Abstract
Background
Explant browning presents a major problem for in vitro culture, and can lead to the death of the explant and failure of regeneration. Considerable work has examined the physiological mechanisms underlying Phalaenopsis leaf explant browning, but the molecular mechanisms of browning remain elusive. In this study, we used whole genome RNA sequencing to examine Phalaenopsis leaf explant browning at genome-wide level.
Methodology/Principal Findings
We first used Illumina high-throughput technology to sequence the transcriptome of Phalaenopsis and then performed de novo transcriptome assembly. We assembled 79,434,350 clean reads into 31,708 isogenes and generated 26,565 annotated unigenes. We assigned Gene Ontology (GO) terms, Kyoto Encyclopedia of Genes and Genomes (KEGG) annotations, and potential Pfam domains to each transcript. Using the transcriptome data as a reference, we next analyzed the differential gene expression of explants cultured for 0, 3, and 6 d, respectively. We then identified differentially expressed genes (DEGs) before and after Phalaenopsis explant browning. We also performed GO, KEGG functional enrichment and Pfam analysis of all DEGs. Finally, we selected 11 genes for quantitative real-time PCR (qPCR) analysis to confirm the expression profile analysis.
Conclusions/Significance
Here, we report the first comprehensive analysis of transcriptome and expression profiles during Phalaenopsis explant browning. Our results suggest that Phalaenopsis explant browning may be due in part to gene expression changes that affect the secondary metabolism, such as: phenylpropanoid pathway and flavonoid biosynthesis. Genes involved in photosynthesis and ATPase activity have been found to be changed at transcription level; these changes may perturb energy metabolism and thus lead to the decay of plant cells and tissues. This study provides comprehensive gene expression data for Phalaenopsis browning. Our data constitute an important resource for further functional studies to prevent explant browning.
Background
Plant tissue culture is an important tool in both basic and applied studies as well as in commercial application, Such as for propagation of orchids, especially in production of Phalaenopsis. The Phalaenopsis genus of popular ornamental plants belongs to the family Orchidaceae and is mainly used in tissue culture because of its rapid propagation. However, explant browning in Phalaenopsis tissue culture presents a major problem in producing regenerated Phalaenopsis in culture. To date, little is known about the browning mechanisms of Phalaenopsis. Therefore, establishing effective and economical methods to reduce explant browning in tissue culture of Phalaenopsis will provide a valuable tool for plant propagation and transgenic manipulation.
Enzymatic oxidation of phenols produces tissue browning [1–3], which involves peroxidase (POD, EC 1.11.1.7), polyphenol oxidase (PPO, EC 1.10.3.1 or EC 1.14.18.1), and phenylalanine ammonia-lyase (PAL, EC 4.3.1.5) [3–7]. There are three hypotheses considered to be responsible for the mechanism of enzymatic browning, namely, phenol-phenolic enzyme regional distribution [8], free radical damage [9], and protective enzyme system [10]. Therefore, these browning enzymes are the important factors and necessary conditions for browning. Our previous studies demonstrated that PAL, PPO, and POD activities increase during browning of Phalaenopsis explants, and PAL and POD transcript levels consistently increase after 3 d of tissue culture [4, 5, 11]. Proteomic studies showed that peroxiredoxin, mitochondrial F-1-ATPase subunit 2, and regulatory protein-like protein increase in Phalaenopsis explants after 3 d of culture [12]. These findings clearly showed that browning of Phalaenopsis explants is tightly regulated at the transcriptome and proteome levels. These studies indicate that simply manipulating the activity of specific enzymes such as PPO may not solve the explant browning problem [13].
Recent advances in sequencing technologies have enabled genomic-scale sequencing projects for many model organisms. These projects include de novo transcriptome analysis and reference mapping of expressed transcripts [14–16]. Using RNA- sequencing (RNA-seq) techniques, Mellidou et al [17] identified different expressed gene (DEGs) mainly involved in lipid metabolism, secondary metabolism, and cell wall modifications in apple fruit browning disorder. The energy-related and stress-related genes were also altered during apple fruit browning development. To investigate pear fruit surface brown (SBS), transcriptome analysis showed that up-regulated the expression of genes related to oxidative phosphorylation, phenolic compound synthesis and PPO [18]. These studies provide a genomics basis for botanists to understand the molecular mechanisms of enzymatic browning.
Recently, OrchidBase (http://lab.fhes.tn.edu.tw/est) has been established from 37,979,342 sequence reads collected from 11 in-house Phalaenopsis orchid cDNA libraries using multiple sequencing techniques [19]. A total of 1,233,823 unique sequences were obtained from Phalaenopsis aphrodite using Roche 454 and Illumina/Solexa high-throughput sequencing technologies [20]. Based on these accomplishments in Phalaenopsis genome research, in the current study, we carried out transcriptome analysis and expression profiling of Phalaenopsis leaf explants during browning.
Here, we used Illumina short-read sequencing for de novo transcriptome assembly and analysis of Phanalenopsis hybrid: Konggangjinli (A red flower cultivar, S1 Fig) explants at the early stage of browning. We constructed a mixed library from 0-d-cultured (control), 3-d-cultured (prior to browning, browning rate 0%), and 6-d-cultured explant (brown rate 100%) (S2 Fig). After sequencing, we used BLAST to compare these reads to the NCBI database and OrchidBase (http://lab.fhes.tn.edu.tw/est) to determine their encoded proteins. The genes and isoforms were annotated and functionally mapped to Gene Ontology (GO) terms. Furthermore, we constructed 3 libraries from the above-mentioned three kinds of explant for expression profile analysis during Phalaenopsis explant browning. The reads were first mapped to the transcriptome and OrchidBase and then analyzed to identify differentially expressed genes (DEGs) among 0 d, 3 d and 6 d. DEGs were categorized into three groups: explants cultured for 3 d vs. 0 d (before explant browning), explants cultured for 6 d vs. 0 d (after explant browning), and explants cultured for 6 d vs. 3 d. Next, we confirmed the differential expressions of interesting genes by quantitative real-time polymerase chain reaction (qRT-PCR). This study sheds fresh light on explant browning at a genome-wide scale and will facilitate the functional analysis of browning-related genes in Phalaenopsis.
Materials and Methods
Plant material
Leaves of Phalaenopsis sp. plants were cut into 0.5 cm × 0.5 cm segments and cultured on Murashige & Skoog medium (Murashige & Skoog, 1962) solidified with 0.8% agar (w/v) at pH 5.8 and supplied with 3 mg/L 6-BA. All cultures were maintained under cool white fluorescent light (approximately 35 μmol m–2 s–1 photon flux density and 16 h photoperiod) at 24 ± 2°C. Explants were collected from leaves cultured for 0, 3, and 6 d, respectively. All explants were immediately frozen in liquid nitrogen and stored at -80°C.
RNA isolation and Illumina PE library preparation for transcriptome analysis
De novo assembly of the transcriptome was performed by short-read sequencing (Illumina). Total RNAs were isolated from explants that were cultured for 0, 3, and 6 d, and then mixed for cDNA library construction. TRIzol Reagent (Invitrogen, CA, USA) was used to isolate total RNA following the manufacturer’s protocol. RNA quality was assessed using the Agilent 2100 Bioanalyzer with 260/280 and 260/230 absorbance ratios.
In this study, transcriptome libraries were constructed by Shanghai Majorbio Bio-pharm Biotechnology Co., Ltd. (Shanghai, China). The Truseq RNA sample prep kit (Illumina, San Diego, CA, USA) was used to construct the cDNA library according to the manufacturer’s protocol. Subsequently, the insert size of the libraries was selected for cDNA target fragments of 300 bp to 500 bp on 2% Low-range Ultra Agarose, followed by PCR amplification using Phusion DNA polymerase (NEB) for 15 PCR cycles. After being quantified by TBS380, libraries were paired-end sequenced using Illumina HiSeq 2000 (2 × 100 bp read length).
Bioinformatics analysis of Phalaenopsis transcriptome
Prior to analyses, all adapter nucleotides were trimmed and the sequences were de novo assembled using SeqPrep, condetri v 2.0.pl and Trinity software (http://trinitynaseq.sourceforge.net) with default parameters respectively. We then collected equestris of Phalaenopsis from the OrchidBase (http://lab.fhes.tn.edu.tw/est.), which contained 37,979,342 sequence reads, to improve the assembly quality. Finaly, BLASTp /BLASTx alignment (E- value of ≤10-5) was performed between isogenes and protein databases such as NCBI’s non-redundant (nr), string. Homologous protein domains from translated transcriptomic sequences of Phalaenopsis were identified by searching against the Pfam database using Hmmscan (http://pfam.sanger.ac.uk/) [21]. BLAST2GO was used to assign putative functionalities, GO terms, and KEGG (Kyoto Encyclopedia of Genes and Genomes) based metabolic pathways [22]. Furthermore we conducted sequence similarity analysis of protein sequences between the assembled transcripts and Arabidopsis thaliana, Nicotiana tabacum, Glycine max, Lycopersicon esculentum, Triticum aestivum, Oryza sativa, and equestris of the Phalaenopsis database. In this analysis, the Phalaenopsis equestris database was downloaded from the NCBI database (http://www.ncbi.nlm.nih.gov/bioproject/53345), by BLASTn search with a cut-off (E- value of ≤10-5). Arabidopsis and Oryza were chosen as they represent dicot and monocot model species, respectively.
Illumina PE library preparation for expression profile analysis and transcriptome mapping for bioinformatics analysis
To examine expression profiles, we then constructed and sequenced three independent libraries from 0 d, 3 d, and 6 d explants. Each cDNA library was constructed as described above and then sequenced by Illumina HiSeq 2000 (1 × 50 bp read length). After removing adaptor sequences, empty reads, and low-quality sequences with unknown sequences “N”, the high quality clean reads were mapped to our Phalaenopsis transcriptome reference database using the Bowtie software with default parameters. Then the transcript abundances were quantified by SEM (http://dewelabbiostat.wise.edu/rsem). The differentially expressed genes (DEGs) between the three samples were identified using the number of mapped reads as EdgeR inputs (http://bioconductor.org/packages/release/bioc/html/edgR.html). In this algorithm, we used FDR (False Discovery Rate) ≤0.05 and the absolute value of |log2FC|>1 as the threshold to judge the significance of differential gene expression. Here, FDR was used to determine the threshold p-value in multiple tests and analysis through manipulating the FDR value, where p-value corresponded to the differential gene expression test.
Three comparisons were conducted: explants cultured for 3 d vs. explants cultured for 0 d (3 d vs. 0 d), explants cultured for 6 d vs. explants cultured for 0 d (6 d vs. 0 d), and explants cultured for 6 d vs. explants cultured for 3 d (6 d vs. 3d). A Venn diagram was constructed using the Venny tool software [23].
The three groups of DEGs were mapped to GO terms. The DEGs were aligned to the GO database for GO functional enrichment analysis using GO tools software with FDR corrections (http://githb.com/tanghaibao/goatools). Statistical significance was considered at P<0.05. We also mapped all DEGs to the KEGG database and searched for significantly enriched KEEG pathways at P<0.05 level using KOBAS software (http://kobas.cbi.pku.edu.cn/home.do). The sequence similarity analysis of protein sequences between the DEGs with Arabidopsis thaliana and Oryza sativa were also conducted as mentioned above. Homologous protein domains for DEGs were identified by searching against the Pfam database using Hmmscan
Validation of gene expression by quantitative real-time PCR (qRT-PCR)
qRT-PCR was performed to confirm the RNA-seq data. Total RNA was extracted for cDNA preparation as described above. Total RNA (1 μg) was reverse transcribed into single-stranded cDNA using the Primescript RT reagent kit (TaKaRa, Dalian, China). qRT-PCR was implemented using the SYBR premix Ex Taq kit (TaKaRa, Dalian, China), with first-strand cDNA as the template. The actin gene from Phalaenopsis was used as an internal control. The relative quantitative method (2-ΔΔCt) was used to calculate the fold change of the target genes. The primers employed in the qRT-PCR are listed in S1 Table. The results of these reference genes were compared to fragments per kilobase of exon model per millions mapped reads (FPKM) estimates.
Date availability
The filtered reads for Phalaenopsis spp were deposited in the NCBI Sequence Read Archive (SRA) under the accession number SRX386023 and SRS506991 (http://www.ncbi.nlm.nih.gov/sra/?term=SRS506991, http://www.ncbi.nlm.nih.gov/sra/?term=SRX386023) (data unpublic)
Results
Phalaenopsis transcriptome assembly and annotation
To examine the Phalaenopsis transcriptome, we collected RNA from 0-d-cultured, 3-d-cultured, and 6-d-cultured explants and constructed one mixed cDNA library. We used the tagged cDNA library for 2×100 bp paired-end sequencing on a single lane of the Illumina Hiseq2000 and generated 118,996,000 raw paired-end reads resulting in 12 gigabases of sequence. After cleaning and quality control, this resulted in 79,434,350 reads (7,544,398,171 bp) (Table 1). Using the RNA seq de novo assembler Trinity, we obtained 31,708 isogenes with a mean size of 1108.07 bp and lengths from 351 bp to 9,265 bp (S2 Table). Using BLASTn sequence similarity search with cut-off E-value≤10-5, about 90% of isogenes (28,267/31,708) have significant sequence similarity to the Phalaenopsis EST database (data not shown). Only 10% of isogenes (3441) were not hit, and were thought to be specific sequence in our study. We used this reference transcriptome for our subsequent analysis of expression profiles during explant browning.
Table 1. Assembly of transcriptome of Phalaenopsis.
| Number | |
|---|---|
| Total number of raw reads | 118,996,000/12,018,596,000(bp) |
| Total number of reads assembled | 79, 434, 350/7 544, 398, 171(bp) |
| Total genes | 21, 348 |
| Total isogenes | 31, 708 |
| Total residues | 35, 134, 614 |
| Smallest isogene | 351 |
| Largest iosgene | 9, 265 |
| average | 1, 108.07 |
About 84% assembled transcripts (26,565/31,708, S3 Table) were annotated by using BLASTp and BLASTn algorithms to search against the NR, gene, string and Nt databases (E-value≤10-5). A BLASTx sequence similarity search with cut-off E-value≤10-5, showed that Phalaenopsis proteins have significant sequence similarity to the proteomes of other plants (Table 2). For example the Phalaenopsis proteome showed substantial amino acid sequence identity to Arabidopsis thaliana (61.71%) and Oryza sativa (63.78%) proteomes. The function of the Phalaenopsis sequences hit in Arabidopsis thaliana and Oryza sativa were shown as supporting information in S4 and S5 Tables. Most of the transcripts had most-significant hits to the Phalaenopsis equestris database, and only 14.34% of the sequences did not show significant identity in the Phalaenopsis equestris database. These do not found homologous sequences suggesting that they may novel proteins in our study.
Table 2. Homology analysis of Phalaenopsis proteome with other plants.
| Proteome of Phalaenopsis | Sequences number(identity≧30) | Sequences number(identity≧70) | Sequences number(identity≧80) | Average Sequences identity(≧30) | |
|---|---|---|---|---|---|
| Arabidopsis thaliana | 26565 | 1120(4.22%) | 350(1.32%) | 121(0.46%) | 61.71% |
| Nicotiana tabacum | 26565 | 2304(8.67%) | 688(2.59%) | 253(0.95%) | 60.96% |
| Glycine max | 26565 | 4386(16.51%) | 1555(5.85%) | 639(2.41%) | 63.45% |
| Lycopersicon esculentum | 26565 | 2237(8.42%) | 731(2.75%) | 267(1.01%) | 62.35% |
| Triticum aestivum | 26565 | 1197(4.51) | 382(1.44%) | 142(0.53%) | 61.37% |
| Oryza sativa | 26565 | 4735(17.82%) | 1734(6.53%) | 631(0.24%) | 63.78% |
| Phalaenopsis equestris | 26565 | 23050(86.78%) | 17942(67.54%) | 16951(63.81%) | 85.66% |
Phalaenopsis expression profile changes during explant browning
To examine the expression of Phalaenopsis genes in browning, we next constructed three cDNA libraries and used 2×100 bp paired-end sequencing to obtain sets of expression reads for explants cultured for 0, 3, and 6 d. After removing the adaptor sequences and low-quality sequences, we mapped the clean reads for each sample (0, 3, and 6 d) to the assembled reference transcriptome of Phalaenopsis (Table 3).
Table 3. Assembly of Phalaenopsis expression profiles and mapping to transcriptome.
| Sample | Number of sequences | Bases (bp) | Number of sequences mapped to transcriptome | Ratio of sequences hit in transcriptome | Total number ofgenes expressed |
|---|---|---|---|---|---|
| 0 d | 7, 750, 687 | 741, 453, 474 | 5, 405, 788/6, 249, 646 | 86.50% | 19,156 |
| 3 d | 9, 085, 070 | 868, 764, 389 | 5, 560, 155/7, 379, 172 | 75.35% | 22,185 |
| 6 d | 9, 529, 153 | 912, 039, 691 | 5, 841, 384/7, 755, 354 | 75.32% | 22,902 |
We used Bowtie (http://bowtie-bio.sourceforge.net/index.shtml) to map reads, used RSEM to assign reads to genes and isoforms, and used edgeR to standardize the reads. We refer to the normalized reads in fragments per kilobase of exon model per millions mapped reads (FPKM), and we used the number of genes with FPKM values ≥0.5 to estimate the total number of genes expressed during Phalaenopsis explant browning(S6 Table). We found 19,156 expressed genes in the explants cultured for 0 d, representing almost 60% of the annotated transcriptome. We also found 22,185 genes (70%) were expressed in 3 d, and 22,902 (72%) in 6 d (Table 3).
Identification of genes differentially expressed during Phalaenopsis explant browning
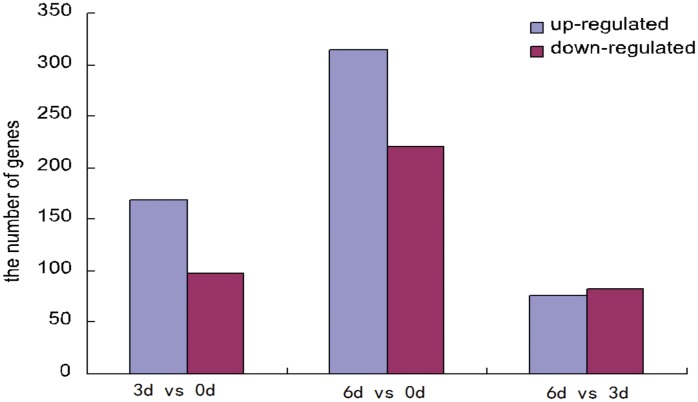
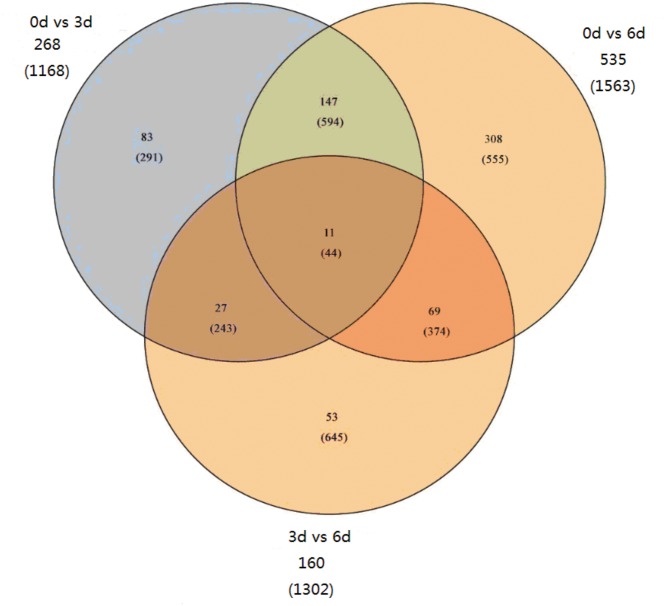
We compared our three time points (0, 3, and 6 d) and identified 698 differentially expressed genes (DEGs) (Fig 1, S7 Table) with FDRs of <0.05. We identified 267 DEGs before Phalaenopsis explant browning in the comparison of 3 d vs. 0 d, 169 (644 isoforms) up-regulated and 98 (523 isoforms) down-regulated. We identified 534 DEGs after explant Phalaenopsis browning in the comparison of 6 d vs. 0 d, 314 (921 isoforms) up-regulated and 220 (641 isoforms) down-regulated. Finally, we only identified 159 DEGs in the comparison of 6 d vs. 3 d, 76 (748 isoforms) up-regulated and 83 (553 isoforms) down-regulated. These results showed that many more genes increased in expression before and after explant browning. Also, the proportion of DEGs was highest in the 0 d vs. 6 d comparison and lowest in the 3 d vs. 6 d comparison. To identify shared and unique DEGs during explant browning, we generated a Venn diagram based on the three comparisons (Fig 2). Only 11(44 isoforms) DEGs were shared in the three groups.
Fig 1. The number of up(blue)- and down(red)- regulate genes in three groups.
Fig 2. Venn diagram showing the number DEGs for each comparison and the overlaps between three comparison groups.
The number in the parenthesis presents the number of isoforms.
Furthermore, DEGs that have the best homolog in Arabidopsis thaliana and Oryza sativa were also studied (S8, S9 and S10 Tables). In the comparison of 3 d vs. 0 d, 6 d vs. 0 d 6 d vs. 3 d, there were 31, 49and 38 isoforms shown sequences identity in Arabidopsis thaliana, and 173, 239,173 isoforms hit in Oryza sativa respectively. The number of homolog in three groups showed low identity in Arabidopsis thaliana comparison with Oryza sativa. Based on the homologs in Arabidopsis thaliana, we found multiple DEGs involved in biosynthesis of secondary metabolites, nucleotide metabolism, carbohydrate metabolism, and amino acids metabolism, such as: NAD (P)- binding rossmann-fold superfamily protein, nucleoside diphosphate kinase family protein, protein kinase superfamily protein, glucose-6-phosphate dehydrogenase 2, caffeoyl-CoA 3-O-methyltransferase.
The highly expression of DEGs before and after Phalaenopsis explant browning was shown in Table 4. The results showed that the expression of pathogenesis-related protein 10c was in higher level and was significantly elevated after explant browning. Notably, glutathione S-transferase (comp16940_0, comp23612_0, comp18470_0, comp23433_0), which involved in Glutathione metabolism, increased by 10–700 times after explant browning, It was also worth mentioning that the expressions of nonsymbiotic hemoglobin (comp23970_0), bibenzyl synthase (chalcone synthase, comp22408_0), 2-hydroxyacyl-CoA lyase (comp22023_0), S-adenosyl-L-methionine synthase (comp19689_0), cytochrome P450 71D11-like (comp20309_0), which appear to be involved in peroxisome, cysteine and methionine metabolism and secondary metabolism, respectively, were very low in the control samples, but increased significantly after Phalaenopsis explant browning occurred, the expression increase by 30–600 folds, of which nonsymbiotic hemoglobin expression increase more than 300 folds. These results demonstrated that there is a close relationship between metabolism and browning. The expression of RNA-dependent RNA polymerase (comp23180_c0), was in high level in control explant and was significantly decreased before and after explant browning. Three DEGs involved in energy metabolism, such as V-type proton ATPase 16 kDa proteolipid subunit-like (comp 16560_c0), which take part in oxidative phosphorylation, putative chlorophyll a/b-binding protein (comp 23175_c0), type III chlorophyll a/b-binding protein (comp20543_c0) which relate to photosynthesis expression reduced before and after explant browning, respectively.
Table 4. The highly expressed DEGs before and after Phalaenopsis explant browning.
| gene id | FPKM | Gene Description | ||
|---|---|---|---|---|
| 0d | 3d | 6d | ||
| Up-regulation | ||||
| comp24400_c0 | 601.4 | 13221.09 | 15931.33 | pathogenesis-related protein 10c [Elaeis guineensis] |
| comp23960_c0 | 243.1 | 8625.85 | 36472.73 | gastrodianin-4A [Gastrodia elata] |
| comp23734_c0 | 874.74 | 4548.3 | 2667.81 | lectin [Cymbidium hybrid cultivar] |
| comp16940_c0 | 48.41 | 2552.83 | 1778.27 | tau glutathione S-transferase [Allium cepa] |
| comp4775_c0 | 58.2 | 2416.77 | 253.37 | S-like RNase[Triticum aestivum] |
| comp22420_c0 | 195.86 | 1665.28 | 1454.83 | PREDICTED: formate dehydrogenase, mitochondrial [Vitis vinifera] |
| comp24492_c0 | 430.54 | 1643.42 | 332.69 | RecName: Full = Probable linoleate 9S-lipoxygenase 5; AltName: Full = Leaf lipoxygenase lipoxygenase [Solanum tuberosum] |
| comp22045_c0 | 45.2 | 836.7 | 672.12 | hypothetical protein SORBIDRAFT_04g034160 [Prunus persica] |
| comp24644_c0 | 47.75 | 660.62 | 319.99 | PREDICTED: L-ascorbate peroxidase 2, cytosolic [Vitis vinifera] |
| comp18704_c0 | 67.83 | 641.47 | 2141.54 | phi class glutathione transferase GSTF7 [Populus trichocarpa] |
| comp22866_c0 | 55.1 | 615.43 | 265.49 | PREDICTED: ferritin-3, chloroplastic [Vitis vinifera] |
| comp23433_c0 | 0.84 | 610.48 | 630.83 | glutathione-S-transferase Cla47 [Elaeis guineensis] |
| comp23612_c0 | 11.87 | 597.78 | 511.05 | tau glutathione S-transferase [Allium cepa] |
| comp19147_c0 | 124.53 | 622.35 | 13177.75 | pathogenesis-related protein 10c [Elaeis guineensis] |
| comp23970_c0 | 1.54 | 483.33 | 1032.45 | nonsymbiotic hemoglobin [Raphanus sativus] |
| comp22408_c0 | 1.21 | 25.66 | 692.42 | bibenzyl synthase [Phalaenopsis sp. chalcone synthase] |
| comp22023_c0 | 17.61 | 217.69 | 655.67 | PREDICTED: 2-hydroxyacyl-CoA lyase [Vitis vinifera] |
| comp22263_c0 | 131.9 | 101.1 | 611.8 | RecName: Full = Adenosylhomocysteinase; Short = AdoHcyase; AltName: Full = S-adenosyl-L-homocysteine hydrolase S-adenosylhomocysteine hydrolase |
| comp23263_c0 | 41.16 | 101.1 | 595.65 | acyl CoA ligase [Linum usitatissimum] |
| comp20309_c0 | 3.53 | 46.43 | 528.32 | PREDICTED: cytochrome P450 71D11-like [Glycine max] |
| comp19689_c0 | 8.72 | 80.93 | 519.67 | S-adenosyl-L-methionine synthase [Musa acuminata AAA Group] |
| comp19151_c0 | 91.85 | 403.39 | 506.52 | glutathione S-transferase [Elaeis guineensis] |
| Down-regulation | ||||
| comp14578_c0 | 5435.85 | 2150.44 | 459.24 | putative chlorophyll a/b-binding protein [Phalaenopsis hybrid cultivar] |
| comp24412_c0 | 3644.04 | 297.48 | 192.06 | ORF170[Phalaenopsis aphrodite subsp. formosana] |
| comp24467_c0 | 2403.94 | 725.68 | 240.26 | PREDICTED: uncharacterized protein LOC100244671 [Vitis vinifera] |
| comp19366_c0 | 1852.48 | 556.8 | 382.63 | hypothetical protein SORBIDRAFT_07g023340 [Sorghum bicolor] |
| comp24193_c0 | 1682.55 | 38.19 | 34.45 | PREDICTED: BTB/POZ and TAZ domain-containing protein 2-like[Glycine max] |
| comp18801_c0 | 1226.98 | 360.29 | 72.21 | type 1 non-specific lipid transfer protein precursor [Triticum aestivum] |
| comp16560_c0 | 1059.9 | 352.26 | 190.68 | PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit-like [Glycine max] |
| comp20543_c0 | 976.56 | 303.2 | 175.84 | type III chlorophyll a/b-binding protein [Lycoris aurea] |
| comp23175_c0 | 873.94 | 122.29 | 96.53 | RecName: Full = Pyruvate, phosphate dikinase, chloroplastic; AltName: Full = Pyruvate, orthophosphate dikinase; Flags: Precursor pyruvate,ortho |
| comp21796_c0 | 764.9 | 259.27 | 134.58 | Os11g0171300[Oryza sativa Japonica Group] |
| comp24776_c0 | 713.77 | 138.06 | 117.99 | Mitochondrial protein, putative [Medicago truncatula] |
| comp20608_c0 | 638.52 | 123.21 | 180.07 | PREDICTED: dnaJ protein homolog 2 [Vitis vinifera] |
| comp20205_c1 | 542.53 | 428.03 | 113.87 | uncharacterized protein LOC100527231 [Glycine max] |
| comp17995_c0 | 541.63 | 295.51 | 51.11 | HAP3-like protein [Citrus sinensis] |
| comp15000_c1 | 531.85 | 71.35 | 60.87 | PREDICTED: 60S ribosomal protein L51, mitochondrial-like [Glycine max] |
| comp21138_c0 | 511.4 | 819.76 | 104.11 | cationic peroxidase 2 precursor [Glycine max] |
The gene were selected at the highest of the three FPKM >500 and were stored by p-value<0.05.
To identified DEGs that were induced before and after Phalaenopsis explant browning, the result showed that there were 36 and 69 DEGs induced before and after Phalaenopsis explant browning, respectively (S11 Table). The top 10 DEGs were showed in Tables 5 and 6. We found that cytokinin oxidase(comp21576), probable mannitol dehydrogenase(comp21741),4-coumarate:CoA ligase(comp20466), which involved in zeatin biosynthesis, phenylpropanoid biosynthesis, fatty acid elongation, respectively, were highly induced before and after Phalaenopsis explant browning.
Table 5. The TOP10 of DEGs induced before Phalaenopsis explant browning.
| Gene id | FPKM | Gene Description | |
|---|---|---|---|
| 0d | 3d | ||
| comp24050_c0 | 0 | 179.85 | PREDICTED: pentatricopeptide repeat-containing protein At4g02750 [Vitis vinifera] |
| comp20452_c0 | 0 | 111.27 | hypothetical protein SORBIDRAFT_ 07g023110 [Sorghum bicolor] |
| comp21576_c0 | 0 | 108.91 | cytokinin oxidase [Dendrobium hybrid cultivar] |
| comp15076_c0 | 0 | 61.13 | trypsin inhibitor 3 [Oncidium Gower Ramsey] |
| comp21640_c0 | 0 | 60.97 | Peroxidase 52 precursor, putative [Ricinus communis] |
| comp21741_c0 | 0 | 46.64 | PREDICTED: probable mannitol dehydrogenase [Vitis vinifera] |
| comp19746_c0 | 0 | 38.15 | RecName: Full = Probable mannitol dehydrogenase; AltName: Full = NAD-dependent mannitol cinnamyl alcohol dehydrogenase [Fragaria] |
| comp20287_c0 | 0 | 36.41 | PREDICTED: uncharacterized protein LOC100243770 [Vitis vinifera] |
| comp11152_c0 | 0 | 34.58 | putative UDP-glucose dehydrogenase 1 [Nicotiana tabacum] |
| comp18523_c0 | 0 | 33.57 | extensin-like protein [Vigna unguiculata] |
The gene were selected at p-value<0.05
Table 6. The TOP10 of DEGs induced after Phalaenopsis explant browning.
| Gene id | FPKM | Gene Description | |
|---|---|---|---|
| 0d | 6d | ||
| comp24050_c0 | 0 | 274.5 | PREDICTED: pentatricopeptide repeat-containing protein At4g02750 [Vitis vinifera] |
| comp20466_c0 | 0 | 184.49 | 4-coumarate:CoA ligase [Petunia x hybrida] |
| comp13345_c0 | 0 | 147.5 | MYB transcription factor MYB92 [Elaeis guineensis] |
| comp18691_c1 | 0 | 144.65 | trans-2-enoyl-CoA reductase [Phalaenopsis amabilis] |
| comp21576_c0 | 0 | 127.96 | cytokinin oxidase [Dendrobium hybrid cultivar] |
| comp21741_c0 | 0 | 123.9 | PREDICTED: probable mannitol dehydrogenase [Vitis vinifera] |
| comp20452_c0 | 0 | 102.5 | hypothetical protein SORBIDRAFT_07g023110 [Sorghum bicolor] |
| comp22222_c0 | 0 | 70.1 | PREDICTED: probable serine/threonine-protein kinase At1g18390 [Vitis vinifera] |
| comp29276_c0 | 0 | 68.39 | glycolate oxidase |
| comp20432_c1 | 0 | 65.15 | hypothetical protein slr1753 [Synechocystis sp. PCC 6803] |
The gene were selected at p-value<0.05
Gene ontology (GO) analysis for Phalaenopsis transcriptome and DEGs
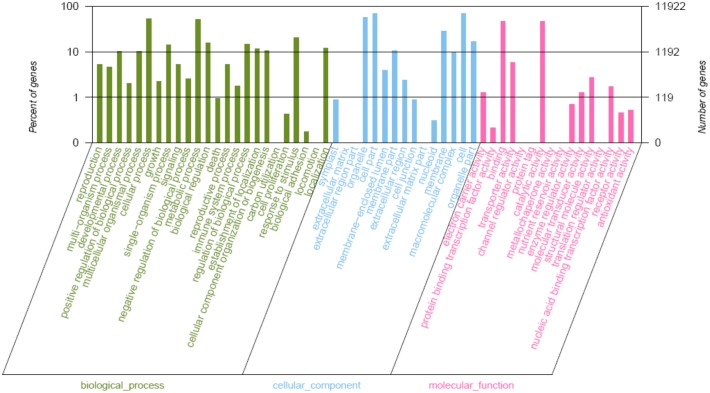
To characterize the Phalaenopsis transcriptome, we used GO assignments to classify the functions of the predicted Phalaenopsis genes. Using Blast2GO, we grouped the 21,384 annotated sequences into 45 sub-categories within the three main GO categories of biological process, cellular component and molecular function (Fig 3). Based on the biological processes, we established 15 categories. We found 6,318 genes (52.99%) involved in cellular processes, 6,167 genes (51.72%) in metabolic processes and 2, 420 genes (20.29%) in responding to stimulus. In addition, we found 14 clusters of genes with roles in cell formation, including cell parts (8,275 genes, 69.41%), organelles (6,885 genes, 57.75%), and membrane (3,431 genes, 28.79%). Based on their putative molecular functions, we also found 23 categories, including genes involved in catalytic activity (5,642 genes, 47.32%) and binding activity (5, 534 genes, 46.42%).
Fig 3. Distribution of the GO categories assigned to the Phalaenopsis transcriptome.
21, 384 genes were grouped into 45 sub-categories, which were divided into three categories: cellular component, molecular function, biological process.
Once we had characterized the transcriptome, we next assigned GO categories to the DEGs and compared them with those in the transcriptome. We found a similar distribution of GO-annotation function of DEGs in the three comparisons (S3 Fig). For biological processes, before and after explant browning, most DEGs were assigned to metabolic process, followed by cellular process and responding to stimulus. For cellular component, the top three categories for DEGs were cell part, cell, and organelles. For molecular functions, the top categories for DEGs were catalytic activity and binding.
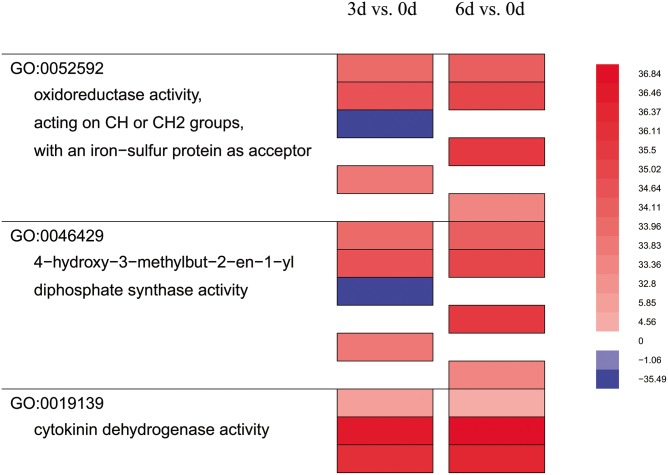
Furthermore, GO enrichment analyses showed that before Phalaenopsis explant browning, the DEGs for 3 d vs. 0 d mainly function in the protein–DNA complex, and nucleosome organization. However, after Phalaenopsis explant browning, the DEGs for 6 d vs. 0 d mainly function in metabolic processes, such as small molecule metabolism, oxoacid metabolism, and carboxylic acid metabolism (S12 Table). We also analyzed the GO enriched functional categories in each of the three pairwise comparisons, which condensed and compressed the genes by removing categories that did not show as significantly different, and uses a false color heat-map to indicate up- or down-regulated classes (Fig 4). Genes encoding enzymes for cytokinin dehydrogenase, oxidoreductase, acting on CH or CH2 groups, with an iron-sulfur protein as acceptor, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase activity were strongly up-regulated in 3 d and 6 d explants compared to control (0 d) (Fig 4). Also, we found no significant change for 6 d vs. 3 d in GO function enrichment analyses.
Fig 4. Enriched functional categories in each of the pair-wise comparison.
Each vertical column represented the genes that were dramatically up- (red) or down- regulated (blue) comparing the two groups indicated at the top. Lines to the left showed functional category and GO number. Each color bar represents an individual locus.
Pathway analysis for Phalaenopsis transcriptome and DEGs
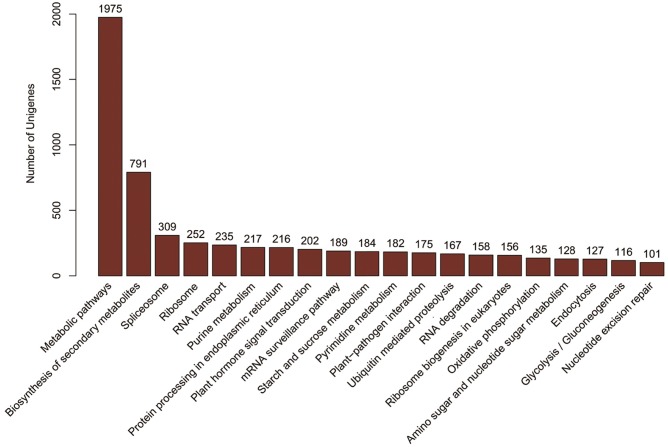
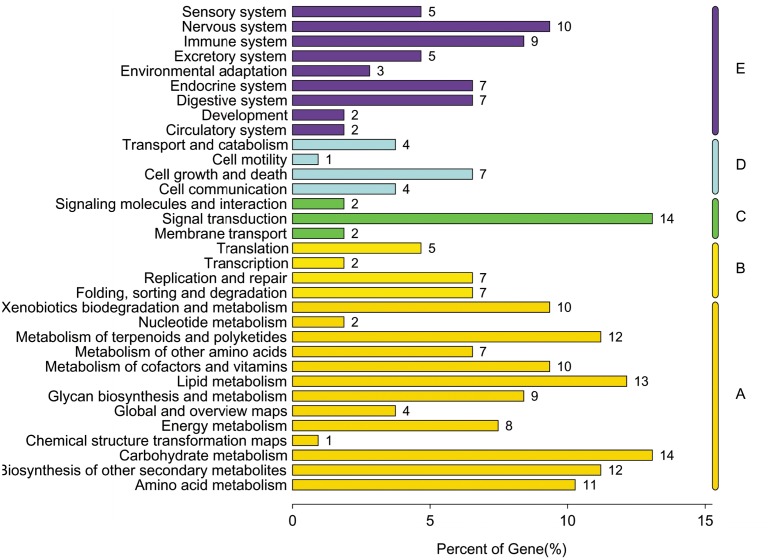
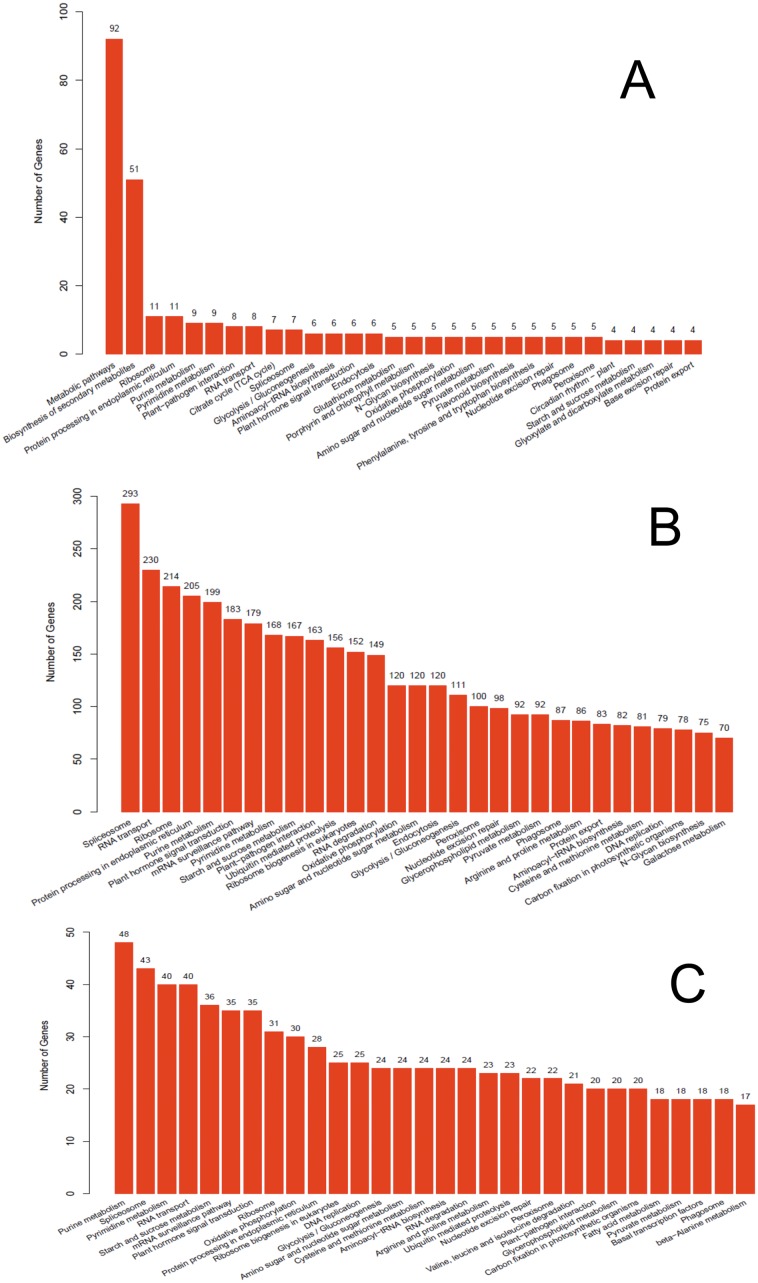
Pathway-based analysis can help elucidate the biological functions of genes. To identify the biological pathways that are active in Phalaenopsis, we mapped all the unigenes to the reference canonical pathways in KEGG. A total of 14,099 unigenes were assigned to 123 KEGG pathways (S13 Table), where ‘Metabolic pathways’ (1,975 unigenes, 14%) is dominant, followed by ‘Biosynthesis of secondary metabolites ’(791 unigenes, 5%) and ‘Spliceosome ’(309 unigenes, 2%) (Fig 5). KEGG classification analysis identified transcripts involved in signal transduction, carbohydrate metabolism and lipid metabolism have a high degree in Phalaenopsis (Fig 6). The distribution of KEGG pathway of Phalaenopsis genes was shown in Fig 7. The top 30 pathways of Phalaenopsis transcripts mapped to the Arabidopsis thaliana and Oryza sativa KEGG were showed that the purine metabolism, spliceosome, pyrimidine metabolism and RNA transport were high hit in Oryza sativa, while metabolic pathways and biosynthesis of secondary metabolites were high mapped in Arabidopsis thaliana. About 293,230 and 214 transcripts mapped in the spliceosome, RNA transport and ribosome in Phalaenopsis were tooken over the top 3 (Fig 7).
Fig 5. Distribution of Phalaenopsis transcriptome sequences among KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways.
The top 20 most highly represented pathways are shown. Analysis was performed using Blast2GO and the KEGG database.
Fig 6. Classification of Phalaenopsis genes involved in KEGG pathways.
(A: Metabolism, B: Genetic Information Processing, C: Environmental Information Processing, D: Cellular Processes, E: Organismal Systems)
Fig 7. Distribution of Phalaenopsis genes and distribution of Phalaenopsis genes mapped in mapped in Arabidopsis thaliana and Oryza sativa among KEGG pathways.
The top 20 most highly represented pathways are shown. A: Phalaenopsis genes mapped in Arabidopsis thaliana, B: Distribution of Phalaenopsis genes in KEGG pathways, C: Phalaenopsis genes mapped in Oryza sativa
To characterize the functional consequences of gene expression changes associated with explant browning, we performed pathway enrichment analysis on DEGs based on the KEGG database with p <0.05 as the threshold. We found that 18 of 90 pathways demonstrated significant changes in 3 d vs. 0 d, 25/102 pathways in 6 d vs. 0 d, and 11/98 pathways in 6 d vs. 3 d (S14 Table). Comparison of the three groups of DEGs revealed that phenylpropanoid biosynthesis, flavonoid biosynthesis, phenylalanine metabolism and terpenoid backbone biosynthesis demonstrated significant changes in all three comparisons. There were seven pathways of carbohydrate metabolism, seven pathways of amino acid metabolism, five of lipid metabolism, four pathways of secondary metabolism significantly changed before and after Phalaenopsis explant browning (Table 7).
Table 7. List of important KEGG pathways enrichment analysis of DEGs before and after Phalaenopsis explant browning.
| Pathway Id | Pathway name | Background.number | Sample number | |
|---|---|---|---|---|
| 3d vs.0d | 6d vs.0d | |||
| Carbohydrate metabolism | ||||
| ko00010 | Glycolysis / Gluconeogenesis | 116 | 12 | |
| ko00040 | Pentose and glucuronate interconversions | 46 | 6 | |
| ko00620 | Pyruvate metabolism | 93 | 10 | |
| ko00640 | Propanoate metabolism | 41 | 6 | |
| ko00630 | Glyoxylate and dicarboxylate metabolism | 55 | 10 | |
| ko00030 | Pentose phosphate pathway | 55 | 11 | |
| ko00051 | Fructose and mannose metabolism | 67 | 12 | |
| Amino acid metabolism | ||||
| ko00250 | Alanine, aspartate and glutamate metabolism | 52 | 9 | |
| ko00270 | Cysteine and methionine metabolism | 84 | 14 | |
| ko00360 | Phenylalanine metabolism | 50 | 13 | |
| ko00400 | Phenylalanine, tyrosine and tryptophan biosynthesis | 49 | 8 | |
| ko00480 | Glutathione metabolism | 67 | 10 | |
| ko00460 | Cyanoamino acid metabolism | 30 | 7 | |
| ko00350 | Tyrosine metabolism | 34 | 5 | |
| Lipid metabolism | ||||
| ko00564 | Glycerophospholipid metabolism | 97 | 10 | |
| ko00565 | Ether lipid metabolism | 28 | 5 | |
| ko00062 | Fatty acid elongation | 37 | 9 | |
| ko00071 | Fatty acid metabolism | 66 | 13 | |
| ko00592 | alpha-Linolenic acid metabolism | 62 | 13 | |
| Energy metabolism | ||||
| ko00910 | Nitrogen metabolism | 41 | 17 | 10 |
| ko00195 | Photosynthesis | 71 | 17 | |
| ko00710 | Carbon fixation in photosynthetic organisms | 83 | 16 | |
| Biosynthesis of other secondary metabolites | ||||
| ko00940 | Phenylpropanoid biosynthesis | 75 | 16 | 19 |
| ko00941 | Flavonoid biosynthesis | 35 | 7 | 8 |
| ko00945 | Stilbenoid, diarylheptanoid and gingerol biosynthesis | 16 | 4 | |
| ko00960 | Tropane, piperidine and pyridine alkaloid biosynthesis | 20 | 4 | 7 |
| Translation | ||||
| ko03008 | Ribosome biogenesis in eukaryotes | 156 | 15 | |
| ko03010 | Ribosome | 252 | 23 | |
| Signal transduction | ||||
| ko04075 | Plant hormone signal transduction | 202 | 18 | |
| Metabolism of terpenoids and polyketides | ||||
| ko00900 | Terpenoid backbone biosynthesis | 46 | 7 | 9 |
| ko00903 | Limonene and pinene degradation | 25 | 5 | |
| ko00908 | Zeatin biosynthesis | 13 | 3 | 4 |
| Transport and catabolism | ||||
| ko04146 | Peroxisome | 100 | 14 | |
Pathway enrichment of DEGs was analysis at P-value<0.05.
Protein domain analysis for Phalaenopsis transcriptome and DEGs
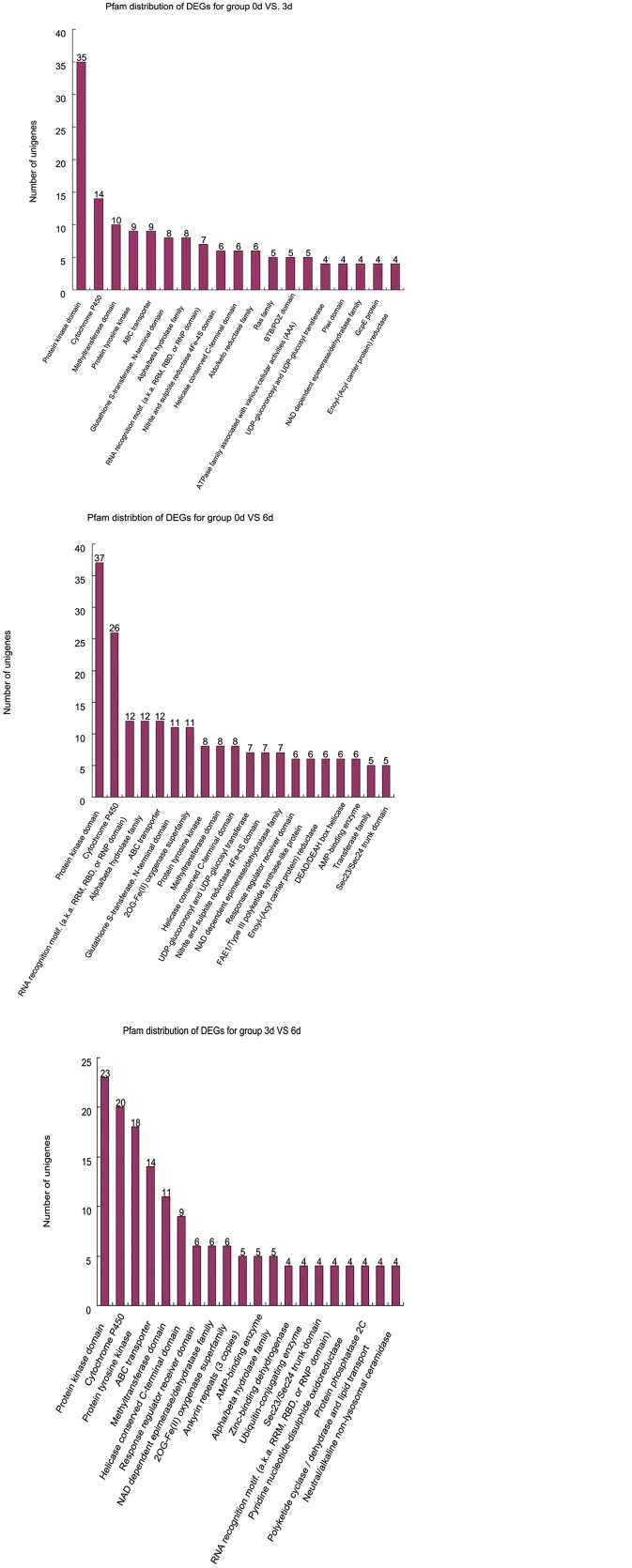
Conserved protein domains were identified in the Phalaenopsis unigenes against the Pfam database by using Hmmscan software with cutoff E-value≤0.1, 12,341 of 26,565 assembled unigenes matched entries that are corresponding to 3,021 different domains/families (S15 Table). The 20 most abundant domains identified are provided in Table 7. The top 3 abundant domains were identified included Pkinase (protein kinase domain) with 568 unigenes, followed by Pkinase_Tyr (protein tyrosine kinase) with 284 unigenes and RRM_1(RNA recognition motif) with 202 unigenes. The PPR domain proteins and cytochrome P450 families were also highly represented.
To specifically identify DEGs protein domains in the Pfam database, comparison of Pfam searches between three groups of DEGs indicated an overall similarity of identified domains. The top 20 domains were shown in Fig 8. Protein kinase domain, cytochrome P450, methyltransferase domain, Protein tyrosine kinase and ABC transporter were most highly represented in three groups of DEGs.
Fig 8. Distribution of the top 20 Pfam domains identified in three groups of DEGs.
Validation of the expression profile data using real-time PCR
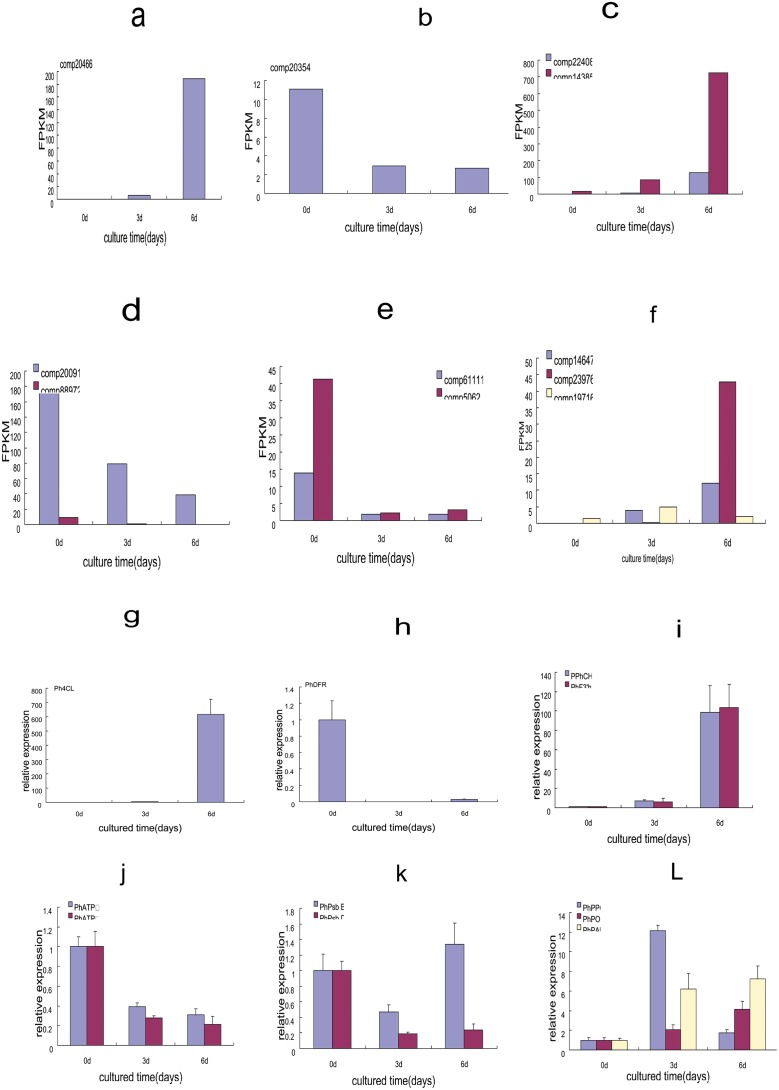
Based on KEGG pathway enrichment analysis and previous studies [4–6, 11–12], we selected eleven DEGs and subjected them to quantitative real-time PCR (qRT-PCR) to validate the results of the expression profile analysis (Fig 9). We found a high correlation (R2 = 0.8841) between RNA-seq and qRT-PCR results (S1 Table). For example, the transcript levels of CHS(chalcone synthase), 4CL(4-coumaroyi·CoA ligasc), and F3’H (flavonoid3’-hydroxylase)were high in 6 d explants, and DFR (dihy- droflavonol 4-reductase)decreased in 3 and 6 d explants (Fig 9h and 9i). ATPase γ subunit, ATPase α subunit, psbB, and psbD were down-regulated in the 3 and 6 d explants, consistent with the results of expression profile analysis. Only the psbB transcript level increased in the 6 d explants, in contrast to the sequence data (Fig 9e and 9k). As expected [4, 11], the transcripts of PPO, POD, and PAL were up-regulated during browning (Fig 9I). Our results indicate that our RNA-seq data reliably identified browning-related genes in Phalaenopsis.
Fig 9. Transcript profile(a-f) and qPCR (g-l) results of selected genes during Phalaenopsis explant browning.
The genes and transcriptome number: comp 20466 (4CL), comp 20354 (DFR), comp 22408 (CHS), comp 14385 (F3'H), comp 20091 (ATP γ subunite), comp 88972 (ATP α subunite); comp 61111 (psbB), comp 5062 (psbD), comp 14647 (POD), comp 23976 (PAL), comp 19716 (PPO). The left y-axis indicates the FPKM (fragments per kilobase of exon modle per millions mapped reads). In a-f and the relative expression level by qPCR(2ΔΔ ct), is presented as fold- changes in gene expression normalized to the actin gene in g-l.
Discussion
Transcriptome and expression profile analysis of DEGs during Phalaenopsis explant browning
Using the Illumina HiSeq 2000 next generation sequencing technology, we obtained transcriptome sequences from explants that were cultured for 0, 3, and 6 d. We assembled more than 79,434,350 reads into approximately 31,708 isogenes and annotated 26,565 proteins. Homology searches showed that 63.78 and 61.71% isogenes have significant similarities to Oryza sativa and Arabidopsis thaliana protein sequences and about 85.66% of isogenes show identity with the publicly available Phalaenopsis equestris. Our data show more sequences with significant identity to known sequences in the plant proteome than the sequences identified by Hsu et al. [24]. Also, we annotated and functionally mapped 21, 348 sequences to GO terms. Gene annotation can help predict potential genes and their functions at the transcriptome level. In this study, we found predominant gene clusters involved in the cellular and metabolic processes of biological processes, the binding and catalytic activities of molecular functions, as well as the cell, cell part, and organelles categories of cellular components. Our results were different from those obtained in Phalaenopsis aphrodite, in which the identified genes were predominantly involved in the biosynthesis of macromolecules and in nitrogen, protein, and nucleic acid metabolism [20]. This difference may be due to the fact that we constructed our cDNA library only from Phalaenopsis leaf tissue.
The DEGs among the explants cultured for 0, 3, and 6 d probably relate to their metabolism and functions. We found that 267 and 534 genes exhibited significant differences in expression before and after (3d and 6d) Phalaenopsis explant browning, respectively. Among them 36 and 69 DEGs were induced before and after explant browning, respectively. The three comparisons shared only 11 genes in common, indicating that the patterns of gene expression differed over time during explant browning. The identified DEGs were involved in biological processes, cellular components, and molecular functions GO terms. Pfam search showed that protein kinase domain, cytochrome P450, methyltransferase domain, protein tyrosine kinase and ABC transporter were most highly represented in three groups of DEGs. Protein kinase and its subcategories tyrosine-protein kinase were known to regulate the majority of cellular pathways. Cytochrome P450 is a large plant protein family that might contribute to extensive modifications of various secondary compounds. These results revealed DEGs with multiple functions, suggesting that 3 d and 6 d were critical stages for explant browning, when many substances related to browning were produced.
DEGs involved in polyphenol synthesis and phenylpropanoid pathways before and after Phalaenopsis explant browning
Previous studies indicated that phenol oxidase activity results in explant browning and the degree of explant browning positively correlates with the contents of phenolic compounds [2–5, 25]. The accumulation and oxidation of phenolic compounds causes enzymatic browning. Also, reduction of polyphenol synthesis and inhibition of the phenylpropanoid pathway repressed browning [26–28]. PAL is the first dedicated enzyme in the phenylpropanoid pathway and converts phenylalanine into trans-cinnamic acid, providing a substrate for further synthesis of phenolic compounds. PPO and POD also function in enzymatic browning. Both enzymes have common diphenolic substrates leading to brown products. In our previous studies, we found that tannin filled in the vascular bundles of browning tissues [29]. Based on previous work and this study, we selected seven genes for qPCR: PhCHS, Ph4CL, PhF3’H, and PhDFR, which function in flavonoid biosynthesis, and PhPPO, PhPOD, and PhPAL, which function in phenylalanine metabolism, and which also showed significant changes in the KEGG pathway enrichment analysis in this study. Consistent with our expectations, PhPPO, PhPOD, and PhPAL were up-regulated in Phalaenopsis explant browning [2, 4, 8, 30] and qRT-PCR confirmed the expression profiling obtained in this study.
Anthocyanins, a class of flavonoids that is responsible for the colors in fruits and most flowers of higher plants, are major water soluble pigments [31,32] and exhibit important physiological functions, such as antioxidative [33,34]. The degradation of anthocyanins and/or the oxidation of phenolics caused by polyphenol oxidase (PPO) results in an enzymatic browning reaction of fruits and vegetables [35,36]. Genes in anthocyanin biosynthesis such as PhPAL, PhCHS, Ph4CL, PhF3’H were up-regulated in 6 d explants of Phalaenopsis indicates that anthocyanin may play important roles in the early stages of Phalaenopsis explant browning. Previous studies have shown that disruptions of oxidation and reduction may induce tissue browning, leading to reactive oxygen species (ROS) bursts, which would affect the membrane integrity [37–39]. ROS which were induced in tissue culture is eliminated by biosynthesis of anthocyanin. DFR is a key enzyme in the catalysis of the stereospecific reduction of dihydroflavonols to leucoanthocyanidins in anthocyanin biosynthesis. Over-expression DFR in the sweet potato enhanced scavenging of reactive oxygen radicals in plants under stressful conditions [40]. PhDFR showed opposite expression profile compared to that of PhPAL, Ph4CL, PhCHS and PhF3’H in the early stages of Phalaenopsis explant, so anthocyanin biosynthesis may be effected. This may lead to imbalance of ROS metabolism and membrane lipid peroxidation, and destroy of cell membrane integrity.
Meanwhile, Cytochrome P450 plays an important role in flavone biosynthesis and has been well characterized. About 115 different cytochrome P450 isogenes were detected in our RAN sequencing data. The cytochrome P450 71D11-like was highly expression after Phalaenopsis explant browning (Tables 4 and 8). Cytochrome P450 expressed only after lotus tissue browning [41], indicating that browning was beneficial for secondary metabolite production.
Table 8. Distribution of the top 20 Pfam domains identified in translated Phalaenopsis sequences.
| Pfam_entry_name | description of Pfam entry | annotated isogenes number |
|---|---|---|
| Pkinase | Protein kinase domain | 568 |
| Pkinase_Tyr | Protein tyrosine kinase | 284 |
| RRM_1 | RNA recognition motif. (a.k.a. RRM, RBD, or RNP domain) | 202 |
| PPR_3 | Pentatricopeptide repeat domain | 138 |
| PPR_2 | PPR repeat family | 117 |
| p450 | Cytochrome P450 | 115 |
| DEAD | DEAD/DEAH box helicase | 88 |
| ABC_tran | ABC transporter | 84 |
| Helicase_C | Helicase conserved C-terminal domain | 78 |
| PPR_1 | PPR repeat | 70 |
| AAA | ATPase family associated with various cellular activities (AAA) | 66 |
| PP2C | Protein phosphatase 2C | 66 |
| Abhydrolase_6 | Alpha/beta hydrolase family | 57 |
| Abhydrolase_5 | Alpha/beta hydrolase family | 54 |
| Epimerase | NAD dependent epimerase/dehydratase family | 53 |
| Myb_DNA-binding | Myb-like DNA-binding domain | 52 |
| TPR_14 | Tetratricopeptide repeat | 52 |
| Methyltransf_11 | Methyltransferase domain | 51 |
| 2OG-FeII_Oxy | 2OG-Fe(II) oxygenase superfamily | 47 |
| RRM_6 | RNA recognition motif (a.k.a. RRM, RBD, or RNP domain) | 47 |
Analysis was performed using HMMER with Pfam database.
DEGs involved in photosynthesis, energy synthesis and other metabolism before and after Phalaenopsis explant browning
Photosystem II (PSII) is a multi-subunit, pigment-protein complex localised in the chloroplast thylakoid membranes. The PSII complex is composed of a number of proteins including D1 (PsbA), D2 (PsbD), CP43 (PsbC), CP47 (PsbB), apoproteins of cytochrome b559, and the extrinsic manganese- stabilizing protein, along with a number of small proteins [42]. In our KEGG pathway enrichment analysis, we found that DEGs involved in photosynthesis also changes significantly in browning explants, so we selected PhpsbB, and PhpsbD for qRT-PCR. F-ATPases (also known as F1F0-ATPase, or H(+)-transporting two-sector ATPase) (EC:3.6.3.14) function as the prime producers of ATP in mitochondria, chloroplasts and bacterial plasma membranes, using the proton gradient generated by oxidative phosphorylation (mitochondria) or photosynthesis (chloroplasts). Thus, we also selected the genes encoding the PhATPase γ subunit, and APhTPase α subunit for qRT-PCR. Our qRT-PCR results were in accordance with the RNA-seq expression profiles in which the transcript levels decreased in the 3 and 6 d explants. By contrast, our qRT-PCR analysis showed that the transcript levels of PhpsbB increased in the 6 d explants. We have found that during explant browning, chloroplasts shrink to dark-colored structures and chlorophyll b content declines [25,29]. In this study, we found that the abundance of PhpsbB and PhpsbD declined before and after Phalaenopsis explant browning. Consistent with this results, two kinds of chlorophyll a/b binding protein were found to be decreased significantly during explant browning(Table 4). To our knowledge, this is the first report to show that photosynthesis genes are associated with explant browning. Liu et al [18] also confirmed that photosynthesis changes during pear fruit browning. These metabolic processes will disrupt the energy metabolism providing a molecular context for the formation of ROS in browning tissue. We conceive that the wound-induced natural defense response leads to accumulation of toxic compounds that impair photosynthesis in browning plant tissues and ultimately damage or kill plant cells.
In plant tissues, ATP content is important for regulating anti-oxidase expression, such as superoxide dismutase, catalase, and ascorbate peroxidase [43]. Jiang et al. [40] also found that ATPase was differentially expressed in Lotus tuber browning tissue using 2-DE and MALDI-TOF-TOF for protein profile analysis. Proteins related to nucleotide metabolism: adenosine kinase, and nucleoside diphosphate kinase were down-expressed after Lotus tissue browning. Their down-regulations indicated that substance metabolism was out of control after browning and the abilities of energy synthesis and utilization declined, accompanied by disorder of ATP energy metabolism. Senescence and stress can also lead to lower ATP production and higher accumulation of reactive oxygen species, resulting in membrane system disorder and triggering browning [42]. Moreover, using LCQ MS/MS for protein expression analysis in explants cultured for 3 d, we linked ATPase with explant browning [12]. In line with this, we herein verified that the transcript levels of the genes encoding the PhATPase γ and PhATPase α subunits decreased in the 3 d and 6 d explants, implying that PhATPase has important functions during explant browning. The expression of V-type proton ATPase 16 kDa proteolipid subunit-like also decreased significantly before and after explant browning (Table 4). Down-regulation of genes encoding ATPase subunits indicated that energy synthesis and utilization declined. Less energy means incapacity to repair membranes, in turn, loosening of cellular compartmentalization allows the release of PPOs and phenols to the cytosol, triggering the actual discolouration reactions. Thus, we consider that ATP metabolism might affect explant browning in Phalaenopsis.
Before and after Phalaenopsis explant browning, DEGs involved in amino acid metabolism, carbohydrate metabolism, lipid metabolism were significantly changed (Table 7). A total of 22 and 44 DEGs involved in amino acid metabolism before and after explant browning, indicating that expressions of some proteins resulted in promotion of the browning reaction. After Lotus tissue browning, Jiang et al found the expression of proteins relating to amino acid metabolism: cysteine synthase and glutamate-ammonia ligase reduced, respectively [40]. Related to lipid metabolism the expression type 1 non-specific lipid transfer protein reduced during explant browning, implying that lipid might be related to plant stress tolerance. Many DEGs involved in lipid degradation pathways were significantly up-regulated in apple fruit browning disorder [17]. It is understandable that changes in these metabolic processes will disrupt the energy metabolism and cell compartments, resulting oxidation of phenolic compounds and influence the explant browning.
Conclusions
Our RNA-seq analysis of changes in gene expression substantially increases the number of available equestris in Phalaenopsis and provides comprehensive information on the mechanism of Phalaenopsis explant browning at a genome-wide scale. Most of the DEGs we identified were up-regulated during Phalaenopsis explant browning. More importantly, transcripts of the genes that control secondary metabolism, energy metabolism, carbohydrate metabolism, and lipid metabolism were differentially expressed in explant browning. In addition, in this study, we identified many new browning-related genes. More detailed and extensive analyses of the identified genes will be essential to further decipher their possible roles in explant browning. Thus, our data provide genome-wide information that enables further exploration of explant browning.
Supporting Information
(EPS)
(EPS)
Each set of 3 vertical bars (blue, red, yellow) indicated the percent of DEGs in the three categories: molecular functions(A), cellular components (B), and biological processes (C).
(TIF)
(DOC)
(DOC)
(XLS)
(XLS)
(XLS)
(XLS)
FDR <0.05 and the absolute value of log2Ratio ≤1 were used as the threshold to judge the significance of gene expression difference.
(XLS)
(XLS)
(XLS)
(XLS)
(XLSX)
Fisher’s exact test with FDR <0.05
(DOC)
A totalof 14,099 sequences were assigned to 123 KEGG pathways.
(XLS)
(XLS)
(XLS)
Acknowledgments
All authors are grateful to Dr. Jinxiang Wang (College of Natural Resources and Environment, South China Agricultural University) for his help in manuscript writing.
Data Availability
All data were deposited in the NCBI Sequence Read Archive (SRA) under the accession number SRX386023 and SRS506991 (http://www.ncbi.nlm.nih.gov/sra/?term=SRS506991, http://www.ncbi.nlm.nih.gov/sra/?term=SRX386023).
Funding Statement
This work was supported by Natural Science Foundation in China (31070618), http://www.nsfc.gov.cn/ (CJX).
References
- 1. Ye M, Wang BC, Duan CR. Advances of explant browning in plant tissue culture. Lett. Biotec. 2004, 15: 426–428. [Google Scholar]
- 2. Laukkanen H, Häggman H, Kontunen-Soppela S, Hohtola A. Tissue browning of in vitro cultures of Scots pine: role of peroxidase and polyphenol oxidase. Physiol. Plant. 1999,106:337–343. [Google Scholar]
- 3. Bhat SR, Chandel KPS. A novel technique to overcome browning in tissue culture. Plant Cell Rep.1991,10: 358–361. 10.1007/BF00193159 [DOI] [PubMed] [Google Scholar]
- 4. Xu CJ, Li L. Changes of total phenol content and the activities of PPO, POD and PAL during the browning in Phalaenopsis explant in vitro. Acta Horti. Sinica. 2006,33(3):671–674. [Google Scholar]
- 5. Xu CJ, Li H, Li L. Phenylalanine ammonia lyase gene expression correlated with Phalaenopsis sp. leaf explants browning. J. Trop. Subtrop. Bot 2007,5: 50–54. [Google Scholar]
- 6. Zhou WL, Tan RF, Xu CJ, Lai YY, Chen DY, Li L. Gibberellic acid Iinhibits browning, enzyme activity and gene expression of phenylalanine ammonia-lyase in Phalaenopsis leaf explants Genes, Genomes and Genomics,2009, 3: 68–71. [Google Scholar]
- 7. Krishna H, Sairam RK, Singh SK, Patel VB, Sharma RR, Grover M, et al. Mango explant browning: Effect of ontogenic age, mycorrhization and pre-treatments. Scientia Horticulturae. 2008,118(2): 132–138. [Google Scholar]
- 8. Mager AM, Harel E. Polyphenoloxidases in plants. Phytochemistry 1979, 8 (2), 193–215. [Google Scholar]
- 9. Parkin KL, Marangoni A, Jackman RL, Yada RY, Stanley DW. Chilling injury: a review of possible causes. J. Food Biochem. 1989, 13 (2), 127–153. [Google Scholar]
- 10. Mitter R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7 (3), 405–410. [DOI] [PubMed] [Google Scholar]
- 11. Xu CJ, Sun XZ, Chen DY, Lai YY, Li L. Clong and charcterizationof peroxidase gene in Phalaenopsis. Pak. J. Bot.2011, 43(4): 2161–2165. [Google Scholar]
- 12. Chen G, Chen DY, Wang T, Xu CJ, Li L. Analysis of the proteins related to browning in leaf culture of Phalaenopsis . Scientia horticulturae. 2012,141:17–22. [Google Scholar]
- 13. Chen DY, Lai YY, Xu CJ, Li L. Influuences of PPO Inhibitors On Browning in Phalaenopsis Explants . Subtropical plant science. 2009,38(12):15–18 (in chinese) [Google Scholar]
- 14. Shin H, Hirst M, Bainbridge MN, Magrini V, Mardis E, Moerman DG. et al. Transcriptome analysis for Caenorhabditis elegans based on novel expressed sequence tags. BMC Biol. 2008,6: 30 10.1186/1741-7007-6-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Meyer E, Aglyamova GV, Wang S, Buchanan-Carter J, Abrego D, Colbourne JK, et al. Sequencing and de novo analysis of a coral larval transcriptome using 454 GSFlx. BMC Genomics. 2009,10: 219 10.1186/1471-2164-10-219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008,320: 1344–1349. 10.1126/science.1158441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mellidou I, Buts K, Hatoum D, Ho QT, Johnston JW, Watkins CB, et al. Transcriptomic events associated with internal browning of apple during postharvest storage. BMC Plant Biology 2014, 14:328–345. 10.1186/s12870-014-0328-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu P, Xue C, Wu T-t, Heng W, Jia B, Ye Z, et al. Molecular Analysis of the Processes of Surface Brown Spot (SBS) Formation in Pear Fruit (Pyrus bretschneideri Rehd. cv. Dangshansuli) by De Novo Transcriptome Assembly. Plosone, 2013, 8(9): e74217 10.1371/journal.pone.0074217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fu CH, Chen YW, Hsiao YY, Pan ZJ, Liu ZJ, Huang YM, et al. OrchidBase: A collection of sequences of transcriptome derived from orchids. Plant Cell Physiol. 2011,52:238–243. 10.1093/pcp/pcq201 [DOI] [PubMed] [Google Scholar]
- 20. Su CL, Chao YT, Alex Chang YC, Chen WC, Chen CY, Lee AY, et al. De novo assembly of expressed transcripts and global analysis of the Phalaenopsis aphrodite transcriptome. Plant Cell Physiol. 2011,52(9):1501–1514 10.1093/pcp/pcr097 [DOI] [PubMed] [Google Scholar]
- 21. Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, et al. The Pfam protein families database. Nucleic Acids Res. 2010,38:D211–D222. 10.1093/nar/gkp985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, et al. Blast2GO:a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics, 2005,21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- 23.Venny OJ. An interactive tool for comparing lists with Venn Diagrams. 2007, Available: http://bioinfogp.cnb.csic.es/tools/venny/index.html.
- 24. Hsu CC, Chung YL, Chen TC, Lee YL, Kuo YT, Tsai WC, et al. An overview of the Phalaenopsis orchid genome through BAC end sequence analysis. BMC Plant Biol. 2011,11(3): [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xu CJ, Li L, Li H, Zhang MG. Preliminary Studies on the Elements of Browning and the Changes in Cellular Texture of Leaf Explant Browning in Phalaenopsis . Acta Hortieulturae Sinica. 2005,32(6): 1111–1113. [Google Scholar]
- 26. Tanaka E, Okumura S, Takamiya R, Hosaka H, Shimamura Y, Murata M. Cinnamaldehyde inhibits enzymatic browning of cut lettuce by repressing the induction of phenylalanine ammonia-lyase without promotion of microbial growth. J Agric Food Chem. 2011,59(12): 6705–6709. 10.1021/jf200382e [DOI] [PubMed] [Google Scholar]
- 27. Hisaminato H, Murata M, Homma S. Relationship between enzymatic browning of cut lettuce and phenylalanine ammonia-lyase activity, and prevention of browning by inhibitors of polyphenol biosynthesis. Biosci. Biotechnol. Biochem. 2001,65:1016–1021 [DOI] [PubMed] [Google Scholar]
- 28. Jones AM, Saxena PK. Inhibition of Phenylpropanoid Biosynthesis in Artemisia annua L.: A Novel Approach to Reduce Oxidative Browning in Plant Tissue Culture. PLoS One. 2013,8(10): 1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu CJ, Tan RF, Chen DY, Lai YY, Li L. Ultrastructure and distribution of phenol in Phalaenopsis browning leaf explants. Northern Horticulture. 2010,(21): 90–92. [Google Scholar]
- 30. Ru Zhiwei, Lai Yanyan, Xu Chuangjun, Li Ling. Polyphenol Oxidase (PPO) in Early Stage of Browning of Phalaenopsis Leaf Explants. Journal of Agricultural Science,2013,5(9):57–64. [Google Scholar]
- 31. Andersen M, Jordheim M, Anthocyanins In: Encyclopedia of Life Sciences (ELS). Chichester: John Wiley and Sons Ltd; 2010,1–12. [Google Scholar]
- 32. Gould K, Davies K, Winefield C. Anthocyanins: biosynthesis, functions, and application. New York: Springer; 2009. [Google Scholar]
- 33. Butelli E, Titta L, Giorgio M, Mock HP, Matros A, et al. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat Biotechnol, 2008, 26: 1301–1308. 10.1038/nbt.1506 [DOI] [PubMed] [Google Scholar]
- 34. Hong MJ, Kim JB, Yoon YH, Kim SH, Ahn JW, Jeong IY, et al. The effects of chronic gamma irradiation on oxidative stress response and the expression of anthocyanin biosynthesis-related genes in wheat (Triticum aestivum). Int J Radiat Biol. 2014,17:1–31. [DOI] [PubMed] [Google Scholar]
- 35. Ruenroengklin N, Sun J, Shi J, Xue SJ, Jiang Y. Role of endogenous and exogenous phenolics in litchi anthocyanin degradation caused by polyphenol oxidase Food Chemistry, 2009,115(4):1253–1256. [Google Scholar]
- 36. Jiang Y. Role of anthocyanins, polyphenol oxidase and phenols in lychee pericarp browning.Journal of the Science of Food and Agriculture.2000, 80, 305–310. [Google Scholar]
- 37. Franck C, Lammertyn J, Ho QT, Verboven P, Verlinden B, et al. Browning disorders in pear fruit. Postharvest Biol Technol 2007,43: 1–13. [Google Scholar]
- 38. Veltman RH, Lentheric I, Van der Plas LHW, Peppelenbos HW. Internal browning in pear fruit (Pyrus communis L. cv Conference) may be a result of a limited availability of energy and antioxidants. Postharvest Biol Technol 2003,28:295–302. [Google Scholar]
- 39. Cascia G, Bulley SM, Punter M, Bowen J, Rassam M, et al. Investigation of ascorbate metabolism during inducement of storage disorders in pear. Physiol Plantarum,2013, 147: 121–134. 10.1111/j.1399-3054.2012.01641.x [DOI] [PubMed] [Google Scholar]
- 40. Jiang J, Jiang L, Zhang L, Luo H, Opiyo AM, Yu Z. Changes of protein profile in fresh-cut lotus tuber before and after browning. J Agric Food Chem. 2012,60(15):3955–3965. 10.1021/jf205303y [DOI] [PubMed] [Google Scholar]
- 41. Wang H, Fan W, Li H, Yang J, Huang J, Zhang P. Functional characterization of Dihydroflavonol-4-reductase in anthocyanin biosynthesis of purple sweet potato underlies the direct evidence of anthocyanins function against abiotic stresses. PLoS One. 2013, 4;8(11): e78484 10.1371/journal.pone.0078484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yu JJ, Vermaas WFJ. Transcript Levels and Synthesis of Photosystem II Components in Cyanobacterial Mutants with lnactivated Photosystem II Genes. The Plant Cell. 1990,2, 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Veltman RH, Lenthéric I, Van der Plas LHW, Peppelenbos HW. Internal browning in pear fruit (Pyrus communis L. cv Conference) may be a result of a limited availability of energy and antioxidants. Postharvest Biology and Technology. 2003,28: 295–302. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(EPS)
(EPS)
Each set of 3 vertical bars (blue, red, yellow) indicated the percent of DEGs in the three categories: molecular functions(A), cellular components (B), and biological processes (C).
(TIF)
(DOC)
(DOC)
(XLS)
(XLS)
(XLS)
(XLS)
FDR <0.05 and the absolute value of log2Ratio ≤1 were used as the threshold to judge the significance of gene expression difference.
(XLS)
(XLS)
(XLS)
(XLS)
(XLSX)
Fisher’s exact test with FDR <0.05
(DOC)
A totalof 14,099 sequences were assigned to 123 KEGG pathways.
(XLS)
(XLS)
(XLS)
Data Availability Statement
All data were deposited in the NCBI Sequence Read Archive (SRA) under the accession number SRX386023 and SRS506991 (http://www.ncbi.nlm.nih.gov/sra/?term=SRS506991, http://www.ncbi.nlm.nih.gov/sra/?term=SRX386023).