Abstract
Purpose
Previous studies have shown that basal breast cancers, which may have an inherent “BRCAness” phenotype and sensitivity to inhibitors of poly (ADP-Ribose) polymerase (PARP), express elevated levels of PARP1. Our lab recently reported that HER2+ breast cancers also exhibit sensitivity to PARP inhibitors (PARPi) by attenuating the NF-kB pathway. In this study, we assessed PARP1 and phospho-p65, a marker of activated NF-kB levels in human breast cancer tissues.
Methods
PARP1 and PARP2 copy number, mRNA, and protein expression was assessed by interrogating the PAM-50 defined breast cancer patient set from the TCGA using the cBioPortal. PARP1 and phospho-p65 immunohistochemistry and correlation to clinical parameters was conducted using 307 primary breast cancer specimens (132 basal, 82 luminal, 93 HER2+) through univariate and multivariate analyses.
Results
In the PAM50 breast cancer data set, PARP1 and 2 expression was altered in 24/58 (41%) HER2+, 32/81 (40%) basal, and 75/324 (23%) luminal A/B breast cancer patients. This correlated with a statistically significant increase in PARP1 protein levels in HER2+ and basal but not luminal breast cancers (p=0.003, p=0.027, p=0.289, respectively). No change in PARP2 protein level was observed. Interestingly, using breast cancer specimens from 307 patients, HER2 positivity correlated with elevated PARP1 expression (p<0.0001) and was three times more likely than HER2 negative breast cancers to exhibit high PARP1 levels. No significant differences were noted between race, ER status, or PR status for PARP1 expression. Additionally, we found a significant correlation between HER2 status and phospho-p65 expression (p<0.0001). Lastly, a direct correlation between PARP1 and phospho-p65 (p<0.0001) was noted.
Conclusions
These results indicate a potential connection between HER2, PARP1, and phospho-p65. Furthermore, these data suggest that the PARPi sensitivity we previously observed in HER2+ breast cancer cells may be due to elevated PARP1 expression.
Keywords: PARP1, p65, HER2+, NF-kB, breast cancer
INTRODUCTION
Analysis of the genetic variation found in primary breast cancer has led to their classification into five distinct subtypes: basal, luminal A and B, HER2-enriched, and normal breast-like[1]. Although sharing numerous features recognized as ‘hallmarks of cancer[2],’ these subtypes possess vastly differing biology, which drastically alter their natural histories and response to treatment. Thus, greater understanding of how these subtypes behave and how they respond clinically is paramount to determining effective treatment for patients.
One therapy generating much excitement in breast cancer clinical studies are inhibitors of the DNA repair enzyme, poly(ADP) ribose polymerase (PARP). PARP inhibitors (PARPi) have shown to be effective in breast tumors with defective homologous recombination (HR) DNA repair pathways, such as those with BRCA1/2 mutations[3-7]. Recent evidence suggests that basal breast cancers possess a “BRCA-ness” phenotype[8] and sensitivity to PARPi[9]. Interestingly, a recent report identified high immunoreactivity to nuclear PARP1 in both BRCA-associated and non-BRCA-associated basal breast cancer[9]. These results suggest that high levels of nuclear PARP1 may correlate with tumor sensitivity to PARPi.
Recently, our laboratory discovered that HER2+ breast cancer cells are susceptible to PARPi alone in vitro and in vivo despite having proficient HR[10]. This susceptibility to PARPi was due to attenuation of NF-kB signaling. Because of the reported correlation between elevated nuclear PARP1 and susceptibility to PARP inhibitors in basal breast cancers[11, 12], we also hypothesized that elevated PARP1 would correspond to increased markers of NF-kB signaling.
In this study, we report that HER2+ breast cancers exhibit increased expression of PARP1 and phospho-p65, a key subunit of the NF-kB heterodimer and marker of NF-kB activity. First, analysis of the TCGA PAM50--defined subtype patient sets demonstrated increased PARP1 mRNA and protein expression in HER2 enriched and basal breast cancers, compared to luminal breast cancers. To verify these intriguing findings, we evaluated levels of PARP1 by immunohistochemistry (IHC) in 307 breast tumors (132 basal, 82 luminal, 93 HER2+) from multiple institutions comprising the BMaP3 (Minority Biospecimen/Biobanking Geographic Management Program, Region 3) consortium. Interestingly, HER2+ tumors express higher baseline levels of PARP1 and phospho-p65 compared to HER2- tumors. Additionally, a direct correlation between PARP1 and phospho-p65 expression was observed. Taken together these results suggest elevated PARP1 levels correspond to increased NF-kB signaling in HER2+ breast cancer patients and that high nuclear PARP1 may explain the PARPi sensitivity we previously observed in HER2+ breast cancer cells.
MATERIALS AND METHODS
Patient characteristics and clinical methodology
We obtained breast cancer tissue from a total of 307 patients. Tissue from 41 HER2+ and 32 HER2-invasive breast cancer patients diagnosed at The University of Alabama at Birmingham (UAB) between the years of 1999 and 2012 were identified from pre-existing databases. An additional 234 patients’ tissue (132 basal, 50 luminal, and 52 HER2+) was acquired as tissue microarrays from the Minority Biospecimen/Biobanking Geographic Management Program (BMaP). Institutional Review Board approval was obtained prior to initiation of this study (IRB#: X101214005).
UAB tissue was obtained from either biopsy or definitive surgery with pre-treatment specimens utilized when at all possible. HER2, estrogen receptor (ER), and progesterone receptor (PR) status as well as other pathologic variables (grade, etc.) were determined at the time of initial diagnosis by the UAB Department of Pathology and recorded in the electronic medical record. The following patient and tumor characteristics were recorded: age at diagnosis, race, pathologic stage, and ER/PR/HER2 receptor status. Treatment details regarding chemotherapy and endocrine therapy were also obtained. In those patients receiving neoadjuvant chemotherapy, response at the time of definitive surgery was assessed. Patients with metastatic disease were not included in the UAB subset. For the BMaP subset, de-identified patient data provided age at diagnosis, race, pathologic stage, ER/PR/HER2 receptor status, and administration of either neoadjuvant or adjuvant systemic treatment. Clinicopathologic parameters were then correlated with expression of PARP1 and phospho-p65.
The median age at diagnosis for the entire cohort was 53.6 years old (range 21-89). When analyzing by subtype, the average age of patients with basal, luminal, and HER2+ breast cancer averaged 54.1, 53.2, and 53.0 years old, respectively. There were a total of 93 HER2+ cases (30%) and 214 HER2- cases (70%). Of the 307 total cases, 117 (38%) were estrogen receptor (ER) + and 190 (62%) ER-, while 124 (40%) were progesterone receptor (PR) + and 183 (60%) PR-. Forty-nine percent of patients were white and 51% African American. Pathologic staging was as follows: 87 (28%) stage I, 152 (50%) stage II, 54 (18%) stage III, and 5 (2%) stage IV. Patient and tumor characteristics for the entire cohort are displayed in Table I.
Table I.
Demographics and clinical information for patients whose breast cancers were included in analysis (n = 307).
| Basal | Luminal | HER2+ | TOTAL | ||
|---|---|---|---|---|---|
| Number | Patients | 132 (43) | 82 (27) | 93 (30) | 307 (100) |
| Age | Mean | 54.08 | 53.25 | 53.01 | --- |
| Range | 27-87 | 28-80 | 21-89 | --- | |
| HER | HER2+ | 0 (0) | 0 (0) | 93 (100) | 93 (30) |
| HER2- | 130 (100) | 84 (100) | 0 (0) | 214 (70) | |
| ER | ER+ | 0 (0) | 83 (97) | 37 (40) | 120 (38) |
| ER- | 130 (100) | 2 (3) | 55 (60) | 187 (62) | |
| PR | PR+ | 0 (0) | 81 (96) | 45 (49) | 126 (40) |
| PR- | 130 (100) | 4 (4) | 47 (51) | 181 (60) | |
| Race | White | 79 (59) | 36 (44) | 37 (40) | 152 (49) |
| Black | 54 (41) | 46 (56) | 57 (62) | 157 (51) | |
| Stage | 1 | 46 (33) | 14 (17) | 27 (29) | 87 (28) |
| 2 | 64 (48) | 48 (56) | 40 (43) | 152 (50) | |
| 3 | 18 (14) | 17 (21) | 19 (21) | 54 (18) | |
| 4 | 1 (1) | 1 (1) | 3 (3) | 5 (2) | |
| NA | 4 (12) | 3 (4) | 3 (3) | 10 (3) | |
| Therapy | Neoadjuvant | 30 (22) | 13 (16) | 23 (25) | 66 (21) |
| Adjuvant | 74 (56) | 61 (74) | 63 (68) | 198 (64) | |
| Neither | 29 (22) | 8 (10) | 6 (7) | 43 (14) | |
Immunohistochemistry
Formalin-fixed, paraffin-embedded tissue blocks of identified patients were obtained from UAB Department of Pathology and tissue microarrays were acquired from the BMaP. For the UAB specimens, presence of tumor within the blocks was verified by a pathologist prior to sectioning into 5 μm slices and mounting on slides. All specimens were deparaffinized and rehydrated using standard techniques. Antigen retrieval was performed with pH 9 EDTA buffer at high temperature. Hydrogen peroxide and 3% goat serum were added to quench peroxidases and block nonspecific binding. Primary antibody was then applied for one hour at room temperature. Primary antibodies consisted of the following: anti-PARP-1 (Cat.#: 1072-1, Epitomics, Burlingame, CA) at a dilution of 1:750 and anti-phospho-p65 (Cat.#: 73745, Epitomics, Burlingame, CA) at a dilution of 1:2000. HRP-conjugated goat anti-rabbit secondary antibody (Cat.#: 111-035-144, Jackson Immuno Research, West Grove, PA) at a dilution of 1:200 was then applied. After DAB chromagen (Scy Tek Laboratories, Logan, UT) was added for seven minutes, the tissues were counterstained with hematoxylin. Sections were then dehydrated and the cover slip mounted with Permount. A multi-specimen breast tumor mircroarray with known staining for the two proteins of interest was used as a positive control and serum without primary antibody was used as a negative control in each run.
All slides were reviewed by two physicians, including a board-certified pathologist, blinded to clinical information. Intensity of staining was graded as 0 (no staining), 1+ (weak), 2+ (moderate), or 3+ (strong). The percent of cells (0-100%) staining for each intensity was determined. Staining was analyzed using three different methodologies. First, an H-score was calculated by multiplying the staining intensity (0-3+) by the percent of cells of each intensity (0-100%) and then adding these together for a final score of 0-300. In addition to H-score, the total percentage of cells with ≥2+ staining was determined. Lastly, any staining (yes or no) of ≥2+ was recorded in order to analyze the correlation with clinicopathologic features using a dichotomized variable. Nuclear and cytosolic staining was graded separately for each of the two proteins.
Statistical analysis
We have evaluated PARP1 and phospho-p65 data from a total of five institutions. Each patient had two or three replicates of PARP1 and phospho-p65 measurements, and we used mean value in the data analysis. PARP1 and phospho-p65 levels are presented as H-score, % (0-100) with ≥2+ staining, and dichotomized as binary outcome as > 2+ based on IHC staining intensity. Descriptive analyses with mean, standard deviation, count, and proportion are presented for each outcome. We compared PARP1 and phospho-p65 level by patient characteristics using two–group student t-test for continuous data and chi-square statistics for binary data. Pearson correlation was used to assess correlations between continuous variables, and Chi-Square statistics was used to assess association between binary variables. We used a linear regression model with analysis of variance method (ANOVA) and a logistics regression model to explore the association between each outcome with race, ER, PR, HER2, and tumor stage in univariate analysis and multivariable analysis. The variation among institutions was controlled in the models. The least square mean is presented and the Odds Ratio and its two sided 95% confidence intervals were presented.
RESULTS
HER2+ breast tumors express elevated levels of PARP1 mRNA and protein
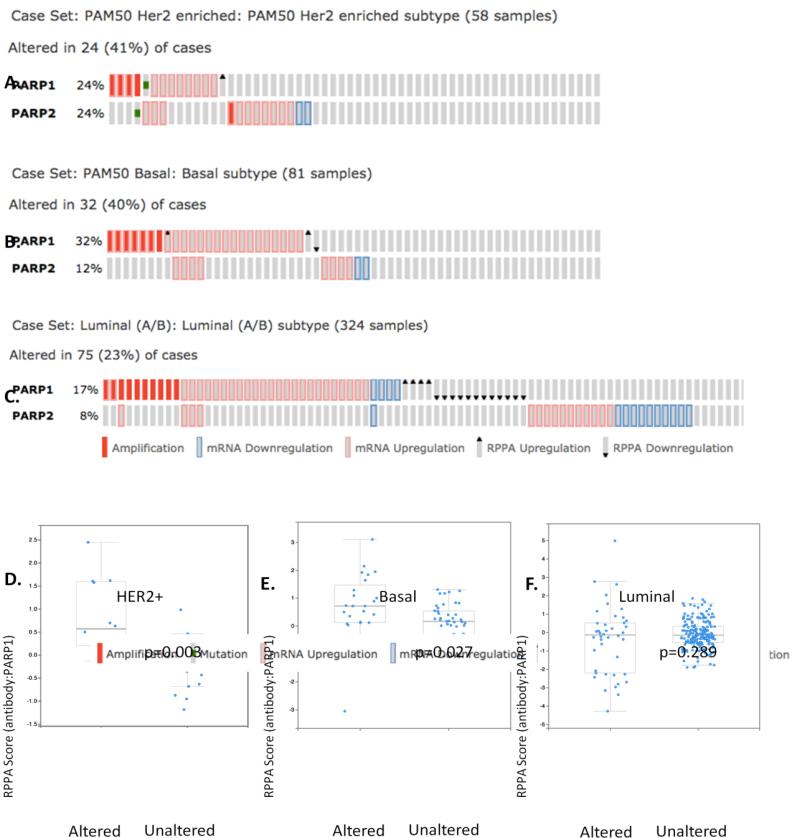
Prior studies have demonstrated that BRCA-associated and some non-BRCA-associated basal breast cancers are sensitive to PARPi[7, 13, 14]. These cancers also expressed high levels of nuclear PARP1, suggesting a potential link between PARP1 levels and sensitivity to PARPi. We have previously shown that HER2+ breast cancer cells have an exquisite sensitivity to PARPi in vitro and in vivo. One possible hypothesis could be that HER2+ breast cancers express high levels of PARP1 similar to the basal subtype. To test this hypothesis, we first interrogated the PAM50-defined subtype patient sets from the TCGA[15]. Utilizing cBioPortal (http://www.cbioportal.org/public-portal/), we examined PARP1 gene copy number, mRNA, and protein expression in 463 breast tumors consisting of 58 HER2-enriched (HER2+), 81 basal, and 324 luminalA/B breast cancers[15]. As PARP inhibitors target both PARP1 and its related family member PARP2, we also examined PARP2 mRNA and protein expression in these tumors. Interestingly, consistent with our hypothesis, PARP1 and/or PARP2 were altered in 41% (24/58) of HER2+ breast cancer cases. These alterations included gene amplification, mutation, and/or mRNA upregulation (Figure 1A). Similar to previously reported data[9], basal breast cancer also exhibited altered PARP1/2 mRNA (40%; 32/81 cases) (Figure 1B). In contrast, this trend was not observed in the luminal A/B subtypes (Figure 1C). Thus, basal and HER2-enriched breast cancers have elevated PARP1/2 gene expression compared to luminal breast cancers.
Figure 1. PARP1 expression is altered at the genomic and protein levels in HER2+ breast cancer patients from the TCGA PAM50 set.
a) cBioPortal analysis of PARP1/PARP2 mRNA expression in HER2+-overexpressing breast cancer patients. b) cBioPortal analysis of PARP1/PARP2 mRNA expression in basal breast cancer patients. c) cBioPortal analysis of PARP1/PARP2 mRNA expression in luminal A/B breast cancer patients. d) PARP1 protein expression is elevated in HER2+-overexpressing breast cancer patients with genomically altered PARP1 (p = 0.003). e) PARP1 protein expression is elevated in basal breast cancer patients with genomically altered PARP1 (p = 0.027). f) PARP1 protein expression is not altered in luminal A/B breast cancer patients (p = 0.289).
We next assessed whether PARP1/2 protein expression was similarly altered amongst the breast cancer subtypes. As shown in Figure 1D and 1E, a statistically significant upregulation in PARP1 protein was found in HER2+ and basal, but not luminal (Figure 1F), breast cancer subtypes using the RPPA analysis as reported on cBioPortal. The average abundance of PARP1 in HER2+, expressed as Z-score, was −0.05 in PARP1/2 unaltered cases versus 0.83 in altered cases (p=0.003) (Figure 1D). Additionally, no alteration in PARP2 protein level was identified by RPPA analysis (data not shown). Based on these intriguing data, we further pursued PARP1 protein levels in a larger, multi-institutional verification set.
PARP1 protein expression correlates with HER2 status, but not ER status, PR status, or race
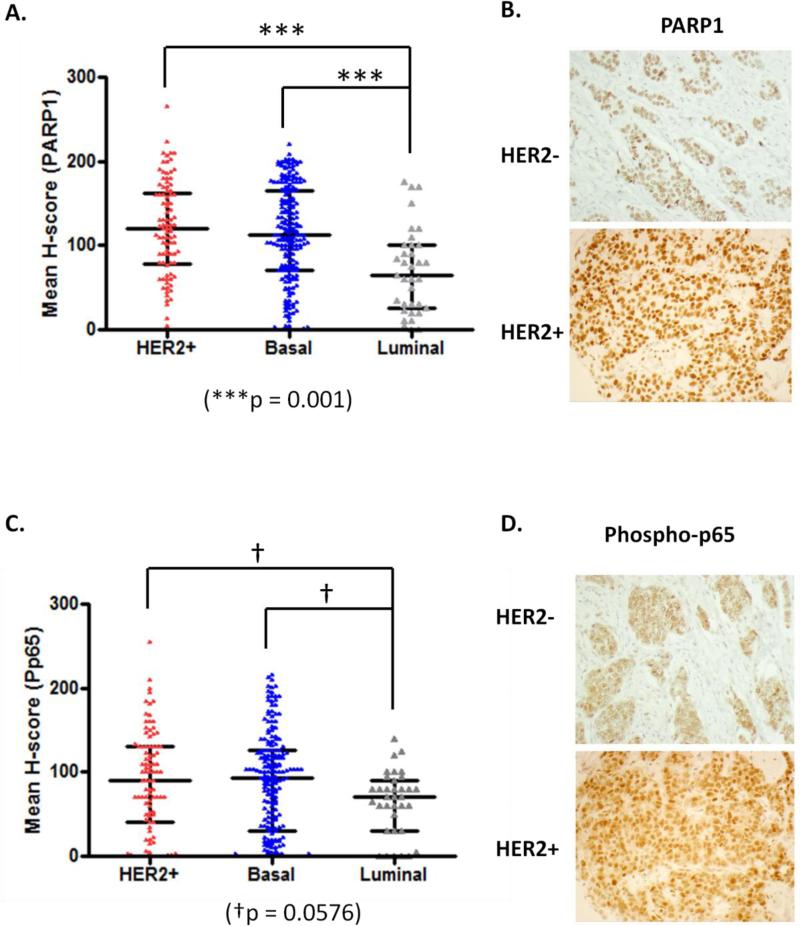
As PARP1 levels were altered in HER2-enriched patients within the PAM50 portion of the TCGA invasive breast cancer data set, we aimed to verify this in a larger data set consisting of 307 primary breast cancers (93 HER2+, 132 basal, 82 luminal) from UAB and the BMaP consortium tissue microarray (see materials and methods). Patient characteristics are summarized in Table I. Utilizing immunohistochemistry (IHC) and H-score analyses of staining intensity, we evaluated PARP1 staining in 307 patient primary tumors. In univariate analysis, we analyzed mean H-score by breast cancer subtype and observed both HER2+ and basal breast cancer had significantly elevated PARP1, as compared to luminal breast cancer specimens (p<0.001, Figure 2A). A representative image of PARP1 staining in both HER2+ and luminal breast cancer tissues is shown in Figure 2B. Of note, PARP1 staining was primarily nuclear in all specimens.
Figure 2. Mean H-score protein levels of PARP1 and phospho-p65 are elevated in HER2+ and triple negative breast cancer patients.
a) Graphical representation of the distribution of mean H-score for PARP staining by breast cancer subtype. Of note, triple negative and HER2+ subtypes had significantly higher expression of PARP1 (p < 0.001) compared to luminal subtypes. b) Representative immunohistochemical staining of HER2+ and HER2- breast cancer samples for PARP1 protein. PARP1 protein expression was almost exclusively nuclear. c) Graphical representation of the distribution of mean H-score for phospho-p65 staining by breast cancer subtype. Of note, basal and HER2+ subtypes had high expression of phospho-p65, but the difference was not significant (p = 0.0576) compared to luminal subtypes. d) Representative immunohistochemical staining of HER2+ and HER2- breast cancer samples for phospho-p65. Phospho-p65 displayed not only weaker staining in HER2- samples but also differed in subcellular location where the majority was perinuclear and cytoplasmic as compared to mainly nuclear in HER2+.
Interestingly, multivariable analysis revealed a significant difference with PARP1 H-scores among the institutions (p<0.0001). The average PARP1 H-scores for Emory University, Moffitt Cancer Institute, Tulane University, UAB, and the University of Mississippi were 77.61, 118.82, 113.15, 76.55, and 71.70, respectively. After controlling for the variation between institutions, we continued to observe a strong association between PARP1 score and HER2 status (p<0.0001). HER2+ patients had a significantly higher level of PARP1 with a least square mean of 97.95 compared to 65.05 in HER2- patients (Table IIA).
Table IIA.
PARP1 mean H-score analyzed according to patient and tumor characteristics (Race, ER status, PR status, HER2 status).
| Number | Mean | Std Dev | Median | Minimum | Maximum | LSMean | P-Value | ||
|---|---|---|---|---|---|---|---|---|---|
| Race | White | 152 | 118 | 56.60 | 120 | 0 | 265 | 104.12 | 0.5057 |
| Black | 154 | 97.87 | 62.07 | 100 | 0 | 223.33 | 99.98 | ||
| Other | 1 | 60 | --- | 60 | 60 | 60 | 40.40 | ||
| ER | ER+ | 138 | 96.38 | 62.40 | 95.83 | 0 | 265 | 75.32 | 0.3959 |
| ER- | 169 | 116.96 | 56.74 | 116.67 | 0 | 223.33 | 87.68 | ||
| PR | PR+ | 128 | 96.47 | 62.52 | 95.83 | 0 | 265 | 82.14 | 0.9300 |
| PR- | 179 | 115.76 | 57.20 | 115 | 0 | 255 | 80.86 | ||
| HER2 | HER2+ | 93 | 117.29 | 59.70 | 116.67 | 0 | 265 | 97.95 | <0.0001 |
| HER2- | 214 | 103.55 | 59.98 | 104.17 | 0 | 220 | 65.05 | ||
| Stage | 0 | 1 | 125 | --- | 125 | 125 | 125 | 54.16 | 0.7014 |
| 1 | 77 | 124.07 | 55.57 | 120 | 0 | 210 | 93.89 | ||
| 2 | 143 | 104.46 | 61.15 | 103.33 | 0 | 265 | 90.60 | ||
| 3 | 74 | 98.55 | 61.44 | 100.83 | 0 | 220 | 80.96 | ||
Similarly, when analyzing by percentage of cells with >2+ PARP1 staining, PARP1 was also significantly different among institutions (p=0.0035) and by HER2 status (p=0.0095) after controlling for institute. A higher percentage of HER2+ breast cancers had >2+ PARP1 staining compared to HER2-, 20.25% and 8.47% respectively (Table IIB).
Table IIB.
Percent of cells with ≥2 PARP1 staining analyzed according to patient and tumor characteristics (Race, ER status, PR status, HER2 status).
| Number | Mean | Std Dev | Median | Minimum | Maximum | LSMean | P-value | ||
|---|---|---|---|---|---|---|---|---|---|
| Race | White | 152 | 33.51 | 33.79 | 20 | 0 | 100 | 26.41 | 0.5577 |
| Black | 154 | 24.92 | 31.55 | 8.33 | 0 | 100 | 24.32 | ||
| Other | 1 | 0 | --- | 0 | 0 | 0 | −7.65 | ||
| ER | ER+ | 138 | 24.23 | 31.04 | 5 | 0 | 100 | 11.00 | 0.4212 |
| ER- | 169 | 32.92 | 33.81 | 20 | 0 | 100 | 17.72 | ||
| PR | PR+ | 128 | 24.25 | 31.00 | 5.83 | 0 | 100 | 15.50 | 0.7863 |
| PR- | 179 | 32.47 | 33.78 | 20 | 0 | 100 | 13.21 | ||
| HER2 | HER2+ | 93 | 31.52 | 30.52 | 20 | 0 | 98.33 | 20.25 | 0.0095 |
| HER2- | 214 | 28.06 | 33.90 | 10 | 0 | 100 | 8.47 | ||
| Stage | 0 | 1 | 30 | --- | 30 | 30 | 30 | −1.64 | 0.6423 |
| 1 | 77 | 36.93 | 36.46 | 20 | 0 | 100 | 21.24 | ||
| 2 | 143 | 28.27 | 32.48 | 13.33 | 0 | 100 | 18.95 | ||
| 3 | 74 | 23.33 | 29.62 | 10 | 0 | 100 | 13.10 | ||
| 4 | 5 | 30 | 39.65 | 23.33 | 0 | 96.67 | 20.16 | ||
Lastly, with logistic regression analysis, HER2+ breast cancers were more likely to have staining >2+, after controlling for variation among institutions (p=0.0019). HER2+ tumors were 3 times more likely to possess higher PARP1 than HER2- tumors, with an odds ratio of 3.081 (95% confidence interval of 1.607-5.906) (Table IIC).
Table IIC.
Tumors with presence or absence of ≥2+ PARP1 staining analyzed according patient and tumor characteristics (Race, ER status, PR status, HER2 status).
| Yes | No | Total | Odds Ratio | CI | P-value | ||
|---|---|---|---|---|---|---|---|
| Race | White | 108 (52.94) | 44 (42.72) | 152 | >999.999 | <0.001->999.999 | 0.9315 |
| Black | 96 (47.06) | 58 (56.31) | 154 | ||||
| Other | 0 (0) | 1 (0.97) | 1 | ||||
| ER | ER+ | 78 (38.24) | 60 (58.25) | 138 | 0.532 | 0.160-1.773 | 0.3640 |
| ER- | 126 (61.76) | 43 (41.75) | 169 | ||||
| PR | PR+ | 72 (35.29) | 56 (18.24) | 128 | 0.979 | 0.296-3.237 | 0.7193 |
| PR- | 132 (64.71) | 47 (45.63) | 179 | ||||
| HER2 | HER2+ | 68 (33.33) | 25 (24.27) | 93 | 3.081 | 1.607-5.906 | 0.0019 |
| HER2- | 136 (66.67) | 78 (75.73) | 214 | ||||
| Stage | 0 | 1 (0.50) | 0 (0) | 1 | >999.999 | <0.001->999.9 | 0.9127 |
| 1 | 55 (27.64) | 22 (21.78) | 77 | 1.095 | 0.128-9.392 | ||
| 2 | 94 (47.24) | 49 (48.51) | 143 | 1.366 | 0.176-10.610 | ||
| 3 | 46 (23.12) | 28 (27.72) | 74 | 0.969 | 0.120-7.813 | ||
No significant differences were observed between race, ER status, or PR status for PARP1 expression after controlling for institute (Table IIA-C). Consistent with TCGA data, HER2 status correlated with PARP1 expression in all methods of analysis (H-score, % cells >2+ staining, and presence of > 2+ staining).
Phospho-p65 protein expression correlates with HER2 status, but not ER status, PR status, or race
We have previously reported that PARP inhibition attenuates NF-kB signaling and corresponds to increased cell death in HER2+ breast cancer cells, suggesting a potential link between HER2, PARP1, and NF-kB. Several studies have also shown the importance of the NF-kB pathway as a downstream effector of HER2+ breast tumorigenesis [16-20]. We thus further interrogated our patient samples for expression of phospho-p65, a marker of active NF-kB signaling.
In univariate analysis, we analyzed mean H-score by breast cancer subtype and observed both HER2+ and basal breast cancers there was elevated phospho-p65, although not significant, as compared to luminal breast cancer specimens (p = 0.0576) (Figure 2C). An example of HER2+ and luminal breast cancer tissue staining for phospho-p65 is displayed in Figure 2D. We appreciated phospho-p65 displayed not only weaker staining in HER2- samples, but also altered location where the majority was perinuclear and cytoplasmic as compared to mainly nuclear in HER2+ group.
In multivariable analyses, we again observed that there is a significant difference among institutions (p<0.0001). The average phospho-p65 scores for Emory University, Moffitt Cancer Center, Tulane University, UAB, and University of Mississippi were 106.68, 110.39, 54.52, 76.28, and 66.19, respectively. HER2+ samples had a significantly higher level of phospho-p65 (p< 0.0322) with a least square mean of 95.04 in HER2+ and 79.03 in HER2- breast tumors (Table IIIA).
Table IIIA.
Phospho-p65 mean H-score statistics analyzed according to patient and tumor characteristics (Race, ER status, PR status, HER2 status).
| Number | Mean | Std Dev | Median | Minimum | Maximum | LSMean | P-value | ||
|---|---|---|---|---|---|---|---|---|---|
| Race | White | 152 | 97.05 | 54.62 | 100 | 0 | 213.33 | 87.74 | 0.9992 |
| Black | 154 | 67.66 | 58.42 | 65 | 0 | 255 | 87.53 | ||
| Other | 1 | 90 | --- | 90 | 90 | 90 | 85.83 | ||
| ER | ER+ | 138 | 71.82 | 58.93 | 70 | 0 | 255 | 89.55 | 0.7153 |
| ER- | 169 | 90.83 | 56.47 | 100 | 0 | 213.33 | 84.52 | ||
| PR | PR+ | 128 | 70.69 | 59.14 | 113.33 | 0 | 223.33 | 78.83 | 0.2381 |
| PR- | 179 | 90.58 | 56.35 | 96.67 | 0 | 213.33 | 95.25 | ||
| HER2 | HER2+ | 93 | 84.24 | 58.26 | 90 | 0 | 255 | 95.04 | 0.0322 |
| HER2- | 214 | 81.44 | 58.37 | 82.5 | 0 | 216.67 | 79.03 | ||
| Stage | 0 | 1 | 115 | --- | 115 | 115 | 115 | 76.99 | 0.0038 |
| 1 | 77 | 108.53 | 60.11 | 110 | 0 | 255 | 98.17 | ||
| 2 | 143 | 73.31 | 55.45 | 73.33 | 0 | 203.33 | 72.79 | ||
| 3 | 74 | 68.18 | 51.46 | 70 | 0 | 190 | 66.08 | ||
| 4 | 5 | 121.33 | 83.15 | 160 | 16.67 | 216.67 | 121.16 | ||
Similarly, the percentage of cells with >2+ phospho-p65 staining was significantly different among institutions (p=0.0010) and by HER2 status (p=0.0027) after controlling for institute. There was also a higher percentage of cells with >2+ staining in HER2+ compared to HER2- samples, 26.61% and 16.37%, respectively (Table IIIB).
Table IIIB.
Percent of cells with ≥2 phospho-p65 staining analyzed according to patient and tumor characteristics (Race, ER status, PR status, HER2 status).
| Number | Mean | Std Dev | Median | Minimum | Maximum | LSMean | P-value | ||
|---|---|---|---|---|---|---|---|---|---|
| Race | White | 152 | 21.78 | 29.04 | 7.5 | 0 | 100 | 21.81 | 0.9931 |
| Black | 154 | 14.63 | 23.07 | 2.08 | 0 | 96.67 | 21.29 | ||
| Other | 1 | 10 | --- | 10 | 10 | 10 | 21.37 | ||
| ER | ER+ | 138 | 15.40 | 25.63 | 0 | 0 | 96.67 | 24.00 | 0.4234 |
| ER- | 169 | 20.39 | 26.82 | 10 | 0 | 100 | 18.98 | ||
| PR | PR+ | 128 | 14.82 | 25.30 | 0 | 0 | 96.67 | 20.03 | 0.6442 |
| PR- | 179 | 20.53 | 26.92 | 10 | 0 | 100 | 22.95 | ||
| HER2 | HER2+ | 93 | 19.73 | 25.58 | 10 | 0 | 95 | 26.61 | 0.0027 |
| HER2- | 214 | 17.46 | 26.74 | 4.17 | 0 | 100 | 16.37 | ||
| Stage | 0 | 1 | 30 | --- | 30 | 30 | 30 | 12.60 | 0.0073 |
| 1 | 77 | 29.98 | 32.03 | 18.33 | 0 | 100 | 25.82 | ||
| 2 | 143 | 14.50 | 24.06 | 1.67 | 0 | 100 | 15.35 | ||
| 3 | 74 | 11.19 | 17.45 | 3.33 | 0 | 90 | 13.29 | ||
| 4 | 5 | 44 | 37.59 | 53.33 | 0 | 93.33 | 40.40 | ||
Lastly, using logistic regression analysis, HER2+ breast cancers were significantly more likely to have >2+ phospho-p65 staining, after controlling for variation among institutions (p=0.0039). HER2+ tumors were approximately 3 times more likely to have >2+ phospho-p65 staining than HER2- with an odds ratio of 2.79 and 95% confidence interval of 1.474-5.3 (Table IIIC).
Table IIIC.
Tumors with presence or absence of ≥2+ phopsho-p65 staining analyzed according patient and tumor characteristics (Race, ER status, PR status, HER2 status).
| Yes | No | Total | Odds Ratio | CI | P-value | ||
|---|---|---|---|---|---|---|---|
| Race | White | 97 (54.49) | 55 (42.64) | 152 | <0.001 | <0.001->999.999 | 0.9974 |
| Black | 80 (44.94) | 74 (57.36) | 154 | ||||
| Other | 1 (0.56) | 0 (0) | 1 | ||||
| ER | ER+ | 66 (37.08) | 72 (55.81) | 138 | 1.019 | 0.328-3.161 | 0.896 |
| ER- | 112 (62.92) | 57 (44.19) | 169 | ||||
| PR | PR+ | 59 (33.15) | 69 (53.49) | 128 | 0.73 | 0.235-2.267 | 0.397 |
| PR- | 119 (66.85) | 62 (46.51) | 179 | ||||
| HER2 | HER2+ | 58 (32.58) | 35 (27.13) | 93 | 2.795 | 1.474-5.3 | 0.0039 |
| HER2- | 120 (67.42) | 94 (72.87) | 214 | ||||
| Stage | 0 | 1 (0.57) | 0 (0) | 1 | >999.999 | <0.001->999.9 | 0.5104 |
| 1 | 57 (32.57) | 20 (16.0) | 77 | 0.457 | 0.039-5.346 | ||
| 2 | 75 (42.86) | 68 (54.4) | 143 | 0.273 | 0.026-2.905 | ||
| 3 | 38 (21.71) | 36 (28.8) | 74 | 0.275 | 0.025-3.045 | ||
| 4 | 4 (2.29) | 1 (0.80) | 5 | ||||
No significant differences were observed between race, ER status, or PR status for phospho-p65 expression after controlling for institute (Table IIIA-C). Similar to PARP1 expression, HER2 status was found to correlate with expression of phospho-p65 in all methods of analysis.
Elevated phospho-p65 correlates with stage IV disease
Utilizing pathological staging data, we also found that stage IV breast cancer had significantly elevated phospho-p65 levels as compared to stage 0 to III breast cancer. The least square mean for H-score was 121.15 in stage IV patients compared to least square means between 66.07 and 98.17 for nonmetastatic patients (p < 0.0038) (Table IIIA). Similarly, in percentage of cells with >2+ phospho-p65 staining the same finding exists with 40.4% of cells in stage IV patients having ≥2+ staining versus 12.6% - 25.81% of cells in stage I-III patients (p<0.0073) (Table IIIB). We did not observe this trend in the binary output (p = 0.5104) (Table IIIC).
Direct correlation between PARP1 and phospho-p65 expression
Based on these intriguing data revealing elevated PARP1 and phospho-p65 in HER2+ breast cancers, we hypothesized that there may be a direct correlation between PARP1 and NF-kB expression within the patient cohort tested. A Pearson's correlation analysis was performed to determine the relationship between the 307 patients’ mean H-scores for PARP1 and phospho-p65 staining intensity. Indeed, a direct correlation between PARP1 and phospho-p65 was observed (r=0.38671, N=307, p<0.0001).
Additionally, we examined correlation between these proteins in our measure of percentage of cells with >2+ staining. Again, a positive correlation was found between PARP1 and phospho-p65 (r=0.29557, N=307, p < 0.0001).
Finally, utilizing the binary measure of presence or absence of >2+ staining, we conducted a Chi-square analysis. We observed a significant relationship between PARP1 and phopsho-p65 levels (X2 (1, N=307) = 36.6463, p < 0.0001). The effect size for this finding, as measured by Cramer's V, was 0.3455. In total, 46.58% of patients were positive for ≥2+ staining of both PARP1 and phospho-p65.
DISCUSSION
In this study, we demonstrated a link between nuclear PARP1 and phospho-p65 expression in primary breast cancer specimens. Specifically, levels of nuclear PARP1 and phospho-p65 are elevated in HER2+ breast cancer as compared to HER2- groups. Previous publications have reported that HER amplification results in increased NF-kB signaling in several cancers[17, 21], including breast cancer where the HER2+ subtype has been shown to harbor increased NF-kB activity[22, 23]. Additionally, HER2 overexpression may be influencing PARP1's nuclear functions such as the regulation of NF-kB-mediated transcription by PARP1 acting as a coactivator[11, 12]. This is supported by our previous studies linking HER2, PARP1, and NF-kB[10]. Our current analysis provides further evidence for this link as we observed a correlation in patient samples between elevated PARP1 and phospho-65. Given these results, this signaling pathway is worth interrogating at the tissue level to yield important information about a patient's cancer.
Our approach of utilizing three scoring methods for evaluation of PARP1 and phospho-p65 expression yielded consistent and significant results. Through analysis of data by continuous (H-score), binned (% cells > 2+), and binary (presence/absence of > 2+) variables we were able to show elevated PARP1 and phospho-p65 in HER2+ breast cancer across all scoring methods. This is important as the clinical utility of this evaluation lies in the ability of pathologists to quickly screen patient specimens using the most time efficient measure, the binary method (presence or absence of >2+ staining). As our data were highly significant across all measures, we hypothesize that this information may be utilized in a time-effective manner through use of binary outcomes.
Interestingly, although prior studies have shown that cytosolic PARP1 does not correlate with HER2 status[24] we have identified a relationship between nuclear PARP1 and HER2+ status. Importantly, in this prior study only 24.6% of HER2+ samples expressed high cytosolic PARP1[24]; whereas, other studies of clinical specimens indicate that between 80-90% of breast cancers display elevated nuclear PARP1[9]. Perhaps this may be an indication that nuclear PARP1 could be a more sensitive marker in breast cancer. We also observed that this effect is specific to PARP1, as PARP2 did not demonstrate significant changes in expression according to our analysis of the TCGA data. Furthermore, previous studies determined PARP2 expression was not significantly different between cancer and normal surrounding tissue[25]. Thus, PARP2 does not appear to play as important a role in tumorigenesis. As our data suggest, similar to the basal subtype, the HER2-enriched breast cancer subtype has elevated expression of nuclear PARP1, indicating the potential utility of nuclear PARP1 as a therapeutic biomarker for PARP inhibitors in this specific subset of breast cancer patients. Alternatively, phospho-p65 could be used as a biomarker based on our data.
We also observed that PARP1 and phospho-p65 had a direct correlation in HER2+ breast cancer specimens. Previous studies, including ours, have also suggested links between these three proteins. In MMTV-neu transgenic mice lacking IKKa, the regulatory subunit of NF-kB, researchers observed a significant decrease in incidence of tumor formation and multiplicity of tumors in MMTV-neu models[26]. Interestingly, our lab recently observed HER2+ breast cancer cells were sensitive to PARP inhibition through the reduction NF-kB signaling[10], suggesting a possible oncogenic addiction to NF-kB signaling. Further evidence suggestive of a link between these proteins identified a nuclear PARP1 signalsome, which is essential to internal activation of the NF-kB pathway in response to IR-induced DNA damage[27]. Furthermore, PARP inhibitors sensitize p65+, but not p65-, cells to radiation by decreasing NF-kB's ability to bind DNA and reducing its transcriptional activity[28]. Together these data argue for an interesting link between HER2, PARP1, and the NF-kB pathway. Currently, in depth mechanistic studies for this link are underway in our laboratory.
There is also increasing evidence correlating tumor aggressiveness to increased NF-kB signaling[29]. Of note, the importance of NF-kB to HER2 transformation[26] corroborates our data in which we observed a correlation between elevated phospho-p65 and stage IV disease. Similarly, others have found nuclear phospho-p65 relates to poor prognosis in other cancers including gastric, pancreatic, hepatocellular, ovarian and cervical[30-34].
In summary, this study provides the first evidence of elevated protein levels and direct correlation between PARP1 and phospho-p65 in HER2+ breast cancer specimens. Whether elevated PARP1 and phospho-p65 levels can serve as a potential marker of PARP inhibitor sensitivity should be tested in future clinical trials.
ACKNOWLEDGEMENTS
This work is supported in part by the Minority Biospecimen/Biobanking Geographic Management Program for Region 3 (BMaP-3) (3U54 CA153509-03S1) from the National Cancer Institute (ESY), AACRGenentech BioOncology Career Development Award for Cancer Research on the HER Family Pathway (12-30-18-YANG, to ESY), Susan G. Komen Career Catalyst Award (CCR12364491, to ESY), NCI/H. Lee Moffitt Cancer Center BMaP Pilot Grant (10-16308-03-17-G1, to ESY), and the UAB MSTP (NIH-NIGMS 5T32GM008361-21 to JAS) and UAB Cell and Molecular Biology Training grant (5T32GM008111-27) (JAS), an intramural pilot grant from the Department of Radiation Oncology (LK). This work was also supported by the Breast SPORE (5P50CA 089019) and Tissue Procurement Shared Facility (P30CA-13148-41) of the Comprehensive Cancer Center at UAB.
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
ETHICAL STANDARDS
The experiments in this manuscript comply with the current laws of the country in which they were performed.
RESEARCH INVOLVING HUMAN PARTICIPANTS AND/OR ANIMALS
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This article does not contain any studies with animals performed by any of the authors.
REFERENCES
- 1.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Robert A, Weinberg Hallmarks of Cancer: The Next Generation. Cell. 144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434(7035):913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 4.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 5.Fong PC, et al. Inhibition of Poly(ADP-Ribose) Polymerase in Tumors from BRCA Mutation Carriers. New England Journal of Medicine. 2009;361(2):123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 6.Audeh MW, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010;376(9737):245–51. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- 7.Tutt A, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. The Lancet. 2010;376(9737):235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- 8.Turner NC, Reis-Filho JS. Basal-like breast cancer and the BRCA1 phenotype. Oncogene. 2006;25(43):5846–5853. doi: 10.1038/sj.onc.1209876. [DOI] [PubMed] [Google Scholar]
- 9.Domagala P, et al. PARP-1 expression in breast cancer including BRCA1-associated, triple negative and basal-like tumors: possible implications for PARP-1 inhibitor therapy. Breast Cancer Research and Treatment. 2011;127(3):861–869. doi: 10.1007/s10549-011-1441-2. [DOI] [PubMed] [Google Scholar]
- 10.Nowsheen S, et al. HER2 overexpression renders human breast cancers sensitive to PARP inhibition independently of any defect in homologous recombination DNA repair. Cancer Res. 2012;72(18):4796–4806. doi: 10.1158/0008-5472.CAN-12-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hassa PO, Hottiger MO. A role of poly (ADP-ribose) polymerase in NF-kappaB transcriptional activation. Biol Chem. 1999;380(7-8):953–9. doi: 10.1515/BC.1999.118. [DOI] [PubMed] [Google Scholar]
- 12.Hassa P, Hottiger M. The functional role of poly (ADP-ribose) polymerase 1 as novel coactivator of NF-κB in inflammatory disorders. Cellular and Molecular Life Sciences CMLS. 2002;59(9):1534–1553. doi: 10.1007/s00018-002-8527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Shaughnessy J, et al. Iniparib plus Chemotherapy in Metastatic Triple-Negative Breast Cancer. New England Journal of Medicine. 2011;364(3):205–214. doi: 10.1056/NEJMoa1011418. [DOI] [PubMed] [Google Scholar]
- 14.Rottenberg S, et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proceedings of the National Academy of Sciences. 2008;105(44):17079–17084. doi: 10.1073/pnas.0806092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biswas DK, et al. NF-kappa B activation in human breast cancer specimens and its role in cell proliferation and apoptosis. Proc Natl Acad Sci U S A. 2004;101(27):10137–42. doi: 10.1073/pnas.0403621101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biswas DK, Iglehart JD. Linkage between EGFR family receptors and nuclear factor kappaB (NF-kappaB) signaling in breast cancer. J Cell Physiol. 2006;209(3):645–52. doi: 10.1002/jcp.20785. [DOI] [PubMed] [Google Scholar]
- 18.Izzo JG, et al. Pretherapy nuclear factor-kappaB status, chemoradiation resistance, and metastatic progression in esophageal carcinoma. Mol Cancer Ther. 2006;5(11):2844–50. doi: 10.1158/1535-7163.MCT-06-0351. [DOI] [PubMed] [Google Scholar]
- 19.Braunstein S, Formenti SC, Schneider RJ. Acquisition of stable inducible up-regulation of nuclear factor-kappaB by tumor necrosis factor exposure confers increased radiation resistance without increased transformation in breast cancer cells. Mol Cancer Res. 2008;6(1):78–88. doi: 10.1158/1541-7786.MCR-07-0339. [DOI] [PubMed] [Google Scholar]
- 20.Prasad S, Ravindran J, Aggarwal BB. NF-kappaB and cancer: how intimate is this relationship. Mol Cell Biochem. 2010;336(1-2):25–37. doi: 10.1007/s11010-009-0267-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merkhofer EC, Cogswell P, Baldwin AS. Her2 activates NF-kappaB and induces invasion through the canonical pathway involving IKKalpha. Oncogene. 2010;29(8):1238–48. doi: 10.1038/onc.2009.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh S, et al. Nuclear factor-kappaB activation: a molecular therapeutic target for estrogen receptor-negative and epidermal growth factor receptor family receptor-positive human breast cancer. Mol Cancer Ther. 2007;6(7):1973–82. doi: 10.1158/1535-7163.MCT-07-0063. [DOI] [PubMed] [Google Scholar]
- 23.Shapochka D, et al. Relationship between NF-κB, ER, PR, Her2/neu, Ki67, p53 expression in human breast cancer. Exp Oncol. 2012;34(4):358–353. [PubMed] [Google Scholar]
- 24.von Minckwitz G, et al. Cytoplasmic Poly(Adenosine Diphosphate–Ribose) Polymerase Expression Is Predictive and Prognostic in Patients With Breast Cancer Treated With Neoadjuvant Chemotherapy. Journal of Clinical Oncology. 2011;29(16):2150–2157. doi: 10.1200/JCO.2010.31.9079. [DOI] [PubMed] [Google Scholar]
- 25.Ossovskaya V, et al. Exploring molecular pathways of triple-negative breast cancer. Genes Cancer. 2011;2(9):870–9. doi: 10.1177/1947601911432496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao Y, Luo J.-l., Karin M. IκB kinase α kinase activity is required for self-renewal of ErbB2/Her2-transformed mammary tumor-initiating cells. Proceedings of the National Academy of Sciences. 2007;104(40):15852–15857. doi: 10.1073/pnas.0706728104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stilmann M, et al. A nuclear poly (ADP-ribose)-dependent signalosome confers DNA damage-induced IκB kinase activation. Molecular cell. 2009;36(3):365–378. doi: 10.1016/j.molcel.2009.09.032. [DOI] [PubMed] [Google Scholar]
- 28.Veuger SJ, Hunter JE, Durkacz BW. Ionizing radiation-induced NF-kappaB activation requires PARP-1 function to confer radioresistance. Oncogene. 2009;28(6):832–42. doi: 10.1038/onc.2008.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dolcet X, et al. NF-kB in development and progression of human cancer. Virchows archiv. 2005;446(5):475–482. doi: 10.1007/s00428-005-1264-9. [DOI] [PubMed] [Google Scholar]
- 30.Guo RX, et al. Increased staining for phosphorylated AKT and nuclear factor-κB p65 and their relationship with prognosis in epithelial ovarian cancer. Pathology international. 2008;58(12):749–756. doi: 10.1111/j.1440-1827.2008.02306.x. [DOI] [PubMed] [Google Scholar]
- 31.Weichert W, et al. High expression of RelA/p65 is associated with activation of nuclear factor-κB-dependent signaling in pancreatic cancer and marks a patient population with poor prognosis. British journal of cancer. 2007;97(4):523–530. doi: 10.1038/sj.bjc.6603878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasaki N, et al. Nuclear factor-κB p65 (RelA) transcription factor is constitutively activated in human gastric carcinoma tissue. Clinical Cancer Research. 2001;7(12):4136–4142. [PubMed] [Google Scholar]
- 33.Li W, et al. Constitutive activation of nuclear factor-kappa B (NF-kB) signaling pathway in fibrolamellar hepatocellular carcinoma. International journal of clinical and experimental pathology. 2010;3(3):238. [PMC free article] [PubMed] [Google Scholar]
- 34.Prusty BK, Husain SA, Das BC. Constitutive activation of nuclear factor-kB: preferential homodimerization of p50 subunits in cervical carcinoma. Front Biosci. 2005;10(5):1510–1519. doi: 10.2741/1635. [DOI] [PubMed] [Google Scholar]