Abstract
Implantation requires coordinated interactions between the conceptus and surrounding decidual cells, but the involvement of clock genes in this process is incompletely understood. Circadian oscillations are predicated on transcriptional-translational feedback loops, which balance the activities of the transcriptional activators CLOCK (circadian locomotor output cycles kaput) and brain muscle arnt-like 1 and repressors encoded by PER (Period) and Cryptochrome genes. We show that loss of PER2 expression silences circadian oscillations in decidualizing human endometrial stromal cells (HESCs). Down-regulation occurred between 12 and 24 hours following differentiation and coincided with reduced CLOCK binding to a noncanonical E-box enhancer in the PER2 promoter. RNA sequencing revealed that premature inhibition of PER2 by small interfering RNA knockdown leads to a grossly disorganized decidual response. Gene ontology analysis highlighted a preponderance of cell cycle regulators among the 1121 genes perturbed upon PER2 knockdown. Congruently, PER2 inhibition abrogated mitotic expansion of differentiating HESCs by inducing cell cycle block at G2/M. Analysis of 70 midluteal endometrial biopsies revealed an inverse correlation between PER2 transcript levels and the number of miscarriages in women suffering reproductive failure (Spearman rank test, ρ = −0.3260; P = 0.0046). Thus, PER2 synchronizes endometrial proliferation with initiation of aperiodic decidual gene expression; uncoupling of these events may cause recurrent pregnancy loss.—Muter, J., Lucas, E. S., Chan, Y.-W., Brighton, P. J., Moore, J. D., Lacey, L., Quenby, S., Lam, E. W.-F., Brosens, J. J. The clock protein period 2 synchronizes mitotic expansion and decidual transformation of human endometrial stromal cells.
Keywords: endometrium, circadian rhythm, cell cycle, miscarriage
Mammalian reproduction is dependent on a series of interlocking signals that control the onset of puberty and the timing of ovulation, blastocyst implantation, and parturition (1). The central circadian pacemakers in the suprachiasmatic nucleus (SCN) are responsible for the establishment of daily rhythms entrained by environmental cues (2–4). These SCN pacemakers relay photic information to GnRHs in the hypothalamus, which is cascaded to the ovaries through the release of pituitary gonadotropins and thus control reproductive cyclicity and ovulation (5, 6). In addition, various cell types in the ovary, fallopian tube, and uterus have their own functional molecular clocks that control circadian gene expression (5, 7, 8). At a cellular level, the circadian clockwork is composed of a set of 4 core clock genes and their paralogs that establish robust and stable transcriptional and translational feedback loops (4). BMAL1 [brain muscle arnt-like 1, encoded by aryl hydrocarbon receptor nuclear translocator-like (ARNTL)] and CLOCK (circadian locomotor output cycles kaput) form a heterodimer that binds to specific DNA motifs (E-boxes) in the promoter regions of target genes, including the Period (PER; 1, PER2, and PER3) and the Cryptochrome (CRY; 1 and 2) genes. PER and CRY proteins then accumulate in the cytoplasm and, after a lag period, return to the nucleus to inhibit their own transcription as well as the expression of other genes activated by the CLOCK-BMAL1 heterodimer (9, 10). In addition, clock proteins are subjected to a wide range of posttranslational modifications, including phosphorylation (11), acetylation (12), ubiquitination (13), and sumoylation (14), that act to fine-tune rhythmic oscillations over an ∼24 hour period.
Tissue-specific gene deletions in mice have highlighted the importance of the peripheral clocks in female reproduction. For example, conditional deletion of Bmal1 in pituitary gonadotropes impacts on estrous cycle length (15), whereas in the ovary and myometrium, it perturbs steroidogenesis and the timing of parturition, respectively (16, 17). A key uterine response indispensable for pregnancy is decidualization, a process characterized by the transformation of endometrial stromal cells into specialist secretory cells that provide a nutritive and immune-privileged matrix for the invading blastocyst and subsequent placental formation (18). Previous studies using transgenic rats expressing a destabilized luciferase reporter under the control of the mouse Per2 promoter have shown that decidualization is associated with down-regulation of Per2 and loss of circadian luciferase oscillations (19). Moreover, female mice lacking both Per1 and Per2 reportedly have more implantation sites but fewer live offspring when compared to wild-type animals (20), indicating that these clock proteins are indispensable for optimal utero-placental interactions.
Unlike the rat and other rodents, decidualization of the human endometrium is not under the control of an implanting blastocyst. Instead, this process is driven by the postovulatory rise in progesterone levels and increasing local cAMP production. Consequently, this process is initiated in each ovulatory cycle and enhanced in response to embryonic signals (18, 21). Decidualization is a dynamic and temporally regulated process that commences with proliferative expansion of the stromal cells during the midluteal phase of the cycle (22). Once initiated, differentiating human endometrial stromal cells (HESCs) mount a transient proinflammatory response that renders the endometrium receptive to implantation. This is followed by an anti-inflammatory response, expansion of cytoplasmic organelles, and acquisition of a secretory phenotype that characterizes decidualizing cells during the late-luteal phase of the cycle (23, 24). Disruption of the temporal organization of the decidual response leads to reproductive failure. For example, endometriosis is associated with uterine progesterone resistance, a blunted decidual response, implantation failure, and conception delay (25). Conversely, a disordered proinflammatory decidual response prolongs the window of endometrial receptivity, which in turn increases the risk for out-of-phase implantation and recurrent pregnancy loss (RPL) (23, 24).
This study investigated the role and regulation of clock proteins during decidual transformation of HESCs. As is the case in rodents, we found that circadian oscillations are lost in differentiating HESCs as a consequence of down-regulation of PER2, which occurs between 12 and 24 hours after exposure of a deciduogenic stimulus. Timing of this event is critical because premature loss of PER2 abolishes mitotic expansion of HESCs and predisposes for a highly disorganized decidual response. The importance of this transitional pathway was underscored by analysis of midluteal endometrial biopsies from recurrent miscarriage patients, showing an inverse correlation between PER2 mRNA levels and the number of preceding failed pregnancies.
MATERIALS AND METHODS
Patient selection and endometrial sampling
The study was approved by the National Health Service (NHS) National Research Ethics-Hammersmith and Queen Charlotte’s & Chelsea Research Ethics Committee (1997/5065). Subjects were recruited from the Implantation Clinic, a dedicated research clinic at University Hospitals Coventry and Warwickshire NHS Trust. Written informed consent was obtained from all participants in accordance with the guidelines in The Declaration of Helsinki 2000. Samples were obtained using a Wallach Endocell sampler (Wallach Surgical Devices, Trumbull, CT, USA), starting from the uterine fundus and moving downward to the internal cervical ostium. A total of 57 fresh endometrial biopsies were processed for primary cultures. The average age (±SD) of the participants was 35.1 ± 4.7 years. For analysis of PER2 mRNA expression, 70 additional biopsies stored in RNAlater solution (Sigma-Aldrich, Poole, United Kingdom) were obtained from patients with RPL. Demographic details are summarized in Supplemental Table 1. All endometrial biopsies were timed between 6 and 10 days after the preovulatory LH surge. None of the subjects was on hormonal treatments for at least 3 months before the procedure.
Primary cell culture
HESCs were isolated from endometrial tissues as described previously (26). Purified HESCs were expanded in maintenance medium of DMEM/F-12 containing 10% dextran-coated charcoal-treated fetal bovine serum (DCC-FBS), l-glutamine (1%), and 1% antibiotic-antimycotic solution. Confluent monolayers were decidualized in DMEM/F-12 containing 2% DCC-FBS with 0.5 mM 8-bromoadenosine-cAMP (8-br-cAMP; Sigma-Aldrich) with or without 10−6 M medroxyprogesterone acetate (MPA; Sigma-Aldrich) to induce a differentiated phenotype. For synchronization, dexamethasone (Sigma-Aldrich) was used at 100 nM for 30 minutes. Actinomycin D (Sigma-Aldrich) was used at a final concentration of 2 μM in DMSO. All experiments were carried out before the third cell passage.
Real-time quantitative RT-PCR
Total RNA was extracted from HESC cultures using RNA STAT-60 (AMS Biotechnology, Abingdon, United Kingdom). Equal amounts of total RNA (1 µg) were treated with DNase and reverse transcribed using the QuantiTect Reverse Transcription Kit (Qiagen, Manchester, United Kingdom), and the resulting cDNA was used as template in quantitative RT-PCR (qRT-PCR) analysis. Detection of gene expression was performed with Power SYBR Green Master Mix (Life Technologies, Paisley, United Kingdom) and the 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The expression levels of the samples were calculated using the ΔCt method, incorporating the efficiencies of each primer pair. The variances of input cDNA were normalized against the levels of the L19 housekeeping gene. All measurements were performed in triplicate. Melting curve analysis confirmed amplification specificity. Primer sequences used are as follows: CLOCK, forward 5′-gac aaa gcg aaa aga gta tct ag-3′ and reverse 5′-cat ctt tct agc att acc agg aa-3′; BMAL1, forward 5′-gac att cct tcc agt ggc cta-3′ and reverse 5′-tac cta tgt ggg ggt tct cac-3′; CRY1, forward 5′-cat cct gga ccc ctg gtt-3′ and reverse 5′-cac tga agc aaa aat cgc c-3′; CRY2, forward 5′-ctg ttc aag gaa tgg gga gtg-3′ and reverse 5′-ggt cat aga ggg tat gag aat tc-3′; PER1, forward 5′-atg gtt cca ctg ctc cat ctc-3′ and reverse 5′-ccg gtc agg acc tcc tc-3′; PER2, forward 5′-gtc cga aag ctt cgt tcc aga-3′ and reverse 5′-gtc cac atc ttc ctg cag tg-3′; and prolactin (PRL), forward 5′-aag ctg tag aga ttg agg agc aaa c-3′ and reverse 5′-tca gga tga acc tgg ctg act a-3′. In the actinomycin D experiments, PER2 mRNA half-life was calculated using t1/2 = 0.693/k, where k is the slope derived from the linear equation lnC = lnC0 − kt, and where C is the relative level of PER mRNA in HESCs (27).
Western blot analysis
Whole-cell protein extracts were prepared by lysing cells in RIPA buffer containing protease inhibitors (cOmplete, Mini, EDTA-free; Roche, Welwyn Garden City, United Kingdom). Protein yield was quantified using the Bio-Rad Protein Assay Dye Reagent Concentrate (Bio-Rad Laboratories, Hemel Hempstead, United Kingdom). Equal amounts of protein were separated by SDS-PAGE before wet transfer onto PVDF membrane (GE Healthcare, Buckinghamshire, United Kingdom). Nonspecific binding sites were blocked by overnight incubation with 5% nonfat dry milk in Tris-buffered saline with 1% Tween 20 [130 mM NaCl, 20 mM Tris (pH 7.6), and 1% Tween 20]. The following primary antibodies were purchased from Abcam (Cambridge, United Kingdom): anti-CLOCK (catalog number ab3517, diluted 1:3000); anti-BMAL1 (ab3350, 1:375); anti-CRY1 (ab54649, 1:500); anti-CRY2 (ab38872, 1:2000); anti-PER1 (ab3443, 1:300); anti-PER2 (ab179813, 1:300); and anti-β-actin (ab8226, 1:10,000). Protein complexes were visualized with ECL Plus chemiluminescence (GE Healthcare). The Western blots are collated in Supplemental Fig. 1.
Transient transfection
Primary HESCs were transfected with small interfering RNA (siRNA) by jetPRIME Polyplus transfection kit (VWR International, Lutterworth, United Kingdom). For gene silencing, undifferentiated HESCs were transiently transfected with 50 nM PER2-siGENOME SMARTpool or siGENOME Non-Targeting siRNA Pool 1 (Dharmacon, GE Healthcare). Transfection studies were performed in triplicate and repeated on primary cultures from 3 subjects.
Chromatin immunoprecipitation
HESCs in 10 cm culture dishes were fixed with 1% formaldehyde for 10 minutes at 37°C. Fixation was stopped with 125 mM glycine, and nuclei were isolated by incubating at 4°C for 10 minutes in 1 ml Swelling buffer [25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, and 0.1% NP40 alternative]. The cells were scraped, homogenized, and centrifuged for 3 minutes at 16,000 × g at 4°C. Pelleted nuclei were resuspended in 500 μl sodium dodecyl sulfate (SDS) lysis buffer [1% SDS, 1% Triton X-100, 0.5% deoxycholate, 10 mM EDTA, and 500 mM Tris-HCl (pH 8.1)] and sonicated for 30 minutes at 4°C on high power in a Diagenode Bioruptor sonicator (Diagenode, Liege, Belgium). The resulting suspension was centrifuged for 15 minutes at 16,000 × g at 4°C and supernatant diluted in immunoprecipitation buffer [0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl (pH 8.1), and 167 mM NaCl] and then precleared at 4°C for 3 hours with Protein A Dynabeads (Life Technologies, Carlsbad, CA, USA). The chromatin was then complexed overnight at 4°C with the appropriate antibody bound to Protein A Dynabeads. Post complexing, samples were washed with the following buffers before eluting the chromatin with 250 μl Elution buffer (1% SDS and 100 mM NaHCO3) and incubating at room temperature for 15 minutes: low-salt buffer [0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl (pH 8.1), and 150 mM NaCl]; high-salt buffer [0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl (pH 8.1), and 500 mM NaCl]; LiCl buffer [250 mM LiCl, 1% NP40 alternative, 1% deoxycholate, 1 mM EDTA, and 10 mM Tris-HCl (pH 8.1)]; and Tris-EDTA buffer [10 mM Tris-HCl (pH 8) and 1 mM EDTA]. A total of 200 mM NaCl was added to reverse cross-link the proteins and the DNA. After an overnight incubation at 65°C, 10 mM EDTA, 40 mM Tris-HCl (pH 8), and 40 μg/ml protease K (Sigma-Aldrich) were added, and the sample was incubated for a further hour at 55°C before proceeding with the DNA purification using the QIAquick PCR purification kit (Qiagen). Buffers were supplemented with protease and phosphatase inhibitor cocktails (Sigma-Aldrich) and 10 mM sodium butyrate (28). The following antibodies were used in the chromatin immunoprecipitation (ChIP) experiments: CLOCK (Abcam), and as negative control, the rabbit polyclonal anti-mouse IgG (M7023; Sigma-Aldrich). The purified DNA was amplified by qRT-PCR using the following primers: PER2 E-box, forward 5′-cag at gaga cgg agt cgc-3′ and reverse 5′-ccc aca gct gca cgt atc-3′; and PER1 E-box, forward 5′-cac gtg cgc ccg tgt gt-3′ and reverse 5′-ccg att ggc tgg gga tct c-3′.
RNA sequencing and data analysis
Total RNA was extracted using RNA STAT-60 from primary HESC cultures first transfected with either PER2 or nontargeting (NT) siRNA and then decidualized with 8-br-cAMP and MPA for 24 hours. There were 3 biologic repeat experiments performed. RNA quality was analyzed on an Agilent 2100 Bioanalyzer (Agilent Technologies, Wokingham, United Kingdom). RNA integrity number score for all samples was ≥8.9. Transcriptomic maps of single-end reads were generated using Bowtie 2.2.3 (29), SAMtools 0.1.19, and TopHat 2.0.12 (30) against the University of California, Santa Cruz, hg19 reference transcriptome (2014) from the Illumina iGenomes resource using the fr-firststrand setting. Gene counts were estimated using HTSeq 0.6.1 (www-huber.embl.de/users/anders/HTSeq/). Transcripts per million (TPM) were calculated as recently described (31). Count data from the TopHat-HTSeq pipeline were analyzed using 2 different methods for differential expression detection, i.e., DESeq and edgeR (32, 33). Expression was considered to be significantly different if the false discovery rate value (edgeR) or adjusted P value (DESeq) was <0.01. Differentially expressed genes were retained if they were detected by at least 2 of the methods used. Expression data have been submitted to the Gene Expression Omnibus (GEO) repository (GSE62854).
In vitro colony-forming assay
Transfected HESCs were seeded at a clonal density of 50 cells/cm2 (to ensure equal loading) onto fibronectin-coated 60 mm culture dishes and cultured in growth medium: DMEM/F-12 containing 10% DCC-FBS, 1% l-glutamine (Invitrogen, Paisley, United Kingdom), 1% antibiotic-antimycotic solution (Invitrogen), insulin (2 µg/ml) (Sigma-Aldrich), estradiol (1 nM) (Sigma-Aldrich), and basic fibroblast growth factor (10 ng/ml) (Merck Millipore, Watford, United Kingdom). The first medium change was after 7 days. Colonies were monitored microscopically to ensure that they were derived from single cells. Cultures were terminated at 10 days and stained with hematoxylin.
Cell cycle analysis, viability, and proliferation assays
For cell cycle analysis, HESCs in suspension were washed with PBS, fixed with cold 70% ethanol, ribonuclease-A (Qiagen) treated, stained with propidium iodide (Sigma-Aldrich), and subjected to flow cytometry analysis. Cell cycle distribution was assessed using FlowJo software (Ashland, OR, USA).
Real-time adherent cell proliferation was determined by the label-free xCELLigence Real-Time Cell Analyzer (RTCA) DP instrument (Roche Diagnostics Gesellschaft mit beschränkter Haftung, Basel, Switzerland), which utilizes specialized microtiter culture plates containing an interdigitized gold microelectrode on which cells attach and proliferate. Cell contact with the electrode increases the impedance across these gold arrays and reported as an arbitrary “cell index” value as an indication of confluency and adherence (34). HESCs were seeded into 16-well plates (E-plate-16; Roche Diagnostics Gesellschaft mit beschränkter Haftung) at a density of 10,000 cells per well and cultured in 10% DCC-FBS until ∼80% confluent. The RTCA DP instrument was placed at 37°C in a humidified environment with 95% air and 5% CO2, and cells were decidualized following transient transfection as per standard protocols. Individual wells within the E-plate-16 were referenced immediately and monitored first every 15 minutes for 3 hours and then hourly for 4 days. Changes in cell index were captured and analyzed using the RTCA Software v1.2 supplied with the instrument.
Statistical analysis
Data were analyzed with the statistical package GraphPad Prism 6 (GraphPad Software Incorporated, La Jolla, CA, USA). Unpaired Student’s t test, Mann–Whitney U test, Spearman rank correlation, and 1-way ANOVA with post hoc Tukey’s test were used when appropriate. Statistical significance was assumed when P < 0.05.
RESULTS
Loss of circadian oscillations upon decidualization of HESCs
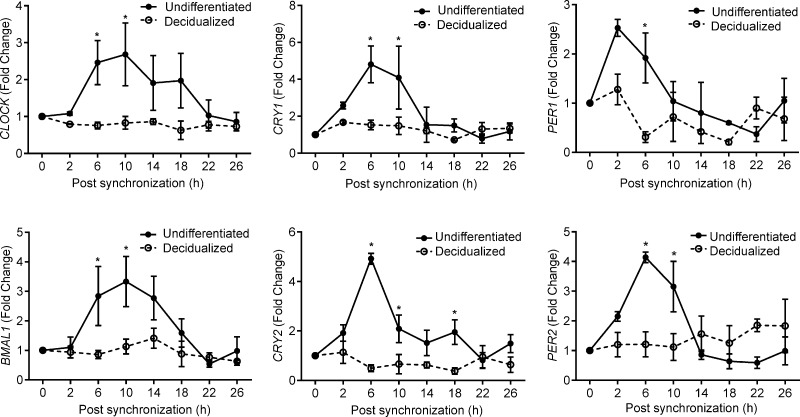
Decidualization of stromal cells in the rat uterus is associated with loss of circadian rhythms (19). We speculated that this phenomenon may aid in synchronizing maternal and embryonic gene expression at implantation and, thus, be conserved. To test this hypothesis, we measured the transcript levels of 6 core clock genes, i.e., CLOCK, BMAL1 (ARNTL), CRY1, CRY2, PER1, and PER2, in primary undifferentiated HESCs and cells decidualized for 4 days. Following dexamethasone synchronization, all 6 clock genes exhibited circadian regulation in undifferentiated cultures with the amplitude of gene expression varying up to 5-fold over a 26 hour period (Fig. 1). By contrast, expression was uniformly aperiodic in decidualizing cultures, confirming that differentiation of HESCs is also associated with loss of rhythmicity.
Figure 1.
Decidualization silences circadian oscillations in HESCs. Primary undifferentiated HESCs and cultures first decidualized with 8-br-cAMP and MPA for 4 days were treated with dexamethasone for 30 minutes, and transcript levels of 6 core clock genes were measured at the indicated time points. The data show relative change (mean ± sem) in transcript levels in 3 independent primary cultures. *P < 0.05.
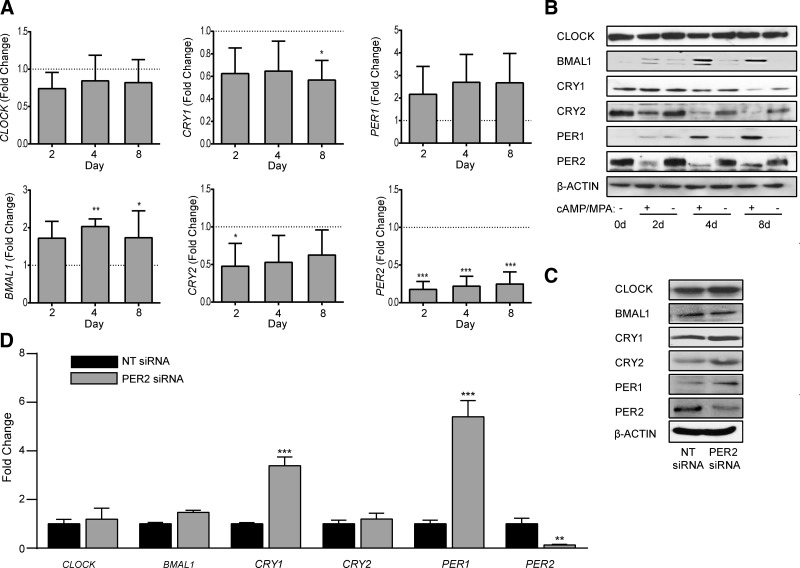
To investigate the underlying mechanism, we profiled the expression of the same clock genes in undifferentiated HESCs and cells decidualized for 2, 4, or 8 days. Decidualization elicited modest but consistent changes in the expression of several transcripts, including up-regulation of BMAL1 mRNA levels and down-regulation of CRY1 and CRY2 expression (Fig. 2A). The changes at transcript level were also apparent at protein level (Fig. 2B). Although up-regulation of PER1 transcripts did not reach statistical significance, expression of the protein gradually increased as the decidual process unfolded. By contrast, CLOCK expression remained constant over the 8 day time course. The most striking observation, however, was the rapid and profound inhibition of PER2 expression with transcript levels falling by 80% within 2 days of differentiation (Fig. 2A). Western blot analysis confirmed the dramatic decline in PER2 levels upon decidualization (Fig. 2B). Furthermore, PER2 mobility on SDS-PAGE became more focused and noticeably enhanced, suggesting that decidualization also impacts on the posttranslational modification status of this component of the circadian machinery.
Figure 2.
Expression of clock genes in decidualizing HESCs. A) The transcripts levels of 6 core clock genes were measured in undifferentiated HESCs and cells decidualized with 8-br-cAMP and MPA for 2, 4, or 8 days. The data show expression (mean ± sem) relative to that in undifferentiated cells (dotted line) in 3 independent primary cultures. B) Total protein lysates from parallel cultures were subjected to Western blotting. β-Actin served as a loading control. C) Western blot analysis of total cell lysates obtained 48 hours following transfection of primary cultures with NT or PER2 siRNA. D) mRNA levels of core clock genes were also determined 48 hours following transfection of primary cultures with NT or PER2 siRNA. At 2 days after transfection, the efficacy of siRNA-mediated knockdown of PER2 was 87 and 53% at mRNA and protein level, respectively. The data show relative change (mean ± sem) in transcript levels in 3 independent primary cultures. *P < 0.05; **P < 0.01; ***P < 0.001.
Because circadian oscillations are predicated on autoregulatory feedback loops, we postulated that PER2 knockdown in undifferentiated HESCs would recapitulate the changes in core clock components associated with decidualization. To test this hypothesis, primary undifferentiated HESCs were transfected with either PER2 or NT siRNA and harvested after 4 days. Although PER2 knockdown resulted in a reciprocal up-regulation of PER1, it did not recapitulate the other decidual changes, suggesting that multiple clock regulators are modulated in response to HESC differentiation (Fig. 2C, D).
Mechanism of PER2 inhibition
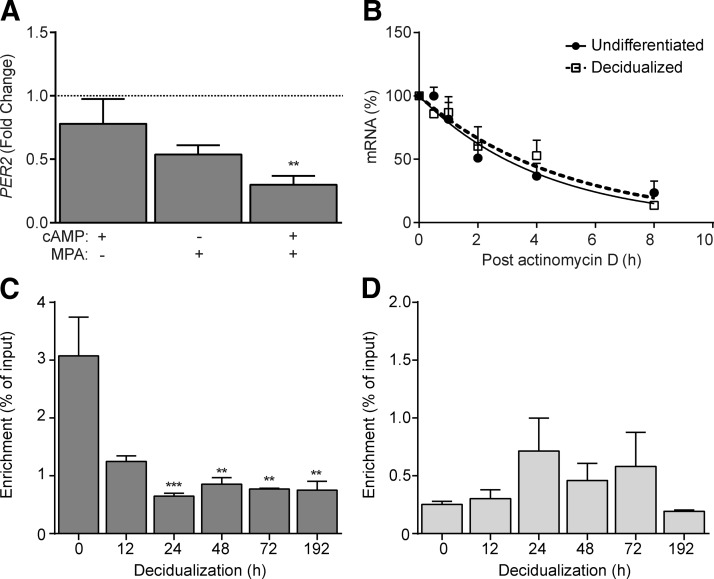
To provide insight into the mechanism of PER2 down-regulation, we first treated primary HESCs with 8-br-cAMP, MPA, or a combination. The decline in PER2 expression was more pronounced with MPA than 8-br-cAMP (Fig. 3A), although the level of inhibition was not statistically significant with either treatment. By contrast, combined treatment had an additive effect, reducing PER2 expression by 70% after 48 hours when compared to vehicle-treated control (P < 0.01). Mining of available ChIP data sets revealed no changes in the levels of trimethylated lysine 27 or lysine 4 of histone 3 (H3K27me3 and H3K4me3, respectively) at the PER2 promoter upon decidualization (data not shown). In the absence of obvious epigenetic changes, we speculated that PER2 could be regulated at the level of RNA stability. To test this hypothesis, undifferentiated and decidualized HESCs were treated with actinomycin D, a potent transcription inhibitor, for 0.5, 1, 2, 4, or 8 hours. As shown in Fig. 3B, the half-life of PER2 transcripts was comparable in undifferentiated and decidualizing cells (2.93 versus 3.39 hours, respectively; P > 0.05).
Figure 3.
Regulation of PER2 in decidualizing HESCs. A) Primary HESC cultures were treated with 8-br-cAMP, MPA, or a combination for 48 hours and PER2 transcript levels measured. The data show relative change (mean ± sem) in mRNA levels compared to vehicle-treated undifferentiated cultures established from 3 different biopsies. B) Primary cultures remained undifferentiated or were decidualized for 48 hours prior to treatment with 5 μg/ml actinomycin D. RNA was extracted at the indicated time points and subjected to qRT-PCR analysis. C) Binding of CLOCK to E2, a noncanonical E-box enhancer in the PER2 promoter, was assessed by ChIP in 3 independent primary cultures, either undifferentiated (0 hour) or decidualized for the indicated time points. The data show relative enrichment compared to input. D) In parallel, CLOCK binding to a regulatory E-box element (E5) in the PER1 promoter was determined. The data show the mean ± sem. **P < 0.01; ***P < 0.001.
In the mouse, Per2 expression is critically dependent on constitutive binding of CLOCK to a noncanonical 5′-CACGTT-3′ E-box enhancer, termed E2, located 20 base pairs (bp) upstream of the transcriptional start site (35). The E2 enhancer and the extended CLOCK:BMAL1 M34 core binding site are highly conserved in humans, raising the possibility that disruption of CLOCK binding disables PER2 transcription in decidualizing cells. ChIP analysis using a CLOCK antibody showed that decidualization was associated with a rapid and sustained loss of CLOCK binding at this locus (amplicon −301 to −162 bp). In 3 independent time course cultures, treatment with MPA and 8-br-cAMP for 24 hours was sufficient to reduce CLOCK binding to E2 in PER2 promoter by 59%, and this level of repression was maintained over an 8 day time course (Fig. 3C). By contrast, CLOCK binding to the E-box (E5; amplicon −142 to −54 bp) in the PER1 promoter remained constant throughout the time course (Fig. 3D).
PER2 knockdown silences circadian oscillations and disrupts HESC decidualization
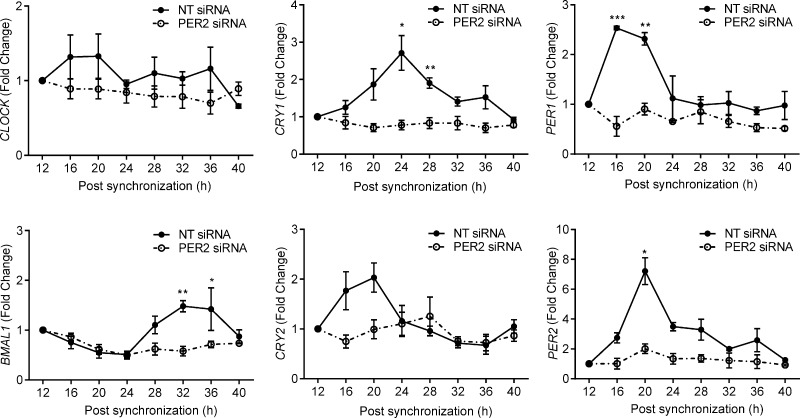
Next, we examined whether PER2 knockdown in undifferentiated HESCs would suffice to disrupt circadian rhythm generation. Paired primary cultures transfected with either NT or PER2 siRNA were synchronized with dexamethasone, and total RNA was harvested at 4 hours intervals over a 28 hour period. Cells transfected with NT siRNA demonstrated robust circadian oscillations in the 6 core clock genes. PER2 knockdown resulted in a nonoscillating expression profile in undifferentiated HESCs (Fig. 4), indicating that down-regulation of this clock protein accounts for the loss of autonomous circadian oscillations in decidual cells.
Figure 4.
PER2 knockdown in HESCs causes loss of rhythmic expression in core clock genes. Primary HESC cultures were transfected with either NT or PER2 siRNA. After 48 hours, the cultures were synchronized with dexamethasone, and total RNA was harvested at indicated time points. Transcript levels of core clock genes were analyzed by qRT-PCR in 3 independent experiments. The data show the mean ± sem. *P < 0.05; **P < 0.01; ***P < 0.001.
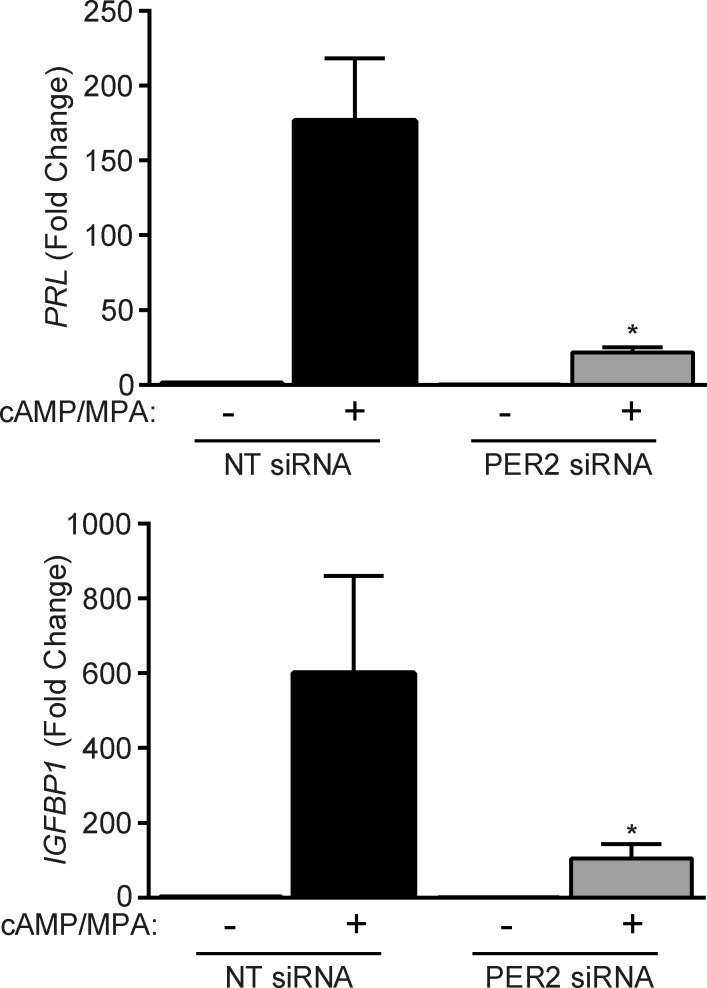
We speculated that loss of PER2-dependent circadian oscillations may sensitize undifferentiated HESCs to deciduogenic cues. However, this was not the case. Instead, PER2 knockdown severely compromised the induction of PRL and IGFBP1 (IGF-binding protein-1), 2 cardinal decidual marker genes, in response to cAMP and MPA signaling (Fig. 5). Thus, whereas PER2 down-regulation is a striking feature of decidual cells, this clock protein seems nevertheless essential for the initial responsiveness of HESCs to differentiation signals.
Figure 5.
PER2 is required for the induction of decidual marker genes. Primary HESCs were transfected with NT or PER2 siRNA. The cultures remained untreated or were decidualized for 48 hours. The data show relative induction (mean ± sem) of the decidual marker genes PRL and IGFBP1 in 3 independent primary cultures. *P < 0.05.
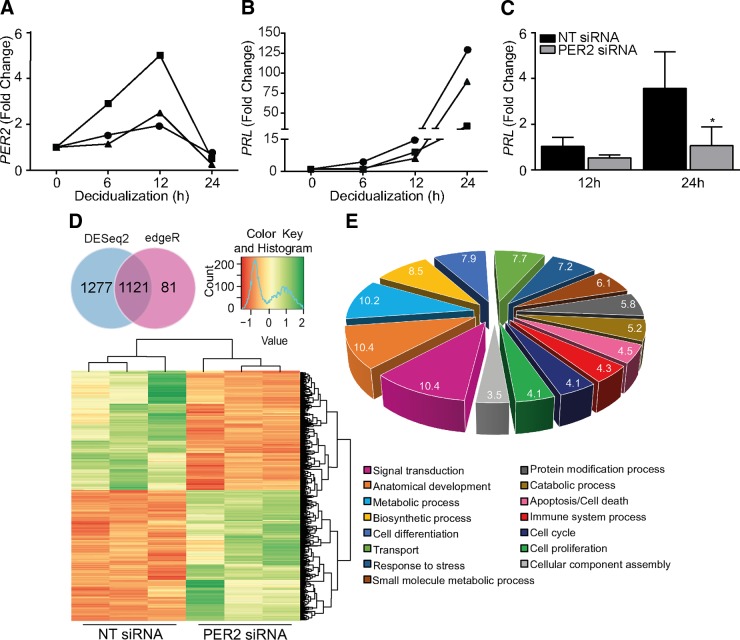
Based on the kinetics of cAMP-dependent induction of the decidual PRL promoter activity, HESC differentiation has been shown to be a biphasic process, characterized by an initial rapid but modest response, which is followed by an accelerated rise in promoter activity after 12 hours of stimulation (36). Hence, we examined the kinetics of PER2 inhibition and PRL induction in a short time course. PER2 transcript levels transiently increased in response to 8-br-cAMP and MPA, with levels peaking at 12 hours, which was followed by a sharp drop by 24 hours (Fig. 6A). As anticipated, the rise in PRL mRNA was modest within the first 12 hours of stimulation but then accelerated in concert with the drop in PER2 transcript levels (Fig. 6B). Intriguingly, PER2 knockdown in HESCs had no significant impact on the expression of PRL transcripts in the first 12 hours of stimulation but inhibited the accelerated induction of this decidual marker between 12 and 24 hours (Fig. 6C).
Figure 6.
Premature PER2 down-regulation leads to a disorganized decidual response. A) The kinetics of PER2 expression in response to 8-br-cAMP and MPA treatment were monitored in 3 independent cultures at the indicated time points by qRT-PCR. PER2 transcript levels were normalized to those in undifferentiated cells. B) Kinetics of PRL induction in the same short time course. C) Primary HESCs were transfected with NT or PER2 siRNA. After 48 hours, the cultures were treated with 8-br-cAMP and MPA for 12 or 24 hours. PRL mRNA levels were normalized to those in undifferentiated cells. The data show mean fold change (±sem) in 3 independent cultures. *P < 0.05. D) Venn diagram comparison of differentially expressed transcripts, identified by DESeq and edgeR, in primary HESCs decidualized for 24 hours following transfection with either NT or PER2 siRNA. Numbers represent differentially expressed transcripts with nonzero counts in 3 independent experiments. Clustered heat map of top-ranked differentially expressed transcripts is also shown. E) Graphic representation of the top 15 GO slim annotations of differentially expressed genes.
To investigate further the consequences of PER2 knockdown on activation of the decidual transcriptome, total RNA harvested from 3 independent unsynchronized HESC cultures, first transfected with either PER2 or NT siRNA and then decidualized for 24 hours, was subjected to RNA sequencing. On average, 25 million single-end reads were sequenced per sample. By combining 2 different analysis tools, DESeq and edgeR, we identified 1121 genes that were significantly altered upon PER2 knockdown, 572 (51%) of which were up-regulated and 549 (49%) down-regulated. To assess further the relatedness of the cultures, we calculated Z scores of the TPM values for the differentially expressed genes. A heat map of this association is depicted in Fig. 6D.
Among the down-regulated genes were PRL and IGFBP1, confirming the initial qRT-PCR analysis. More puzzling, however, was the observation that PER2 knockdown up-regulated various other genes essential for decidual transformation in HESCs, including genes encoding key transcription factors [e.g., CREM, CEBPβ, CEBPα, and NURR1 (37, 38)], kinases and phosphatases (e.g., SGK1 and MKP1) (39, 40), the cell surface receptor for IL33 (IL1RL1, also known as ST2) (23), and BMP2, a key decidual morphogen (41). Thus, rather than preventing or attenuating differentiation, premature down-regulation of PER2 predisposes for a disordered decidual program. Also striking was the induction of genes coding metabolic regulators, including peroxisome proliferator-activated receptor γ (PPARG) and PPARG coactivator 1-α, following siRNA-mediated PER2 inhibition.
PER2 prevents clonal expansion of HESCs by inducing G2/M arrest
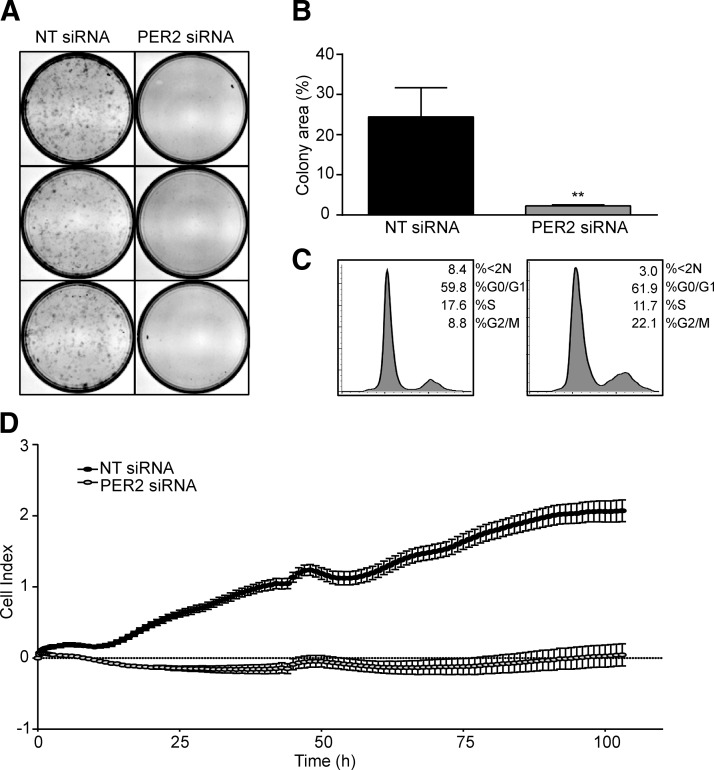
Gene ontology (GO) analysis (GO slim) identified “cell cycle” and “proliferation” among the biologic processes prominently affected by PER2 knockdown (Fig. 6E). It is well established that stromal cells must undergo mitotic expansion prior to full decidualization (22). Based on the sequencing data, we speculated that premature PER2 inhibition deregulates decidual gene expression by interfering with the proliferative potential of HESCs. In agreement, the ability of HESCs to form colonies when plated at low density was severely compromised upon PER2 knockdown (Fig. 7A, B). Flow cytometry analysis of 3 independent primary cultures revealed accumulation of HESCs in G2/M phase of the cycle following transfection with PER2 siRNA when compared to NT siRNA (mean ± sem, 22.1 ± 1.4% versus 8.8% ± 2.3, respectively; P = 0.03), which was accompanied by a reduction of cells in S phase (11.7 ± 1.1% versus 17.6 ± 1.0%, respectively; P = 0.03) (Fig. 7C). Interestingly, the apoptotic cell fraction (<2 N) tended to be lower upon transfection with PER2 siRNA when compared to NT siRNA. Real-time monitoring of cell proliferation over 100 hours using microelectronic sensor technology (xCELLigence) confirmed that siRNA-mediated PER2 knockdown results in complete growth inhibition of HESC cultures (Fig. 7D). These results show that the lack of mitotic expansion observed upon PER2 knockdown is, at least in part, due to imposition of cell cycle block at G2/M. This observation fits well with the RNA-sequencing data, showing that 52 of the 73 cell cycle-related genes perturbed upon PER2 knockdown are involved in G2/M transition (Supplemental Table 2).
Figure 7.
PER2 knockdown blocks mitotic expansion of HESCs. A) Colony-forming assays of 3 independent primary cultures first transfected with either NT or PER2 siRNA. B) Total colony area as measured by ImageJ analysis (U.S. National Institutes of Health, Bethesda, MD, USA), and the data represent mean colony area (±sem). C) Representative gated cell cycle histograms obtained 48 hours after transfection of primary HESCs with either NT or PER2 siRNA. D) Real-time monitoring of cell growth over 100 hours following transfection with NT or PER2 siRNA. **P < 0.01.
Midluteal endometrial PER2 expression in recurrent miscarriage
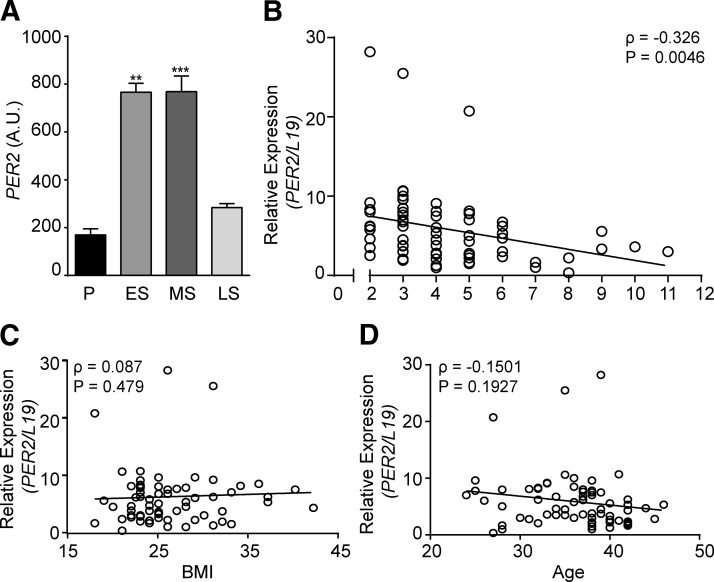
A search of the GEO database revealed that endometrial PER2 transcript levels [GEO profiles; identification (ID) 24460199] increase 4-fold between the proliferative and early-secretory phase of the cycle (Fig. 8A). Elevated levels are maintained during the midluteal phase of the cycle but then fall in concert with a sharp increase in the expression of decidual marker genes, including IGFBP1 (ID 24460250) (42). Next, we examined PER2 expression in midluteal endometrial biopsies obtained from 70 women with ovulatory cycles attending a dedicated miscarriage clinic. All patients suffered consecutive miscarriages, ranging between 2 and 11 (median ± SD, 4 ± 2). Most losses occurred in the first trimester of pregnancy. Within this cohort, endometrial PER2 transcript levels correlated inversely with the number of previous miscarriages (Spearman rank test, ρ = −0.3260; P = 0.0046) (Fig. 8B). By contrast, no association was found between PER2 expression and other demographics relevant to miscarriages such as age (ρ = 0.01070; P = 0.9) or body mass index (ρ = −0.1501; P = 0.2) (Fig. 8C, D).
Figure 8.
Endometrial PER2 expression in RPL. A) PER2 transcripts, expressed in arbitrary units (A.U.), in proliferative (P), early-secretory (ES), midsecretory (MS), and late-secretory (LS) human endometrium. Expression levels were derived from in silico analysis of publicly available microarray data (GEO profiles; ID 24460199) (42). B) Correlation between PER2 transcript levels in midluteal endometrial biopsies and the number of preceding miscarriages in 70 subjects with RPL. C) Correlation between PER2 transcript levels in midluteal endometrial biopsies and body mass index (BMI) in the RPL cohort. D) Correlation between PER2 transcript levels in midluteal endometrial biopsies and age of cohort subjects. **P < 0.01; ***P < 0.001.
DISCUSSION
Circadian rhythms permeate a vast array of physiologic processes by maintaining tissue homeostasis in anticipation of environmental change (43). We show that oscillations of the core clock machinery are halted, or potentially “paused,” upon decidualization of HESCs. Decidualization is a tightly spatiotemporally controlled process that commences with proliferative expansion of stromal cells in the superficial endometrial layer during the midluteal phase of the cycle. Differentiating cells then transit through defined phenotypic changes that sequentially control endometrial receptivity, embryo selection, and, ultimately, either menstrual shedding or resolution of pregnancy (18). Human preimplantation embryos do not express circadian genes apart from maternal transcripts, but these are degraded prior to the implantation-competent blastocyst stage (1). A parsimonious explanation for the silencing of circadian oscillations in both conceptus and surrounding decidualizing cells is that it synchronizes embryo-maternal interactions. Although PER2 levels drop significantly, all components of the core clock machinery remain expressed in nonoscillatory decidual cells, suggesting that these cells are poised to resume rhythmicity, possibly entrained by embryonic cues.
Down-regulation of PER2 in the endometrium is precisely timed. In the rat, it marks the transition from oscillatory receptivity to the nonoscillatory decidual (postreceptive) endometrium (19). This pattern of expression is recapitulated in the human uterus with the decline in PER2 transcript levels heralding the progression from mid-to-late-luteal endometrium. In primary culture, down-regulation of PER2 mRNA occurred between 12 and 24 hours following exposure to a standard deciduogenic treatment. Again, inhibition of this clock gene marked the onset of an important transitional phase, characterized by increases in reactive oxygen production, altered redox signaling, and accelerated expression of decidual marker genes (26, 44). Previous studies reported that PER2 is acutely responsive to hormonal and other signals that converge onto a cAMP-response element in its promoter region (45–47). This pathway provides a likely explanation for the initial transient rise in PER2 transcript levels in differentiating HESCs. However, sustained expression and circadian oscillations in peripheral tissues require constitutive binding of a transcriptional complex containing CLOCK to the highly conserved E2 enhancer in the proximal PER2 promoter. In agreement, we found that loss of PER2 expression in decidualizing HESCs coincided with attenuated binding of CLOCK to the E2 enhancer. This was not accounted for by a general reduction in the DNA-binding activity of the CLOCK:BMAL1 heterodimer as exemplified by the ChIP analysis of the PER1 promoter. The precise mechanism of selective silencing of PER2 in differentiating HESCs remains to be defined. One possibility is that PER2 repression in decidualizing cells reflects accumulation of p53 (48), which in other cell systems has been shown to disrupt the binding of CLOCK to the E2 enhancer (49).
PER2 differs from other core clock proteins in that it exhibits several structural features of steroid receptor coregulators (50), including 2 conserved nuclear receptor-binding motifs (LXXLL). Furthermore, PER2 has been shown to bind estrogen receptor-α (ERα) and antagonize estrogen-dependent proliferation of breast cancer cell lines, at least in part by enhancing receptor degradation (51, 52). It acts as a transcriptional corepressor by recruiting histone deacetylases (e.g., HDAC2) and components of the polycomb repressor complex (e.g., EZH2 and SUZ12) to promoter regions of target genes (53). The kinetics of PER2 down-regulation in differentiating HESCs coupled to the enhanced activation of marker genes, such as PRL and IGFBP1, suggest that this clock protein is a major repressor of decidual gene expression, either through an epigenetic mechanism, as a putative corepressor of the progesterone receptor, or possibly through a combination of these mechanisms. Yet, PER2 knockdown did not sensitize HESCs to deciduogenic signals but resulted in a grossly disordered differentiation response. These seemingly contradictory findings are explained by the imposition of G2/M block upon PER2 knockdown, which prevented the obligatory mitotic expansion of stromal cells prior to the onset of decidual gene expression. This observation is itself intriguing because PER2 is widely regarded to be a tumor suppressor (54, 55). As mentioned previously, PER2 knockdown accelerates proliferation of ERα-positive breast cancer cells (51). Notably, endometrial PER2 transcript levels are also low during the proliferative phase of the cycle. In leukemia cell lines, PER2 overexpression induces growth arrest in the G2/M phase of the cell cycle by inhibiting c-MYC and cyclin B1 and up-regulating p53 (54). Thus, the ability of PER2 to promote or inhibit cell cycle progression seems to be dependent on hormonal inputs within a cell-specific context.
Miscarriage is the most common complication of pregnancy. One in 7 recognized pregnancies ends in miscarriage during the first trimester, and 1–2% fail between 13 and 24 weeks gestation (18). The American Society for Reproductive Medicine defines RPL as ≥2 consecutive miscarriages before the fetus reaches viability. Affected couples are routinely screened for various anatomic, endocrine, immunologic, thrombophilic, and genetic risk factors, although the value of these investigations is highly contentious. In a majority of patients, no underlying associations are found, and conversely, many subclinical disorders or risk factors perceived to cause miscarriages are also prevalent in women with uncomplicated pregnancies. Embryonic chromosomal imbalances are estimated to account for approximately 50% of all miscarriages; but importantly, the incidence of euploidic fetal loss increases with each additional miscarriage, whereas the likelihood of a future successful pregnancy decreases (56–58). In other words, the likelihood of a causal maternal factor increases with each additional pregnancy loss.
Several lines of evidence from experimental as well as epidemiologic studies suggest that an aberrant decidual response predisposes for subsequent pregnancy failure (18, 23, 39). The observation of a significant inverse correlation between midluteal PER2 transcript levels and the number of previous miscarriages strongly infers that deregulation of this core clock gene increases the likelihood of persistent miscarriages. Additional studies are warranted to assess the role of PER2 in the endometrial epithelial cells and to examine the tissue distribution of this clock protein in patients with RPL and control subjects. Interestingly, a recent systematic review and meta-analysis reported a significant association between night shifts and miscarriages (adjusted odds ratio, 1.41; 95% confidence interval, 1.22–1.63) (59). Taken together, these observations demonstrate that disruption of both central as well as peripheral circadian outputs predisposes for reproductive failure.
Supplementary Material
Acknowledgments
This work was supported by the Biomedical Research Unit in Reproductive Health, a joint initiative between the University Hospitals Coventry and Warwickshire National Health Service Trust and Warwick Medical School. J.M. is the recipient of a Warwick Chancellor’s studentship. The authors declare no conflicts of interest.
Glossary
- 8-br-cAMP
8-bromoadenosine-cAMP
- ARNTL
aryl hydrocarbon receptor nuclear translocator-like
- BMAL1
brain muscle arnt-like 1
- bp
base pair
- ChIP
chromatin immunoprecipitation
- CLOCK
circadian locomotor output cycles kaput
- CRY
cryptochrome
- DCC-FBS
dextran-coated charcoal-treated fetal bovine serum
- ERα
estrogen receptor-α
- GEO
Gene Expression Omnibus
- GO
gene ontology
- HESC
human endometrial stromal cell
- ID
identification
- IGFBP1
IGF-binding protein-1
- MPA
medroxyprogesterone acetate
- NHS
National Health Service
- NT
nontargeting
- PER1
period 1
- PER2
period 2
- PPARG
peroxisome proliferator-activated receptor γ
- PRL
prolactin
- qRT-PCR
quantitative RT-PCR
- RPL
recurrent pregnancy loss
- RTCA
real-time cell analyzer
- SCN
suprachiasmatic nucleus
- SDS
sodium dodecyl sulfate
- siRNA
small interfering RNA
- TPM
transcripts per million
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
REFERENCES
- 1.Boden M. J., Varcoe T. J., Kennaway D. J. (2013) Circadian regulation of reproduction: from gamete to offspring. Prog. Biophys. Mol. Biol. 113, 387–397 [DOI] [PubMed] [Google Scholar]
- 2.Markson J. S., O’Shea E. K. (2009) The molecular clockwork of a protein-based circadian oscillator. FEBS Lett. 583, 3938–3947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morin L. P. (2013) Neuroanatomy of the extended circadian rhythm system. Exp. Neurol. 243, 4–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reppert S. M., Weaver D. R. (2002) Coordination of circadian timing in mammals. Nature 418, 935–941 [DOI] [PubMed] [Google Scholar]
- 5.Dolatshad H., Davis F. C., Johnson M. H. (2009) Circadian clock genes in reproductive tissues and the developing conceptus. Reprod. Fertil. Dev. 21, 1–9 [DOI] [PubMed] [Google Scholar]
- 6.Chappell P. E., White R. S., Mellon P. L. (2003) Circadian gene expression regulates pulsatile gonadotropin-releasing hormone (GnRH) secretory patterns in the hypothalamic GnRH-secreting GT1-7 cell line. J. Neurosci. 23, 11202–11213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kennaway D. J., Varcoe T. J., Mau V. J. (2003) Rhythmic expression of clock and clock-controlled genes in the rat oviduct. Mol. Hum. Reprod. 9, 503–507 [DOI] [PubMed] [Google Scholar]
- 8.Karman B. N., Tischkau S. A. (2006) Circadian clock gene expression in the ovary: Effects of luteinizing hormone. Biol. Reprod. 75, 624–632 [DOI] [PubMed] [Google Scholar]
- 9.Ko C. H., Takahashi J. S. (2006) Molecular components of the mammalian circadian clock. Hum. Mol. Genet. 15, R271–R277 [DOI] [PubMed] [Google Scholar]
- 10.Fu L., Lee C. C. (2003) The circadian clock: pacemaker and tumour suppressor. Nat. Rev. Cancer 3, 350–361 [DOI] [PubMed] [Google Scholar]
- 11.O’Neill J. S., Maywood E. S., Hastings M. H. (2013) Cellular mechanisms of circadian pacemaking: beyond transcriptional loops. Handb. Exp. Pharmacol. 217, 67–103 [DOI] [PubMed] [Google Scholar]
- 12.Grimaldi B., Nakahata Y., Kaluzova M., Masubuchi S., Sassone-Corsi P. (2009) Chromatin remodeling, metabolism and circadian clocks: the interplay of CLOCK and SIRT1. Int. J. Biochem. Cell Biol. 41, 81–86 [DOI] [PubMed] [Google Scholar]
- 13.Stojkovic K., Wing S. S., Cermakian N. (2014) A central role for ubiquitination within a circadian clock protein modification code. Front. Mol. Neurosci. 7, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee J., Lee Y., Lee M. J., Park E., Kang S. H., Chung C. H., Lee K. H., Kim K. (2008) Dual modification of BMAL1 by SUMO2/3 and ubiquitin promotes circadian activation of the CLOCK/BMAL1 complex. Mol. Cell. Biol. 28, 6056–6065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu A., Zhu L., Blum I. D., Mai O., Leliavski A., Fahrenkrug J., Oster H., Boehm U., Storch K. F. (2013) Global but not gonadotrope-specific disruption of Bmal1 abolishes the luteinizing hormone surge without affecting ovulation. Endocrinology 154, 2924–2935 [DOI] [PubMed] [Google Scholar]
- 16.Ratajczak C. K., Boehle K. L., Muglia L. J. (2009) Impaired steroidogenesis and implantation failure in Bmal1-/- mice. Endocrinology 150, 1879–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ratajczak C. K., Asada M., Allen G. C., McMahon D. G., Muglia L. M., Smith D., Bhattacharyya S., Muglia L. J. (2012) Generation of myometrium-specific Bmal1 knockout mice for parturition analysis. Reprod. Fertil. Dev. 24, 759–767 [DOI] [PubMed] [Google Scholar]
- 18.Gellersen B., Brosens J. J. (2014) Cyclic decidualization of the human endometrium in reproductive health and failure. Endocr. Rev. 35, 851–905 [DOI] [PubMed] [Google Scholar]
- 19.Uchikawa M., Kawamura M., Yamauchi N., Hattori M. A. (2011) Down-regulation of circadian clock gene period 2 in uterine endometrial stromal cells of pregnant rats during decidualization. Chronobiol. Int. 28, 1–9 [DOI] [PubMed] [Google Scholar]
- 20.Pilorz V., Steinlechner S. (2008) Low reproductive success in Per1 and Per2 mutant mouse females due to accelerated ageing? Reproduction 135, 559–568 [DOI] [PubMed] [Google Scholar]
- 21.Jones M. C., Fusi L., Higham J. H., Abdel-Hafiz H., Horwitz K. B., Lam E. W. F., Brosens J. J. (2006) Regulation of the SUMO pathway sensitizes differentiating human endometrial stromal cells to progesterone. Proc. Natl. Acad. Sci. USA 103, 16272–16277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W., Li Q., Bagchi I. C., Bagchi M. K. (2010) The CCAAT/enhancer binding protein beta is a critical regulator of steroid-induced mitotic expansion of uterine stromal cells during decidualization. Endocrinology 151, 3929–3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salker M. S., Nautiyal J., Steel J. H., Webster Z., Sućurović S., Nicou M., Singh Y., Lucas E. S., Murakami K., Chan Y. W., James S., Abdallah Y., Christian M., Croy B. A., Mulac-Jericevic B., Quenby S., Brosens J. J. (2012) Disordered IL-33/ST2 activation in decidualizing stromal cells prolongs uterine receptivity in women with recurrent pregnancy loss. PLoS One 7, e52252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salker M., Teklenburg G., Molokhia M., Lavery S., Trew G., Aojanepong T., Mardon H. J., Lokugamage A. U., Rai R., Landles C., Roelen B. A. J., Quenby S., Kuijk E. W., Kavelaars A., Heijnen C. J., Regan L., Macklon N. S., Brosens J. J. (2010) Natural selection of human embryos: impaired decidualization of endometrium disables embryo-maternal interactions and causes recurrent pregnancy loss. PLoS One 5, e10287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al-Sabbagh M., Lam E. W. F., Brosens J. J. (2012) Mechanisms of endometrial progesterone resistance. Mol. Cell. Endocrinol. 358, 208–215 [DOI] [PubMed] [Google Scholar]
- 26.Brosens J. J., Hayashi N., White J. O. (1999) Progesterone receptor regulates decidual prolactin expression in differentiating human endometrial stromal cells. Endocrinology 140, 4809–4820 [DOI] [PubMed] [Google Scholar]
- 27.Ross J. (1995) mRNA stability in mammalian cells. Microbiol. Mol. Biol. Rev. 59, 423–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grimaldi G., Christian M., Steel J. H., Henriet P., Poutanen M., Brosens J. J. (2011) Down-regulation of the histone methyltransferase EZH2 contributes to the epigenetic programming of decidualizing human endometrial stromal cells. Mol. Endocrinol. 25, 1892–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langmead B., Trapnell C., Pop M., Salzberg S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trapnell C., Pachter L., Salzberg S. L. (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagner G. P., Kin K., Lynch V. J. (2012) Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 131, 281–285 [DOI] [PubMed] [Google Scholar]
- 32.Anders S., Huber W. (2010) Differential expression analysis for sequence count data. Genome Biol. 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson M. D., McCarthy D. J., Smyth G. K. (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lal S., Allan A., Markovic D., Walker R., Macartney J., Europe-Finner N., Tyson-Capper A., Grammatopoulos D. K. (2013) Estrogen alters the splicing of type 1 corticotropin-releasing hormone receptor in breast cancer cells. Sci. Signal. 6, ra53. [DOI] [PubMed] [Google Scholar]
- 35.Yoo S. H., Ko C. H., Lowrey P. L., Buhr E. D., Song E. J., Chang S., Yoo O. J., Yamazaki S., Lee C., Takahashi J. S. (2005) A noncanonical E-box enhancer drives mouse Period2 circadian oscillations in vivo. Proc. Natl. Acad. Sci. USA 102, 2608–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Telgmann R., Gellersen B. (1998) Marker genes of decidualization: activation of the decidual prolactin gene. Hum. Reprod. Update 4, 472–479 [DOI] [PubMed] [Google Scholar]
- 37.Lamas M., Monaco L., Zazopoulos E., Lalli E., Tamai K., Penna L., Mazzucchelli C., Nantel F., Foulkes N. S., Sassone-Corsi P. (1996) CREM: a master-switch in the transcriptional response to cAMP. Philos. Trans. R. Soc. Lond. B Biol. Sci. 351, 561–567 [DOI] [PubMed] [Google Scholar]
- 38.Mantena S. R., Kannan A., Cheon Y. P., Li Q., Johnson P. F., Bagchi I. C., Bagchi M. K. (2006) C/EBPbeta is a critical mediator of steroid hormone-regulated cell proliferation and differentiation in the uterine epithelium and stroma. Proc. Natl. Acad. Sci. USA 103, 1870–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salker M. S., Christian M., Steel J. H., Nautiyal J., Lavery S., Trew G., Webster Z., Al-Sabbagh M., Puchchakayala G., Föller M., Landles C., Sharkey A. M., Quenby S., Aplin J. D., Regan L., Lang F., Brosens J. J. (2011) Deregulation of the serum- and glucocorticoid-inducible kinase SGK1 in the endometrium causes reproductive failure. Nat. Med. 17, 1509–1513 [DOI] [PubMed] [Google Scholar]
- 40.Leitao B., Jones M. C., Fusi L., Higham J., Lee Y., Takano M., Goto T., Christian M., Lam E. W. F., Brosens J. J. (2010) Silencing of the JNK pathway maintains progesterone receptor activity in decidualizing human endometrial stromal cells exposed to oxidative stress signals. FASEB J. 24, 1541–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Q., Kannan A., Wang W., Demayo F. J., Taylor R. N., Bagchi M. K., Bagchi I. C. (2007) Bone morphogenetic protein 2 functions via a conserved signaling pathway involving Wnt4 to regulate uterine decidualization in the mouse and the human. J. Biol. Chem. 282, 31725–31732 [DOI] [PubMed] [Google Scholar]
- 42.Talbi S., Hamilton A. E., Vo K. C., Tulac S., Overgaard M. T., Dosiou C., Le Shay N., Nezhat C. N., Kempson R., Lessey B. A., Nayak N. R., Giudice L. C. (2006) Molecular phenotyping of human endometrium distinguishes menstrual cycle phases and underlying biological processes in normo-ovulatory women. Endocrinology 147, 1097–1121 [DOI] [PubMed] [Google Scholar]
- 43.Ripperger J. A., Brown S. A. (2010) Transcriptional regulation of circadian clocks. In Circadian Clock, Vol. 12, (Albrecht U., ed.), pp. 37–78, Springer, New York [Google Scholar]
- 44.Al-Sabbagh M., Fusi L., Higham J., Lee Y., Lei K., Hanyaloglu A. C., Lam E. W. F., Christian M., Brosens J. J. (2011) NADPH oxidase-derived reactive oxygen species mediate decidualization of human endometrial stromal cells in response to cyclic AMP signaling. Endocrinology 152, 730–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakamura T. J., Moriya T., Inoue S., Shimazoe T., Watanabe S., Ebihara S., Shinohara K. (2005) Estrogen differentially regulates expression of Per1 and Per2 genes between central and peripheral clocks and between reproductive and nonreproductive tissues in female rats. J. Neurosci. Res. 82, 622–630 [DOI] [PubMed] [Google Scholar]
- 46.Koyanagi S., Hamdan A. M., Horiguchi M., Kusunose N., Okamoto A., Matsunaga N., Ohdo S. (2011) cAMP-response element (CRE)-mediated transcription by activating transcription factor-4 (ATF4) is essential for circadian expression of the Period2 gene. J. Biol. Chem. 286, 32416–32423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O’Neill J. S., Maywood E. S., Chesham J. E., Takahashi J. S., Hastings M. H. (2008) cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science 320, 949–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pohnke Y., Schneider-Merck T., Fahnenstich J., Kempf R., Christian M., Milde-Langosch K., Brosens J. J., Gellersen B. (2004) Wild-type p53 protein is up-regulated upon cyclic adenosine monophosphate-induced differentiation of human endometrial stromal cells. J. Clin. Endocrinol. Metab. 89, 5233–5244 [DOI] [PubMed] [Google Scholar]
- 49.Miki T., Matsumoto T., Zhao Z., Lee C. C. (2013) p53 regulates Period2 expression and the circadian clock. Nat. Commun. 4, 2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Albrecht U., Bordon A., Schmutz I., Ripperger J. (2007) The multiple facets of Per2. Cold Spring Harb. Symp. Quant. Biol. 72, 95–104 [DOI] [PubMed] [Google Scholar]
- 51.Gery S., Virk R. K., Chumakov K., Yu A., Koeffler H. P. (2007) The clock gene Per2 links the circadian system to the estrogen receptor. Oncogene 26, 7916–7920 [DOI] [PubMed] [Google Scholar]
- 52.Nakamura T. J., Sellix M. T., Menaker M., Block G. D. (2008) Estrogen directly modulates circadian rhythms of PER2 expression in the uterus. Am. J. Physiol. Endocrinol. Metab. 295, E1025–E1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hwang-Verslues W. W., Chang P. H., Jeng Y. M., Kuo W. H., Chiang P. H., Chang Y. C., Hsieh T. H., Su F. Y., Lin L. C., Abbondante S., Yang C. Y., Hsu H. M., Yu J. C., Chang K. J., Shew J. Y., Lee E. Y., Lee W. H. (2013) Loss of corepressor PER2 under hypoxia up-regulates OCT1-mediated EMT gene expression and enhances tumor malignancy. Proc. Natl. Acad. Sci. USA 110, 12331–12336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun C. M., Huang S. F., Zeng J. M., Liu D. B., Xiao Q., Tian W. J., Zhu X. D., Huang Z. G., Feng W. L. (2010) Per2 inhibits k562 leukemia cell growth in vitro and in vivo through cell cycle arrest and apoptosis induction. Pathol. Oncol. Res. 16, 403–411 [DOI] [PubMed] [Google Scholar]
- 55.Thoennissen N. H., Thoennissen G. B., Abbassi S., Nabavi-Nouis S., Sauer T., Doan N. B., Gery S., Müller-Tidow C., Said J. W., Koeffler H. P. (2012) Transcription factor CCAAT/enhancer-binding protein alpha and critical circadian clock downstream target gene PER2 are highly deregulated in diffuse large B-cell lymphoma. Leuk. Lymphoma 53, 1577–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ogasawara M., Aoki K., Okada S., Suzumori K. (2000) Embryonic karyotype of abortuses in relation to the number of previous miscarriages. Fertil. Steril. 73, 300–304 [DOI] [PubMed] [Google Scholar]
- 57.Rai R., Regan L. (2006) Recurrent miscarriage. Lancet 368, 601–611 [DOI] [PubMed] [Google Scholar]
- 58.Brosens J. J., Salker M. S., Teklenburg G., Nautiyal J., Salter S., Lucas E. S., Steel J. H., Christian M., Chan Y. W., Boomsma C. M., Moore J. D., Hartshorne G. M., Sućurović S., Mulac-Jericevic B., Heijnen C. J., Quenby S., Koerkamp M. J. G., Holstege F. C. P., Shmygol A., Macklon N. S. (2014) Uterine selection of human embryos at implantation. Sci. Rep. 4, 3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stocker L. J., Macklon N. S., Cheong Y. C., Bewley S. J. (2014) Influence of shift work on early reproductive outcomes: a systematic review and meta-analysis. Obstet. Gynecol. 124, 99–110 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.