Abstract
Recent advances in medical imaging are beginning to allow us to quantify brain tissue maturation in the growing human brain prior to normal term age, and are beginning to shed new light on early human brain growth. These advances compliment the work already done in cellular level imaging in animal and post mortem studies of brain development. The opportunities for collaborative research that bridges the gap between macroscopic and microscopic windows on the developing brain are significant. The aim of this paper is to provide a review of the current research into MR imaging of the living fetal brain with the aim of motivating improved interfaces between the two fields. The review begins with a description of faster MRI techniques that are capable of freezing motion of the fetal head during the acquisition of a slice, and how these have been combined with advanced post-processing algorithms to build 3D images from motion scattered slices. Such rich data has motivated the development of techniques to automatically label developing tissue zones within MRI data allowing their quantification in 3D and 4D within the normally growing fetal brain. These methods have provided the basis for later work that has created the first maps of tissue growth rate and cortical folding in normally developing brains in-utero. These measurements provide valuable findings that compliment those derived from post-mortem anatomy, and additionally allow for the possibility of larger population studies of the influence of maternal environmental and genes on early brain development.
Keywords: MRI, In-Utero Imaging, Brain Morphometry
1. Introduction
This article reviews some of the recent advances that are beginning to allow us to quantifying brain tissue maturation in the growing human brain prior to normal term age. This work has been driven by the increased clinical use of magnetic resonance imaging to study both premature neonatal brain injury and to study fetal brain growth in-utero. Such techniques now provide an opportunity not only for improved clinical research but for basic neuroscience studies that can compliment the significant body of work on fetal brain growth derived from post-mortem data. In particular, cellular level studies have already provided dramatic new insights into the migration of cells in the formation of the cortex. However, the next step is to provide stronger links between these microscale observations and the formation of macroscale anatomy. The opportunities for collaborative research that bridges between these microscopic and macroscopic windows on the developing brain are significant. The aim of this paper is therefore partly to provide a review of the current research into macroscopic imaging of the living human fetal brain and partly to act as a motivation for improved links between the two complimentary fields of endeavour.
2. Advances in MR Imaging and Motion Correction
Although modern ultrasound imaging provides sensitive, high resolution images of the growing fetus that have become essential for clinical evaluation of pregnancy, its use in studying the brain has been limited by the presence of the skull and the ability of ultrasound to distinguish subtle differences in brain tissues. As a result, challenging clinical cases have driven (Weinreb et al., 1985; Powell et al., 1988; Horton and Tempany, 1995) the increased use of MRI of the human fetus, particularly in studying the brain (Williamson et al., 1989; Wilde et al., 2005). However, this clinically driven development has also opened up the possibility of improving our basic knowledge of brain development in-utero, with the opportunity to study the process of brain tissue growth in detail as it happens. This capability can potentially compliment information that has already been gained from many postmortem studies of the human fetal brain. MRI however also has significant limitations for fetal brain studies that arise from the relatively low signal strength. This necessitates the use of longer imaging times and increases susceptibility to unavoidable fetal movements during studies. This has lead to the use of multi-slice imaging rather than the acquisition of full 3D volumetric images to study the fetal brain. Developments in so called snapshot imaging techniques such as half-Fourier turbo spin echo (HASTE) (Kiefer et al., 1994) such as those shown in figure 1, and single shot fast spin echo (SSFSE) (Busse et al., 2005), require a second or less for a single slice. These techniques have opened a route to reducing the effect of fetal head motion within each slice acquisition and provide the clinical radiologist with vital tissue contrast at relatively high in-plane spatial resolution of 1mm or better. The occurrence of motion between slices remains both a clinical challenge (preventing accurate interpretation of the anatomy observed within a slice) and a challenge for more quantitative studies that seek to measure volume or distance within the anatomy.
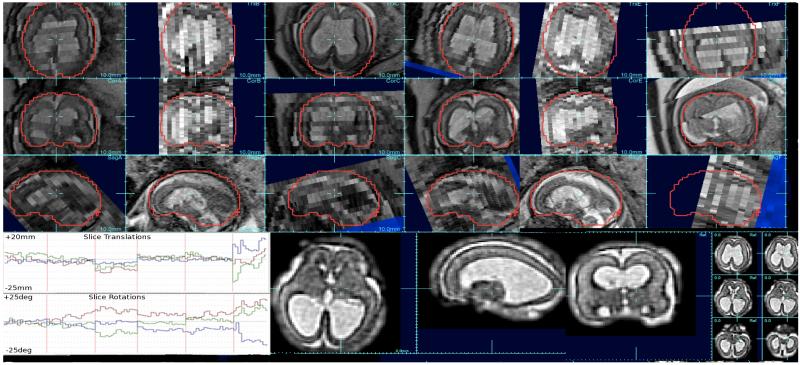
Figure 1.
Example clinically acquired multi-slice study (upper 3 rows) where 6 stacks (A-F) of 1×1mm slices with thicknesses varying between 3mm and 4mm were acquired of a fetus with enlarged ventricles: note the motion between odd and even slices (data acquired with an interleave of 2), differences in slice intensity due to through plane motion and intensity bias, and low through-plane resolution. Between slice motion was estimated using SIMC (Kim et al., 2010) (see bottom left traces of rotation and translation corrections of each slice pose) and signal differences were corrected using SIBC ((Kim et al., 2011). A 3D image reconstruction was created using a robust iterative scheme with iterative slice profile deconvolution (Fogtmann et al., 2012) to create a 1×1×1mm resolution voxel size (bottom right).
Since 2005 there has been a series of developments that have sought to address these limitations in fetal MRI by combining methods from computer vision with those of fast multi-slice MR imaging (Studholme, 2011). The majority of approaches make use of a two step reconstruction-alignment methodology that forms a putative 3D volume from the scattered slices and then refines the alignment of the slice data to this 3D imaging using slice to volume techniques, and then repeating these two steps in an iterative framework to form a consistent description of the fetal head anatomy. In particular, the work of our group using this approach (Rousseau et al., 2005) showed that it was possible to recover between slice motion by assuming rigid head motion and by employing a hierarchical estimation of the fetal head trajectory Rousseau et al. (2006), that incorporated information about when slices were acquired. Later work by other groups have made use of such a two step scheme (Jiang et al., 2007; Gholipour et al., 2010; Limperopoulos and Clouchoux, 2009) to develop a series of computational approaches to the estimation of slice to slice fetal head motion that can recover the full 3D anatomical positioning of each slice. However, such approaches are not inherently guaranteed to converge as the two steps of 3D reconstruction and slice matching are not directly linked via a single criteria that can lead, for example, to problems where slice data for a given region of anatomy are missing due to motion. An alternative scheme developed by our group (Kim et al., 2010) seeks to completely separate the slice motion and 3D reconstruction steps by making use of the acquisition of intersecting slices, and enforcing their agreement or match where they intersect in 3D. This basic ideas allows the creation of collective slice alignment approaches that can be compared to the simpler case of photographic image mosaicing in 2D.
Such approaches to accurate between slice motion estimation have been complimented by the refinement of techniques to build accurate 3D images from motion scattered slice data, where the acquired slices have high in plane resolution but provide only low resolution in the through plane direction. Algorithms have been developed to provide a consistent high isotropic spatial resolution by building on early interpolation based techniques Rousseau et al. (2006), making use of an iterative estimation of the 3D image. This process seeks to deconvolve the through-plane point spread function Gholipour et al. (2010); Rousseau et al. (2010) from the data as a 3D image is formed from scattered slices. Most recently, methods have been proposed Fogtmann et al. (2012) to unify iterative between-slice-motion estimation and 3D reconstruction, where both alignment and reconstruction problems are solved by minimizing the same robustly fit model to enforce a consistent optimization framework. As well as the problem of resolution, accurate fusion of many repeated acquisitions requires the removal of any inconsistencies in MRI signal level that occur as the fetus moves with respect to the MRI imaging coils. Such a problem can be formulated in a similar way to slice alignment Kim et al. (2011) and has been shown to improve the contrast of subtle developing tissues zones such as the sub-plate and germinal matrix. An example of such a 3D image derived from a clinical study made up of many multi-slice acquisitions is shown in figure 1. Recent work has also looked at the use of robust estimators the exclude or reduce the influence of corrupted data rather than explicitly modelling the signal artefact Kuklisova-Murgasova et al. (2012).
3. MRI Contrast and Automated Tissue Delineation
One of the initial basic steps in any MRI study of brain anatomy, either in the adult or pediatric population, is to assign tissue classes to the observed MRI intensities, in order to impose a tissue model on the data that can be used to quantify anatomy. In adult imaging the most common task is to divide the MRI data into grey and white tissue compartments (Kapur et al., 1996; Shattuck et al., 2001; Van Leemput et al., 2003). Techniques that make use of a statistical model based labelling of the observed data (where the maximum a posteriori estimate is usually found by applying the Expectation-Maximization (EM) algorithm) have achieved significant success in automated tools (Leemput et al., 1999; Ashburner and Friston, 2005) and often make heavy use of a spatial statistical prior for the tissue occurrence (Ashburner and Friston, 2009). These rely on the hard assumption of the fixed tissue classes present, or introduce model extensions to extract additional pathological classes (Leemput et al., 2001). In the case of brain development seen in the earliest stages of human fetal imaging, these classes and models of structure are less appropriate as the brain structure is not made up of static mature tissues, but consists of developing tissue zones or layers Bystron et al. (2008) that have distinctly different MRI properties Corbett-Detig et al. (2011). In addition, these zones have a highly transient nature in the brain that is reflected in the relative strength of the MRI signal. A key point to notice in this context is the fact that the appearance of fetal brain tissues in MR images is changing rapidly with gestational age. However, the ability to accurately quantify the state of these tissue zones within the brain, particularly around the age of the first clinical scan at around 20 gestational weeks, may be critically important in providing a basic measure of brain growth. Given the limited information available in the T2 weighed (T2W) MRI contrast, any labelling of a given voxel in the MRI must take into account the anatomical location of that voxel as well as its MRI intensity. Therefore research on automated atlas based tissue labelling has made heavy use of manual tracing from which to learn a model of tissue shape and appearance. The first methods specifically developed for fetal brain anatomy applied a single age specific model of tissue zones (Habas et al., 2008, 2010b) derived from careful manual delineation of brain anatomy in reference images. These approaches were then extended (Habas et al., 2010a) to create a continuous computational atlas of the developing fetal brain. This mathematical representation of the changing brain is capable of forming an age specific prior estimate or template at any given age, as illustrated in figure 2. These priors templates can then be used to initialize automated tissue labelling schemes that adapt the ideal average created for the age of a new fetal scan to be labelled, to the observed anatomy in the subjects MRI data as illustrated in Figure 3.
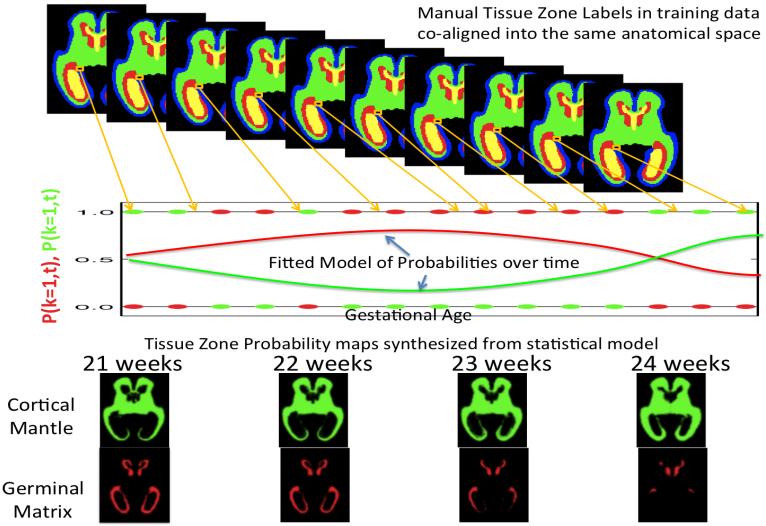
Figure 2.
Using Manual Tracing data to learn the probability of developing tissues zones visible in MRI scans of fetuses of different ages: For each voxel, each manually marked dataset mapped to a common fetal coordinate system contributes a binary decision at a given age (red and green dots on graph). A constrained mathematical model is fitted to these binary observations to form a continuous estimate of the probability of observing a tissue class (red and green curves) at any given age at that point in the anatomy. The formation of such a complete computational model that also captures age related changes in fetal brain shape and appearance (contrast) in MRI allows the formation of age specific templates and tissue priors for any given gestational age (bottom row). These age specific templates can be used as a starting estimate for automated techniques that label tissues in new MRI scans (Habas et al., 2010a) by adapting the average model to the new scan of an individual.
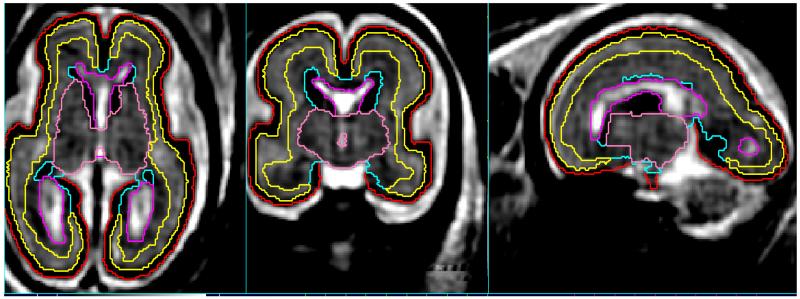
Figure 3.
Automated tissue delineation in a 3D fetal brain image using an age specific statistical prior (Habas et al., 2010a) to drive an expectation maximization labelling scheme.
Such approaches have achieved accurate automated results as illustrated in 3 and have been shown to be consistent with manual expert definitions of boundaries (Habas et al., 2010a). The techniques have since been applied to study both normally growing anatomy (Scott et al., 2011) and to automatically delineate abnormally developing fetal neuroanatomy (Scott et al., 2012).
Another atlas-based segmentation approach has recently been studied for such problem. It is based on the use of multiple atlases and the core principle is to propagate labels from manually annotated images to new images based on overall similarity of the new subject to one of the manually traced subjects. This label propagation can be applied using non-rigid registration methods (Heckemann et al., 2006) to bring a new subject into alignment with a set of different manually marked anatomies, and selecting the manually marked anatomy whose MRI scan best matches the new subjects MRI scan after non-rigid mapping. Alternatively, methods that do not depend on accurate registration of the anatomy can be applied by selecting regional matches derived from a dictionary of local images using so-called patch-based techniques (Rousseau et al., 2011; Coupé et al., 2011). Such label propagation has recently been used as an initialization step for a multi-shape technique for ventricle volumetry (Gholipour et al., 2012). Atlas based labelling has been developed specifically in a form to make use of the inherent laminar structure and ordering of the younger fetal brain in constructing a statistical prior (Habas et al., 2009). Finally, atlas-free techniques have been recently proposed to segment specific brain tissues by including local topological constraints (Caldairou et al., 2011). Many of these newer techniques show promise but have not been evaluated or applied on larger scale neuroscience studies as that of age specific EM segmentation (Scott et al., 2011, 2012)
4. Atlases and Tissue Growth Rates
One of the key initial motivations for computational modelling of fetal brain growth in MRI studies has been to assist in tissue segmentation, through the ability to create an age specific prior map of tissue composition. This requires a model of both how MRI contrast changes with age, how brain shape changes and how the probability of tissues at given locations in the brain vary with age. Figure 4 illustrates an instantiation of such a statistical MRI growth model for three gestational weeks showing the dramatic changes apparent. This represents the shape average anatomy that is extracted from a set of individual anatomies covering an age range, via the construction of a minimum deformation shape average followed by general linear modelling of the shape deformations and MRI contrasts over time (Habas et al., 2010a). One of the key challenges in constructing this shape average is the problem of aligning MRI scans acquired at different gestational ages in a way that ensure a continuous mapping over time and enables a voxel by voxel modelling of growth. For the equivalent adult models where tissue classification remains consistent over time (i.e. the same tissues occur at all ages), MRI image registration can be directly employed (Rohlfing and Maurer, 2005). However for the fetal case, since tissues zones such as the germinal matrix and sub-plate are transient in their appearance with age in MRI (Corbett-Detig et al., 2011), accurate non-rigid correspondence between one anatomy and another cannot be driven directly by MRI intensities. In response to this problem, new approaches have employed automated age specific tissue labelling of subject MRI data, prior to accurate spatial normalization, to abstract the process of spatial normalisation away from changes in tissue properties, to ensure that only age consistent tissue boundaries are used in the alignment process (Rajagopalan et al., 2011a). Accurate spatial normalisation enables the application of techniques than map and analyze patterns of tissue growth across the brain with age. allowing the creation of maps of tissue growth rate. When combined with voxel wise linear modelling this has enabled the investigation of the emergence of distinct patterns of tissue growth rates associated with the formation of primary sulci by statistically detecting those regions that are increasing in volume at a rate that is statistically greater or lesser than the brain as a whole (Rajagopalan et al., 2011a). This scalar growth analysis clearly opens up the possibility for cross comparison and evaluation against post-mortem growth and migration measurements being made at a cellular level in different brain tissue regions.
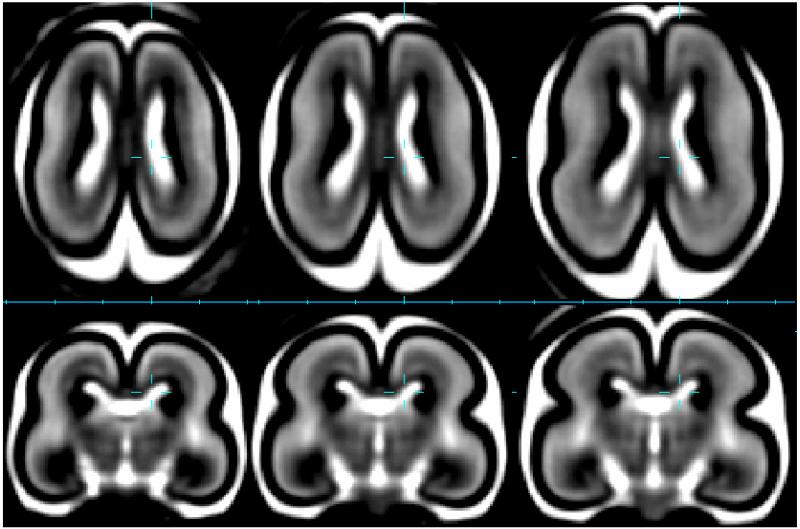
Figure 4.
Average brain tissue contrast and shape captured at 20GW (left), 21GW (middle) and 22GW (right) in a T2W MRI growth model (Habas et al., 2010a) derived from in-utero data of 40 healthy subjects. Outer surface consists of dark cortical plate below which a bright region of subplate is visible. Around the ventricles is the dark region of germinal matrix.
Most recently these methods have been extended to examine not only scalar expansion of tissue but the directional components of that expansion (Rajagopalan et al., 2012, 2010), and to look at the relationship in growth rates across different tissue zone boundaries (Rajagopalan et al., 2011b). This is of particular importance in attempting to understand the role of transient layers such as the sub-plate in cortex formation (Corbett-Detig et al., 2011), allowing the identification of temporally or spatially related events in different brain layers that may not be directly accessible to post-mortem histology studies.
5. Mapping and Ordering of Cortical Folding Events
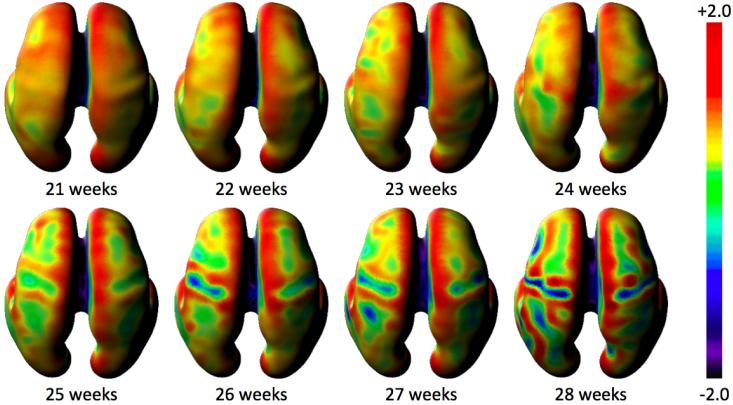
The availability of high resolution 3D images of the fetal brain with accurate tissue labelling enables not only internal volumetric studies of growth, but studies of tissue boundary shape changes. In particular they have enabled the first quantitative maps of brain folding in the human brain in utero (Habas et al., 2012). Rather than basing the evaluation of folding on simple 2D inspection of brain slices, such methods can employ spatial modelling and surface based curvature analysis, as illustrated in Figure 5, to quantify average surface folding patterns for the fetus. When this is combined with statistical testing, we can map locations on a common anatomical coordinate system where folding at a consistent location can be examined in all subjects. In particular, (Habas et al., 2012) used temporally localized regression across a range of fetal brain shapes to map those regions of the brain where there was a statistically detectable change in brain folding occurring over time as illustrated in Figure 6. These techniques, first used to investigate normal brain growth, have since been used to examine and confirm abnormal cortical folding in ventriculomegaly, the most common clinically identified fetal brain abnormality (Scott et al., 2012).
Figure 5.
Quantifying Average Brain Folding Patterns: Inner cortical plate curvature (mean curvature (H) at each surface point) derived from a T2W MRI growth model (Habas et al., 2012) of in-utero MRI data of 40 healthy subjects. These colour maps are displayed in common coordinate system (mean surface shape of the group) so that each point is in correspondence allowing the formation of temporal model at each location on the surface.
Figure 6.
Detecting Significant Changes in Brain Folding: Mapping dynamic changes in curvature detectable in a temporal and spatially consistent location across a population of fetal brain anatomies of different ages.
Making use of minimum deformation, template free, normalisation it is also possible to construct left-right symmetric fetal brain models to investigate the emergence of asymmetry in brain surface curvature over time (Habas et al., 2012). This work was built on that of Dubois and co-authors (Dubois et al., 2009) who confirmed the presence of early asymmetries in premature neonates, by showing that folding asymmetries emerge at the earliest stages of primary sulcation in the fetus. In practice such asymmetries may be more statistically significant at these stages because of the lack of individualized brain folding in the form of secondary and tertiary gyri. As a result these asymmetry patterns may also be useful as a powerful marker for early abnormal development.
6. Diffusion Imaging of Tissue Microstructure in the Fetal Brain
Clinically, the use of slice based measurements of scalar diffusion properties of the fetal brain has allowed the mapping of the developmental trajectory of white matter tissue properties in-utero (Schneider et al., 2007, 2009; Bui et al., 2006; Kim et al., 2008). In challenging clinical conditions, in utero diffusion weighted MRI has been shown to provide a valuable marker for acute hypoxic ischemic fetal brain lesions (Baldoli et al., 2002; Girard et al., 2003). More recently it has been used in the mapping of abnormalities of the laminar structure of the fetal brain in Cobbelstone complex (Widjaja et al., 2009). Berman (Agid et al., 2006) also reported higher diffusivity in parietal white matter and the thalamus in fetuses with congenital heart defects when compared to controls. In addition to scalar microstructural properties, from post-mortem studies of the fetal brain, full 3D diffusion direction and tractography measurements have illustrated the possibility of mapping the development of white matter connectivity using MRI (Huang et al., 2009). This research has motivated the first attempts at in utero diffusion tensor based tractography studies (Zanin et al., 2011; Kasprian et al., 2008; Koob et al., 2011) that have shown the possibility of mapping the emergence of white matter connections in utero in cases of limited fetal head motion. However, for practical clinical applications more robust approaches that can deal with fetal head motions are required to allow reliable estimation of tissue microstructure. Over the last 5 years, following the work on motion correction of fast multi-slice T2W structural imaging (Rousseau et al., 2006; Jiang et al., 2007), there has been a significant interest in developing techniques for improving DTI acquisition and post-processing for challenging adult, pediatric and fetal cases.
The key challenge is then to estimate the changes in position and orientation of the diffusion weighted slices with respect to the underlying anatomy, and to then form a regularly sampled estimate of the diffusion profile across the fetal brain volume from the motion scattered slice data. Algorithms have been developed that estimate individual slice locations and orientations in the anatomical frame, and reorientate the diffusion measurements based on this mapping, before fitting or interpolating a diffusion tensor or general orientation distribution function (ODF) to the scattered diffusion measurements. The first work to address between slice fetal motion during DTI (Jiang et al., 2009) extended our work of slice to volume reconstruction based motion estimation (Rousseau et al., 2006) to that of diffusion measurements by explicitly considering the re-orientation of individual slice diffusion geometry into a consistent coordinate frame (as opposed to simply volume orientation (Aksoy et al., 2010) in adults).
In adult brain tractography studies it has been shown that the rank 2 diffusion tensor is not su cient in some regions of anatomy to model all possible configurations of white matter fibre bundles (for example in the case of crossing fibres). For the later stages of fetal brain development this may be a similar concern. Oubel et al (Oubel et al., 2010, 2012) have proposed a reconstruction method of dMRI sequences from scattered data that is independent of the diffusion model, and that can be used with more complex diffusion models such as the Gaussian mixture model or high-order diffusion tensors. The approach consists of first registering jointly the DW images and then using a derived image for registration with a T2-weighted reference image. Diffusion measurements are finally interpolated in the spherical domain. It has been shown that using these techniques in case studies that in-utero tractography can now be used in conjunction with an analysis of pathology data to assist in the identification of abnormal tract development (Koob et al., 2011).
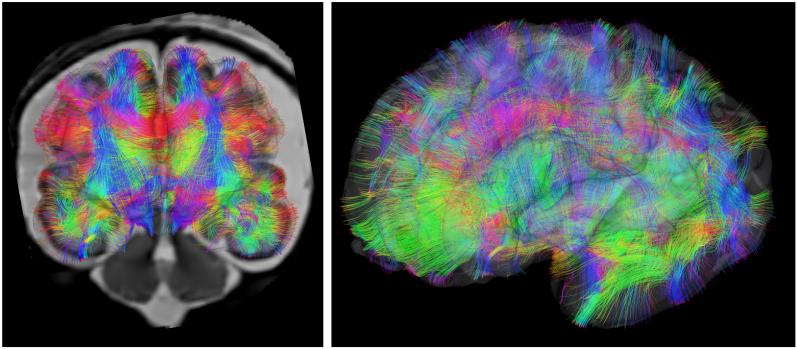
The most recent work in this area seeks to unify slice reconstruction and ODF estimation within a single framework. This importantly seeks to explicitly provide diffusion orientation sensitivity within the slice matching process. Such approaches also incorporate a deconvolution framework to optimally fuse spatial information Fogtmann et al. (2014) from thick slices acquired with different orientations. Such recent work has shown the power of the approach on challenging, clinically acquired diffusion data. An illustration of whole brain tractography map from a motion corrected DTI study of human a fetus acquired in utero is shown in Figure 7.
Figure 7.
In utero connectome mapping: Motion correction of diffusion weighted MRI measurements that account for changes in anatomical position and orientation with respect to the scanner permit the construction of maps of white matter connectivity in the developing fetus.
The ability to image whole brain diffusion patterns in a large fraction of fetuses opens a route to studying the early development of the human connectome (Tymofiyeva et al., 2012; Sporns, 2011) before birth. However, another challenge in extending adult connectivity studies to the rapidly developing fetus is the problem of consistently defining anatomical correspondence over time, as an anatomical parcellation is the starting point for many connectivity analysis techniques. Recent work in this area has examined alternative approaches that use graph based measures of connectivity derived from generic parcellation schemes Cheng et al. (2013) that seek to avoid a one to one correspondence over time.
7. Conclusions
In this paper we have covered some of the main advances which are beginning to enable us to study the human fetal brain in the same way that MRI has allowed to us to study the adult and pediatric brain. The nature of the developing brain tissues and motion of the anatomy being imaged pose fundamental limitations to the use of conventional MRI studies on all but a fraction of fetuses that are not moving, and have therefore historically limited fetal research to slice based measurements. By combining advances in slice based snapshot MRI with post acquisition correction techniques, it is now possible to form true 3D images even in studies with significant fetal head motion. This enables the formation of 3D images in more than just a small fraction of fetal studies and raises the feasible yield of a study of fetal brain growth to a practically useful level, and may also permits the use of 3D imaging within a clinical setting. For such studies, the highly transient nature of fetal brain tissues zones pose a significant second challenge to the extraction of quantitative anatomical measurements using automated methods. However, new techniques that can adapt tissue classification schemes to the developmental stage being studied are a key step in enabling automated delineation and quantification of transient tissue zones, such as the germinal matrix and sub-plate. This work has enabled the first 3D in utero morphometric studies of the growth of such transient tissue zones (Corbett-Detig et al., 2011; Rajagopalan et al., 2011a, 2012). The next step will be translational studies linking these findings more closely to findings in microscopic studies of cell growth and migration (Lui et al., 2011; Kriegstein et al., 2006) that could provide both a validation of in-utero MRI measurements, but also new insights into macroscopic patterning of brain structure over time.
Highlights.
Description of the context for the development of fetal MRI techniques
A review and classification of motion correction techniques for fetal MRI
A review of the challenges for automated tissue labeling in fetal MRI and a description of the current state of the art approaches in fetal MRI tissue labeling.
A description of the early papers in computational morphometry of fetal brain growth including volume based and surface based models of normal growth.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agid R, Lieberman S, Nadjari M, Gomori J. Prenatal MR diffusion-weighted imaging in a fetus with hemimegalencephaly. Pediatr. Radiol. 2006;36:138–140. doi: 10.1007/s00247-005-0003-3. [DOI] [PubMed] [Google Scholar]
- Aksoy M, Skare S, Holdsworth S, Bammer R. Effects of motion and b-matrix correction for high resolution dti with short-axis propeller-epi. NMR in Biomedicine. 2010;23:794–802. doi: 10.1002/nbm.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J, Friston K. Unified segmentation. NeuroImage. 2005;26:839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston K. Computing average shaped tissue probability templates. NeuroImage. 2009;45:333–341. doi: 10.1016/j.neuroimage.2008.12.008. [DOI] [PubMed] [Google Scholar]
- Baldoli C, Righini A, Parazzini C, Scotti G, Triulzi F. Demonstration of acute ischemic lesions in the fetal brain by diffusion magnetic resonance imaging. Ann. Neurol. 2002;52:243–246. doi: 10.1002/ana.10255. [DOI] [PubMed] [Google Scholar]
- Bui T, Daire J, Chalard F, Zaccaria I, Alberti C, Elmaleh M, Garel C, Luton D, Blanc N, Sebag G. Microstructural development of human brain assessed in utero by diffusion tensor imaging. Pediatr. Radiol. 2006;36:1133–1140. doi: 10.1007/s00247-006-0266-3. [DOI] [PubMed] [Google Scholar]
- Busse R, Zaharchuk G, Glenn O. A rapid two shot t1-measurement and its applicability to oxygen partial pressure (po2) measurement in fluid; Proceedings of ISMRM 2005; Miami, Fl., USA. 2005. [Google Scholar]
- Bystron I, Blakemore C, Rakic P. Development of the human cerebral cortex: Boulder committee revisited. Nature Reviews Neuroscience. 2008;9:110–122. doi: 10.1038/nrn2252. [DOI] [PubMed] [Google Scholar]
- Caldairou B, Passat N, Habas P, Studholme C, Koob M, Dietemann J, Rousseau F. Segmentation of the cortex in fetal mri using a topological model; Biomedical Imaging: From Nano to Macro, 2011 IEEE International Symposium on; IEEE. 2011.pp. 2045–2048. [Google Scholar]
- Cheng X, Wilm J, Seshamani S, Fogtmann M, Kroenke C, Studholme C. Adapting parcellation schemes to study fetal brain connectivity in serial imaging studies; Proceedings IEEE EMBS; IEEE. 2013; [DOI] [PubMed] [Google Scholar]
- Corbett-Detig JM, Habas PA, Scott JA, Kim K, Rajagopalan V, McQuillen PS, Barkovich AJ, Glenn OA, Studholme C. 3-D global and regional patterns of human fetal subplate growth determined in utero. Brain Struct. Funct. 2011;215:255–263. doi: 10.1007/s00429-010-0286-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coupé P, Manjón JV, Fonov V, Pruessner J, Robles M, Collins DL. Patch-based segmentation using expert priors: Application to hippocampus and ventricle segmentation. NeuroImage. 2011;54:940–954. doi: 10.1016/j.neuroimage.2010.09.018. [DOI] [PubMed] [Google Scholar]
- Dubois J, Hertz-Pannier L, Cachia A, Mangin J, Le Bihan D, Dehaene-Lambertz G. Structural asymmetries in the infant language and sensori-motor networks. Cerebral Cortex. 2009;19:414–423. doi: 10.1093/cercor/bhn097. [DOI] [PubMed] [Google Scholar]
- Fogtmann M, Chapman T, Kim K, Seshamani S, Studholme C. A unified approach for motion-estimation and super-resolution reconstruction from structural magnetic resonance on moving subjects; Proceedings of the MICCAI 2012 Workshop on Perinatal and Paediatric Imaging: PaPI 2012; 2012. [Google Scholar]
- Fogtmann M, Seshamani S, Kroenke C, Cheng X, Chapman T, Wilm J, Rousseau F, Koob M, Dietemann JL, Studholme C. A unified approach to diffusion direction sensitive slice registration and 3d dti reconstruction from moving fetal brain anatomy. IEEE Transactions on Medical Imaging N, N. 2014 doi: 10.1109/TMI.2013.2284014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gholipour A, Akhondi-Asl A, Estro JA, Warfield SK. Multiatlas multi-shape segmentation of fetal brain mri for volumetric and morphometric analysis of ventriculomegaly. Neuroimage. 2012;60:1819–1831. doi: 10.1016/j.neuroimage.2012.01.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gholipour A, Estro J, Warfield S. Robust super-resulution volume reconstruction from slice acquisitions: Application to fetal brain mri. IEEE transactions on Medical Imaging. 2010;29:1739–1758. doi: 10.1109/TMI.2010.2051680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard N, Gire C, Sigaudy S, Porcu G, d’Ercole C, Figarella-Branger D, Raybaud C, Confort-Gouny S. Mr imaging of acquired fetal brain disorders. Childs Nerv Syst. 2003;19:490–500. doi: 10.1007/s00381-003-0761-x. [DOI] [PubMed] [Google Scholar]
- Habas P, Scott J, Roosta A, Rajagopalan V, Kim K, Rousseau F, Barkovich A, Glenn O, Studholme C. Early folding patterns and asymmetries of the normal human brain detected from in utero mri. Cerebral Cortex. 2012;22:13–25. doi: 10.1093/cercor/bhr053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habas PA, Kim K, Chandramohan D, Rousseau F, Glenn OA, Studholme C. Statistical model of laminar structure for atlas-based segmentation of the fetal brain from in-utero MR images; Proceedings SPIE Medical Imaging 2009: Image Processing; 2009.pp. 17:1–17:8. [Google Scholar]
- Habas PA, Kim K, Corbett-Detig JM, Rousseau F, Glenn OA, Barkovich AJ, Studholme C. A spatiotemporal atlas of MR intensity, tissue probability and shape of the fetal brain with application to segmentation. Neuroimage. 2010a;53:460–470. doi: 10.1016/j.neuroimage.2010.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habas PA, Kim K, Rousseau F, Glenn OA, Barkovich AJ, Studholme C. Medical Image Computing and Computer-Assisted Intervention. LNCS; 2008. Atlas-based segmentation of the germinal matrix from in utero clinical MRI of the fetal brain; pp. 351–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habas PA, Kim K, Rousseau F, Glenn OA, Barkovich AJ, Studholme C. Atlas-based segmentation of developing tissues in the human brain with quantitative validation in young fetuses. Human Brain Mapping. 2010b;31:1348–1358. doi: 10.1002/hbm.20935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckemann R, Hajnal J, Aljabar P, Rueckert D, Hammers A, et al. Automatic anatomical brain mri segmentation combining label propagation and decision fusion. NeuroImage. 2006;33:115–126. doi: 10.1016/j.neuroimage.2006.05.061. [DOI] [PubMed] [Google Scholar]
- Horton K, Tempany C. MRI in pregnancy. In: Tempany C, editor. MR and imaging of the female pelvis. Mosby, Mosby; 1995. pp. 235–260. [Google Scholar]
- Huang H, Xue R, Zhang J, Ren T, Richards L, Yarowsky P, Miller M, Mori S. Anatomical characterization of human fetal brain development with diffusion tensor magnetic resonance imaging. The Journal of Neuroscience. 2009;29:4263–4273. doi: 10.1523/JNEUROSCI.2769-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Xue H, Counsell S, Anjari M, Allsop J, Rutherford M, Rueckert D, Hajnal J. Diffusion tensor imaging (dti) of the brain in moving subjects: Application to in-utero fetal and ex-utero studies. Magnetic Resonance in Medicine. 2009;62:645–655. doi: 10.1002/mrm.22032. [DOI] [PubMed] [Google Scholar]
- Jiang S, Xue H, Glover A, Rutherford M, Rueckert D, Hajnal J. MRI of moving subjects using multislice snapshot images with volume reconstruction (SVR): application to fetal, neonatal, and adult brain studies. IEEE Trans Med Imaging. 2007;26:967–980. doi: 10.1109/TMI.2007.895456. [DOI] [PubMed] [Google Scholar]
- Kapur T, Grimson W, Wells W, Kikinis R. Segmentation of brain tissue from magnetic resonance images. Med. Image Anal. 1996:1. doi: 10.1016/S1361-8415(96)80008-9. [DOI] [PubMed] [Google Scholar]
- Kasprian G, Brugger PC, Weber M, Krssak M, Krampl E, Herold C, Prayer D. In utero tractography of fetal white matter development. neuroimage. 2008;43:213–224. doi: 10.1016/j.neuroimage.2008.07.026. [DOI] [PubMed] [Google Scholar]
- Kiefer B, Grassner J, Hausmann R. Image acquisition in a second with half-fourier-acquisition single-shot turbo spin echo 4; Proceedings of first society for magnetic resonance; 1994. [Google Scholar]
- Kim D, Chung S, Vigneron D, Barkovich A, Glenn O. Diffusion-weighted imaging of the fetal brain in vivo. Magnetic Resonance in Medicine. 2008;59:216–220. doi: 10.1002/mrm.21459. [DOI] [PubMed] [Google Scholar]
- Kim K, Habas P, Rajagopalan V, Scott J, Corbett-Detig J, Rousseau F, Barkovich A, Glenn O, Studholme C. Bias field inconsistency correction of motion-scattered multislice mri for improved 3d image reconstruction. IEEE Transactions on Medical Imaging. 2011;30:1704. doi: 10.1109/TMI.2011.2143724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Habas PA, Rousseau F, Glenn OA, Barkovich AJ, Studholme C. Intersection based motion correction of multislice MRI for 3-d in utero fetal brain image formation. IEEE Trans. Med. Imaging. 2010;29:146–158. doi: 10.1109/TMI.2009.2030679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob M, Weingertner AS, Gasser B, Oubel E, Dietemann JL. Thick corpus callosum: a clue to the diagnosis of fetal septopreoptic holo-prosencephaly? Pediatr Radiol. 2011 doi: 10.1007/s00247-011-2260-7. [DOI] [PubMed] [Google Scholar]
- Kriegstein A, Noctor S, Martínez-Cerdeño V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nature Reviews Neuroscience. 2006;7:883–890. doi: 10.1038/nrn2008. [DOI] [PubMed] [Google Scholar]
- Kuklisova-Murgasova M, Quaghebeur G, Rutherford M, Hajnal J, Schnabel J. Reconstruction of fetal brain mri with intensity matching and complete outlier removal. Medical Image Analysis. 2012 doi: 10.1016/j.media.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leemput KV, Maes F, Vandermeulen D, Colchester A, Suetens P. Automated segmentation of multiple sclerosis lesions by model outlier detection. IEEE Transactions on Medical Imaging. 2001;20:667–688. doi: 10.1109/42.938237. [DOI] [PubMed] [Google Scholar]
- Leemput KV, Maes F, Vandermeulen D, Suetens P. Automated model-based bias field correction of MR images of the brain. IEEE Transactions on Medical Imaging. 1999;18:885–896. doi: 10.1109/42.811268. [DOI] [PubMed] [Google Scholar]
- Limperopoulos C, Clouchoux C. Advancing fetal brain mri: targets for the future. Semin Perinatol. 2009;33:289–298. doi: 10.1053/j.semperi.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Lui J, Hansen D, Kriegstein A. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oubel E, Koob M, Studholme C, Dietemann JL, Rousseau F. Reconstruction of scattered data in fetal diffusion MRI; Proceedings MICCAI 2010; Beijing, Springer. 2010; pp. 574–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oubel E, Koob M, Studholme C, Dietemann JL, Rousseau F. Reconstruction of scattered data in fetal diffusion MRI. 2012;16:28–37. doi: 10.1016/j.media.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell M, Worthington B, Buckley J, et al. Magnetic resonance imaging (MRI) in obstetrics. ii. fetal anatomy. Br J Obstet Gynaecol. 1988;95:38–46. doi: 10.1111/j.1471-0528.1988.tb06478.x. [DOI] [PubMed] [Google Scholar]
- Rajagopalan V, Scott J, Habas P, Kim K, Corbett-Detig J, Rousseau F, Barkovich A, Glenn O, Studholme C. Local tissue growth patterns underlying normal fetal human brain gyrification quantified in utero. The Journal of Neuroscience. 2011a;31:2878–2887. doi: 10.1523/JNEUROSCI.5458-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan V, Scott J, Habas P, Kim K, Rousseau F, Glenn O, Barkovich A, Studholme C. Mapping directionality specific volume changes using tensor based morphometry: An application to the study of gyrogenesis and lateralization of the human fetal brain. NeuroImage. 2012;63:947–958. doi: 10.1016/j.neuroimage.2012.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan V, Scott JA, Habas PA, Kim K, Rousseau F, Glenn OA, Barkovich AJ, Studholme C. Medical Image Computing and Computer Assisted Intervention. spinger; 2010. Measures for characterizing directionality specific volume changes in TBM of brain growth; pp. 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan V, Scott JA, Habas PA, Kim K, Rousseau F, Glenn OA, Barkovich AJ, Studholme C. Spatiotemporal morphometry of adjacent tissue layers with application to the study of sulcal formation; Proceedings of MICCAI 2011; Springer Verlag, LNCS. 2011b; pp. 476–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlfing T, Maurer C. Multi-classifier framework for atlas-based image segmentation. Pattern Recognition Letters. 2005;26:2070–2079. [Google Scholar]
- Rousseau F, Glenn O, Iordanova B, Rodriguez-Carranza C, Vigneron D, Barkovich J, Studholme C. In: Duncan J, Gerig G, editors. Reconstructing high resolution in vivo fetal MR brain images; Proceedings of Medical Image Computing and Computer Assisted Intervention (MICCAI); Springer Verlag. 2005; pp. 548–555. [DOI] [PubMed] [Google Scholar]
- Rousseau F, Glenn O, Iordanova B, Rodriguez-Carranza C, Vigneron D, Barkovich J, Studholme C. Registration-based approach for reconstruction of high-resolution in utero fetal MR brain images. Academic radiology. 2006;13:1072–1081. doi: 10.1016/j.acra.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Rousseau F, Habas PA, Studholme C. A supervised patch-based approach for human brain labeling. IEEE Trans Med Imaging. 2011;30:1852–1862. doi: 10.1109/TMI.2011.2156806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau F, Kim K, Studholme C, Koob M, Dietemann J. On super-resolution for fetal brain mri; Medical Image Computing and Computer-Assisted Intervention-MICCAI 2010; Springer Verlag, LNCS. 2010; pp. 353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J, Confort-Gouny S, Le Fur Y, Viout P, Bennathan M, Chapon F, Fogliarini C, Cozzone P, Girard N. Diffusion-weighted imaging in normal fetal brain maturation. European radiology. 2007;17:2422–2429. doi: 10.1007/s00330-007-0634-x. [DOI] [PubMed] [Google Scholar]
- Schneider M, Berman J, Baumer F, Glass H, Jeng S, Jeremy R, Esch M, Biran V, Barkovich A, Studholme C, et al. Normative apparent diffusion coe cient values in the developing fetal brain. American Journal of Neuroradiology. 2009;30:1799–1803. doi: 10.3174/ajnr.A1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JA, Habas PA, Kim K, Rajagopalan V, Hamzelou KS, Corbett-Detig JM, Barkovich AJ, Glenn OA, Studholme C. Growth trajectories of the human fetal brain tissues estimated from 3D reconstructed in utero mri. Int J Dev Neurosci. 2011;29:529–536. doi: 10.1016/j.ijdevneu.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JA, Habas PA, Rajagopalan V, Kim K, Barkovich AJ, Glenn OA, Studholme C. Volumetric and surface-based 3d mri analyses of fetal isolated mild ventriculomegaly: Brain morphometry in ventriculomegaly. Brain Struct Funct. 2012 doi: 10.1007/s00429-012-0418-1. [DOI] [PubMed] [Google Scholar]
- Shattuck D, Sandor-Leahy S, Schaper K, Rottenberg D, Leahy R. Magnetic resonance image tissue classification using a partial volume model. Neuroimage. 2001;13:856–876. doi: 10.1006/nimg.2000.0730. [DOI] [PubMed] [Google Scholar]
- Sporns O. The human connectome: a complex network. Ann N Y Acad Sci. 2011;1224:109–125. doi: 10.1111/j.1749-6632.2010.05888.x. [DOI] [PubMed] [Google Scholar]
- Studholme C. Mapping fetal brain development in utero using magnetic resonance imaging: The big bang of brain mapping. Annual Review of Biomedical Engineering. 2011;13:345–368. doi: 10.1146/annurev-bioeng-071910-124654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tymofiyeva O, Hess CP, Ziv E, Tian N, Bonifacio SL, McQuillen PS, Ferriero DM, Barkovich AJ, Xu D. Towards the “baby connectome”: mapping the structural connectivity of the newborn brain. PLoS One. 2012:7. doi: 10.1371/journal.pone.0031029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leemput K, Maes F, Vandermeulen D, Suetens P. A unifying framework for partial volume segmentation of brain mr images. Medical Imaging, IEEE Transactions on. 2003;22:105–119. doi: 10.1109/TMI.2002.806587. [DOI] [PubMed] [Google Scholar]
- Weinreb J, Lowe T, Santos-Ramos R, et al. Magnetic resonance imaging in obstetric diagnosis. Radiology. 1985;154:157–161. doi: 10.1148/radiology.154.1.3880601. [DOI] [PubMed] [Google Scholar]
- Widjaja E, Geibprasert S, Blaser S, Rayner T, Shannon P. Abnormal fetal cerebral laminar organization in cobblestone complex as seen on post-mortem MRI and DTI. Pediatric radiology. 2009;39:860–864. doi: 10.1007/s00247-009-1255-0. [DOI] [PubMed] [Google Scholar]
- Wilde JD, Rivers A, Price D. A review of the current use of magnetic resonance imaging in pregnancy and safety implications for the fetus. Prog Biophys Mol Bio. 2005;87:335–353. doi: 10.1016/j.pbiomolbio.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Williamson R, Weiner C, Yuh W, Abu-yousef M. Magnetic resonance imaging of anomalous fetuses. Obstetrics and Gynecology. 1989;73:952–956. doi: 10.1097/00006250-198906000-00009. [DOI] [PubMed] [Google Scholar]
- Zanin E, Ranjeva J, Confort-Gouny S, Guye M, Denis D, Cozzone P, Girard N. White matter maturation of normal human fetal brain. an in vivo diffusion tensor tractography study. Brain and Behavior. 2011 doi: 10.1002/brb3.17. [DOI] [PMC free article] [PubMed] [Google Scholar]