s−1) | d-mantADP | 3.46 ± 0.11 | ∼4.95c | 0.72 | 2.72 ± 0.16 |
| k−AD (s−1) | d-mantADP a | 5.19 ± 0.55 | n.d. | n.d. | n.d. |
| k−AD (s−1) | Pyrene-actinb | 5.53 ± 0.08 | 28.9 ± 0.07 | 15 | n.d. |
| k−AD (s−1) | Light scatteringb | 4.27 ± 0.05 | 29.67 ± 0.15 | n.d. | n.d. |
| k−AD (s−1) | d-mantADPb | 5.06 ± 0.05 | 31.43 ± 0.49 | n.d. | 2.68 ± 0.30 |
| KAD (µM) | k−AD/k+AD | 1.5 ± 0.17 | 5.99 ± 0.88 | 58 | 1.4 ± 0.4 |
| Coupling | KAD/KD | 0.32 ± 0.05 | 1.30 ± 0.24 | ∼36 | 0.7 |
| Coupling | k−AD/k−D | 2.14 ± 0.25 | ∼17.6 | ∼9.4 | 2.8 |
| Actin interaction | |||||
| k+A (µM−1 s−1) | Light scattering | 1.14 ± 0.03 | 10.88 ± 0.22 | 1.34 | 0.73 ± 0.03 |
| k−A (s−1) | Pyrene-actin | ∼0.07 | ∼0.001 | 0.0068 | <0.007 |
| KA (nM) | k−A/k+A | ∼60 | ∼0.1 | ∼5.1 | <10 |
| k+DA (µM−1 s−1) | Light scattering | 0.26 ± 0.01 | 1.17 ± 0.04 | n.d. | 0.19 ± 0.02 |
| k−DA (s−1) | Pyrene-actin | ∼0.01 | ∼0.014 | n.d. | ∼0.004 |
| KDA (nM) | k−DA/k+DA | ∼38 | ∼12 | n.d. | ∼20 |
For comparison, the kinetic parameters from selected nonmuscle myosin-2s are listed. n.d., not determined. aFrom y-intercept. bFrom chasing experiment. cCalculation from k−AD/KAD.
ADP interaction
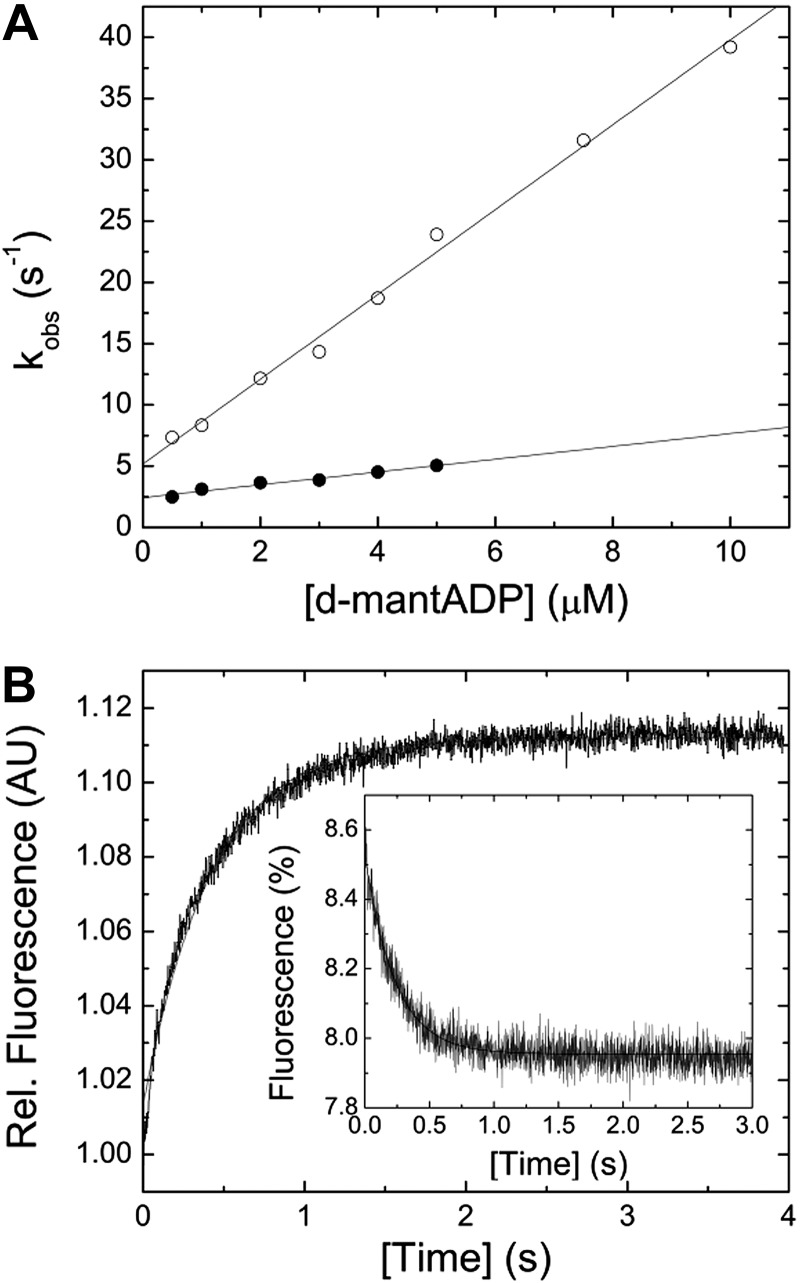
The bimolecular interaction between myosin and the fluorescent nucleotide analog d-mantADP results in a single-exponential transient fluorescence increase, indicating a 1-step binding mechanism. Plotting of kobs vs. [d-mantADP] results in a linear dependence (Fig. 3A). Fitting the data to a straight line yields a second-order binding rate constant for ADP, k+D, of 0.52 ± 0.04 µM−1 s−1. Extrapolating the data to [d-mantADP] = 0 yields the ADP release rate constant k−D of 2.42 ± 0.13 s−1. For comparison, direct measurement of the respective constant in ATP-chasing experiments yields a k−D of 2.92 ± 0.03 s−1 or 2.35 ± 0.01 s−1 with the extrinsic (d-mantADP) or intrinsic (tryptophan) fluorescence signal (Fig. 3B). Calculation of the dissociation equilibrium constant, based on the mathematical relationship k−D/k+D, results in a value of 4.65 ± 0.44 µM.
Figure 3.
Interaction between myosin and actomyosin with ADP. A) [d-mantADP] dependence of the kobs for myosin (closed circles) or actomyosin (open circles) binding to the fluorescent ADP analog. Linear fits to the data sets yield the second-order binding rate constants k+D of 0.52 ± 0.04 µM−1 s−1 and k+AD of 3.46 ± 0.1 s−1. Extrapolating the data to [d-mantADP] = 0 yields the ADP release rate constant k−D of 2.42 ± 0.13 s−1 and k−AD of 5.19 ± 0.55 s−1, in the absence and presence of F-actin, respectively. B) Representative transients for the ADP release from myosin and actomyosin (inset). Assay conditions were as follows in the absence of F-actin: 1.5 mM ATP versus 0.25 µM myosin and 15 µM ADP. The fluorescence was exited at a wavelength of 297 nm and the fluorescence emission detected after the passage of a 320 nm cutoff filter. The obtained transient was best described by a single exponential function yielding a kobs of 2.35 s−1. The inset shows a light-scattering signal upon the dissociation of a 0.25 µM pyrene-actomyosin complex in the presence of 10 µM ADP with 4 mM ATP. Single-exponential fit of the data yields a kobs of 4.27 s−1.
Mixing myosin with increasing [d-mantADP] in the presence of F-actin results in a 7-fold acceleration of the second-order ADP binding rate constant to k+AD = 3.46 ± 0.11 µM−1 s−1 (Fig. 3A). Extrapolating the linear fit to [d-mantADP] = 0 yields the apparent ADP release rate constant, k−AD, of 5.19 ± 0.55 s−1. The ADP release rate from actomyosin was independently measured by chasing d-mantADP or ADP from the actomyosin complex with excess [ATP]. Fluorescence emission transients of d-mantADP, pyrene, or a change in the light-scattering signal result in observed dissociation constants of k−AD = 5.06 ± 0.05 s−1, k−AD = 5.53 ± 0.08 s−1, or k−AD = 4.27 ± 0.05 s−1, respectively (Fig. 3B, inset). Calculation of the dissociation equilibrium constant KAD (KAD = k−AD/k+AD) gives 1.5 ± 0.17 µM. Interestingly, the actin-activated ADP release rate is not significantly altered in the presence of varying concentrations of free [Mg2+] in the physiologic range (0.2–2 mM; data not shown). All kinetic constants describing the interaction between myosin and actomyosin with ADP are listed in Table 2.
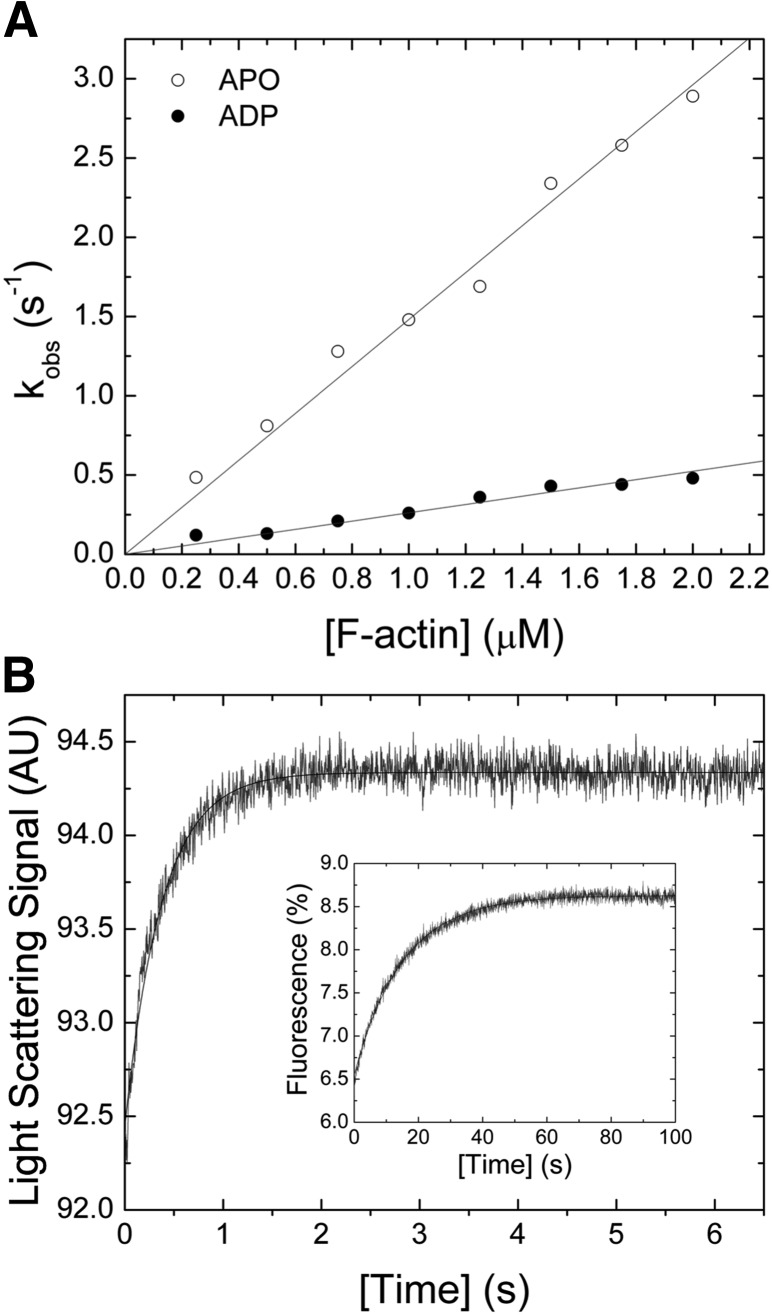
F-actin interaction
The bimolecular interaction between myosin and F-actin was probed in light-scattering assays in the presence and absence of saturating [ADP]. Mixing of both proteins results in an increase of the light-scattering signal (Fig. 4B). A single exponential fit to the data set indicates a 1-step binding event. Plotting of kobs as a function of [F-actin] results in a linear dependence, yielding the second-order rate constants for F-actin binding k+A of 1.14 ± 0.03 µM−1 s−1 and k+DA of 0.26 ± 0.01 µM−1 s−1 in the absence or presence of saturating [ADP] (Fig. 4A). The corresponding release rate constants k−A ∼0.07 s−1 and k−DA ∼0.01 s−1 were obtained from chasing experiments (Fig. 4B, inset). From the ratio of the release and binding rate constants (k−A/k+A and k−DA/k+DA), the dissociation equilibrium constants KA and KAD were calculated to ∼60 and ∼38 nM. The kinetic constants describing the interaction between F-actin and Drosophila nonmuscle myosin-2 are listed in Table 2.
Figure 4.
Interaction between myosin and F-actin. A) [F-actin] dependence of kobs for the reaction between myosin and filamentous actin. The second-order binding rate constants k+A of 1.14 ± 0.03 µM−1 s−1 (open circle) and k+DA of 0.26 ± 0.01 µM−1 s−1 (closed circle) in the absence or presence of 100 µM [ADP] were obtained from the linear fits to the data. B) Representative light-scattering signal of the interaction between 0.3 µM myosin and 1.5 µM F-actin. Single-exponential fit of the data yields an amplitude of 1.89% and a kobs of 2.65 s−1. The inset shows an observed F-actin release rate constant kobs of 0.07 s−1 that was obtained after fitting the transient, obtained from chasing 0.25 µM pyrene-actomyosin with 10 µM F-actin, to a single exponential function.
Interaction with blebbistatin
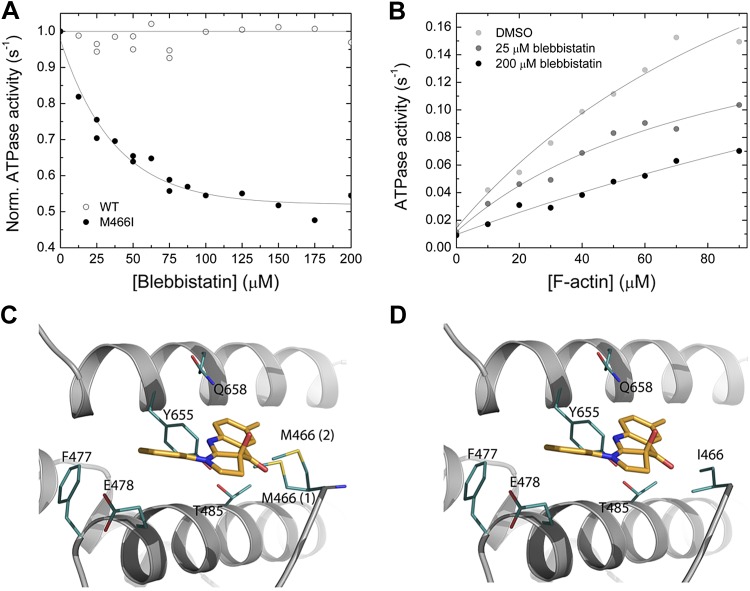
The interaction between myosin and the commonly used myosin-2 inhibitor blebbistatin was tested in the actin-activated ATPase assay. At constant [F-actin] of 30 µM, increasing [blebbistatin] up to 200 µM does not significantly inhibit the steady-state ATPase activity of Drosophila nonmuscle myosin-2 (Fig. 5A). Structural analysis of the Dictyostelium nonmuscle myosin-2/blebbistatin complex shows that the inhibitor binds to a pocket at the apex of the 50 kDa cleft, at a distance of ∼15 Å from the nucleotide binding pocket (29). Superimposition with the Drosophila nonmuscle myosin-2 motor domain homology model indicates that all key myosin/blebbistatin binding site residues, as defined by Allingham et al. (29), are conserved (Table 3). A notable feature of the Drosophila nonmuscle myosin-2 blebbistatin binding site is that amino acid Met466 substitutes for a consensus isoleucine in switch-2, as outlined in greater detail above. Switch-2 is not only an essential part of the nucleotide binding region but also flanks the rear of the blebbistatin binding site. Structural analysis of the Drosophila nonmuscle myosin-2 motor domain homology model in the prepower stroke state shows that the large side chain of Met466 occupies the blebbistatin binding site and restricts the free conformational space around the switch-2 loop. Out of the 13 possible rotamers for Met466 backbone-dependent side-chain conformations, 2 of the major rotamers, representing the energetically most favorable (17 and 13%) and the most likely side-chain conformations, occlude the inhibitor binding site (Fig. 5C). In contrast, the consensus isoleucine side-chain rotamer is positioned normally and allows free conformational space for blebbistatin to bind properly (Fig. 5D). We hypothesize that Met466 sterically hinders blebbistatin binding and interferes with the communication between the nucleotide and the allosteric blebbistatin binding site and thus could partially explain the insensitive nature of Drosophila nonmuscle myosin-2 toward blebbistatin.
Figure 5.
Interaction between myosin and blebbistatin. A) Normalized (Norm.) actin-activated steady-state ATPase activity of Drosophila nonmuscle myosin-2 (open circles) and M466I (closed circles) as a function of [blebbistatin]. [F-actin] was kept constant at 30 µM, and the data were normalized to the starting value of the ATPase activity in the absence of inhibitor. Single-exponential fit of the data set gives an IC50 of 36.3 ± 4.1 µM for M466I, whereas no inhibition is observed for the native protein [wild-type (WT)]. B) Blebbistatin-mediated decrease of the steady-state actin-activated ATPase activity. Increasing [blebbistatin] leads to a concentration-dependent decrease in the actin-activated ATP turnover. At 90 µM [F-actin], the ATPase activity is reduced >2-fold. For comparison, the uninhibited control was supplemented with 1.8% DMSO to account for the solvent effect. C, D) Close-up views of the blebbistatin binding site in the Drosophila nonmuscle myosin-2 and M466I motor domain homology models in prepower stroke state (ADP·Pi). Key residues are shown as sticks in teal, blebbistatin in mango color. There are 2 major side-chain rotamers [17% (1), and 13% (2)] of Met466 in the active-site loop switch-2 shown for nonmuscle myosin-2 (C). In silico substitution of switch-2 residue methionine 466 by “isoleucine” increases the conformational space available for blebbistatin binding in mutant M466I (D).
TABLE 3.
Comparison of IC50 values and blebbistatin binding site residues of nonmuscle myosin-2s from selected model organisms
| Drosophila nonmuscle myosin-2 | Drosophila nonmuscle myosin-2 M466I | Acanthamoeba nonmuscle myosin-2 | Dictyostelium nonmuscle myosin-2 | Human nonmuscle myosin-2A | Blebbistatin interaction | |
|---|---|---|---|---|---|---|
| IC50 (µM) | >>200 | 36.3 | 83 (14) | 4.9 (14) | 5.1 (14) | |
| Residue | Leu271 | Leu271 | Leu262 | Leu262 | Leu258 | Benzyl ring |
| Residue | Phe477 | Phe477 | Phe468 | Phe466 | Phe464 | Benzyl ring |
| Residue | Glu478 | Glu478 | Glu469 | Glu467 | Glu465 | Benzyl ring |
| Residue | Val651 | Val651 | Val652 | Val630 | Val646 | Benzyl ring |
| Residuea | Ala467 | Ala467 | Ser458 | Ser456 | Ala454 | Tetrahydropyrrolo ring |
| Residue | Ile482 | Ile482 | Ile473 | Ile471 | Ile469 | Tetrahydropyrrolo ring |
| Residue | Thr485 | Thr485 | Thr476 | Thr474 | Thr472 | Methylquinolinone |
| Residue | Tyr655 | Tyr655 | Tyr656 | Tyr634 | Tyr650 | Methylquinolinone |
| Residue | Gln658 | Gln658 | Gln659 | Gln637 | Gln653 | Methylquinolinone |
| Residue | Leu662 | Leu662 | Leu663 | Leu641 | Leu657 | Methylquinolinone |
| Residuea | Met466 | Ile466 | Ile459 | Ile457 | Ile455 | None |
Reference binding site residues were previously described for the Dictyostelium nonmuscle myosin-2/blebbistatin complex by Allingham et al. (29). Residues corresponding to Drosophila nonmuscle myosin-2 Met466 are listed for completeness. aDeviations in the amino acid sequence.
To test for the potential impact of the nonconsensus switch-2 residue Met466 on the blebbistatin insensitivity of Drosophila nonmuscle myosin-2, we overproduced the switch-2 mutant M466I, in which methionine Met466 is replaced by a consensus isoleucine. M466I has a basal ATPase activity (kbasal) of 0.021 s−1 under steady-state conditions. The kcat of 0.58 ± 0.1 s−1 is similar to the respective value of Drosophila nonmuscle myosin-2, whereas the Kapp of 299 ± 88 µM is higher (data not shown). Supporting the prediction from the in silico modeling approach, M466I is blebbistatin sensitive. Blebbistatin inhibits the steady-state actin-activated ATPase of M466I by a maximum of 50% and an IC50 of 36.3 ± 4.1 at 30 µM F-actin (Fig. 5A). The effect of blebbistatin on the steady-state ATPase turnover of M466I in the concentration range up to 90 µM [F-actin] was examined (Fig. 5B). It is not possible to measure the Kapp of M466I with great accuracy given its very high value and the need for extremely high actin concentrations and, thus, is not possible to draw a firm conclusion whether Kapp, kcat, or both parameters are altered by [blebbistatin]. However, direct comparison of the ATPase activities at 90 µM [F-actin] indicates a 50% inhibition between 0 and 200 µM. The same trend is also observed for the kbasal (Fig. 5B).
DISCUSSION
Actomyosin ATPase cycle
Drosophila nonmuscle myosin-2 has a nonconsensus amino acid (Met466) in switch-2 instead of isoleucine, which is normally present in myosins. Isoleucine substitutions at this site are extremely rare in the myosin superfamily but have been reported for mammalian myosin-18A and both myosin-18 and myosin-20 from Drosophila. Both myosin classes have additional nonconsensus amino acids in the nucleotide binding region and are enzymatically inactive pseudoenzymes (30–32). However, our studies demonstrate that Drosophila nonmuscle myosin-2 is a bona fide motor as would be predicted by genetic studies in Drosophila and tissue culture models (11, 33).
Signatures include a slow steady-state ATPase activity and a low coupling ratio kcat/Kapp (Table 1). M466I has a similar kcat but a 50% reduced coupling efficiency due to an increased Kapp. The kbasal is slightly increased in M466I compared with the wild-type motor domain. Overall, the observed effects on the steady-state ATPase activity resemble alanine scanning results at the respective site in Dictyostelium nonmuscle myosin-2, indicating that an isoleucine to alanine substitution does not impact the overall in vitro and in vivo motor function but results in a slightly higher kbasal (33). Drosophila nonmuscle myosin-2 has a very high affinity for ADP, and the ADP release rate is only marginally activated by F-actin (k−AD/k−D, ∼2). The actin-activated ADP release k−AD (∼5 s−1) is around 10 times faster than the kcat, making it unlikely to rate limit the catalytic cycle under physiologic conditions. In agreement with other reports on class-2 myosins, the release of inorganic phosphate (Pi) is the kinetic step limiting the actomyosin ATPase cycle (23, 25). The fast ADP release rate compared with the kcat leads to a low duty ratio of 0.1, calculated from
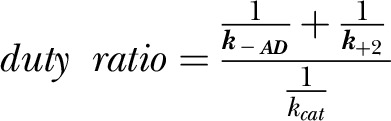 |
Contractile forces exerted by nonmuscle myosin-2 derive from bipolar minifilamentous structures in which the myosin motor domains are radially distributed at opposing filament ends. As described by Kiehart and Feghali (34), native Drosophila nonmuscle myosin-2 bipolar filaments contain on average 16 myosins, resulting in 16 heads on each side of the filament. Although Drosophila nonmuscle myosin-2 would not be processive as a single molecule, such a filament composed of multiple myosin-2 molecules could have a high effective duty ratio and therefore act as a processive unit. Based on the filament geometry, the effective duty ratio of a Drosophila nonmuscle myosin-2 filament equals 0.81 using the following equation:
with n equaling the number of heads per filament side. The high effective duty ratio would promote a continuous, processive interaction between a Drosophila nonmuscle myosin-2 filament and neighboring F-actin, as it has been reported for human nonmuscle myosin-2B (35). The duty ratio of Drosophila nonmuscle myosin-2 is comparable to the duty ratios for isoforms of vertebrate nonmuscle myosins-2B and -2C (16, 36), whereas amoeboid nonmuscle myosins from unicellular eukaryotes have slightly lower duty ratios (23).
Kinetic comparison of nonmuscle myosin-2 paralogs from model organisms
In comparison to previously characterized nonmuscle myosin-2s from model organisms, the overall kinetic signatures of Drosophila nonmuscle myosin-2 resemble those of the mammalian paralogs, underlining the closer phylogenetic relationship (Table 2). Its steady-state ATPase activity and the actin-activated ADP release rate constants are similar to those reported for human class-2 nonmuscle myosins (Table 2) (16, 25, 36). In comparison, the reported values for class-2 myosins from Acanthamoeba and Dictyostelium (15–31 s−1) are significantly higher (Table 2) (23, 26). We speculate that alternatively spliced variants of fly nonmuscle myosin-2 with insertions of either 8 or 40 amino acids in loop-1 might have faster actin-activated ADP release rates, as it has been described for human nonmuscle myosin-2C and smooth muscle myosin-2 splice variants (36–39). Possible functional consequences include alterations in duty ratio and mechanoenzymatic properties. Transient kinetic signatures of nonmuscle myosin-2s from amoeba to humans include reduced second-rate binding constants for ATP in the presence of F-actin and fast second-order binding rate constants for ADP in the presence of F-actin (k+AD/K1k+2). The resulting product inhibition impacts the duty ratio and the in vitro sliding velocity of the proteins (12). For the cytoplasmic myosin-2 from Drosophila, the k+AD/K1k+2 ratio of ∼10 is identical to the number calculated for human nonmuscle myosin-2A and slightly higher when compared to Acanthamoeba (k+AD/K1k+2, ∼7) and Dictyostelium nonmuscle myosin-2 (k+AD/K1k+2, ∼3) (Table 2). Kinetic (k−AD/k−D) and thermodynamic (KAD/KD) coupling ratios of ∼2 and ∼0.3 are low for the Drosophila paralog and similar to those reported for nonmuscle myosin-2s from human. The respective amoeboid myosins have higher kinetic coupling ratios between 9.4 and 17.6, indicating a significant increase in the ADP release rate by F-actin. The thermodynamic coupling ratio for Dictyostelium is around 10-fold higher than the one reported for Drosophila nonmuscle myosin-2.
Although nonmuscle myosin-2s from Drosophila and human are kinetically similar, the actin-activated ADP release of Drosophila cytoplasmic myosin-2 is magnesium insensitive in the physiologic range of free [Mg2+]. [Mg2+] sensitivity has been described as a modulatory mechanism for the ADP release rate constants, ADP affinities, and duty ratios of mammalian nonmuscle myosin-2s (36). This behavior suggests a different allosteric communication within the Drosophila nonmuscle myosin-2 motor domain, which resembles more the pattern of amoeboid myosins or rabbit skeletal muscle myosin-2.
Extrinsic regulation
Blebbistatin is a class-2–specific, allosteric, noncompetitive myosin inhibitor, which was identified as a small molecule compound that inhibits cytokinesis in various cells types from model organisms, with the exception of Drosophila (15). Previous in vitro studies demonstrated that blebbistatin inhibited the actin-activated ATPase activity of vertebrate and amoeboid nonmuscle myosin-2s by >80%. The IC50 values, reflecting the half-maximal inhibitory blebbistatin concentration, range from 1.8 to 5.1 µM (14, 15). The Acanthamoeba paralog shows only 60% inhibition and a high IC50 with 83 µM (14). Consistent with the findings of the in vivo studies, the in vitro actin-activated steady-state ATPase activity of Drosophila nonmuscle myosin-2 is blebbistatin insensitive up to an inhibitor concentration of 200 µM (Fig. 5A) (15). This does not exclude a blebbistatin-mediated inhibition of the enzymatic activity at higher inhibitor concentration but suggests an IC50 outside the experimentally accessible concentration for in vitro and in vivo studies.
Mechanistically, switch-2 undergoes a conformational change during the ATP hydrolysis cycle and alternates between an open and a closed conformation including the main-chain rotations at its consensus isoleucine (33, 40). Blebbistatin decreases the rate-limiting phosphate release step of the catalytic cycle and increases the lifetime of the weakly bound ADP·Pi intermediate by stabilizing the closed switch-2 conformation (41–43). In the Drosophila nonmuscle myosin-2 homology model in the prepower stroke state, Met466 rotamers occupy the free conformational space of the blebbistatin binding site, thereby interfering with compound binding. We further predict that an open switch-2 conformation results in an even more pronounced decrease of the inhibitor binding pocket. The switch-2 mutant M466I is blebbistatin sensitive with an IC50 value of 36.3 ± 4.1 µM, which lies between those reported for Acanthamoeba nonmuscle myosin-2 and the paralogs from Dictyostelium and mammals (Table 3) (14, 44). As reported previously for Acanthamoeba nonmuscle myosin-2, blebbistatin only leads to a partial inhibition of the steady-state ATPase activity of M466I. Despite the substantial increase in blebbistatin sensitivity for M466I when compared to the wild-type protein, the solely 2-fold inhibition and the high IC50 make the mutant protein not suitable for in vivo cell biologic applications. However, it underlines the proposed link between switch-2 and blebbistatin inhibition, which contributes to the understanding of the inhibitor-mediated myosin inhibition. Introducing further mutations at switch-2 in Drosophila nonmuscle myosin-2 in addition to isoleucine could possibly increase the conformational space at this region and increase the potency of blebbistatin. It may also lead to a new generation of blebbistatin derivatives that increase the inhibitory potency. Furthermore, the in vitro blebbistatin insensitivity of Drosophila nonmuscle indicates that the myosin/inhibitor interaction is more than an induced-fit mechanism. Nonbinding site residues within the myosin motor domain are less conserved in the class-2 myosins, allowing different degrees of protein motion and conformers, which could favor or constrain compound binding (45). Furthermore, the myosin/inhibitor complex formation must be thermodynamically favorable and long lived enough in terms of complex stability so that the induced fit can take place within a reasonable time. A third explanation for the reduced in vitro and in vivo sensitivity might be a different allosteric communication pathway that operates between the blebbistatin binding site and the active site. Blebbistatin has been occasionally used to probe nonmuscle myosin-2 motor function in Drosophila cells and tissues, and some small effects were observed (46). This would not be predicted by the in vitro results of the present study and the initial in vivo study (15), suggesting that there might be off-target effects as were noted in Dictyostelium myosin-2 null cells where blebbistatin inhibits cell streaming and other cellular processes (44).
CONCLUSIONS
Drosophila cytoplasmic myosin-2 is a low duty ratio molecular motor with kinetic signatures that resemble those of mammalian rather than amoeboid nonmuscle myosin-2s. This is consistent with the diverse roles that nonmuscle myosin-2s play in multicellular organisms. The nonconsensus amino acid Met466 in switch-2 is not crucial for Drosophila nonmuscle myosin-2 motor function but serves as blebbistatin desensitizer. It is unfortunate for those studying the function of nonmuscle myosin-2 in Drosophila that blebbistatin cannot be used as a pharmacologic tool. Our demonstration that the M466I mutation partially sensitizes nonmuscle myosin-2 to blebbistatin is the first step toward engineering a Drosophila nonmuscle myosin-2 that would be fully inhibited by this reagent. If such a mutant myosin could be engineered into the Drosophila genome using state-of-the-art gene-editing methodology, replacing the endogenous nonmuscle myosin-2 with a blebbistatin-sensitive version would greatly facilitate studies on the role of this myosin in fly development and homeostasis.
The overall kinetic similarities between Drosophila and mammalian nonmuscle myosin-2s suggest that Drosophila may be a good model system for testing the function of MYH9 disease mutations. However, we do not recommend this approach. Mutations responsible for most human genetic diseases involving myosin-2s lie in regions that are not directly associated with nucleotide or F-actin binding and are sometimes associated with subtle kinetic phenotypes (47, 48). The overall sequence identity between the Drosophila and mammalian nonmuscle myosin-2s motor domain is around 72%, which means that there will be many other amino acid substitutions, in addition to the residue that is intentionally mutated, which could alter the impact of the chosen mutation.
Supplementary Material
Acknowledgments
The authors thank Adam C. Martin (Massachusetts Institute of Technology, Cambridge, MA, USA) for the Drosophila nonmuscle myosin-2 cDNA and the polycistronic expression vector encoding for Sqh and Mlc-c. They also thank Fang Zhang for the preparation of F-actin. J.R.S. was funded by the Intramural Research Program of the U.S. National Institutes of Health National Heart, Lung, and Blood Institute.
Glossary
- d-mant
2′-deoxy-3′-O-(N′-methylanthraniloyl)
- IC50
half-maximal inhibitory concentration
- kbasal
basal ATPase activity
- kcat
maximum catalytic activity
- kobs
observed rate constant
- MOPS
3-(N-morpholino) propanesulfonic acid
- PDB ID
Protein Data Bank identification
- Pi
inorganic phosphate
- S1
subfragment 1
- Sf9
Spodoptera frugiperda
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
REFERENCES
- 1.Young P. E., Richman A. M., Ketchum A. S., Kiehart D. P. (1993) Morphogenesis in Drosophila requires nonmuscle myosin heavy chain function. Genes Dev. 7, 29–41 [DOI] [PubMed] [Google Scholar]
- 2.Sellers J. R. (2000) Myosins: a diverse superfamily. Biochim. Biophys. Acta 1496, 3–22 [DOI] [PubMed] [Google Scholar]
- 3.Bloor J. W., Kiehart D. P. (2001) zipper Nonmuscle myosin-II functions downstream of PS2 integrin in Drosophila myogenesis and is necessary for myofibril formation. Dev. Biol. 239, 215–228 [DOI] [PubMed] [Google Scholar]
- 4.Okumura T., Fujiwara H., Taniguchi K., Kuroda J., Nakazawa N., Nakamura M., Hatori R., Ishio A., Maeda R., Matsuno K. (2010) Left-right asymmetric morphogenesis of the anterior midgut depends on the activation of a non-muscle myosin II in Drosophila. Dev. Biol. 344, 693–706 [DOI] [PubMed] [Google Scholar]
- 5.Aldaz S., Escudero L. M., Freeman M. (2013) Dual role of myosin II during Drosophila imaginal disc metamorphosis. Nat. Commun. 4, 1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franke J. D., Montague R. A., Kiehart D. P. (2010) Nonmuscle myosin II is required for cell proliferation, cell sheet adhesion and wing hair morphology during wing morphogenesis. Dev. Biol. 345, 117–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferreira T., Prudêncio P., Martinho R. G. (2014) Drosophila protein kinase N (Pkn) is a negative regulator of actin-myosin activity during oogenesis. Dev. Biol. 394, 277–291 [DOI] [PubMed] [Google Scholar]
- 8.Wheatley S., Kulkarni S., Karess R. (1995) Drosophila nonmuscle myosin II is required for rapid cytoplasmic transport during oogenesis and for axial nuclear migration in early embryos. Development 121, 1937–1946 [DOI] [PubMed] [Google Scholar]
- 9.Mahajan-Miklos S., Cooley L. (1994) Intercellular cytoplasm transport during Drosophila oogenesis. Dev. Biol. 165, 336–351 [DOI] [PubMed] [Google Scholar]
- 10.Martin A. C., Kaschube M., Wieschaus E. F. (2009) Pulsed contractions of an actin-myosin network drive apical constriction. Nature 457, 495–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vasquez C. G., Tworoger M., Martin A. C. (2014) Dynamic myosin phosphorylation regulates contractile pulses and tissue integrity during epithelial morphogenesis. J. Cell Biol. 206, 435–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heissler S. M., Manstein D. J. (2013) Nonmuscle myosin-2: mix and match. Cell. Mol. Life Sci. 70, 1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franke J. D., Montague R. A., Rickoll W. L., Kiehart D. P. (2007) An MYH9 human disease model in flies: site-directed mutagenesis of the Drosophila non-muscle myosin II results in hypomorphic alleles with dominant character. Hum. Mol. Genet. 16, 3160–3173 [DOI] [PubMed] [Google Scholar]
- 14.Limouze J., Straight A. F., Mitchison T., Sellers J. R. (2004) Specificity of blebbistatin, an inhibitor of myosin II. J. Muscle Res. Cell Motil. 25, 337–341 [DOI] [PubMed] [Google Scholar]
- 15.Straight A. F., Cheung A., Limouze J., Chen I., Westwood N. J., Sellers J. R., Mitchison T. J. (2003) Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science 299, 1743–1747 [DOI] [PubMed] [Google Scholar]
- 16.Wang F., Kovacs M., Hu A., Limouze J., Harvey E. V., Sellers J. R. (2003) Kinetic mechanism of non-muscle myosin IIB: functional adaptations for tension generation and maintenance. J. Biol. Chem. 278, 27439–27448 [DOI] [PubMed] [Google Scholar]
- 17.Lehrer S. S., Kerwar G. (1972) Intrinsic fluorescence of actin. Biochemistry 11, 1211–1217 [DOI] [PubMed] [Google Scholar]
- 18.Criddle A. H., Geeves M. A., Jeffries T. (1985) The use of actin labelled with N-(1-pyrenyl)iodoacetamide to study the interaction of actin with myosin subfragments and troponin/tropomyosin. Biochem. J. 232, 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Millar N. C., Geeves M. A. (1983) The limiting rate of the ATP-mediated dissociation of actin from rabbit skeletal muscle myosin subfragment 1. FEBS Lett. 160, 141–148 [DOI] [PubMed] [Google Scholar]
- 20.Sali A., Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 21.Dunbrack R. L., Jr (2002) Rotamer libraries in the 21st century. Curr. Opin. Struct. Biol. 12, 431–440 [DOI] [PubMed] [Google Scholar]
- 22.Yang Z., Lasker K., Schneidman-Duhovny D., Webb B., Huang C. C., Pettersen E. F., Goddard T. D., Meng E. C., Sali A., Ferrin T. E. (2012) UCSF Chimera, MODELLER, and IMP: an integrated modeling system. J. Struct. Biol. 179, 269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heissler S. M., Liu X., Korn E. D., Sellers J. R. (2013) Kinetic characterization of the ATPase and actin-activated ATPase activities of Acanthamoeba castellanii myosin-2. J. Biol. Chem. 288, 26709–26720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furch M., Geeves M. A., Manstein D. J. (1998) Modulation of actin affinity and actomyosin adenosine triphosphatase by charge changes in the myosin motor domain. Biochemistry 37, 6317–6326 [DOI] [PubMed] [Google Scholar]
- 25.Kovács M., Wang F., Hu A., Zhang Y., Sellers J. R. (2003) Functional divergence of human cytoplasmic myosin II: kinetic characterization of the non-muscle IIA isoform. J. Biol. Chem. 278, 38132–38140 [DOI] [PubMed] [Google Scholar]
- 26.Ritchie M. D., Geeves M. A., Woodward S. K., Manstein D. J. (1993) Kinetic characterization of a cytoplasmic myosin motor domain expressed in Dictyostelium discoideum. Proc. Natl. Acad. Sci. USA 90, 8619–8623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furch M., Remmel B., Geeves M. A., Manstein D. J. (2000) Stabilization of the actomyosin complex by negative charges on myosin. Biochemistry 39, 11602–11608 [DOI] [PubMed] [Google Scholar]
- 28.Kuhlman P. A., Bagshaw C. R. (1998) ATPase kinetics of the Dictyostelium discoideum myosin II motor domain. J. Muscle Res. Cell Motil. 19, 491–504 [DOI] [PubMed] [Google Scholar]
- 29.Allingham J. S., Smith R., Rayment I. (2005) The structural basis of blebbistatin inhibition and specificity for myosin II. Nat. Struct. Mol. Biol. 12, 378–379 [DOI] [PubMed] [Google Scholar]
- 30.Cao Y., White H. D., Li X. D. (2014) Drosophila myosin-XX functions as an actin-binding protein to facilitate the interaction between Zyx102 and actin. Biochemistry 53, 350–360 [DOI] [PubMed] [Google Scholar]
- 31.Guzik-Lendrum S., Heissler S. M., Billington N., Takagi Y., Yang Y., Knight P. J., Homsher E., Sellers J. R. (2013) Mammalian myosin-18A, a highly divergent myosin. J. Biol. Chem. 288, 9532–9548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guzik-Lendrum S., Nagy A., Takagi Y., Houdusse A., Sellers J. R. (2011) Drosophila melanogaster myosin-18 represents a highly divergent motor with actin tethering properties. J. Biol. Chem. 286, 21755–21766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sasaki N., Shimada T., Sutoh K. (1998) Mutational analysis of the switch II loop of Dictyostelium myosin II. J. Biol. Chem. 273, 20334–20340 [DOI] [PubMed] [Google Scholar]
- 34.Kiehart D. P., Feghali R. (1986) Cytoplasmic myosin from Drosophila melanogaster. J. Cell Biol. 103, 1517–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagy A., Takagi Y., Billington N., Sun S. A., Hong D. K., Homsher E., Wang A., Sellers J. R. (2013) Kinetic characterization of nonmuscle myosin IIb at the single molecule level. J. Biol. Chem. 288, 709–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heissler S. M., Manstein D. J. (2011) Comparative kinetic and functional characterization of the motor domains of human nonmuscle myosin-2C isoforms. J. Biol. Chem. 286, 21191–21202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lauzon A. M., Tyska M. J., Rovner A. S., Freyzon Y., Warshaw D. M., Trybus K. M. (1998) A 7-amino-acid insert in the heavy chain nucleotide binding loop alters the kinetics of smooth muscle myosin in the laser trap. J. Muscle Res. Cell Motil. 19, 825–837 [DOI] [PubMed] [Google Scholar]
- 38.Spudich J. A. (1994) How molecular motors work. Nature 372, 515–518 [DOI] [PubMed] [Google Scholar]
- 39.Sweeney H. L., Rosenfeld S. S., Brown F., Faust L., Smith J., Xing J., Stein L. A., Sellers J. R. (1998) Kinetic tuning of myosin via a flexible loop adjacent to the nucleotide binding pocket. J. Biol. Chem. 273, 6262–6270 [DOI] [PubMed] [Google Scholar]
- 40.Murphy C. T., Rock R. S., Spudich J. A. (2001) A myosin II mutation uncouples ATPase activity from motility and shortens step size. Nat. Cell Biol. 3, 311–315 [DOI] [PubMed] [Google Scholar]
- 41.Kovács M., Tóth J., Hetényi C., Málnási-Csizmadia A., Sellers J. R. (2004) Mechanism of blebbistatin inhibition of myosin II. J. Biol. Chem. 279, 35557–35563 [DOI] [PubMed] [Google Scholar]
- 42.Zhao F. Q., Padrón R., Craig R. (2008) Blebbistatin stabilizes the helical order of myosin filaments by promoting the switch 2 closed state. Biophys. J. 95, 3322–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramamurthy B., Yengo C. M., Straight A. F., Mitchison T. J., Sweeney H. L. (2004) Kinetic mechanism of blebbistatin inhibition of nonmuscle myosin IIb. Biochemistry 43, 14832–14839 [DOI] [PubMed] [Google Scholar]
- 44.Shu S., Liu X., Korn E. D. (2005) Blebbistatin and blebbistatin-inactivated myosin II inhibit myosin II-independent processes in Dictyostelium. Proc. Natl. Acad. Sci. USA 102, 1472–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teague S. J. (2003) Implications of protein flexibility for drug discovery. Nat. Rev. Drug Discov. 2, 527–541 [DOI] [PubMed] [Google Scholar]
- 46.Seabrooke S., Stewart B. A. (2011) Synaptic transmission and plasticity are modulated by nonmuscle myosin II at the neuromuscular junction of Drosophila. J. Neurophysiol. 105, 1966–1976 [DOI] [PubMed] [Google Scholar]
- 47.Moore J. R., Leinwand L., Warshaw D. M. (2012) Understanding cardiomyopathy phenotypes based on the functional impact of mutations in the myosin motor. Circ. Res. 111, 375–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma X., Adelstein R. S. (2014) The role of vertebrate nonmuscle Myosin II in development and human disease. BioArchitecture 4, 88–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.