Abstract
N6-methyladenosine (m6A) is an abundant internal modification in eukaryotic mRNA and plays regulatory roles in mRNA metabolism. However, methods to precisely locate the m6A modification remain limited. We present here a photo-crosslinking-assisted m6A sequencing strategy (PA-m6A-seq) to more accurately define sites with m6A modification. Using this strategy, we obtained a high-resolution map of m6A in a human transcriptome. The map resembles the general distribution pattern observed previously, and reveals new m6A sites at base resolution. Our results provide insight into the relationship between the methylation regions and the binding sites of RNA-binding proteins.
Keywords: N6-methyladenosine, photo-crosslinking, RNA modification, transcriptome sequencing
Post-transcriptional modifications are important features of RNA molecules.[1] Particularly, N6-methyladenosine (m6A) is a ubiquitous modification found within eukaryotic messenger RNA and various nuclear noncoding RNAs.[2] m6A formation in the nucleus is catalyzed by a complex containing methyltransferase like 3 (METTL3), methyltransferase like 14 (METTL14), and Wilms’ tumor 1-associating protein (WTAP).[3] Recent discoveries indicate that two human AlkB family proteins, fat mass and obesity-associated protein (FTO) and ALKBH5, serve as RNA demethylases to remove m6A in mammalian poly(A)-tailed RNA, indicating that RNA methylation is reversible and plays dynamic roles in related biological processes.[4] A “reader” protein of m6A, YTHDF2, has been recently shown to specifically recognize thousands of mRNA methylation sites and mediates a methylation-dependent mRNA decay, thus demonstrating a significant role of m6A in mRNA metabolism.[5]
Precise knowledge of m6A locations within the mammalian transcriptome is essential to understanding its biological function. The recently developed high-throughput method, termed m6A-seq or MeRIP-seq (m6A-specific methylated RNA immunoprecipitation with next-generation sequencing), utilizes anti-m6A antibodies for the capture and enrichment of the m6A-containing RNA fragments, followed by high-throughput sequencing to profile m6A distributions in mammalian transcriptomes. This modification was shown to accumulate at 3’-UTR around stop codons and within exons.[6] The resolution of these maps hovers around 200 nt and therefore cannot pinpoint the precise locations of the m6A.[6] A higher-resolution map of yeast m6A methylome has been generated with an improved approach of m6A-seq using shorter fragments to identify m6A sites.[7] A ligation-based detection and SCARLET (site-specific cleavage and radioactive-labeling followed by ligation-assisted extraction and thin-layer chromatography) were also developed to precisely determine methylation sites with single-nucleotide resolution.[8] The SCARLET method, based on site-specific RNase H or DNAzyme cleavage, is effective but also time-consuming, and is not yet feasible for high-throughput applications.[9]
Photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) is a photo-crosslinking-based method to identify binding sites of RNA-binding proteins with high resolution.[10] A photoactivatable ribonucleoside, 4-thiouridine (4SU) or 6-thioguanosine (6SG), is incorporated into messenger RNA and covalently crosslinks with nearby aromatic amino acid residues in RNA-binding proteins upon 365 nm UV irradiation. Inspired by PAR-CLIP, we applied a similar approach, named photo-crosslinking-assisted m6A-sequencing (PA-m6A-seq), which efficiently improves the accuracy of the methylation site assignments, and provides a high-resolution transcriptome-wide mammalian m6A map (~ 23 nt) [GEO: GSE54921]
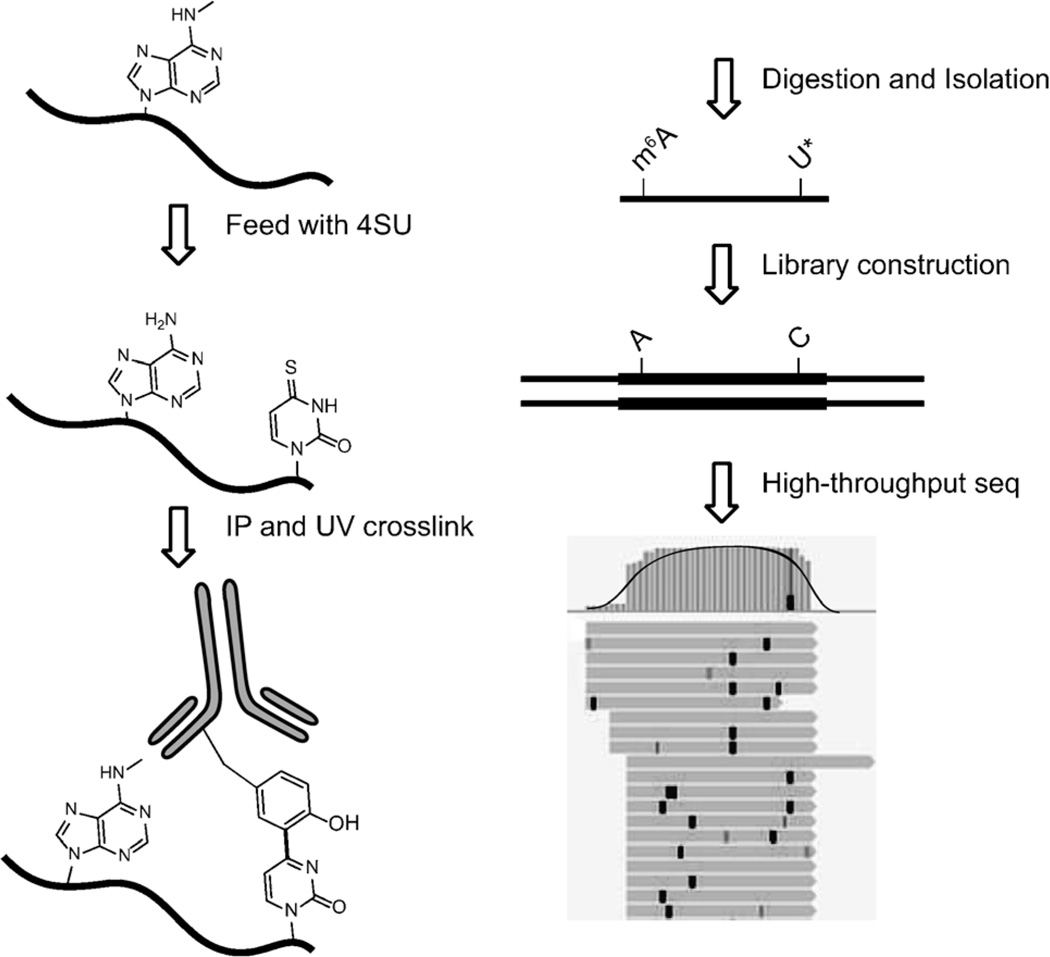
The photo-crosslinking-assisted m6A-seq strategy is shown in Scheme 1.[6a, 10, 11] HeLa cells readily uptake and incorporate 4-thiouridine (4SU) into RNA when 4SU is added to growth medium. The 4SU-containing mRNA is purified by oligo-dT-conjugated magnetic beads. Similar to the procedure of m6A-seq, an immunoprecipitation (IP) step is performed, in which we use full-length rather than fragmented mRNA molecules. After the IP step, the sample is irradiated by 365 nm UV light to initiate crosslinking. Crosslinked RNA is digested to around 30 nt using RNase T1 and further processed to possess a 5′ phosphate group and a 3′ hydroxy group. RNA fragments are washed and extracted with TRIzol reagent after proteinase K digestion to remove covalently bonded peptides. Libraries are prepared from purified RNA by using Illumina TruSeq Small RNA Prep Kit.
Scheme 1.
The strategy of photo-crosslinking-assisted m6A-seq (PA-m6A-seq). Covalently crosslinked 4SU is labeled as U*, which is read as C in RT-PCR. The example of the high-throughput sequencing result is shown on the bottom right. Black bold vertical bars represent T-to-C transition induced by 4SU and covalent crosslinking, compared to reference genome hg19.
4SU, in which oxygen at the 4′ position is substituted by sulfur, forms a thioketone structure. The effect of substitution of sulfur, similar to the effect of substitution of bromine in 5-bromouridine, significantly decreases the bond dissociation energy, facilitating the homolysis of the carbon–sulfur bond and the formation of a radical. The rearrangement and deprotonation of 4-thiouridine leads to the T-to-C transition then the base-pair reading changes in PCR step.[12]
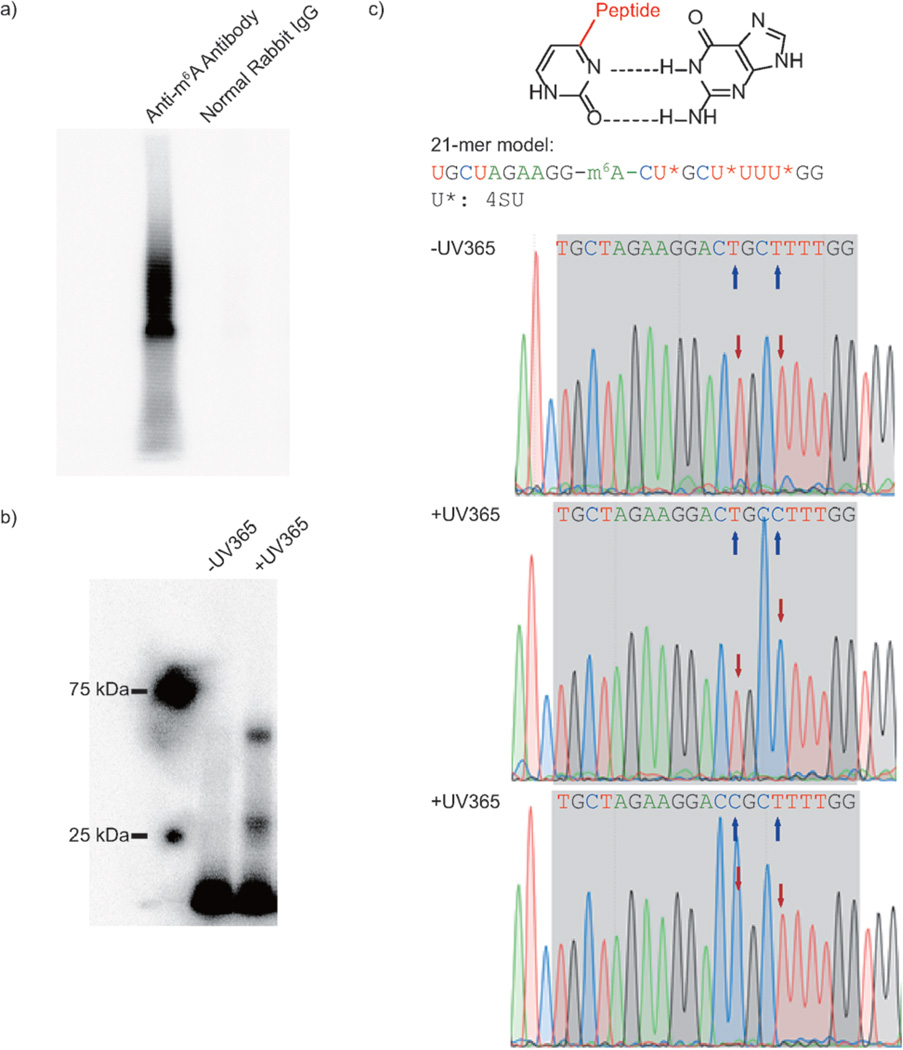
The specificity and immunoprecipitation capability of the anti-m6A antibody has been well documented in previously published works.[4a, 13] However, it is still necessary to confirm that crosslinking comes from the specific recognition of m6A by the antibody. Two parallel immunoprecipitation reactions were established, one with anti-m6A polyclonal rabbit IgG, the other with normal rabbit IgG. With the same treatment, only the anti-m6A antibody afforded visible radioactive signals, demonstrating that specific m6A recognition by the selected antibody is critical for crosslinking (Figure 1 a).
Figure 1.
Model study using PA-m6A-seq. a) Control study to confirm crosslinking is based on recognition of antibody against m6A-containing RNA. b) Control study to prove UV365 is the trigger of covalent crosslinking by using a synthesized 21-mer RNA oligonucleotide. c) T-to-C transition introduced by 4SU and UV irradiation. Proposed mechanism on how the 4SU crosslinked with protein changes the Watson–Crick base pair (top). The sequence of 21-mer model is shown in the middle. Sanger sequencing results of 21-mer model with and without UV365 proved the crosslinked 4SU near m6A was read as C. The blue arrows indicate the 4SU sites on the model, whereas red arrows point out the chromatogram reads of corresponding 4SU with or without crosslinking.
To further confirm that 365 nm UV irradiation triggers 4SU-based crosslinking, a 21-mer RNA oligonucleotide containing 4SU and m6A was synthesized as a model substrate (Figure S1; Supporting Information, SI). Along with the irradiation, the radioactive signal also appeared, representing the crosslinked complex, which confirms that the 4SU-mediated crosslinking is light-dependent (Figure 1 b).
Furthermore, crosslinked 21-mer RNA oligonucleotide was isolated and inserted into a vector for Sanger sequencing, showing that the covalent crosslinking between 4SU and antibody is indeed able to introduce a T-to-C transition nearby m6A, as in the in vivo PAR-CLIP (SI). The T-to-C transition in model work ensures the reliability of this strategy and allows for the use of PAR-CLIP algorithms to analyze PA-m6A-seq data (Figure 1c). With the model work above demonstrating the effectiveness of the strategy, we proceeded with two biological replicates of 4SU-incorporated HeLa cell mRNA for high-throughput sequencing.
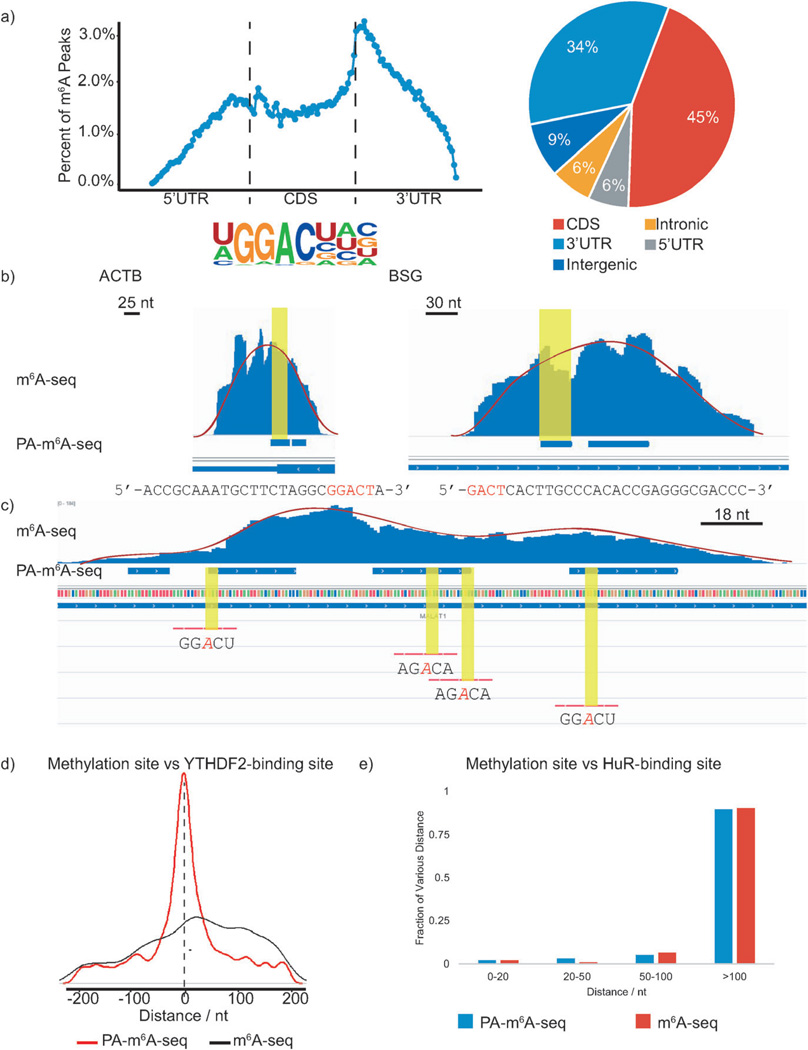
The libraries were constructed following the procedure shown in Scheme 1, and were subjected to high-throughput sequencing. We identified 13 486 m6A peaks within the human transcriptome, with an average length of 23 nt; much shorter than previous published results (Figure S2, SI).[6] We further classified our reads into five different segments: 5′-untranslated region (5′UTR), coding DNA sequence (CDS), 3′UTR, intergenic region, and intronic region. The distribution of our data confirmed that m6A is significantly enriched near the stop codon and mainly localized in CDS and 3′UTR, consistent with previously published results (Figure 2 a).[6]
Figure 2.
PA-m6A-seq applied to poly(A)-tailed RNA purified from HeLa cells. In following figures, blue bars represent methylation sites identified by PA-m6A-seq, whereas blue ‘peaks’ above those bars are from normal m6A-seq. a) Validation of PA-m6A-seq strategy. Metagene profile and pie-chart of the enrichment of RNA segments are consistent with previous reported distribution of m6A, and the motif search yielded GGACU as the predominant one, which was the same as the result from normal m6A-seq. b) Comparison of predicted methylation sites in β-actin (ACTB) and homo sapien basigin (BSG) from PA-m6A-seq with peaks from normal m6A-seq and single sites by SCARLET. Sequences of predicted sites are shown below, consensus motif containing m6A in red. All input background of normal m6A-seq has been subtracted. Both sites were verified by SCARLET. c) Multiple methylation sites in MALAT1 transcript. The methylation sites confirmed by SCARLET, which were covered by peaks from normal m6A-seq, were also identified by PA-m6A-seq with higher resolution. Yellow regions indicate the RRACH motif. Red lines are the probes used to verify these sites with SCARLET. d) Distance of predicted methylation sites using PA-m6A-seq versus peaks obtained from normal m6A-seq to YTHDF2-binding sites. e) Distance of predicted methylation sites obtained from PA-m6A-seq versus peaks obtained from normal m6A-seq to HuR-binding sites.
The consensus sequence for the m6A modification is known as R1R2-m6A-CH (R = G or A; H = A, C, or U; R2 : G > A).[13c, 14] The unbiased motif search based on two sets of high-throughput data uncovered a similar consensus.[6] Here, we used the HOMER motif discovery tool to analyze the possible consensus sequence of m6A.[15] Indeed, GGACU is the most enriched motif in our data (Figure 2a).
After validation, we performed further comparison and analyses with our higher-resolution map. The methylation sites identified by PA-m6A-seq can be confirmed by SCARLET and m6A-seq/MeRIP-seq. Blue horizontal bars and blue peaks were chosen to represent PA-m6A-seq and original m6A-seq peaks, respectively. The SCARLET-identified sites were emphasized in red, shown in the same figures. For example, methylation sites on β-actin mRNA (ACTB) and homo sapien basigin mRNA (BSG) were previously identified by m6A-seq and precisely detected using SCARLET.[6a, 8b] We uploaded our new high-resolution map and compared its contents of methylation sites in ACTB and BSG transcripts to the published results, which showed that these SCARLET-identified methylated regions exist in a 30 nt region in PA-m6A-seq map, whereas they were found in the 200 nt region using m6A-seq/MeRIP-seq (Figure 2b).
The higher-resolution map also suggests a “clustering” property of m6A deposition on transcripts, which is similar to the methylation of cytosine on genomic DNA. Multiple methylation sites were identified in transcripts such as MALAT1 by PA-m6A-seq (Figure 2c). These single sites previously confirmed by using SCARLET were also discovered by our PA-m6A-seq strategy.[8b] Intriguingly, the multiplicity of methylation or the “clustering” property of m6A on transcripts implies the likelihood that these transcripts are highly affected by m6A reader proteins as recently suggested, resembling DNA 5-methylcytosine methylation.[5]
The power of PA-m6A-seq lies in its ability to identify single consensus methylation sequences within a ~ 23 base region, enabling single-base resolution detection of the m6A modification. Hence, we used SCARLET as an independent approach to validate new methylation sites found in PA-m6A-seq, which confirmed the ability of PA-m6A-seq to pinpoint methylation site in a transcriptome-wide manner (Figure S3; SI).
Next, we analyzed the spatial relationship between the methylation sites and the binding sites of two RNA-binding proteins shown to recognize m6A. Wang et al. proved that YTHDF2 is a selective binder/reader of m6A, and identified over 3000 RNA targets using PAR-CLIP.[5] The high-resolution methylation sites identified by PA-m6A-seq overlap very well with the binding sites of YTHDF2. The high-resolution map obtained in this study allowed us to conclude that most YTHDF2-binding sites are within 30–50 nt of the m6A sites, strongly supporting direct interactions of YTHDF2 with m6A and the regulatory role of m6A in YTHDF2-mediated RNA decay (Figure 2d).
Together with YTHDF2, another well-studied RNA-binding protein, HuR (ELAVL1), was also pulled down using an m6A-containing bait.[6a] In contrast to YTHDF2, the consensus sequence recognized by HuR is very different from that of the m6A site.[16] A recent work evaluating the binding of HuR to various probes containing m6A suggested interesting spatial constraints that affect potential interactions between HuR and m6A.[3e] We attempted to further probe the binding sites of HuR and the high-resolution m6A sites on mRNA by applying the same analysis shown above for YTHDF2, and plotting the distribution of distances between the high-resolution methylation sites and HuR-binding sites. The results of both PA-m6A-seq and normal m6A-seq analyses indicate that the majority of the HuR-binding sites are further away (100 nt) from the m6A site (Figure 2e). This analysis suggests that HuR may “indirectly” (through other proteins or mRNA structure changes) interact with m6A if it associates with m6A.
In summary, we have established a photo-crosslinking-assisted strategy to improve the resolution of m6A-seq/MeRIP-seq, and report a high-resolution methylation map of the mammalian transcriptome. The covalent crosslinking and effective RNase T1 digestion resulted in a significant resolution improvement on localization of m6A on fragmented RNA. On the whole, the general analyses validated our strategy, and the higher-resolution map showed the ability to reveal new methylation sites at base resolution. Furthermore, more precise methylation location made it possible to explore the distance between m6A peaks and the binding sites of RNA-binding proteins. These analyses confirm that YTHDF2 directly binds m6A, whereas direct interaction of m6A by HuR is less likely, which suggests that HuR may affect m6A through other mechanisms. The PA-m6A-seq approach presented here can be widely applied to study precise m6A sites in various organisms and the UV-crosslinking strategy is compatible with investigation on nucleic acid–protein interactions.
Experimental Section
High throughput sequencing and bioinformatics analysis: Adapters were trimmed by FASTX and also the first and last bases were removed because of the sequencing quality. Then the reads were mapped to the human genome (hg19) using bowtie with two mismatches at most.[17] PARalyzer was used for peak calling.[18] 23880658 and 21456137 reads were mapped to the genome separately for each replicate and 22 396 and 25 509 peaks were called by PARalyzer. We took the 13 486 peaks shared by two replicates as reliable peak sets for the following analysis. Refseq genes were used for peaks annotation. These peaks were mapped to 6176 genes (13 499 isoforms) (see Table in the SI).
Supplementary Material
Footnotes
C. He is supported as an Investigator of the Howard Hughes Medical Institute. D. Dominissini is supported by an HFSP fellowship. Q.D. is supported by the US National Institutes of Health grant 5K01HG006699 (Q.D.). We thank I. Roundtree and T. Shpidel for suggestions and editing.
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.201410647.
Contributor Information
Kai Chen, Department of Chemistry, Institute for Biophysical Dynamics, Howard Hughes Medical Institute, The University of Chicago, 929 East 57th Street, Chicago, IL 60637 (USA).
Zhike Lu, Department of Chemistry, Institute for Biophysical Dynamics, Howard Hughes Medical Institute, The University of Chicago, 929 East 57th Street, Chicago, IL 60637 (USA).
Xiao Wang, Department of Chemistry, Institute for Biophysical Dynamics, Howard Hughes Medical Institute, The University of Chicago, 929 East 57th Street, Chicago, IL 60637 (USA).
Ye Fu, Department of Chemistry, Institute for Biophysical Dynamics, Howard Hughes Medical Institute, The University of Chicago, 929 East 57th Street, Chicago, IL 60637 (USA).
Guan-Zheng Luo, Department of Chemistry, Institute for Biophysical Dynamics, Howard Hughes Medical Institute, The University of Chicago, 929 East 57th Street, Chicago, IL 60637 (USA).
Nian Liu, Department of Biochemistry and Molecular Biology, Institute for Biophysical Dynamics, The University of Chicago, 929 East 57th Street, Chicago, IL 60637 (USA).
Dali Han, Department of Chemistry, Institute for Biophysical Dynamics, Howard Hughes Medical Institute, The University of Chicago, 929 East 57th Street, Chicago, IL 60637 (USA).
Dan Dominissini, Department of Chemistry, Institute for Biophysical Dynamics, Howard Hughes Medical Institute, The University of Chicago, 929 East 57th Street, Chicago, IL 60637 (USA).
Qing Dai, Department of Chemistry, Institute for Biophysical Dynamics, Howard Hughes Medical Institute, The University of Chicago, 929 East 57th Street, Chicago, IL 60637 (USA).
Tao Pan, Department of Biochemistry and Molecular Biology, Institute for Biophysical Dynamics, The University of Chicago, 929 East 57th Street, Chicago, IL 60637 (USA).
Chuan He, Department of Chemistry, Institute for Biophysical Dynamics, Howard Hughes Medical Institute, The University of Chicago, 929 East 57th Street, Chicago, IL 60637 (USA).
References
- 1.He C. Nat. Chem. Biol. 2010;6:863–865. doi: 10.1038/nchembio.482. [DOI] [PubMed] [Google Scholar]
- 2.a) Desrosiers RC, Friderici KH, Rottman FM. Proc. Natl. Acad. Sci. USA. 1974;71:3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Desrosiers RC, Friderici KH, Rottman FM. Biochemistry. 1975;14:4367–4374. doi: 10.1021/bi00691a004. [DOI] [PubMed] [Google Scholar]; c) Jia G, Fu Y, He C. Trends Genet. 2013;29:108–115. doi: 10.1016/j.tig.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Fu Y, Dominissini D, Rechavi G, He C. Nat. Rev. Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 3.a) Bokar JA, Rath-Shambaugh ME, Ludwiczak R, Narayan P, Rottman F. J. Biol. Chem. 1994;269:17697–17704. [PubMed] [Google Scholar]; b) Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. RNA. 1997;3:1233–1247. [PMC free article] [PubMed] [Google Scholar]; c) Liu J, et al. Nat. Chem. Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Ping XL, et al. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. Nat. Cell Biol. 2014;16:191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Jia G, et al. Nat. Chem. Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zheng G, et al. Mol. Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, et al. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Dominissini D, et al. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]; b) Meyer K, Saletore Y, Zumbo P, Elemento O, Mason C, Jaffrey S. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartz S, et al. Cell. 2013;155:1409–1421. doi: 10.1016/j.cell.2013.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Dai Q, Fong R, Saikia M, Stephenson D, Yu YT, Pan T, Piccirilli JA. Nucleic Acids Res. 2007;35:6322–6329. doi: 10.1093/nar/gkm657. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Liu N, Parisien M, Dai Q, Zheng G, He C, Pan T. RNA. 2013;19:1848–1856. doi: 10.1261/rna.041178.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Yu YT, Shu MD, Steitz JA. RNA. 1997;3:324–331. [PMC free article] [PubMed] [Google Scholar]; b) Zhao X, Yu YT. RNA. 2004;10:996–1002. doi: 10.1261/rna.7110804. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Hengesbach M, Meusburger M, Lyko F, Helm M. RNA. 2008;14:180–187. doi: 10.1261/rna.742708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hafner M, et al. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dominissini D, Moshitch-Moshkovitz S, Salmon-Divon M, Amariglio N, Rechavi G. Nat. Protoc. 2013;8:176–189. doi: 10.1038/nprot.2012.148. [DOI] [PubMed] [Google Scholar]
- 12.a) Dietz TM, Koch TH. Photochem. Photobiol. 1987;46:971–978. doi: 10.1111/j.1751-1097.1987.tb04879.x. [DOI] [PubMed] [Google Scholar]; b) Dietz TM, Vontrebra RJ, Swanson BJ, Koch TH. J. Am. Chem. Soc. 1987;109:1793–1797. [Google Scholar]; c) Ito S, Saito I, Matsuura T. J. Am. Chem. Soc. 1980;102:7535–7541. [Google Scholar]
- 13.a) Bringmann P, Lührmann R. FEBS Lett. 1987;213:309–315. doi: 10.1016/0014-5793(87)81512-0. [DOI] [PubMed] [Google Scholar]; b) Dante R, Niveleau A. FEBS Lett. 1981;130:153–157. doi: 10.1016/0014-5793(81)80686-2. [DOI] [PubMed] [Google Scholar]; c) Horowitz S, Horowitz A, Nilsen TW, Munns TW, Rottman FM. Proc. Natl. Acad. Sci. USA. 1984;81:5667–5671. doi: 10.1073/pnas.81.18.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Munns TW, Liszewski MK, Oberst RJ, Sims HF. Biochemistry. 1978;17:2573–2578. doi: 10.1021/bi00606a018. [DOI] [PubMed] [Google Scholar]; e) Munns TW, Liszewski MK, Sims HF. Biochemistry. 1977;16:2163–2168. doi: 10.1021/bi00629a019. [DOI] [PubMed] [Google Scholar]; f) Munns TW, Oberst RJ, Sims HF, Liszewski MK. J. Biol. Chem. 1979;254:4327–4330. [PubMed] [Google Scholar]; g) Munns TW, Sims HF, Liszewski MK. J. Biol. Chem. 1977;252:3102–3104. [PubMed] [Google Scholar]
- 14.a) Canaani D, Kahana C, Lavi S, Groner Y. Nucleic Acids Res. 1979;6:2879–2899. doi: 10.1093/nar/6.8.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Dimock K, Stoltzfus CM. Biochemistry. 1977;16:471–478. doi: 10.1021/bi00622a021. [DOI] [PubMed] [Google Scholar]; c) Harper JE, Miceli SM, Roberts RJ, Manley JL. Nucleic Acids Res. 1990;18:5735–5741. doi: 10.1093/nar/18.19.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Kane SE, Beemon K. Mol. Cell. Biol. 1985;5:2298–2306. doi: 10.1128/mcb.5.9.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Schibler U, Kelley DE, Perry RP. J. Mol. Biol. 1977;115:695–714. doi: 10.1016/0022-2836(77)90110-3. [DOI] [PubMed] [Google Scholar]; f) Wei CM, Moss B. Biochemistry. 1977;16:1672–1676. doi: 10.1021/bi00627a023. [DOI] [PubMed] [Google Scholar]
- 15.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lebedeva S, Jens M, Theil K, Schwanhäusser B, Selbach M, Landthaler M, Rajewsky N. Mol. Cell. 2011;43:340–352. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 17.Langmead B, Trapnell C, Pop M, Salzberg SL. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corcoran DL, Georgiev S, Mukherjee N, Gottwein E, Skalsky RL, Keene JD, Ohler U. Genome Biol. 2011;12:R79. doi: 10.1186/gb-2011-12-8-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.