Abstract
Rhesus and cynomolgus macaques are frequently used in biomedical research, and the availability of their reference genomes now provides for their use in genome-wide association studies. However, little is known about linkage disequilibrium (LD) in their genomes, which can affect the design and success of such studies. Here we studied LD by using 1781 conserved single-nucleotide polymorphisms (SNPs) in 183 rhesus macaques (Macaca mulatta), including 97 purebred Chinese and 86 purebred Indian animals, and 96 cynomolgus macaques (M. fascicularis fascicularis). Correlation between loci pairs decayed to 0.02 at 1146.83, 2197.92, and 3955.83 kb for Chinese rhesus, Indian rhesus, and cynomolgus macaques, respectively. Differences between the observed heterozygosity and minor allele frequency (MAF) of pairs of these 3 taxa were highly statistically significant. These 3 nonhuman primate taxa have significantly different genetic diversities (heterozygosity and MAF) and rates of LD decay. Our study confirms a much lower rate of LD decay in Indian than in Chinese rhesus macaques relative to that previously reported. In contrast, the especially low rate of LD decay in cynomolgus macaques suggests the particular usefulness of this species in genome-wide association studies. Although conserved markers, such as those used here, are required for valid LD comparisons among taxa, LD can be assessed with less bias by using species-specific markers, because conserved SNPs may be ancestral and therefore not informative for LD.
Abbreviations: GWAS, genome-wide association study; LD, linkage disequilibrium; MAF, minor allele frequency
Contributing to the widespread use of nonhuman primates in biomedical research, captive-breeding programs such as those of the National Primate Research Center system in the United States were established initially by using animals imported from Asia. The 2 most commonly used primates are rhesus macaques (Macaca mulatta) and long-tailed or cynomolgus macaques (M. fascicularis fascicularis).
After humans, rhesus macaques are the most widely distributed primate species.37,38 This species is found throughout mainland Asia, ranging from Afghanistan to India and eastward through Thailand and southern China to the Yellow Sea.31,34 In addition to their significant morphological differences,9 rhesus macaques of Indian and Chinese origins have been demonstrated to exhibit significant phenotypic differences that are directly relevant to their use as biomedical models in experimental studies.2,23,42 Cynomolgus macaques are found south of the subtropical and temperate geographic distributions of rhesus macaques, in the south and southeast Indo-Malayan regions.8,10
The 2 species share a common ancestor that lived 1 to 2 million years ago.3,13,25 This ancestral population of rhesus macaques diverged from a fascicularis-like ancestor shared in common with both rhesus and cynomolgus macaques after cynomolgus macaques expanded from their homeland in Indonesia.36 For this reason, genetic markers present in Indian rhesus macaques are either highly derived or are conserved as ancestral markers shared with Chinese rhesus macaques. The interspecific boundaries of rhesus and cynomolgus macaques are delineated by a narrow zone of parapatry in northern Indochina,7,8,10 within which male-biased gene flow37,39 and relatively high, but highly variable, levels of introgression of genes32 have occurred from rhesus to cynomolgus macaque groups.37,39 Because cynomolgus macaques originated in Indonesia36 and because rhesus macaques probably diverged from cynomolgus macaques in southwestern China,11 genetic markers shared between Indonesian cynomolgus macaques and Chinese rhesus macaques comprise a unique set of markers that are conserved in both macaque species.
The wide assortment of morphometric differences8,9 and the broad geographic distribution of these 2 macaque species foster an expectation of high genetic diversity within and between them that could be exploited for mapping genes responsible for phenotypic differences between taxa. A better understanding of linkage disequilibrium (LD) in these nonhuman primate species can lead to a more informed selection of study subjects for, and more efficient conduct of, genome-wide association studies (GWAS) of particular diseases that macaques share in common with humans. LD is the nonrandom association of alleles at 2 or more adjacent loci that descend from single, ancestral chromosomes.29 LD plays a critical role in gene mapping, both as a tool for fine mapping of complex disease genes and in GWAS-based approaches. GWAS facilitate the identification of genes associated with complex and common traits or diseases by examining LD estimates among large numbers of common genetic variants, typically single-nucleotide polymorphisms (SNPs), between pairs of different groups of subjects to determine whether any variant is associated with a trait or disease of interest. LD data make tightly linked variants strongly correlated to produce successful association studies. For instance, LD reduces the number of markers and sample size of study subjects required to map genes influencing phenotypes to the genome because markers in LD are linked and inherited together.13 In addition, differences in LD can be used to identify orthologs for detecting the signatures of selective sweeps,21 as defined by dN/dS ratios obtained through the McDonald–Kreitman neutrality test.24 Furthermore, LD assessments can provide a more complete understanding of genome structure by defining the boundaries of haplotype blocks, within which recombination is rare or absent and which are separated by recombination ‘hotspots,’ in genomes.43
Evidence from a study based on 1476 SNPs identified in ENCODE regions of the Indian rhesus macaque genome13 indicated that the rate of LD decay is higher in Chinese than in Indian rhesus macaques due to an hypothesized genetic bottleneck experienced by Indian rhesus macaques after diverging from the eastern subspecies, and, therefore, that Indian rhesus macaques, having higher LD, may be more useful for GWAS than Chinese rhesus macaques. In that study,13 only 33% of the SNPs were shared in common between the 2 subspecies, with Chinese rhesus macaques contributing to more than 60% of the remaining rhesus SNPs. Conversely, another study41 reported a slower rate of decay of LD in 25 Chinese than in 25 Indian rhesus macaques on the basis of 4040 SNPs, only 2% of which fell in coding regions, but 68% of those SNPs were shared between the 2 subspecies, with Indian rhesus macaques contributing almost 60% of the remaining SNPs. The marked disparity between the 2 studies in the proportions of shared SNPs used, the subspecies with the most genetic diversity, the sample size of Chinese rhesus macaques, the proportions of SNPs located in or near coding regions that are subject to functional constraints, and the greater disparity in LD decay between the 2 subspecies of rhesus macaques might reflect biases in either or both studies. For example, the use of markers whose frequencies are uncharacteristically low in one subspecies relative to the other can underestimate the rate of LD decay because lower frequency alleles, on average, are younger and have experienced less time for recombination.26 To avoid the influence of such ascertainment biases, comparisons of LD between 2 taxa should involve only SNPs conserved in both taxa. Moreover, because 2 points do not provide a phylogenetic or cladistic analysis to assign specific SNPs to origin on one phylogenetic line or another, comparing just the Indian and Chinese rhesus macaques without an additional primate taxon makes it is difficult to establish polarity and distinguish between derived and conserved SNPs. This limitation likely led to the contradictory conclusions of the 2 previously cited studies13,41 regarding the rate of LD decay in Chinese and Indian rhesus macaques.
Because rhesus and cynomolgus macaques share a common fascicularis-like ancestor, a comparison of heterospecific SNPs among cynomolgus, Indian rhesus, and Chinese rhesus macaques would likely be fundamental to inferences regarding genome-wide LD estimates. The objective of the present study was to evaluate the conclusions of previous studies13,41 by using our panel of 1781 autosomal SNPs that are conserved in both rhesus and cynomolgus macaques to estimate the rates at which genome-wide LD decays in Indian and Chinese rhesus macaques and cynomolgus macaques, the species ancestral to rhesus macaques, and to evaluate the suitability of these populations for GWAS.
Materials and Methods
The research reported here adhered with the approved protocols of the UC Davis IACUC and the legal requirements of the United States, where the research took place.
Whole-blood samples were obtained from 189 (100 Chinese; 89 Indian) rhesus macaques maintained at the California National Primate Research Center in housing arrangements as reported previously.16 DNA was extracted and quantified as described previously,19 and the samples were genotyped for 14 short tandem repeat markers,15 to confirm the animals’ geographic origin prior to SNP genotyping. Whole-blood samples from 112 unrelated cynomolgus macaques (M. f. fascicularis) were a gift of Primate Products (Immokalee, FL). All of these animals were captive bred in cynomolgus-macaque-only breeding facilities outside the United States and were imported into this country by Primate Products. All blood samples were drawn into EDTA-treated vacuum phlebotomy tubes by veterinary staff at Primate Products according to standard operating procedures. Any macaque that showed unexpected adverse effects, such as stress or trauma, was treated according to the standard operating procedures of Primate Products.
The 1781 SNP used in this study represent a subset of the 2808 rhesus macaque SNPs previously identified in cynomolgus macaques.19 SNPs, especially those with low minor allele frequencies (MAF), will have been lost in the more derived populations of rhesus (non-Chinese populations) and cynomolgus (non-Indonesian populations) macaques, a feature that might bias results by overestimating rates of LD decay in both species, but such bias is not anticipated to favor one of the 2 species over the other. Therefore, to minimize the bias of ascertainment resulting from the use of the 2808 markers that were first identified in rhesus macaques, we used only those 1781 SNPs that were conserved in both the Chinese rhesus and the Indonesian cynomolgus macaques in this analysis. All samples were processed and genotyped according to previously described methods.19
Because these markers were discovered in rhesus macaques, their mapped positions in the rhesus genome were used for subsequent analyses and to determine intermarker distances. Principal component analysis was completed by using the R package Adegenet 1.4-214 to identify population structure and outliers that could skew results. Any outliers were removed prior to further analysis to improve the results of LD estimation. Observed and expected heterozygosities for the Indian and Chinese rhesus and cynomolgus macaque populations and pairwise fixation indexes were calculated for all population pairs by using Arlequin version 3.5.1.36 (http://cmpg.unibe.ch/software/arlequin35/) and minor allele frequency (MAF) were calculated by using Plink v.1.0727 (http://pngu.mgh.harvard.edu/purcell/plink/). The differences in observed and expected heterozygosity and MAF estimates among the 3 study groups were evaluated for statistical significance by using a Kruskal–Wallis test. LD values were estimated in Haploview 4.21 (www.broad.mit.edu/mpg/haploview/) as r2, the correlation coefficient between the allele frequencies of the 2 SNPs,12 according to previously described methods.41 MAF values for LD pairs were binned in intervals of 0.1 for frequency-matched pairs,4 and only LD pairs with MAF values greater than or equal to 0.01 were used, to ensure that rare alleles did not represent sequencing errors.20,22 The r2 estimates were plotted against the distances (in kilobases) between marker pairs, and a nonlinear least-squares regression line that applies Remington and colleagues's30 equation to estimate linkage decay was fitted by using R script (http://www.rilab.org/code/files/LDit.html). Although LD dissipates completely when the correlation between a pair of loci is zero (r2 = 0),41 we report results as the decay distance as calculated from Remington and colleagues's30 equation when r2 was set at 0.02, to reduce sampling error at greater distances between SNPs.
Results
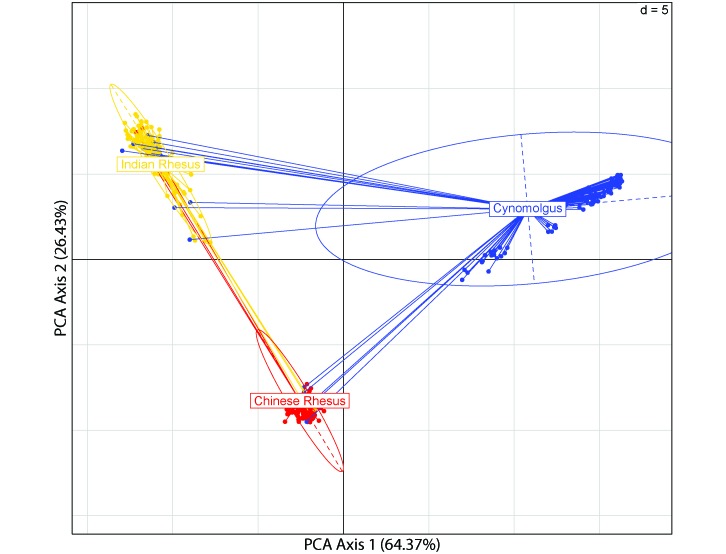
The principal component analysis (Figure 1) revealed 22 outliers among the 3 populations: 3 each in the Chinese and Indian rhesus macaques and 16 in the cynomolgus macaques. These outliers were not interspecific hybrids, given the isolated captive breeding of the cynomolgus macaques prior to their importation into the United States, which precluded any possibility of crossbreeding between rhesus and cynomolgus macaques. Furthermore, pedigree records from the California National Primate Research Center for the rhesus macaques sampled here show no evidence of crossbreeding between the Chinese and Indian rhesus macaques. After omission of the outliers, 97 Chinese rhesus, 86 Indian rhesus, and 96 cynomolgus macaques remained for subsequent analyses. All 1781 SNPs were distributed across the 20 macaque autosomes, and almost all of these SNPs could be genotyped for 90% or more of the members of both rhesus macaque populations (1 marker was 89.69% complete in Chinese rhesus macaques, and 3 Indian rhesus subjects were less than 90% complete, with the lowest at 77.15% completeness). However, 28 of the SNPs provided genotypes for fewer than 90% of the cynomolgus macaque samples, the least complete of which provided genotypes for only 60.42% of the samples.
Figure 1.
Principal component analysis identifying the population structure and outliers in each population. The majority of the variance in the data (90.8%) is explained by the first 2 components, and their respective percentages are given on the axes. Ellipses indicate 95% of the total variance of each study population, and grid intervals show the coordinates of each individual (points) in ±5 units in relation to 0 (black grid lines).
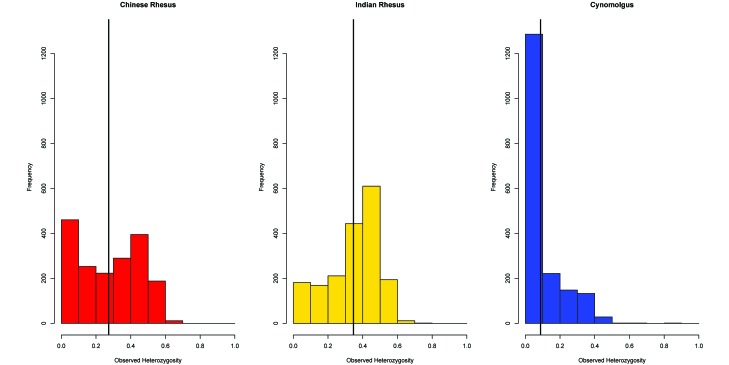
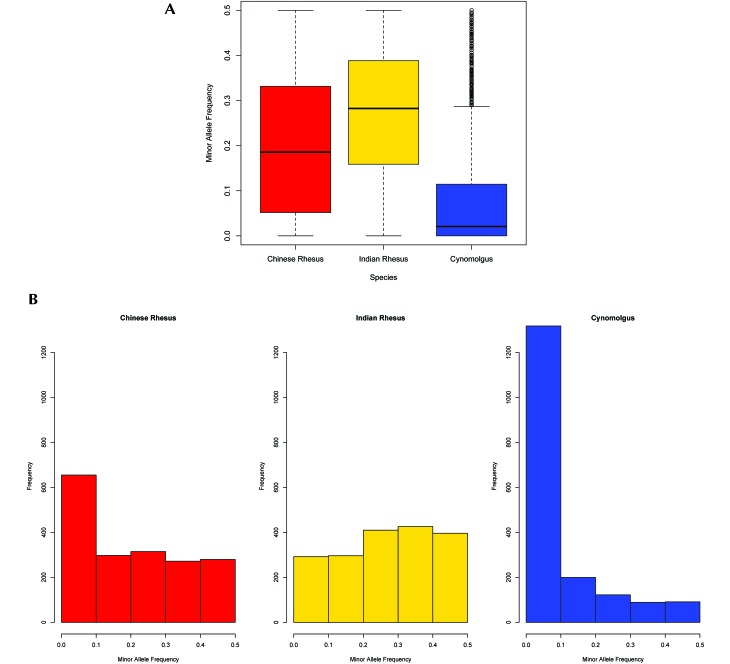
Average observed and expected heterozygosities and MAF for all species are found in Table 1, and their overall distributions appear in Figures 2 and 3A. The distribution of MAF values binned in 0.1 intervals is shown in Figure 3B. Cynomolgus macaques exhibited a pronounced deficiency in estimates of observed heterozygosity (0.088), estimated heterozygosity (0.126), and MAF (0.088) compared with both rhesus subspecies (0.350, 0.355, and 0.271, respectively, for Indian and 0.275, 0.272 and 0.199, respectively, for Chinese rhesus macaques). The Kruskal–Wallis test revealed a statistically significant effect of group on both observed and expected heterozygosity and MAF estimates (P < 0.01 for all comparisons). A posthoc test using Mann–Whitney tests with Bonferroni correction revealed statistically significant differences in heterozygosity and MAF estimates between Indian rhesus and cynomolgus macaques (P < 0.01) and between Chinese rhesus and cynomolgus macaques (P < 0.01). These lower values of all 3 parameters in cynomolgus macaques probably reflect the ascertainment bias of selecting SNPs identified in rhesus macaques that had been conserved during their divergence from cynomolgus macaques, and the higher value of estimated heterozygosity (0.126) relative to that of observed heterozygosity (0.088) in cynomolgus macaques probably reflects the genetic subdivision within this species. Not unexpectedly, genetic differentiation based on pairwise fixation index estimates was higher between cynomolgus and rhesus macaques (0.425 with Indian and 0.358 with Chinese rhesus) than between the 2 rhesus macaques (0.202; Table 2). The greater distance of cynomolgus macaques from Indian than from Chinese rhesus macaques undoubtedly reflects the more highly derived status of Indian rhesus macaques.
Table 1.
Mean heterozygosities and minor allele frequency for each species
| Population | Overall heterozygosity | Estimated heterozygosity | Minor allele frequency |
| Chinese rhesus macaques (n = 97) | 0.275 | 0.272 | 0.199 |
| Indian rhesus macaques (n = 86) | 0.350 | 0.355 | 0.271 |
| Cynomolgus macaques (n = 96) | 0.088 | 0.126 | 0.088 |
Figure 2.
Histograms of observed heterozygosity (OH) for both Macaca species. Mean heterozygosities are significantly different (P < 0.01).
Figure 3.
(A) Average minor allele frequency (MAF) values in each species. The differences between mean MAF are significant (P < 0.01). (B) MAF distributions for each 0.1-MAF bin in each species.
Table 2.
Pairwise fixation indexes
| Chinese (n = 97) | Indian (n = 86) | |
| Indian (n = 86) | 0.202 | |
| Cynomolgus (n = 96) | 0.358 | 0.425 |
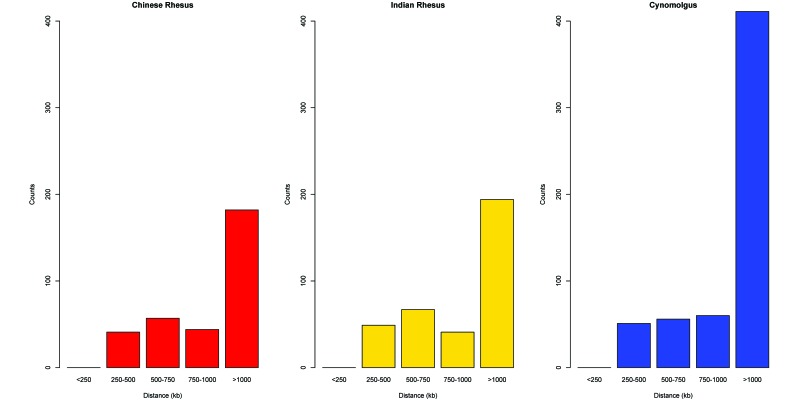
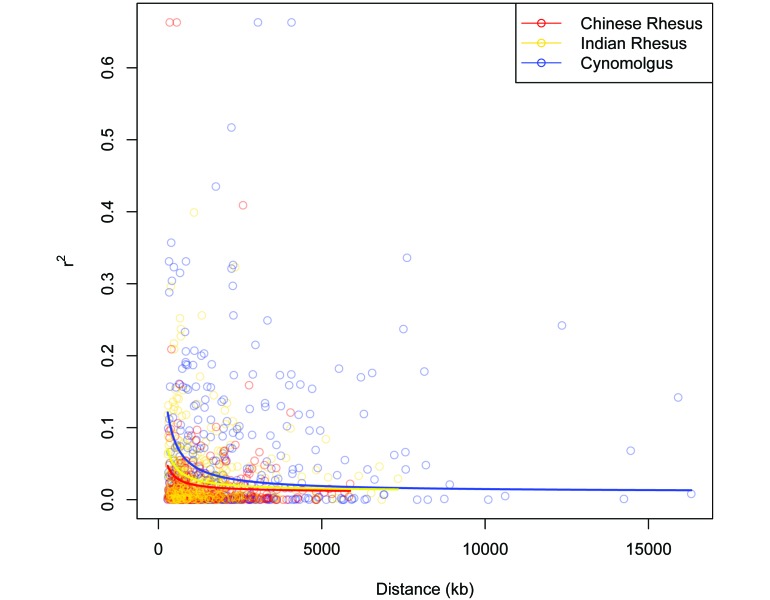
The genomic coverage of these markers was approximately 1 SNP every 1.58 Mb (± 292 kb; Table 3). The marker pair counts and average distances are listed in Table 4, and the distribution of the LD marker pair distances is shown in Figure 4. The average intermarker distances for cynomolgus macaques (2.05 Mb) exceeded that for both Indian (1.45 Mb) and Chinese (1.41 Mb), again reflecting an ascertainment bias in the selection of SNPs. Of the 1781 SNPs, 1753 (98.4%) were polymorphic in all 3 populations (Chinese rhesus, Indian rhesus, and cynomolgus macaques), and 28 (1.6%) were monomorphic, the minor allele having been lost, in the Indian rhesus population. In addition, 324, 351, and 578 marker pairs were frequency matched with MAF values greater than or equal to 0.01 for the Chinese rhesus, Indian rhesus, and cynomolgus macaques, respectively. The r2 values for each LD marker pair were plotted against the intermarker distances for these 3 populations (Figure 5). LD decayed to r2 = 0.02 at 1146.83 kb for Chinese rhesus, 2197.92 kb for Indian rhesus, and 3955.83 kb for cynomolgus macaques.
Table 3.
Gaps and MAF values for each chromosome of each species
| Rhesus chromosome no. | No. of SNPs | Mean gap (kb) | Median gap (kb) | Chinese MAF | Indian MAF | Cynomolgus MAF |
| 1 | 155 | 1441.33 | 1091.03 | 0.219 | 0.268 | 0.085 |
| 2 | 134 | 1402.99 | 1090.18 | 0.207 | 0.270 | 0.074 |
| 3 | 121 | 1629.82 | 991.86 | 0.196 | 0.280 | 0.101 |
| 4 | 98 | 1685.48 | 1352.60 | 0.201 | 0.287 | 0.096 |
| 5 | 142 | 1276.63 | 908.09 | 0.205 | 0.252 | 0.076 |
| 6 | 118 | 1499.34 | 1024.29 | 0.201 | 0.288 | 0.081 |
| 7 | 126 | 1346.33 | 1007.58 | 0.203 | 0.266 | 0.085 |
| 8 | 104 | 1424.05 | 1011.12 | 0.169 | 0.262 | 0.099 |
| 9 | 94 | 1408.11 | 1126.77 | 0.214 | 0.269 | 0.085 |
| 10 | 68 | 1338.31 | 1147.22 | 0.192 | 0.304 | 0.076 |
| 11 | 93 | 1392.85 | 954.30 | 0.209 | 0.274 | 0.085 |
| 12 | 54 | 1863.75 | 1397.68 | 0.215 | 0.270 | 0.068 |
| 13 | 100 | 1361.55 | 850.08 | 0.205 | 0.260 | 0.087 |
| 14 | 73 | 1831.24 | 1404.06 | 0.222 | 0.262 | 0.099 |
| 15 | 68 | 1606.46 | 1355.44 | 0.171 | 0.268 | 0.079 |
| 16 | 46 | 1717.69 | 1496.39 | 0.198 | 0.274 | 0.130 |
| 17 | 74 | 1243.15 | 955.03 | 0.188 | 0.247 | 0.077 |
| 18 | 44 | 1644.88 | 1585.08 | 0.209 | 0.287 | 0.087 |
| 19 | 26 | 2410.40 | 1353.10 | 0.182 | 0.268 | 0.092 |
| 20 | 43 | 2061.54 | 1437.38 | 0.172 | 0.275 | 0.098 |
| Overall | 1579.29 | 1176.96 |
Table 4.
Distance counts for LD marker pairs
| No. of pairs |
|||
| Distance (kb) between markers | Chinese rhesus | Indian rhesus | Cynomolgus |
| <250 | 0 | 0 | 0 |
| 250–500 | 41 | 49 | 51 |
| 500–750 | 57 | 67 | 56 |
| 750–1000 | 44 | 41 | 60 |
| >1000 | 182 | 194 | 411 |
| Average marker pair distance | 1584.66 | 1473.21 | 2467.98 |
Figure 4.
LD marker pair distance distributions for all populations.
Figure 5.
LD r2 plotted against physical distance (kb), with nonlinear least-squares regression lines for the rhesus and cynomolgus macaque populations.
Discussion
The observed and estimated heterozygosities and the MAF values across the 1781 loci we examined were higher for rhesus than for cynomolgus macaques, with Indian rhesus macaques exhibiting more variability than Chinese rhesus macaques. This result is consistent with some previous findings18,19,40,41 but in conflict with results based on studies of mitochondrial DNA sequences,34,35 short tandem repeat loci33 and the expected increasing decline in genetic diversity with increasing distance from the geographic origin of a species.28 The 4040 SNPs from which the 1781 used here were derived are probably biased toward high-frequency SNPs by their method of discovery,41 and the occurrence of low-frequency SNPs among the group of 4040 markers was twice as high in Chinese than in Indian rhesus macaques, suggesting that the 1781 SNPs that we used were representative of the 4040 from which they were selected. The unexpected lower genetic diversity of Chinese, relative to Indian, rhesus macaques, despite the fact that Indian macaques are the derived subspecies, may have been influenced by the method of identification of the original 4040 SNPs or by higher rates of evolution of mitochondrial DNA and short tandem repeats than SNPs, especially in a rapidly expanding population, but there is no reason to expect this effect to influence LD of SNPs that originated in the ancestral rhesus macaque population.
Even though we took precautions, such as using only conserved SNPs, to minimize ascertainment bias, the 1781 SNPs studied here were discovered initially in rhesus macaques,41 a factor that may have led to some underestimates of MAF and heterozygosity in cynomolgus macaques. However, neither that bias nor the loss of some of these SNPs in Indian rhesus macaques or cynomolgus macaques likely significantly influenced estimates of LD in the direction of results described earlier. We assume that all 1781 pairs of alleles analyzed in the present study are ancestral to cynomolgus macaques and derived in rhesus macaques, because rhesus macaques are more derived than are cynomolgus macaques9 and because homoplasy in SNPs is rare owing to their low mutation rate.5
The rates of LD decay in Indian and Chinese rhesus macaques in the present study are more similar than those estimated in earlier studies,13,41 which reported a much faster rate of decay of LD in Chinese than in Indian rhesus macaques. However, our results do not support the previous argument13 that a past genetic bottleneck in Indian rhesus macaques caused them to experience a slower LD decay than Chinese rhesus macaques and that, as a result, Indian rhesus macaques are better suited than the Chinese rhesus macaques for GWAS. Such a genetic bottleneck would, in fact, be inconsistent with the greater level of genetic diversity in Indian than in Chinese rhesus macaques seen in both earlier studies13,41 and the present study. The larger differences in LD estimates of a previous study13 may have resulted from functional constraints on the SNPs used, which would be much more stringent on Indian than Chinese rhesus macaques, and the small sample size of Chinese rhesus macaques used in that study. The proliferation of new, necessarily low-frequency alleles associated with the expansion of rhesus macaques westward to India might have delayed LD decay because these alleles, eliminated from the present analysis by the use of conserved SNPs, would have experienced less time to achieve LD and are functionally constrained by purifying selection. In addition, insufficient population representation of Chinese rhesus macaques13 can fail to capture recombination events and overestimate haplotype block size.43 Another study41 that used large sample sizes of both species yielded a smaller discrepancy between the rates of LD decay between the 2 rhesus macaque subspecies but did not exclude SNPs present in Indian, but not Chinese, rhesus macaques.
In the present study, cynomolgus macaques exhibited a much lower rate of linkage decay compared with both rhesus macaque populations. This higher LD could not have resulted from the same ascertainment bias that caused their lower values of heterozygosity and MAF, because the low-frequency alleles in cynomolgus macaques are ancestral in rhesus macaques and, therefore, have had a greater, not less, opportunity for recombination to dissipate LD than alleles in rhesus macaques.26 The level of population substructure in cynomolgus macaques17 may have influenced the results of this analysis, but such influences have not been studied systematically. Despite being recognized as a single subspecies, that is, M. f. fascicularis, the cynomolgus macaque population we used here is more diversified (fixation index, 0.391) than are both subspecies of rhesus macaques (Chinese, 0.280; Indian, 0.313), because, unlike rhesus macaques, which comprise a single geographically contiguous species, cynomolgus macaques have had fewer opportunities for natural hybridization, having been confined to island populations for much of their evolutionary past.
Because the 2 species studied share a common Pleistocene ancestor,3,13,25 some SNPs screened in the present study are probably derived in both species and may be linked in larger blocks due to ‘hitchhiking’ neighboring SNPs.4 Due to their common ancestral origins, SNPs conserved in multiple populations are prone to ascertainment biases that might obscure inferences of LD at the genomic level in the descendant populations but enable preliminary comparisons of closely related genomes. Species-specific SNPs might be biased by stochastic events in the subdivided cynomolgus macaque populations such that an unbiased estimate of LD might be possible only for Indonesian cynomolgus macaques, still extant in their species’ original homeland. Although the higher LD in cynomolgus macaques suggests that fewer SNPs would be required for disease-association studies than would be necessary for such experiments in the rhesus macaques we studied, additional studies of LD in larger samples of regionally representative populations of cynomolgus macaques are ongoing to assess the potentially complex influences of genetic subdivision and population histories on LD in cynomolgus macaques.
Acknowledgments
This study was supported by the California National Primate Research Center base grant (No. RR000169-48), ARRA supplement grant RR018144-07 to SK, and NIH grants RR005090 and RR025871 to DGS. We thank the staff of the Molecular Anthropology Laboratory at University of California–Davis for their assistance with sample preparation. We are also grateful to the three anonymous reviewers for their insightful comments, which helped to significantly improve this manuscript.
References
- 1.Barrett JC, Fry B, Maller J, Daly MJ. 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265. [DOI] [PubMed] [Google Scholar]
- 2.Cohen J. 2000. Vaccine studies stymied by shortage of animals. Science 287:959–960. [DOI] [PubMed] [Google Scholar]
- 3.Delson E. 1980. Fossil macaques, phyletic relationships, and a scenario of deployment, p 10–30 In: Lindburg DG. The macaques: studies in ecology, behavior, and evolution. New York (NY): Van Nostrand Reinhold. [Google Scholar]
- 4.Eberle MA, Rieder MJ, Kruglyak L, Nickerson DA. 2006. Allele frequency matching between SNP reveals an excess of linkage disequilibrium in genic regions of the human genome. PLoS Genet 2:e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ebersberger I, Metzler D, Schwarz C, Pääbo S. 2002. Genomewide comparison of DNA sequences between humans and chimpanzees. Am J Hum Genet 70:1490–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Excoffier L, Laval G, Schneider S. 2007. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 7.Fittinghoff NA, Jr, Lindburg DG. 1980. Riverine refuging in East Bornean Macaca fascicularis. In: Lindburg DG. The macaques: studies in ecology, behavior, and evolution. New York (NY): Van Nostrand Reinhold. [Google Scholar]
- 8.Fooden J. 1995. Systematic review of Southeast Asian longtail macaques, Macaca fascicularis (Raffles 1821). Fieldiana Zoology 81:1–206. [Google Scholar]
- 9.Fooden J. 2000. Systematic review of the rhesus macaque, Macaca mulatta (Zimmermann 1780). Fieldiana Zoology 96:1–180. [Google Scholar]
- 10.Groves CP. 2001. Primate taxonomy. Washington (DC): Smithsonian Institution Press. [Google Scholar]
- 11.Hasan MK, Feeroz MM, Jones-Engel L, Engel GA, Smith DG. 2014. Origin and dispersal of rhesus macaques. Int J Primatol In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hedrick P, Kumar S. 2001. Mutation and linkage disequilibrium in human mtDNA. Eur J Hum Genet 9:969–972. [DOI] [PubMed] [Google Scholar]
- 13.Hernandez RD, Hubisz MJ, Wheeler DA, Smith DG, Ferguson B, Rogers J, Nazareth L, Indap A, Bourquin T, McPherson J, Muzny D, Gibbs R, Nielsen R, Bustamante CD. 2007. Demographic histories and patterns of linkage disequilibrium in Chinese and Indian rhesus macaques. Science 316:240–243. [DOI] [PubMed] [Google Scholar]
- 14.Jombart T. 2008. Adegenet: an R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405. [DOI] [PubMed] [Google Scholar]
- 15.Kanthaswamy S, Kou A, Satkoski J, Penedo MC, Ward T, Ng J, Gill L, Lerche NW, Erickson BJ, Smith DG. 2010. Genetic characterization of specific pathogen-free rhesus macaque (Macaca mulatta) populations at the California National Primate Research Center (CNPRC). Am J Primatol 72:587–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanthaswamy S, Kou A, Smith DG. 2010. Population genetic statistics from rhesus macaques (Macaca mulatta) in 3 different housing configurations at the California National Primate Research Center. J Am Assoc Lab Anim Sci 49:598–609. [PMC free article] [PubMed] [Google Scholar]
- 17.Kanthaswamy S, Ng J, Satkoski Trask J, George DA, Kou AJ, Hoffman LN, Doherty TB, Houghton P, Smith DG. 2013. The genetic composition of populations of cynomolgus macaques (Macaca fascicularis) used in biomedical research. J Med Primatol 42:120–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanthaswamy S, Satkoski J, Kou A, Malladi V, Glenn Smith D. 2010. Detecting signatures of inter-regional and interspecific hybridization among the Chinese rhesus macaque specific pathogen-free (SPF) population using single-nucleotide polymorphic (SNP) markers. J Med Primatol 39:252–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanthaswamy S, Trask JS, Ross CT, Kou A, Houghton P, Smith DG, Lerche N. 2012. A large-scale SNP-based genomic admixture analysis of the captive rhesus macaque colony at the California National Primate Research Center. Am J Primatol 74:747–757. [DOI] [PubMed] [Google Scholar]
- 20.Kim SY, Lohmueller K, Albrechtsen A, Li Y, Korneliussen T, Tian G, Grarup N, Jiang T, Andersen G, Witte D, Jorgensen T, Hansen T, Pedersen O, Wang J, Nielsen R. 2011. Estimation of allele frequency and association mapping using next-generation sequencing data. BMC Bioinformatics 12:231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim Y, Nielsen R. 2004. Linkage disequilibrium as a signature of selective sweeps. Genetics 167:1513–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimura M. 1983. Rare variant alleles in the light of the neutral theory. Mol Biol Evol 1:84–93. [DOI] [PubMed] [Google Scholar]
- 23.Ling B, Veazey RS, Luckay A, Penedo C, Xu K, Lifson JD, Marx PA. 2002. SIVmac pathogenesis in rhesus macaques of Chinese and Indian origin compared with primary HIV infections in humans. AIDS 16:1489–1496. [DOI] [PubMed] [Google Scholar]
- 24.McDonald JH, Kreitman M. 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351:652–654. [DOI] [PubMed] [Google Scholar]
- 25.Morales JC, Melnick DJ. 1998. Phylogenetic relationships of the macaques (Cercopithecidae: Macaca), as revealed by high-resolution restriction site mapping of mitochondrial ribosomal genes. J Hum Evol 34:1–23. [DOI] [PubMed] [Google Scholar]
- 26.Nielsen R, Signorovitch J. 2003. Correcting for ascertainment biases when analyzing SNP data: applications to the estimation of linkage disequilibrium. Theor Popul Biol 63:245–255. [DOI] [PubMed] [Google Scholar]
- 27.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramachandran S, Deshpande O, Roseman CC, Rosenberg NA, Feldman MW, Cavalli-Sforza LL. 2005. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc Natl Acad Sci USA 102:15942–15947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reich DE, Cargill M, Bolk S, Ireland J, Sabeti PC, Richter DJ, Lavery T, Kouyoumjian R, Farhadian SF, Ward R, Lander ES. 2001. Linkage disequilibrium in the human genome. Nature 411:199–204. [DOI] [PubMed] [Google Scholar]
- 30.Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt SR, Doebley J, Kresovich S, Goodman MM, Buckler ES. 2001. Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc Natl Acad Sci USA 98:11479–11484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rowe N. 1996. The pictorial guide to the living primates. East Hampton (NY): Pogonias Press. [Google Scholar]
- 32.Satkoski Trask JA, Garnica WT, Smith DG, Houghton P, Lerche N, Kanthaswamy S. 2013. Single-nucleotide polymorphisms reveal patterns of allele sharing across the species boundary between rhesus (Macaca mulatta) and cynomolgus (M. fascicularis) macaques. Am J Primatol 75:135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith DG, George D, Kanthaswamy S, McDonough J. 2006. Identification of country of origin and admixture between Indian and Chinese rhesus macaques. Int J Primatol 27:881–898. [Google Scholar]
- 34.Smith DG, McDonough J. 2005. Mitochondrial DNA variation in Chinese and Indian rhesus macaques (Macaca mulatta). Am J Primatol 65:1–25. [DOI] [PubMed] [Google Scholar]
- 35.Smith DG, McDonough JW, George DA. 2007. Mitochondrial DNA variation within and among regional populations of longtail macaques (Macaca fascicularis) in relation to other species of the fascicularis group of macaques. Am J Primatol 69:182–198. [DOI] [PubMed] [Google Scholar]
- 36.Smith DG, Ng J, George DA, Trask JS, Houghton P, Singh B, Villano J, Kanthaswamy S. 2014. A genetic comparison of 2 alleged subspecies of Philippine cynomolgus macaques. Am J Phys Anthropol 155:136–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Southwick CH, Siddiqi MF. 1994. Population status of nonhuman primates in Asia, with emphasis on rhesus macaques in India. Am J Primatol 34:51–59. [DOI] [PubMed] [Google Scholar]
- 38.Southwick CH, Yongzu Z, Haisheng J, Zhenhe L, Wenyuan Q. 1996. Population ecology of rhesus macaques in tropical and temperate habitats in China, p 95–105 In: Fa JE, Lindburg DG. Evolution and ecology of macaque societies. Cambridge (UK): Cambridge University Press. [Google Scholar]
- 39.Tosi AJ, Morales JC, Melnick DJ. 2003. Paternal, maternal, and biparental molecular markers provide unique windows onto the evolutionary history of macaque monkeys. Evolution 57:1419–1435. [DOI] [PubMed] [Google Scholar]
- 40.Trask JA, Malhi RS, Kanthaswamy S, Johnson J, Garnica WT, Malladi VS, Smith DG. 2011. The effect of SNP discovery method and sample size on estimation of population genetic data for Chinese and Indian rhesus macaques (Macaca mulatta). Primates 52:129–138. [DOI] [PubMed] [Google Scholar]
- 41.Trask JS, Garnica WT, Kanthaswamy S, Malhi RS, Smith DG. 2011. 4040 SNP for genomic analysis in the rhesus macaque (Macaca mulatta). Genomics 98:352–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trichel AM, Rajakumar PA, Murphey-Corb M. 2002. Species-specific variation in SIV disease progression between Chinese and Indian subspecies of rhesus macaque. J Med Primatol 31:171–178. [DOI] [PubMed] [Google Scholar]
- 43.Wang N, Akey JM, Zhang K, Chakraborty R, Jin L. 2002. Distribution of recombination crossovers and the origin of haplotype blocks: the interplay of population history, recombination, and mutation. Am J Hum Genet 71:1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]