Abstract
The Protein Kinase C family of enzymes is a group of serine/threonine kinases that play central roles in cell-cycle regulation, development and cancer. A key step in the activation of PKC is translocation to membranes and binding of membrane-associated activators including diacylglycerol (DAG). Interaction of novel and conventional isotypes of PKC with DAG and phorbol esters occurs through the two C1 regulatory domains (C1A and C1B), which exhibit distinct ligand binding selectivity that likely controls enzyme activation by different co-activators.
PKC has also been implicated in physiological responses to alcohol consumption and it has been proposed that PKCα [1, 2], PKCε [3] and PKCδ [4, 5] contain specific alcohol-binding sites in their C1 domains. We are interested in understanding how ethanol affects signal transduction processes through its affects on the structure and function of the C1 domains of PKC. Here we present the 1H, 15N and 13C NMR chemical shift assignments for the Rattus norvegicus PKCδ C1A and C1B proteins.
Keywords: Protein Kinase C Delta, C1A domain, C1B domain, diacyl glycerol, Phorbol ester binding
Biological Context
There is a significant body of evidence that alcohols can directly interact with and modulate PKC activity, including PKCδ [1–4, 6, 7]. PKCδ null mice are less sensitive to ethanol intoxication, indicating that the enzyme contributes to the behavioral response to the drug [7]. The adaptive increase of L-type calcium channels in response to chronic ethanol exposure is also contingent upon PKCδ [8], demonstrating that the enzyme may be involved in neuronal hyperexcitability that occurs during withdrawal and so contribute to a propensity for alcoholism. Understanding the basic mechanisms by which ethanol produces its cellular and physiological effects is important for developing new approaches for treating the disease.
PKC enzymes are separated into three different families based on their domain organization and cofactor requirements. Activation requires a series of phosphorylation steps and conformational changes in the protein that are induced by the binding of co-activators at membrane surfaces. Conventional PKC (cPKC) isozymes (α, βI, βII and γ) ubind calcium ions and phosphatidylserine at their C2 domain in addition to either DAG, or exogenously-applied phorbol esters, through their C1 domains. In contrast, novel PKC (nPKC) isozymes (δ, ε, η and θ) do not require calcium, but can be activated by DAG or phorbol esters alone. This calcium independence is achieved by the presence of C1 domains that have a higher affinity for ligands than those of the conventional isozymes [9].
The structures of the C1B domains from PKCα [10], and PKCγ [11] have been solved by NMR and the structure of the C1B from PKCδ domain has been solved by X-ray crystallography [12]. Further, the crystal structure of the complex between PKCδ C1B and phorbol acetate shows that ligand binding occurs between two juxtaposed hairpin loops [12]. These domains exhibit a similar overall fold that is maintained by the presence of two C3H1 zinc coordination sites. It is highly likely that the C1A domain of PKCδ will have a similar fold as the C1B domain. However, these domains bind selectivity to different ligands [9, 13–18] and are potentially differentially affected by alcohols [1]. It is difficult to explain the mechanistic basis for these difference as there is no structural information available for the C1A domain and structure of the C1B domain suggest very few changes occur upon ligand binding. By solving the structure of the C1A domain and comparing the effects of alcohol on ligand binding to different C1 domains, we will be in a position to better define the mechanism of alcohol action on PKC and how this contributes to cellular changes associated with alcohol consumption. Towards this end, we report the 1H, 15N and 13C NMR chain chemical shift assignments of the PKCδ C1A and C1B domains as a precursor to NMR studies to examine how alcohols influence C1:ligand/membrane interactions.
Methods
Protein Expression and Purification
PKCδ C1A and C1B proteins were cloned, expressed and purified according to standard protocols. Briefly, Rattus norvegicus cDNA was used as a template to separately subclone the C1A and C1B sequences between the BamHI and SalI restriction sites of pGEX-4T1 vector (GE Healthcare) and generate constructs containing an N-terminal GST tag for protein expression. The resulting plasmids were verified by DNA sequencing and transformed into Rosetta 2 E. coli cells (Novagen) for protein expression. The C1A construct contains a total of 80 amino acids (labeled 149–228) derived from PKCδ (residues 154–218) and residues from the pGEX-4T1 vector (residues 149–153 and 219–228). The C1B construct contains 82 amino acids (labeled 214–295) derived from PKCδ (residues 219–285) and residues from the pGEX-4T1 vector (residues 214–218 and 286–295). The solubility of both domains was greatly improved by inclusion of these additional residues, without which the proteins were expressed predominantly into inclusion bodies.
For NMR studies, proteins were expressed from cell cultures grown in minimal media supplemented with 2 g/L 13C D-glucose (>99 atom %) and 1 g/L 15N NH4Cl (> 98 atom %). Cells were grown at 37 °C to an OD600 of 0.3–0.4, and protein expression induced by addition of 1 mM IPTG for 18 hours at 25°C. Cells were disrupted by treatment with lysozyme followed by sonication on ice, and the resulting solution treated with DNAseI. The clarified soluble fraction was applied to glutathione sepharose 4B resin (GE Heathcare) and washed with 20 mM Tris-HCl pH 7.0, 150 mM NaCl, 10 uM ZnCl, 1mM DTT, 25 mM CaCl2 and 0.01% Triton X-100. Soluble protein was eluted in the same buffer following incubation with 50–100 units of thrombin at room temperature for 18 hours and purified by size exclusion chromatography using Superdex-75 resin (Amersham Biosciences) in the same buffer. Protein yields range between 8–13 milligrams per liter of cell growth, and concentrations of 300–500 uM are readily achievable. The identity and integrity of the final proteins was confirmed by electrospray mass spectrometry and SDS-PAGE analysis.
NMR Experiments
NMR experiments were recorded using 15N and 13C labeled C1A and C1B protein (300–500uM) in 20 mM sodium phosphate at pH 6.9, 10% D2O, 100 mM NaCl, 10 uM ZnCl, 1 mM DTT and a trace amount of 2,2,-dimethyl-2-silapentane-5-sulfonic acid (DSS) as the chemical shift reference All experiments were performed at 25°C on Varian INOVA spectrometers operating at 500 MHz, 600 MHz (with cryoprobe) or 900 MHz using the Varian supplied pulse sequences. Assignments of the main-chain atoms were made using: 2D 1H/15N-HSQC, 2D 1H/13C-HSQC, 3D HNCO, 3D HCA(CO)CANH, 3D HNCACB and 3D CBCA(CO)NH and 3D 1H,15N-HSQC-NOESY-HSQC and 15N-edited NOESY-HSQC. While assignments of side chain 13C and 1H resonances were made using 3D HCCH-TOCSY, 3D (H)C(CO)NH-TOCSY, and 2D (HB)CB(CGCD)HD and 2D (HB)CB(CGCDCE)HE for aromatic side chains. Data were processed with NMRpipe [19] and resonance assignments were determined using the Analysis program of the CCPNmr package [20].
Extent of assignments
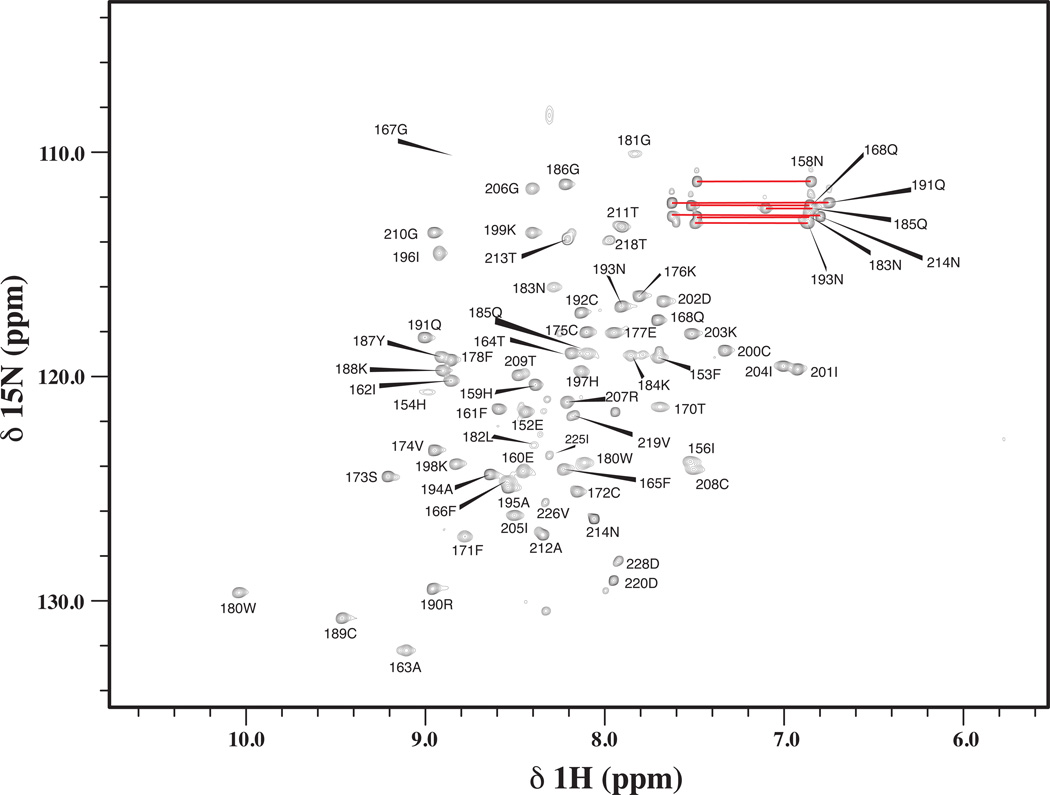
Figure 1 presents the 1H,15N-HSQC spectrum of PKCδ C1A and illustrates backbone amide assignments. The majority of residues from C1A domain (PKCδ residues 159–208) exhibited strong well dispersed NMR peaks, indicating that it is well structured. Several residues outside of the C1A domain (residues 149–153 and 219–228 derived from the pGEX-4T1vector) were absent from the HSQC spectrum or are not involved in long-range NOEs, indicating that they are unstructured. For the C1A domain, backbone resonance assignments have been obtained for 98% of the HN/N pairs (48 out of 49 non-proline residues). All 13Cα, 13CO, and 53 of 54 1Hα backbone atoms have been assigned. In addition, 87% (201 of 232) of 1H and 82% (120 out of 147) of 13C side chain atoms have been assigned. Outside of the C1A domain, 154–156, 209–214 and 218 from PKCδ and residues 152, 219, 220, 225, 226 and 228 from pGEX-4T1 have been assigned. The assignments for the PKCδ C1A have been deposited into the BioMagResBank database under accession number 17113.
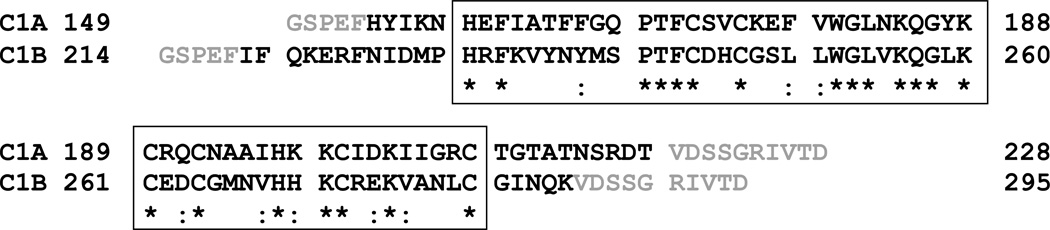
Figure 1.
Alignment of protein constructs used in this study. Amino acids derived from the wild-type C1A and C1B domains are in black and residues derived from the pGEX-4T1 vector are in grey. The extents of the conserved C1 domains are indicated in the boxed region, identical residues are indicated with an asterisk (*) and strongly conserved residues are indicated with a colon (:). Alignment and analysis based on CLUSTAL W algorithm [21].
The 1H, 15N-HSQC spectrum of the C1B domain of PKCδ is presented in Figure 2 and is labeled with backbone amide assignments. Within the C1B domain (residues 231–280), most amide groups exhibit strong and disperse NMR signals, indicative of a structured protein. However, in contrast to the C1A domain, there are several amide resonances within the core C1 domain that are completely missing in the 1H-15N-HSQC spectrum. These resonances are not observed at a range of spectrometer frequencies (500MHz–900MHz) and temperatures, implying that they are in intermediate exchange on the NMR timescale. Because of this, backbone resonance assignments for the C1B protein have been obtained for only 76% of the HN/N pairs (37 out of 49 non-Proline residues). In addition, 86% of 13Cα (43 out of 50), 84% of 13CO (42 out of 50), and 85% of 1Hα (46 out of 54) backbone atoms have been assigned. While assignments have been obtained for 68% (159 out of 235) of the 1H atoms and 67% (96 out of 144) of the 13C side chain atoms. The missing assignments almost completely coincide with residues for which 1HN chemical shifts could not be obtained due to intermediate exchange. Outside of the C1B domain, residues 219–229, 281–285 derived from PKCδ and residues 217, 218 and 286–295 derived from the pGEX-4T1 have also been assigned. The 1H, 13C and 15N assignments for PKCδ C1B have been deposited into the BioMagResBank database under accession number 17112.
Figure 2.
1H,15N-HSQC spectrum of PKCδ C1A recorded at 600 MHz. Each peak labeled with the chemical shift assignments. The side chain amides for Asn and Gln residues are linked by horizontal bars.
The missing assignments within the C1B domain are for residues 237–239, 241–242, 251–254, 256–257 and 258 and these residues are mostly located in the loops of that form the ligand binding site in C1B (Figure 4). The addition of ligands including phorbol acetate, phorbol esters, or DAG does not lead to any improvement in the NMR spectra, and these residues are still not observable, suggesting that there is a significant degree of conformational exchange in these regions of the protein.
Figure 4.
Ribbon diagram of the core C1B domain of PKCδ (based on PDB ID 1PTQ [12]). The location of the two zinc binding sites are shown in orange. Residues that cannot be observed in either 1H,15N-HSQC or 1H,13C-HSQC are colored red. This includes almost all residues in the phorbol acetate binding loops between residues N237-T242 and L251-G258. This directly contrast with the C1A domain, where all these residues can be observed
Discussion
In this work, we have determined the backbone and side chain resonance assignments for the C1A and C1B regulatory domains of PKCδ. These domains are important in binding to regulatory co-factors and have been implicated in the potential alcohol sensitivity of this protein. This study has revealed significant differences in the intrinsic dynamics of the C1A domains compared to the C1B domain. There are a number of residues in the C1B domain that are in intermediate exchange on the NMR timescale and are not observable in the 1H-15N HSQC spectrum of this domain, indicating that they are significantly more dynamic than the same region in the C1A domain (Figure 4) [12]. The majority of these resonances belong to amino acids residing in the two proximal loops that bind phorbol acetate. In contrast, the corresponding residues in the C1A domain of PKCδ are all observable suggesting this region is significantly more ordered. This difference in dynamics may account for previously observed differences in ligand binding specificity of these domains.
The crystal structures of the C1B domain suggest that there are very few if any changes in the structure of the protein upon binding phorbol acetate, and that the ligand binding site is well structured in the absence of ligand [12]. Our studies suggest that this is not the case in solution. However, this does not appear to be a result of any significant difference in the overall solution structure compared to the crystal structure. A number of medium (i>2) and long range (i >4) HN to HN NOEs that are predicted from the crystal structure are clearly observed in the HSQC-NOESY-HSQC and NOESY-HSQC spectra which supports the conclusion that the overall structure observed in the crystal is essentially maintained in solution. We are currently investigating how alcohols affect the structure and dynamics of these domains, and how alcohol influences binding of ligands to these domains.
Figure 3.
1H,15N-HSQC spectrum of PKCδ C1B recorded at 600 MHz. Each observable amide is labeled with its assignments within this construct. The side chain amides for Asn and Gln residues are linked by horizontal bars.
Acknowledgements
We thank Dr. Geoff Armstrong for his assistance with operation of the 900 MHz spectrometer. This work was supported by NIH/NIAAA grant AA013618-06A1 to DNMJ. The operation of the NMR facilities is supported by the University of Colorado Cancer Center Support Grant (NIH 5P30-CA046934) and operation of the 900 MHz NMR by NIH grant 5P41-GM068928.
Literature Cited
- 1.Slater SJ, Kelly MB, Larkin JD, Ho C, Mazurek A, Taddeo FJ, Yeager MD, Stubbs CD. Interaction of alcohols and anesthetics with protein kinase Calpha. J. Biol. Chem. 1997;272:6167–6173. doi: 10.1074/jbc.272.10.6167. [DOI] [PubMed] [Google Scholar]
- 2.Slater SJ, Malinowski SA, Stubbs CD. The nature of the hydrophobic n-alkanol binding site within the C1 domains of protein kinase Calpha. Biochemistry. 2004;43:7601–7609. doi: 10.1021/bi049755z. [DOI] [PubMed] [Google Scholar]
- 3.Das J, Pany S, Rahman GM, Slater SJ. PKC epsilon has an alcohol-binding site in its second cysteine-rich regulatory domain. Biochem J. 2009;421:405–413. doi: 10.1042/BJ20082271. [DOI] [PubMed] [Google Scholar]
- 4.Das J, Addona GH, Sandberg WS, Husain SS, Stehle T, Miller KW. Identification of a general anesthetic binding site in the diacylglycerol-binding domain of protein kinase Cdelta. J Biol Chem. 2004;279:37964–37972. doi: 10.1074/jbc.M405137200. [DOI] [PubMed] [Google Scholar]
- 5.Das J, Zhou X, Miller KW. Identification of an alcohol binding site in the first cysteine-rich domain of protein kinase Cdelta. Protein Sci. 2006;15:2107–2119. doi: 10.1110/ps.062237606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rex EB, Rankin ML, Ariano MA, Sibley DR. Ethanol regulation of D(1) dopamine receptor signaling is mediated by protein kinase C in an isozyme-specific manner. Neuropsychopharm. 2008;33:2900–2911. doi: 10.1038/npp.2008.16. [DOI] [PubMed] [Google Scholar]
- 7.Choi DS, et al. Protein kinase Cdelta regulates ethanol intoxication and enhancement of GABA-stimulated tonic current. J Neurosci. 2008;28:11890–11899. doi: 10.1523/JNEUROSCI.3156-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerstin EH, Jr, McMahon T, Dadgar J, Messing RO. Protein kinase Cdelta mediates ethanol-induced up-regulation of L-type calcium channels. J Biol Chem. 1998;273:16409–16414. doi: 10.1074/jbc.273.26.16409. [DOI] [PubMed] [Google Scholar]
- 9.Giorgione JR, Lin JH, McCammon JA, Newton AC. Increased membrane affinity of the C1 domain of protein kinase Cdelta compensates for the lack of involvement of its C2 domain in membrane recruitment. J Biol Chem. 2006;281:1660–1669. doi: 10.1074/jbc.M510251200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ichikawa S, Hatanaka H, Takeuchi Y, Ohno S, Inagaki F. Solution structure of cysteine-rich domain of protein kinase C alpha. J Biochem (Tokyo) 1995;117:566–574. doi: 10.1093/oxfordjournals.jbchem.a124745. [DOI] [PubMed] [Google Scholar]
- 11.Xu RX, Pawelczyk T, Xia TH, Brown SC. NMR structure of a protein kinase C-gamma phorbol-binding domain and study of protein-lipid micelle interactions. Biochemistry. 1997;36:10709–10717. doi: 10.1021/bi970833a. [DOI] [PubMed] [Google Scholar]
- 12.Zhang G, Kazanietz MG, Blumberg PM, Hurley JH. Crystal structure of the cys2 activator-binding domain of protein kinase C delta in complex with phorbol ester. Cell. 1995;81:917–924. doi: 10.1016/0092-8674(95)90011-x. [DOI] [PubMed] [Google Scholar]
- 13.Ananthanarayanan B, Stahelin RV, Digman MA, Cho W. Activation mechanisms of conventional protein kinase C isoforms are determined by the ligand affinity and conformational flexibility of their C1 domains. J. Biol. Chem. 2003;278:46886–46894. doi: 10.1074/jbc.M307853200. [DOI] [PubMed] [Google Scholar]
- 14.Dries DR, Gallegos LL, Newton AC. A single residue in the C1 domain sensitizes novel protein kinase C isoforms to cellular diacylglycerol production. J. Biol. Chem. 2007;282:826–830. doi: 10.1074/jbc.C600268200. [DOI] [PubMed] [Google Scholar]
- 15.Giorgione J, Hysell M, Harvey DF, Newton AC. Contribution of the C1A and C1B domains to the membrane interaction of protein kinase C. Biochemistry. 2003;42:11194–11202. doi: 10.1021/bi0350046. [DOI] [PubMed] [Google Scholar]
- 16.Johnson JE, Giorgione J, Newton AC. The C1 and C2 domains of protein kinase C are independent membrane targeting modules, with specificity for phosphatidylserine conferred by the C1 domain. Biochemistry. 2000;39:11360–11369. doi: 10.1021/bi000902c. [DOI] [PubMed] [Google Scholar]
- 17.Slater SJ, Ho C, Kelly MB, Larkin JD, Taddeo FJ, Yeager MD, Stubbs CD. Protein kinase Calpha contains two activator binding sites that bind phorbol esters and diacylglycerols with opposite affinities. J. Biol. Chem. 1996;271:4627–4631. doi: 10.1074/jbc.271.9.4627. [DOI] [PubMed] [Google Scholar]
- 18.Stahelin RV, Digman MA, Medkova M, Ananthanarayanan B, Rafter JD, Melowic HR, Cho W. Mechanism of diacylglycerol-induced membrane targeting and activation of protein kinase Cdelta. J Biol Chem. 2004;279:29501–29512. doi: 10.1074/jbc.M403191200. [DOI] [PubMed] [Google Scholar]
- 19.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 20.Vranken WF, et al. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 21.Higgins DG, Bleasby AJ, Fuchs R. CLUSTAL V: Improved software for multiple sequence alignment. Computer Applications in the Biosciences (CABIOS) 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]