Abstract
Drosophila larval neuroblasts are a model system for studying stem cell self-renewal and differentiation. Here we report a novel role for the Drosophila gene Bj1 in promoting larval neuroblast self-renewal. Bj1 is the guanine-nucleotide exchange factor for Ran GTPase, which regulates nuclear import/export. Bj1 transcripts are highly enriched in larval brain neuroblasts (in both central brain and optic lobe), while Bj1 protein is detected in both neuroblasts and their neuronal progeny. Loss of Bj1 using both mutants or RNAi causes a progressive loss of larval neuroblasts, showing that Bj1 is required to maintain neuroblast numbers. Loss of Bj1 does not result in neuroblast apoptosis, but rather leads to abnormal nuclear accumulation of the differentiation factor Prospero, and premature neuroblast differentiation. We conclude that the Bj1 RanGEF promotes Prospero nuclear export and neuroblast self-renewal.
Introduction
Stem cells are defined by their ability to continuously undergo divisions to maintain a stem cell population (self-renewal) while also producing a population of differentiated progeny. However, a stem cell must be able to precisely maintain this balance between self-renewal and differentiation to prevent events like premature cell cycle exit, which could cause a loss of stem cells, or over-proliferation, which could generate tumors. Drosophila larval neural stem cells, called neuroblasts, are an excellent model system for studying self-renewal versus differentiation. They are undifferentiated, generate differentiating progeny, proliferate without forming tumors, and can undergo mitotic quiescence (reviewed in Doe, 2008; Homem and Knoblich, 2012). Furthermore, Drosophila larval neuroblasts and their progeny are genetically tractable, and throughout larval life the neuroblast does not lose its size, replicative ability, or position in the brain. Finally, well-established molecular markers can be used to identify neuroblasts, so one can readily and accurately quantify neuroblast numbers.
Here we investigate the mechanism of self-renewal in larval central brain neuroblasts (Figure 1A). There are two types of Drosophila central brain neuroblasts. Type I neuroblasts divide asymmetrically to give rise to a neuroblast and a ganglion mother cell (GMC), and then GMCs divide once to form two differentiated neurons or glia. In contrast, type II neuroblasts divide to give rise to a neuroblast and a series of self-renewing intermediate neural progenitor (INPs) which themselves divide asymmetrically ~6 times to self-renew and make ~12 progeny (Bayraktar et al., 2010; Bello et al., 2008; Boone and Doe, 2008; Bowman et al., 2008; Izergina et al., 2009; Wang et al., 2014; Yang et al., 2013). Type I neuroblasts have been studied in more detail, and in this work we focus on type I neuroblast divisions.
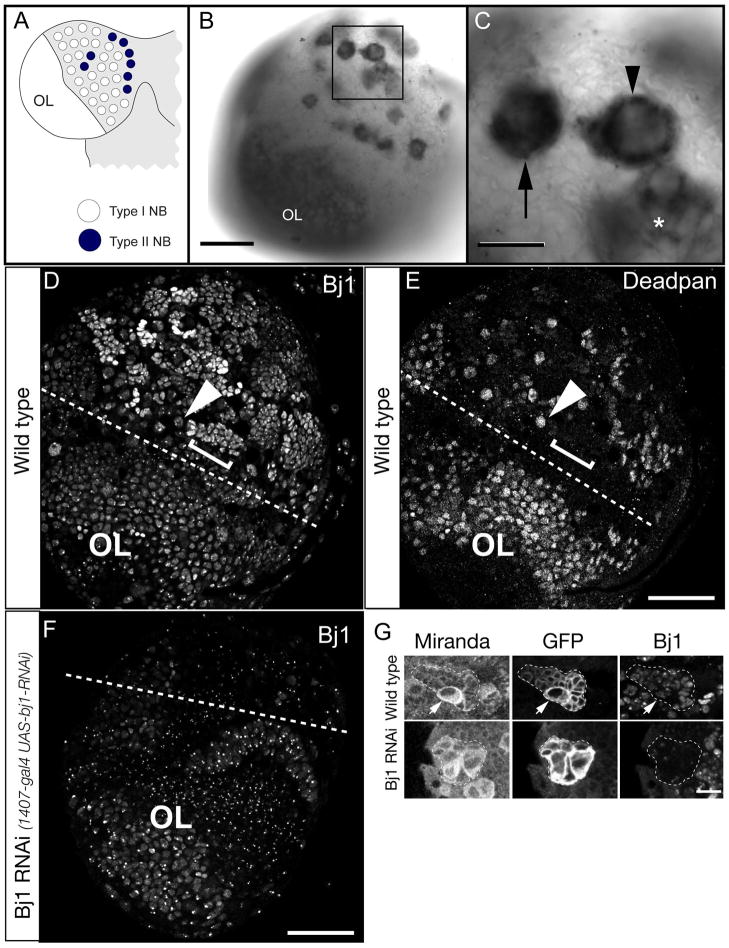
Figure 1. Bj1 RNA is highly enriched in larval type I and II neuroblasts.
(A) Schematic of larval CNS showing one brain lobe. The type I neuroblasts (white) and type II neuroblasts (black); optic lobe neuroblasts are smaller and located more laterally (OL).
(B) Low magnification view of an entire third larval instar brain lobe showing Bj1 RNA enriched in optic lobe neuroblasts (OL) as well as type I and II neuroblasts (boxed).
(C) Enlargement of boxed region in B showing Bj1 RNA is enriched in type I neuroblasts (arrow) and type II neuroblasts (arrowhead), as well as INP progeny of type II neuroblasts (asterisk).
(D,E) Bj1 protein stain (D) and the Deadpan (Dpn) neuroblast/INP marker (E) in a single third instar larval brain lobe. The Dpn+ Bj1+ optic lobe neuroblasts are below the dashed line (OL). A representative type I neuroblast is labeled (arrowhead); neuroblasts show relatively weak Bj1 protein staining. Representative neuroblast progeny, GMCs and neurons, are labeled (bracket); they show relatively strong Bj1 staining.
(F) Bj1 protein in a transgenic Bj1 RNAi third instar larval brain lobe. The central brain is much reduced in size due to Bj1 RNAi causing loss of neuroblasts, whereas the optic lobe is unaffected (because 1407-gal4 is not expressed in this region). Accordingly, Bj1 protein is strongly reduced by only in the central brain. Punctate staining may be background, cross reactivity, or intense Bj1 staining that is less obviously reduced by RNAi.
(G) Bj1 protein staining in larval type II neuroblast lineages (marked with GFP, dashed outlines).
Top row: in wild type, Bj1 is detected in the neuroblast (arrow) and progeny.
Bottom row: Bj1 RNAi reduces Bj1 protein levels.
Scale bar is 50μm in B–F and 10μm in G.
Here we characterize the role of Bj1 in neuroblast self-renewal. Our lab initially identified Bj1 through a functional genomics screen designed to identify genes transcribed at higher levels in neuroblasts than neurons (Carney et al., 2012). A gene was flagged for further study if, when knocked down with RNAi, there were significantly more or less neuroblasts in the central brain, compared to the wild type number (~100 per brain lobe) (Carney et al., 2012). Knockdown of Bj1 in this screen caused a substantial reduction in central brain neuroblasts down to ~50 per brain lobe, implicating Bj1 as a novel regulator of neuroblast self-renewal. The human ortholog of Bj1 is Regulator of Chromosome Condensation 1 (RCC1). RCC1 has a basic N-terminal domain followed by seven RCC1 repeats; the Bj1 protein includes these repeats, plus three additional Bj1 repeats (of different sequence than the seven RCC1 repeats) and an EK-like domain at the C-terminus (Shi and Skeath, 2004). Currently Bj1 is the only known guanine-nucleotide exchange factor (GEF) for the Ran GTPase, which is essential for the translocation of RNA and proteins through the nuclear pore complex (Guttler and Gorlich, 2011) and for assembly of the mitotic spindle (Yudin and Fainzilber, 2009). Previous research has assessed in vivo function of Bj1 during Drosophila embryonic development and in some adult tissues (Shi and Skeath, 2004), but this is the first study of the in vivo function of Bj1 in Drosophila neuroblasts or during larval development.
Materials and Methods
Fly Stocks and genetics
Bj1 TRiP RNAi [Flybase stock 0036067], mCherry TRiP RNAi in attP2, UAS-Dicer2, UAS-p35 (III), 1407-Gal4, y w, and UAS-NLS:GFP::lacZ were all from the Bloomington Drosophila Stock Center (BDSC); the strong hypomorphic Bj1FF32 allele and the UAS-Bj1 stock were courtesy of Jim Skeath (Washington Univ.). RNAi experiments were done at 26–28°C. We also used FRT 2A, Df (3L) exel 7210 (http://www.drosdel.org.uk) and wor-Gal4 (Albertson et al., 2004). Larvae were dissected at the specified hours ALH (where time ALH is reported independent of temperature shifts) or as wandering third instar larvae. When assessing Bj1 alleles over a Bj1 deficiency, virgins were used from the deficiency line and males used from the Bj1 allele line.
In situ RNA hybridization on larval brains
Larval brain in situ RNA hybridization was carried out as previously published (Lim et al., 2007) with minor modifications (0.5% SDS instead of Proteinase K). The Bj1 DNA template was generated from cDNA clone LD22520 by PCR using vector specific primers 5′ GTCGACGTTAGAACGCGGCTAC and 5′ GGGTTAAATTCCCGGGTACTGC. The purified PCR product was transcribed using DIG RNA Labeling Mix (Roche Diagnostics, IN) and Sp6 polymerase according to manufacturer’s instructions. The sense and antisense probes were hydrolyzed to obtain a size around 100bp. Images were taken on a Zeiss LSM 700 and assembled in Illustrator and Photoshop (Adobe, San Jose, CA).
Fixation, antibody staining, and confocal microscopy
Immunohistochemical staining of larval brains was performed with these antibodies: mouse anti-Bj1 (Bj70) 1:50 (Manfred Frasch); rabbit anti-Ase, 1:2000 (Brand et al., 1993); rat anti-Dpn, 1:50 (Doe lab); chicken anti-GFP, 1:2000 (Aves Laboratories, Tigard, OR); guinea pig anti-Mira, 1:1000 (Doe Lab); rabbit anti-PH3, 1:20,000 (Millipore, Billerica, MA); and mouse anti-Pros, 1:1000 (Doe Lab). Antibody staining was performed according to (Carney et al., 2012), but the blocking solution was PBST (phosphate-buffered saline+0.1% Triton-X100; Sigma Aldrich) + 2.5% normal goat serum (Vector Laboratories, Burlingame, CA) + 2.5% normal donkey serum (Jackson ImmunoResearch, West Grove, PA). Images were acquired on a Zeiss700 or Zeiss710 confocal microscope. We took confocal stacks through the entire brain lobe of a larva and counted the number of central brain neuroblasts per brain lobe. Neuroblasts were identified using antibodies against Dpn and Mira, both of which are neuroblast specific proteins in type I lineages. Optic lobe neuroblasts and type II INPs were omitted from these counts based on their much smaller size compared to central brain neuroblasts, their stereotyped position in the brain lobe, and their tight clustering.
Statistics
All error bars are +/− one standard deviation of the mean.
Results
Bj1 RNA is enriched in larval brain neuroblasts, while Bj1 protein is widely detected throughout the larval brain
Bj1 is more highly expressed in the CNS than any other larval tissue (FlyAtlas Anatomical Expression Data; http://flybase.org/reports/FBgn0002638.html). We assessed the specific localization of Bj1 RNA in the larval central nervous system (CNS) using in situ hybridization and found that Bj1 RNA is highly enriched in neuroblasts in the central brain and optic lobe (Figure 1B,C). It is enriched in both type I and type II neuroblasts of the central brain, as well as intermediate neural progenitors (INPs; Figure 1B,C). In contrast, the Bj1 sense probe did not show any specific staining (data not shown). To determine whether Bj1 protein was also enriched in neuroblasts, we used Bj1 antibody (Frasch, 1991) to stain larval brains. Bj1 protein was to be widely expressed in the larval brain, including neuroblasts of the optic lobe (Figure 1D,E labeled “OL”), type I neuroblasts (Figure 1D,E arrowheads) and neuroblast progeny (GMCs and neurons; Figure 1D, E brackets). Although Bj1 mRNA is strongly enriched in neuroblasts compared to their progeny, Bj1 protein is the opposite: low in neuroblasts and high in their progeny, which may indicate translational control. The Bj1 protein staining is specific, however, because it is substantially eliminated by transgenic Bj1 RNAi (Figure 1F). Bj1 protein is also detected in type II neuroblasts and their INP and neuronal progeny (Figure 1G, top row). Here too, expression of transgenic Bj1 RNAi led to a reduction in Bj1 protein levels (Figure 1G, bottom row), further confirming the specificity of Bj1 antibody staining, and the efficacy of Bj1 RNAi.
Bj1 is required for the maintenance of larval brain neuroblasts
We previously showed that neuroblast-specific Bj1 RNAi led to a decrease in the number of larval brain neuroblasts at the wandering third instar stage (Carney et al., 2012), but it was unknown whether this was due to a defect in neuroblast formation, survival, exit from quiescence, or self-renewal. To determine if Bj1 RNAi altered neuroblast formation or exit from quiescence, we quantified neuroblast numbers throughout larval development; a defect in neuroblast formation should result in lower numbers of neuroblasts from the earliest stages of larval life. We used the neuroblast-specific 1407-Gal4 line to drive UAS-Bj1 RNAi to reduce Bj1 levels in neuroblasts. We used the quiescent and proliferating neuroblast marker Deadpan (Dpn) to identify and count neuroblasts (Doe, 2008; Egger et al., 2008; Homem and Knoblich, 2012). We found that wild type and Bj1 RNAi first instar larvae had similar numbers of ~100 Dpn+ neuroblasts, showing that there was no defect in neuroblast formation (Figure 2A–E). We conclude that loss of Bj1 does not affect neuroblast formation or exit from quiescence.
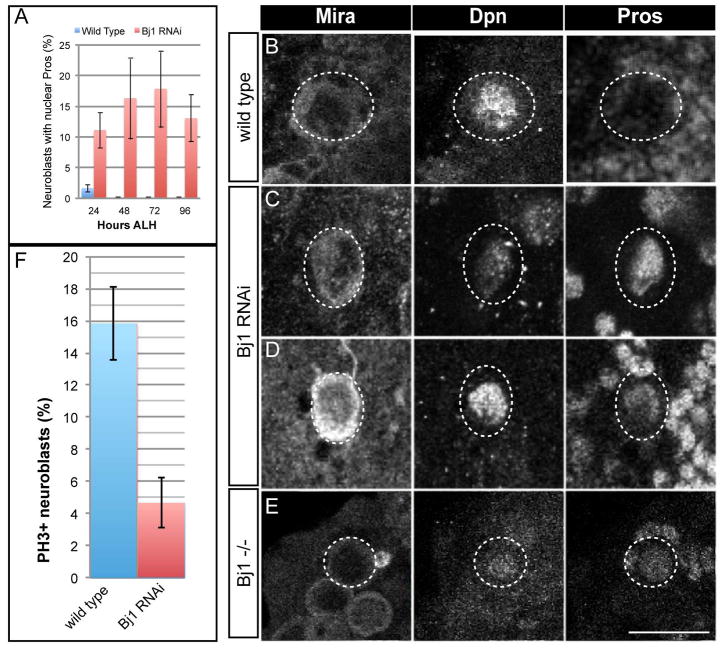
Figure 2. Reducing Bj1 levels leads to loss of larval neuroblasts by an apoptosis-independent mechanism.
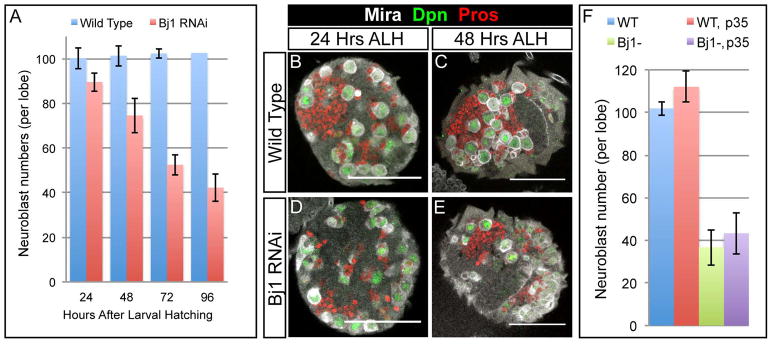
(A) The number of neuroblasts present per brain lobe at four time points after larval hatching (ALH) were quantified for wild type control RNAi and Bj1 RNAi larvae (at 26°C). There were ~100 Dpn+ neuroblasts per brain lobe in the control at each time point, whereas the number of Dpn+ neuroblasts per lobe steadily decreased over time in the Bj1 LOF background.
(B–E) Wild type control and Bj1 LOF brain lobes are shown stained for Mira, Dpn, and Pros (which is weakly expressed in the cytoplasm of wild type neuroblasts and strongly expressed in wild type neurons). (B, D) The neuroblasts shown in the wild type control brain and the Bj1 LOF brain at 24h ALH are equally as large, suggesting the neuroblasts in both genotypes have exited quiescence and have grown in order to re-enter the cell cycle. (C, E) By 48h ALH, the number of neuroblasts has decreased in the Bj1 LOF background compared to wild type, though the neuroblasts present are still as large as the wild type neuroblasts, showing they are not entering quiescence.
(F) Neuroblasts in the Bj1 RNAi background are not undergoing apoptosis because the addition of baculovirus p35 (a potent inhibitor of apoptosis) does not rescue neuroblast number. The first genotype is Wor-Gal4, UAS-Dicer2; UAS-NLS:GFP/TRiP mCherry RNAi (denoted as WT, for wild type) and it is a control that yields wild type larvae; the second genotype is Wor-Gal4, UAS-Dicer2; UAS-p35/TRiP mCherry RNAi (denoted WT, p35) and it is a control using p35; the third genotype is Wor-Gal4, UAS-Dicer2/Bj1 TRiP RNAi; UAS-NLS:GFP (denoted Bj1−) and is a knockdown of Bj1; and the fourth genotype is Wor-Gal4, UAS-Dicer2/Bj1 TRiP RNAi; UAS-p35 (denoted Bj1−, p35) and it is an attempted rescue of Bj1 RNAi neuroblast number by the addition of p35. Each genotype assessed had a total of 4 transgenes, with 2 total transgenes per genotype driven by the neuroblast specific Wor-Gal4 promoter. The UAS-NLS:GFP controls for the UAS-p35, while the TRiP mCherry RNAi is a nonsense RNAi that controls for the Bj1 TRiP RNAi. Scale bars in (B-E) are 50μm. For (A), n = 3, 5, 5, and 3 for WT and n = 5, 5, 4, and 4 for Bj1 RNAi, at 24, 48, 72, and 96 h ALH respectively. For (F), n = 10 for all four genotypes. Error bars are +/- one standard deviation.
Wild type neuroblast number is maintained at ~100 per brain lobe throughout larval life, although progressively more of these neuroblasts enlarge and exit from quiescence over time (Britton and Edgar, 1998). Similarly, many Bj1 RNAi neuroblasts enlarged by 24h ALH (Figure 2D), showing that Bj1 is not required for neuroblast exit from quiescence. However, Bj1 RNAi larval brains showed a progressive reduction in the total number of quiescent and proliferating neuroblasts that was not observed in wild type (Figure 2A). To determine if neuroblasts are lost due to apoptosis, we misexpressed anti-apoptotic caspase inhibitor p35 (Clem, 2001) together with Bj1 RNAi in larval neuroblasts, but found it did not restore neuroblast numbers (Figure 2F). Consistent with this result, previous work has shown that loss of Bj1 dominantly suppresses apoptosis in the Drosophila eye, rather than increasing apoptosis (Shi and Skeath, 2004). We conclude that loss of Bj1 does not lead to neuroblast apoptosis.
Bj1 prevents neuroblast nuclear import of the Prospero differentiation factor
We next tested whether loss of Bj1 might trigger premature neuroblast differentiation. Neuroblasts normally keep the Prospero (Pros) differentiation factor in the cytoplasm during interphase and at the cortex during mitosis (Hirata et al., 1995; Knoblich et al., 1995; Spana and Doe, 1995), and high levels of Pros can lead to its nuclear accumulation and trigger neuroblast differentiation (Bayraktar et al., 2010; Cabernard and Doe, 2009; Choksi et al., 2006). Thus, we tested whether loss of Bj1 resulted in abnormal nuclear accumulation of Pros and neuroblast differentiation. Wild type neuroblasts never show nuclear Pros (Figure 3A,B). In contrast, Bj1 RNAi neuroblasts often showed either high or low levels of nuclear Pros (Figure 3A,C,D). We conclude that loss of Bj1 results in abnormal accumulation of Pros in the neuroblast nucleus and premature neuroblast differentiation.
Figure 3. Reducing Bj1 levels leads to nuclear Pros in larval neuroblasts.
(A) Quantification of neuroblasts with nuclear Pros at the indicated stages (n 3 brains for each timepoint). Error bars are +/- one standard deviation.
(B–E) Larval brain neuroblasts at 48h ALH (circled). (B) Wild type neuroblasts have no detectable nuclear Pros. (C,D) Bj1 RNAi neuroblasts can show nuclear Pros. (E) Bj1 mutant neuroblasts can show nuclear Pros. Scale bar, 10μm. (F) Quantification of mitotic neuroblasts at 48h ALH detected by phosphohistone H3 (PH3) staining in wild type and Bj1 RNAi neuroblasts. Error bars are +/− one standard deviation.
We tested whether the distinctive Bj1 RNAi phenotype can be reproduced in a Bj1 mutant background, as a control for off-target RNAi effects. We examined the strong loss of function allele Bj1FF32 (Shi and Skeath, 2004) in trans to a Bj1 deficiency allele, subsequently called Bj1−/− or Bj1 mutant. Similar to the Bj1 RNAi phenotype, many Bj1 mutant neuroblasts showed nuclear Pros (Figure 3E). In addition, Bj1 mutant neuroblasts are typically much smaller than wild type neuroblasts; the basis for this presumed cell growth defect is unknown. In conclusion, we have shown that reducing Bj1 by two different methods leads to accumulation of Pros in the neuroblast nucleus and premature neuroblast differentiation.
Mislocalization of Pros to the nucleus can occur when neuroblast cell polarity is defective, most notably in miranda mutant neuroblasts (Ikeshima-Kataoka et al., 1997; Schuldt et al., 1998; Shen et al., 1997). We find no defects in aPKC/Bazooka apical crescents or basal Miranda crescents (n=20 mitotic neuroblasts from 10 brain lobes; data not shown), so the nuclear Pros seen following reduction of Bj1 protein levels is not due to altered neuroblast cell polarity. In addition, neuroblast differentiation should be accompanied by cell cycle exit. To determine if Bj1 RNAi leads to reduced cell cycle progression in larval neuroblasts we assayed the mitotic marker phospho-histone H3 (PH3). We found that significantly fewer neuroblasts are in mitosis in Bj1 RNAi brains compared to wild type (Figure 3F), consistent with initiating premature differentiation.
Bj1 does not regulate nuclear import/export of all neuroblast proteins
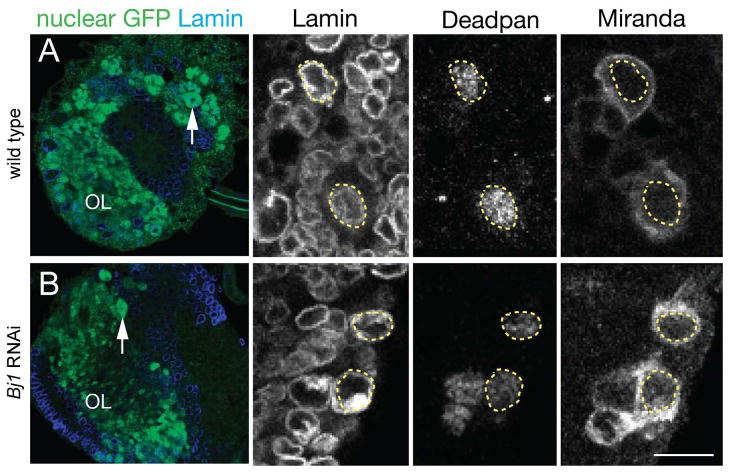
We next asked whether Bj1 has a general role in promoting nuclear export of all proteins within larval neuroblasts, similar to its requirement for Pros nuclear export. In addition, it was previously shown that Bj1 mutant embryos lacked nuclear import of a nuclear localization signal:beta-galactosidase (NLS:βgal) fusion protein (Shi and Skeath, 2004), so we also tested for inappropriate nuclear import. We assayed Bj1 RNAi neuroblasts for three nuclear proteins (Dpn, Ase, and NLS:GFP) to see if they showed nuclear import failure, and one cytoplasmic protein (Miranda) to see if it showed nuclear export failure. As expected, wild type larval neuroblasts had nuclear Dpn, Ase, and NLS:GFP and cytoplasmic Miranda (Figure 4A and data not shown). Similarly, Bj1 RNAi larval neuroblasts had nuclear Dpn, Ase, and NLS:GFP and cytoplasmic Miranda (Figure 4B and data not shown). The observation that Bj1 is required for NLS:βgal import in the embryonic CNS, but not required for nuclear import of NLS:GFP in the larval CNS, may be due to the different stages of development or due to differences between the NLS:βgal and NLS:GFP proteins. We conclude that Bj1 does not have a general role in regulating nuclear import/export in larval neuroblasts.
Figure 4. Reducing Bj1 levels does not lead to general defects in protein nuclear import/export.
(A) Left panel: wild type third instar larval brain lobe showing nuclear NLS:GFP (green) and Lamin nuclear envelope marker (blue); arrow, neuroblast (the dark central spot is the nucleolus); OL, optic lobe. Right panels: Wild type larval neuroblasts have nuclear Deadpan and cytoplasmic Miranda localization; the nucleus is marked by a dashed outline based on Lamin nuclear envelope marker staining. Scale bar, 50μm (left panel), 10μm (right panels).
(B) Left panel: Bj1 RNAi third instar larval brain lobe showing nuclear NLS:GFP (green) and Lamin nuclear envelope marker (blue); arrow, neuroblast (the dark central spot is the nucleolus); OL, optic lobe. Right panels: Bj1 RNAi larval neuroblasts maintain nuclear Deadpan and cytoplasmic Miranda localization; the nucleus is marked by a dashed outline based on Lamin nuclear envelope marker staining. Scale bar, 50μm (left panel), 10μm (right panels).
Bj1 is not sufficient to induce neuroblast identity
The presence of Bj1 protein in GMCs and neurons suggests that Bj1 is not sufficient to promote neuroblast identity. To test this hypothesis, we overexpressed Bj1 within neuroblast lineages (wor-Gal4 UAS-Bj1), but observed no increase in neuroblast numbers (108 +/- 5 neuroblasts per lobe, n = 12; wild type had 104 +/- 3 neuroblasts per lobe, n = 9). We conclude that Bj1 is not sufficient to generate neuroblast identity.
Discussion
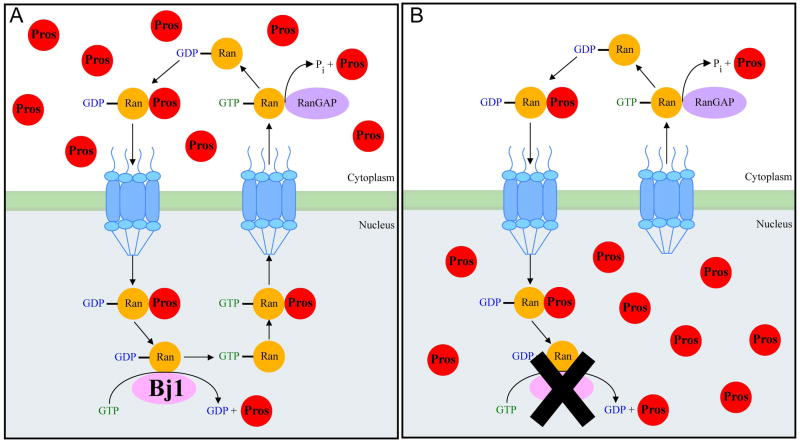
We have shown that Bj1 is an essential gene whose mRNA is highly enriched in larval neuroblasts but whose protein is widely detected throughout the larval brain. Reduced Bj1 levels by RNAi or genetic mutation led to a failure to maintain nuclear export of Pros protein, and concomitant failure to maintain neuroblast self-renewal leading to a progressive loss in neuroblast numbers. The loss of neuroblasts appeared random throughout the brain; we did not identify any specific group of neuroblasts that are reliably unaffected by loss of Bj1. Loss of neuroblasts is not due to failure of neuroblasts to exit quiescence, or apoptosis; we can’t exclude the possibility that neuroblasts disappear by non-apototic pathways such as necrosis (Kuang et al., 2014). We propose a model for Bj1 and Pros in regulating neuroblast self-renewal (Figure 5). In wild type neuroblasts, Pros is transcribed and translated but is kept inactive by NES-dependent transport out of the nucleus. In Bj1 loss of function, there would be less Ran-GTP, leading to reduced nuclear export of Pros, which would trigger neuroblast differentiation.
Figure 5. A model for the role of Bj1 in regulating Prospero nuclear import.
(A) In wild type neuroblasts the Pros NES is dominant over the Pros NLS resulting in cytoplasmic localization of Pros. We propose Bj1 facilitates a GDP to GTP exchange on Ran-GDP to produce a Ran-GTP gradient that would promote Pros nuclear to cytoplasm transport.
(B) Reduction of Bj1 levels leads to decreased Ran-GTP and failure in Pros nuclear export.
Although we have shown that Bj1 is required to prevent premature neuroblast differentiation, it is entirely likely that Bj1 has additional functions. For example, bj1 mutants may survive early embryonic stages due to maternally-deposited RNA that masks an early function. Similarly, our neuroblast-specific RNAi would not affect non-neural tissues, and may not affect early steps of neurogenesis due to incomplete knockdown of Bj1 mRNA/protein levels.
It is remarkable that Bj1 RNA is so highly enriched in larval neuroblasts yet the Bj1 protein is widely distributed. Bj1 protein may have a long half-life (made in neuroblasts, persisting in neural progeny) or neuroblast progeny make low levels of Bj1 transcript which is sufficient to produce the observed Bj1 protein. In addition, the Bj1 RNA may have a neuroblast-specific function that we have not identified.
We have shown that Bj1 does not have a general role in regulating nuclear import/export in larval neuroblasts. Why then is Pros selectively affected? Pros has both a nuclear localization signal (NLS) and a nuclear export signal (NES) (Demidenko et al., 2001). Pros may thus be unusually sensitive to a disruption in the RanGEF/RanGAP nuclear/cytoplasmic gradient. Other Drosophila proteins shuttle between the nucleus and cytoplasm (e.g. Extradenticle, Dishevelled) (Chan et al., 2008; Stevens and Mann, 2007), and it would be interesting to see if they are regulated by Bj1 similar to Pros. Shi and Skeath (2004) propose that Bj1 is required for nuclear import in the embryo, we assessed NLS:GFP import in a Bj1 knockdown in larvae and did not get the same result. The difference could be due to biological differences between embryo and larva, or perhaps between genotypes.
Why do we observe less than 20% of neuroblasts with nuclear Pros? Presumably, this reflects the gradual loss of neuroblasts in Bj1 mutant/RNAi brains. Thus, at any point in time, some neuroblasts have nuclear Pros (these are in the process of differentiating), some neuroblasts had nuclear Pros at earlier stages and have already differentiated and are no longer visible, and some neuroblasts do not yet have nuclear Pros (these may differentiate at a later timepoint). If we add the first and second categories together, it would give a much higher percentage of neuroblasts that – at some point in their life – contained nuclear Pros. It is possible that some neuroblasts are not dependent on Bj1 for self-renewal; however, we have not been able to identify a specific population of neuroblasts that are never affected by loss of Bj1. Rather, it is more likely that different neuroblasts survive in each larval brain, due to the incomplete and variable RNAi knockdown of Bj1.
We would predict that loss of Pros would prevent Bj1-induced neuroblast differentiation, but unfortunately pros homozygotes die in embryogenesis, prior to the appearance of the bj1 mutant phenotype. Similarly, pros mutant clones form aggressive neuroblast tumors (Choksi et al., 2006), which makes it impossible to discern if the removal of Bj1 function has an effect on neuroblast numbers.
Acknowledgments
We thank Jim Skeath (Washington Univ.) for Bj1 flies, Manfred Frasch (Universität Erlangen-Nürnberg) for Bj1 antibody, the Bloomington Drosophila Stock Center for providing fly stocks, Sen-Lin Lai and Kate Walsh for comments, Travis Carney for mentoring, and Janet Hanawalt for copy editing the manuscript. This work was funded by an NIH Developmental Biology Training Grant (T32HD007348-26) and HHMI, where CQD is an Investigator.
References
- Albertson R, Chabu C, Sheehan A, Doe CQ. Scribble protein domain mapping reveals a multistep localization mechanism and domains necessary for establishing cortical polarity. J Cell Sci. 2004;117:6061–6070. doi: 10.1242/jcs.01525. [DOI] [PubMed] [Google Scholar]
- Bayraktar OA, Boone JQ, Drummond ML, Doe CQ. Drosophila type II neuroblast lineages keep Prospero levels low to generate large clones that contribute to the adult brain central complex. Neural Development. 2010;5:26–35. doi: 10.1186/1749-8104-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello BC, Izergina N, Caussinus E, Reichert H. Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Develop. 2008;3:5. doi: 10.1186/1749-8104-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone JQ, Doe CQ. Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Dev Neurobiol. 2008;68:1185–1195. doi: 10.1002/dneu.20648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman SK, Rolland V, Betschinger J, Kinsey KA, Emery G, Knoblich JA. The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Developmental Cell. 2008;14:535–546. doi: 10.1016/j.devcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand M, Jarman AP, Jan LY, Jan YN. asense is a Drosophila neural precursor gene and is capable of initiating sense organ formation. Development. 1993;119:1–17. doi: 10.1242/dev.119.Supplement.1. [DOI] [PubMed] [Google Scholar]
- Cabernard C, Doe CQ. Apical/basal spindle orientation is required for neuroblast homeostasis and neuronal differentiation in Drosophila. Developmental Cell. 2009;17:134–141. doi: 10.1016/j.devcel.2009.06.009. [DOI] [PubMed] [Google Scholar]
- Carney TD, Miller MR, Robinson KJ, Bayraktar OA, Osterhout JA, Doe CQ. Functional genomics identifies neural stem cell sub-type expression profiles and genes regulating neuroblast homeostasis. Developmental biology. 2012;361:137–146. doi: 10.1016/j.ydbio.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CC, Zhang S, Rousset R, Wharton KA., Jr Drosophila Naked cuticle (Nkd) engages the nuclear import adaptor Importin-alpha3 to antagonize Wnt/beta-catenin signaling. Dev Biol. 2008;318:17–28. doi: 10.1016/j.ydbio.2008.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choksi SP, Southall TD, Bossing T, Edoff K, de Wit E, Fischer BE, van Steensel B, Micklem G, Brand AH. Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Developmental Cell. 2006;11:775–789. doi: 10.1016/j.devcel.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Clem RJ. Baculoviruses and apoptosis: the good, the bad, and the ugly. Cell Death Differ. 2001;8:137–143. doi: 10.1038/sj.cdd.4400821. [DOI] [PubMed] [Google Scholar]
- Demidenko Z, Badenhorst P, Jones T, Bi X, Mortin MA. Regulated nuclear export of the homeodomain transcription factor Prospero. Development. 2001;128:1359–1367. doi: 10.1242/dev.128.8.1359. [DOI] [PubMed] [Google Scholar]
- Doe CQ. Neural stem cells: balancing self-renewal with differentiation. Development. 2008;135:1575–1587. doi: 10.1242/dev.014977. [DOI] [PubMed] [Google Scholar]
- Egger B, Chell JM, Brand AH. Insights into neural stem cell biology from flies. Philos Trans R Soc Lond B Biol Sci. 2008;363:39–56. doi: 10.1098/rstb.2006.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch M. The maternally expressed Drosophila gene encoding the chromatin-binding protein BJ1 is a homolog of the vertebrate gene Regulator of Chromatin Condensation, RCC1. Embo j. 1991;10:1225–1236. doi: 10.1002/j.1460-2075.1991.tb08064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttler T, Gorlich D. Ran-dependent nuclear export mediators: a structural perspective. Embo j. 2011;30:3457–3474. doi: 10.1038/emboj.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata J, Nakagoshi H, Nabeshima Y, Matsuzaki F. Asymmetric segregation of the homeodomain protein Prospero during Drosophila development. Nature. 1995;377:627–630. doi: 10.1038/377627a0. [DOI] [PubMed] [Google Scholar]
- Homem CC, Knoblich JA. Drosophila neuroblasts: a model for stem cell biology. Development. 2012;139:4297–4310. doi: 10.1242/dev.080515. [DOI] [PubMed] [Google Scholar]
- Ikeshima-Kataoka H, Skeath JB, Nabeshima Y, Doe CQ, Matsuzaki F. Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature. 1997;390:625–629. doi: 10.1038/37641. [DOI] [PubMed] [Google Scholar]
- Izergina N, Balmer J, Bello B, Reichert H. Postembryonic development of transit amplifying neuroblast lineages in the Drosophila brain. Neural Dev. 2009;4:44. doi: 10.1186/1749-8104-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich JA, Jan LY, Jan YN. Asymmetric segregation of Numb and Prospero during cell division. Nature. 1995;377:624–627. doi: 10.1038/377624a0. [DOI] [PubMed] [Google Scholar]
- Kuang C, Golden KL, Simon CR, Damrath J, Buttitta L, Gamble CE, Lee CY. A novel fizzy/Cdc20-dependent mechanism suppresses necrosis in neural stem cells. Development. 2014;141:1453–1464. doi: 10.1242/dev.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C, Lee J, Choi C, Kim J, Doh E, Choe J. Functional role of CREB-binding protein in the circadian clock system of Drosophila melanogaster. Mol Cell Biol. 2007;27:4876–4890. doi: 10.1128/MCB.02155-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldt AJ, Adams JH, Davidson CM, Micklem DR, Haseloff J, St Johnston D, Brand AH. Miranda mediates asymmetric protein and RNA localization in the developing nervous system. Genes Dev. 1998;12:1847–1857. doi: 10.1101/gad.12.12.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen CP, Jan LY, Jan YN. Miranda is required for the asymmetric localization of Prospero during mitosis in Drosophila. Cell. 1997;90:449–458. doi: 10.1016/s0092-8674(00)80505-x. [DOI] [PubMed] [Google Scholar]
- Shi WY, Skeath JB. The Drosophila RCC1 homolog, Bj1, regulates nucleocytoplasmic transport and neural differentiation during Drosophila development. Dev Biol. 2004;270:106–121. doi: 10.1016/j.ydbio.2004.02.011. [DOI] [PubMed] [Google Scholar]
- Spana EP, Doe CQ. The prospero transcription factor is asymmetrically localized to the cell cortex during neuroblast mitosis in Drosophila. Development. 1995;121:3187–3195. doi: 10.1242/dev.121.10.3187. [DOI] [PubMed] [Google Scholar]
- Stevens KE, Mann RS. A balance between two nuclear localization sequences and a nuclear export sequence governs extradenticle subcellular localization. Genetics. 2007;175:1625–1636. doi: 10.1534/genetics.106.066449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YC, Yang JS, Johnston R, Ren Q, Lee YJ, Luan H, Brody T, Odenwald WF, Lee T. Drosophila intermediate neural progenitors produce lineage-dependent related series of diverse neurons. Development. 2014;141:253–258. doi: 10.1242/dev.103069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Awasaki T, Yu HH, He Y, Ding P, Kao JC, Lee T. Diverse neuronal lineages make stereotyped contributions to the Drosophila locomotor control center, the central complex. J Comp Neurol. 2013;521:2645–2662. Spc2641. doi: 10.1002/cne.23339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudin D, Fainzilber M. Ran on tracks--cytoplasmic roles for a nuclear regulator. J Cell Sci. 2009;122:587–593. doi: 10.1242/jcs.015289. [DOI] [PubMed] [Google Scholar]