Abstract
Great advancements have been made in understanding the basic mechanisms of ictogenesis using single-cell electrophysiology (e.g. patch clamp, sharp electrode), large-scale electrophysiology (e.g. EEG, field potential recording), and large-scale imaging (MRI, PET, calcium imaging of AM dye-loaded tissue). Until recently, it has been challenging to study experimentally how population rhythms emerge from cellular activity. Newly developed optical imaging technologies hold promise for bridging this gap by making it possible to simultaneously record the many cellular elements that comprise a neural circuit. Furthermore, easily accessible genetic technologies for targeting expression of fluorescent protein-based indicators make it possible to study, in animal models of epilepsy, epileptogenic changes to neural circuits over long periods of time. In this review, we summarize some of the latest imaging tools (fluorescent probes, gene delivery methods, and microscopy techniques) that can lead to the advancement of cell- and circuit-level understanding of epilepsy, which in turn may inform and improve development of next generation anti-epileptic and anti-epileptogenic drugs.
Keywords: imaging, WONOEP, probes, microscopy
Background
The nature of epilepsy as a cellular vs. network disease has long been discussed. Of course, the dynamics of a neuronal network depend both on the properties of its constituting elements and the connections among those elements. As an example of “cell-based” epilepsy, mutations related to ion channel function can lead to epilepsy by changing intrinsic properties of neurons, as in the SCN1A mutations of Dravet Syndrome1. Conversely, “network-based” epilepsy can be seen when otherwise normal brain tissue undergoes trauma and the newly re-wired circuit become epileptic, as in post-traumatic epilepsy. Further complicating matters is the array of dynamic interdependencies between micro, macro, and whole system levels. For example, a single protein mutation in one specific neuronal subtype can lead to changes in synaptic plasticity in key neural circuits. In juxtaposition, ion channel expression is dynamically and homeostatically regulated by synaptic network activity levels2. Thus, intrinsic molecular dynamics can affect cellular network connectivity and vice versa. Ultimately, there are many defects at the molecular, cellular and network connectivity levels that can cause a brain to become epileptic and it is important to understand their collective role in epileptogenesis.
With traditional electrophysiological approaches, it has been possible to evaluate intrinsic properties of excitability in single neurons (e.g. action potential threshold, resting membrane potential, chloride reversal potential) and the resulting effects on network dynamics (e.g. field recordings of population activity). Extensive histology of tissue resected from epileptic animals and humans has also revealed aberrant patterns of connectivity. But, until recently, it has been impossible to experimentally bridge the gap between intracellular physiology and population dynamics. Newly developed imaging technologies are enabling, for the first time, the measurement of neural activity at the molecular, cellular, and network levels. The past decades have brought a suite of new imaging probes (highlighted in Table 1), allowing fluorescent protein-based detection of morphology, activity, intracellular ion concentration, voltage fluctuations, neurotransmitter release and regulation, and molecular pathways. Currently, many of these sensors can be targeted to genetically defined populations of cells and imaged over long periods of time to track changes in neuronal networks before and after the emergence of chronic seizures. In this review, inspired by discussions that took place at the WONOEP XII congress in Estérel, Quebec, Canada, we will discuss recent advances in imaging technologies relevant and used to study epilepsy.
Table 1.
Summary of fluorescent probes
| Measured Parameter | Select probe(s) | Ref |
|---|---|---|
| Absolute calcium concentration | YC3.6, Twitch | 12,13 |
| Relative calcium concentration | GCaMP6 | 14 |
| Voltage | ArcLight, ASAP1, QuasAr | 22–24 |
| Absolute chloride concentration | Clomeleon, SuperClomeleon | 29,63 |
| Absolute sodium concentration | SBFI | 64 |
| Relative sodium concentration | Asante Natrium Green | 31 |
| Extracellular glutamate | FLII-E | 34 |
| Vesicular release | synaptopHluorin | 38 |
| Metabolic activity | Intrinsic fluorescence of NADH, FAD | 32 |
Fluorescent-protein based probes
Anatomy
A fluorescent protein in its simplest form is an anatomical probe that can be genetically targeted to a defined population of cells (see next section). eGFP and its numerous spectral variants fill the cytoplasm of a neuron and, depending on expression pattern and imaging modality, can be used to track changes in neuronal morphology associated with epilepsy such as dendritic and axonal branching and spine growth. Other known anatomical features of epilepsy that can be monitored with constitutively fluorescent proteins include astrogliosis, microglia activation and movement, and cell death3–5. Such studies can be enhanced with the use of photoactivatable, photoswitchable, or photoconvertible fluorescent proteins, which upon irradiation become fluorescent (“turn on”), toggle between fluorescent and not fluorescent, and change wavelength respectively6–8 (c.f. 9). These features can be used to track selected neurons over long periods of time and enable the use of super-resolution microscopy such as PALM10 or STORM11.
Cellular activity
The most extensively developed class of genetically encoded sensors of neural activity is the genetically encoded calcium indicator (GECI). GECIs fall broadly into two classes: ratiometric and single wavelength indicators. Ratiometric indicators like those in the Yellow Cameleon and Twitch families are YFP-CFP constructs linked with calmodulin or troponin C, which change FRET efficiency upon calcium binding12,13. These can be calibrated to make absolute measurements of intracellular calcium concentration for recording not only neural activity, but also changes in baseline intracellular calcium. The most popular single wavelength GECIs are the GCaMP family, which consists of calmodulin bound to a circularly permutated GFP molecule. Calcium binding to calmodulin causes a conformational change which transitions GFP from its non-fluorescent, calcium-free conformation, to a brightly fluorescent form. The latest version, GCaMP6, has three variants, s, m, and f, which span from slow kinetics and high signal (s) to fast kinetics, low signal14. Although the latest GECIs are now capable of detecting single action potentials, the fast rise-time kinetics of inorganic dyes make them the best choice for applications requiring more precise identification of spike timing15. GECIs are ideally suited for recording from genetically defined populations of cells or long-term imaging experiments, in which individual neurons are tracked over many days using, for example, two-photon imaging16 or head-mounted, miniaturized microscopes17. While most cells tolerate long term expression of virally delivered GECIs18,19, a small fraction display nuclear filling with the fluorescent protein, a sign of cytotoxicity19, and are often excluded from analysis.
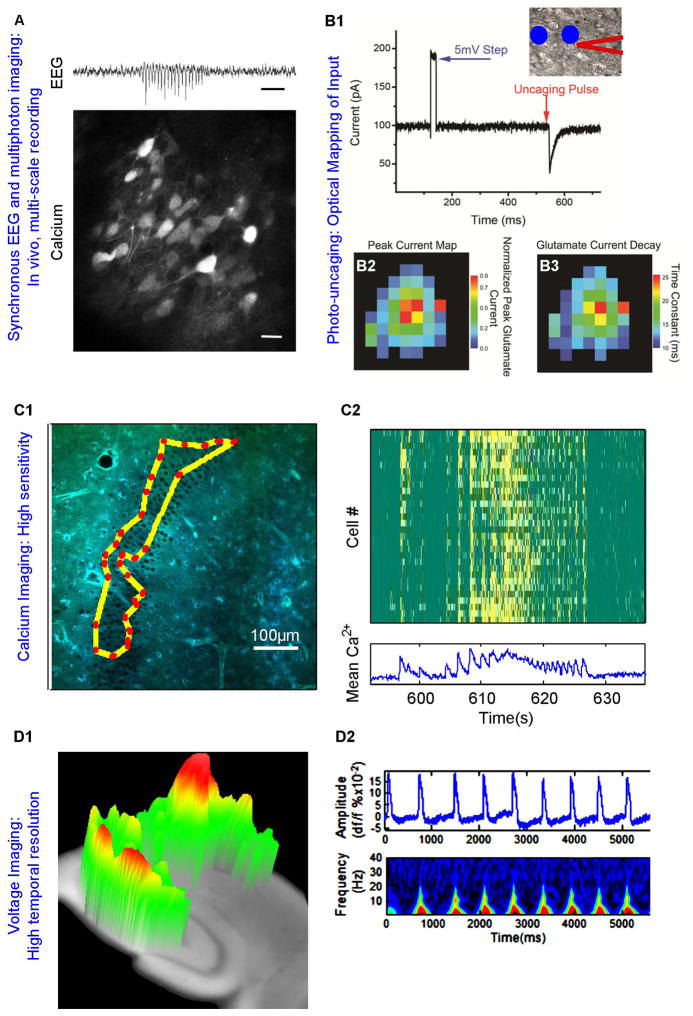
As one illustration of their utility in epilepsy research, GECIs are being used to simultaneously record activity in multiple neurons to evaluate the role of individual cells in generating population rhythms on a larger scale. For example, to examine an AMPA receptor trafficking defect in interneurons of stargazer mice20, neurons were transfected with GCaMP6 AAV vectors. EEG was recorded synchronously with in vivo two-photon imaging of calcium dynamics in an attempt to understand the cellular basis of cortical spike-wave discharges (Figure 1A).
Figure 1. Molecular, cellular, and network imaging at the circuit level.
A) While simultaneously recording intracranial EEG (top, scale = 1 second), calcium activity can be imaged from neurons transfected with GCaMP6 as seen here in layer 2/3 of visual cortex of stargazer, a mouse model of absence epilepsy (bottom, scale = 20 μm; courtesy of Jochen Meyer, Atul Maheshwari, Stelios Smirnakis and Jeffrey Noebels, Baylor College of Medicine). B1) Differential interference contrast of a cortical astrocyte (center) shown with patch-clamp electrode (outlined in red). Photostimulation sites are indicated by blue circles. Whole-cell recording of an astrocyte voltage-clamped at −80 mV reveals robust glutamate transporter current evoked by photostimulation proximal to the soma. This technique can be repeated to generate maps of peak current (B2) and transporter decay time (B3) for an individual astrocyte. C1) Two-photon Targeted Path Scan (TPS) imaging traces the laser along a path that bisects a predefined subset of cells to rapidly sample many cells across a large area. C2) In this example, TPS was used to sample 30 cells at >30Hz during seizure-like activity in an organotypic slice culture (Lillis and Staley, unpublished). D1) High speed (1.2KHz) voltage-sensitive dye imaging of 4-aminopyridine-evoked epileptiform bursts in enables low resolution 3D visualization of travelling waves hippocampal slices. D2) VSD imaging data can be processed to look at spatial, temporal, and spectral characteristics. Adapted from 27.
While GECIs are primarily used as proxies for electrical activity by detecting action potential-induced changes in calcium, voltage sensitive fluorescent proteins (VSFPs) show promise for the direct measurement of both supra- and sub-threshold membrane voltage. The low signal to noise ratios that have plagued fluorescent voltage sensors have improved dramatically in the past decade. In particular, VSFP and hybrid voltage sensors (hVOS) have improved in both sensitivity and membrane targeting (for reduced background fluorescence). hVOS offer increased signal over first-generation protein-based sensors, but depend on the interaction of a membrane-bound fluorescent protein and a voltage sensitive quencher such as the explosive and toxic compound dipicrylamine21. The latest generation of VSFPs, ArcLight, ASAP1, QuasAr, and FRET-based QuasAr fusions offer comparable or superior SNR in a more experimentally practical protein-based format22–25. Although ArcLight is ~80x brighter than QuasArs (which require excitation light of 800W/cm2), QuasArs detect single action potentials with ~5x signal-to-noise ratio and 15x photostability24,25.
Purely synthetic voltage-sensitive dyes (VSDs) are also being used to map changes in circuit activity in epilepsy models, including temporal lobe epilepsy induced by pilocarpine26 and acute models, such as 4-AP27. VSDs do not maintain single cell resolution, but can provide a ‘bird’s eye view’ of the circuit activity, reaching imaging rates over 2KHz (Figure 1D). VSDI is very useful for mapping gross rewiring in epileptic circuits, measuring levels of excitability in normal and epileptogenic tissues28, and for determining how different drugs modulate neural signal propagation velocity and other spatiotemporal dynamics.
Intracellular ion concentration
In addition to the calcium ion indicators described above, fluorescent probes have been developed to measure intracellular concentrations of chloride, sodium, and potassium. The YFP-CFP FRET-based Clomeleon family of chloride sensors29 have already been used to demonstrate transient ictal chloride accumulation30. Similarly, there are a number of dyes that are sensitive to potassium and sodium ions. Most of these dyes, such as Natrium Green, can be loaded into the cells using AM loading or as a salt through individual glass recording pipettes31. The quality of these and other inorganic sodium dyes such as CoroNa and SBFI, are being constantly improved. Sodium dyes have been used to image intracellular sodium fluctuations during seizure-like activity and also during cortical spreading depression; or monitoring changes in resting cellular sodium levels in animal models of epilepsy. Imaging of the ionic fluctuations inside and outside of the cells and networks will be key to revealing ionic dynamics and their modulation during seizures. It is important to add that many of the currently available dyes are improving in quality, lowering toxicity, photo-bleaching, cell leakage, and increasing signal-to-noise ratio. These new imaging probes are far more advantageous than the traditional probes for measuring ionic changes using electrodes, allowing simultaneous dynamic recordings in multiple cells and interconnected networks.
Metabolism
Metabolic activity can be imaged using the intrinsic fluorescence of NAD(P)H (NADH and NADPH)32. The reduced form of NADH and NADPH emits fluorescence at 450 nm when excited with UV light at 360 nm, whereas the oxidized form is less fluorescent. The advantage of fluorescence-based detection of metabolites is the immediate measurement of energy metabolism, as opposed to electrode-based measurements of O2 or metabolite concentrations, which are altered by diffusion within the tissue. Comparison of normal and chronically epileptic tissue from pilocarpine-treated rats and temporal lobe epilepsy (TLE) patients revealed alterations in activity-induced NAD(P)H fluorescence transients, suggesting that chronically epileptic neural tissue is “hypometabolic”33.
Neurotransmitters
As the primary excitatory neurotransmitter in the brain, even modest changes in glutamate signaling can have a wide array of pro-convulsive effects. Recent advances in imaging glutamate have allowed visualization of extracellular glutamate transients on a circuit and cellular level34–36. These same approaches are now being used to investigate the uptake of extracellular glutamate by astrocytes. Impairment of glutamate reuptake can have a dramatic effect on the dynamics of a neural network. To gain a better understanding of this phenomenon, a CFP-YFP FRET-based glutamate biosensor was developed to assay regulation of extracellular glutamate34. In the freeze lesion model of cortical dysplasia, distinct alterations in peri-lesion glutamate transport capacity were recorded by measuring extracellular glutamate concentration37. In addition, photo-uncaging of compounds, like MNI-glutamate and Rubi-GABA, can be used to map the spatial organization of functional neurotransmitter input to cells in normal and epileptic tissue (Figure 1B).
Vesicular neurotransmitter release is commonly measured using the pH-sensitive GFP mutant synaptopHluorin38, which is quenched in the acidic vesicle, but fluoresces brightly when released into the neutral extracellular space. In the pilocarpine model of epilepsy, this has been used to demonstrate higher rates of vesicular glutamate release and endocytosis in the mossy fibers39.
Genetic targeting of fluorophores
Concomitant with the great expansion in the toolbox of stably expressing indicators of neurological activity have been advancements in techniques for targeting expression of these proteins to genetically defined populations of cells.
Cre-lox system for targeted expression
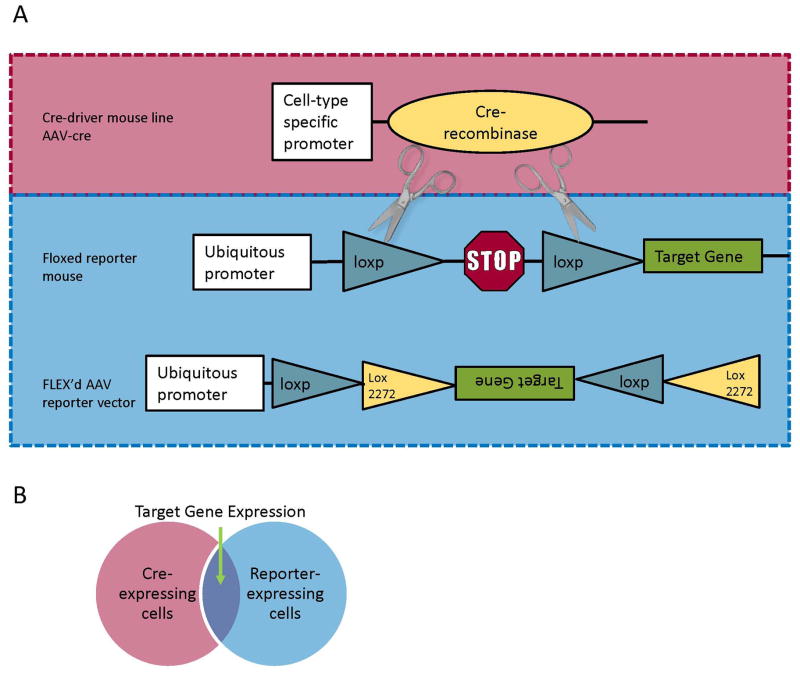
The discovery of cre recombinase-mediated deletion or inversion of loxP-flanked DNA sequences in eukaryotic cells40 has led to a remarkable streamlining in the generation of transgenic mice with targeted expression of new fluorescent indicators. Typically, when a new indicator is developed, a “reporter” mouse line is generated, which contains, at a ubiquitously expressed locus in the genome, a loxP-flanked stop codon followed by the gene for the indicator. Cells that express cre-recombinase at some point in development will have the stop codon deleted, resulting in constitutive expression of the gene of interest (schematized in Figure 2). By crossing a reporter mouse with a mouse selected from a continually growing library of cre “driver” mice, researchers can target expression of the new indicator to a subset of cells defined by cre expression in the driver mouse. For example, somatostatin-cre driver mice were crossed channelrhodopsin+YFP reporter mice to demonstrate anatomical and functional ectopic sprouting of stratum oriens interneurons to the molecular layer of the dentate gyrus in the pilocarpine model of epilepsy41. Cre drivers, however, should be validated by independent investigators since it has become clear that some drivers may not be as specific as originally intended42. Since most cells types are not best defined by expression of a single promoter, complementary recombinase systems Flp-Frt and Dre-rox can be used in combination with Cre-lox to find intersectional populations of cells, co-expressing two genes. For example, parvalbumin-cre driver mice crossed with somatostatin-Flp driver mice have been infected with cre- and Flp- dependent viral constructs to target expression in interneurons co-expressing PV and SOM. While the number of Flp and Dre driver lines is currently quite limited, the toolbox is continually growing.
Figure 2. Cre-mediated expression.
A) A typical scheme for targeted expression of a genetically encoded fluorescent probe involves cre recombinase-mediated deletion of a loxp-flanked stop codon to achieve expression of the target gene. Cre can be delivered in the form of a transgenic “cre-driver” animal that expresses cre in a subset of cells or an AAV virus. Similarly the “reporter” construct can be a transgenic “floxed” mouse or a FLEX’d virus, both of which constitutively express the target gene following cre-mediated recombination. The “floxed” construct snips out a stop codon upon cre-mediated recombination. The FLEX construct first flips the transgene to the forward direction, and then excises one each of the lox sites to prevent further recombination. B) Target gene expression is achieved only in cells co-expressing cre-recombinase and reporter construct. Typically expression is directed to a set of cells genetically defined by cre expression. When viral constructs are used, spatial organization of expression can also be determined by the site of injection.
AAV vectors for transgene delivery
Perhaps the simplest method for delivering genetically encoded fluorescent indicators is the administration of AAV vectors. While viral transduction is somewhat more invasive than the use of transgenic animals, there are many characteristics which make it a desirable expression system. And, there is mounting evidence that AAV2 can produce long term, stable expression of a transgene, with minimal negative impact on infected cells43. AAV vectors have the added advantage of being able to delivering genes in a variety of species for which mutant animals are less readily available (e.g. rats, non-human primates) or in combination with genetic models of disease to produce animals with multiple transgenes.
AAV can be targeted to a desired population of cells through the appropriate selection of capsid serotype, injection site, promoter and/or cre-mediated expression. The proper capsid serotype must be chosen for the targeted tissue (AAV1, 2, 5, 8 and 9 are commonly used in the CNS, but c.f.44). Expression can be spatially defined by site of injection to either target cell bodies located at the injection site or to infect axons and retrogradely label cells projecting to the injection site. Small promoters or promoter fragments such as human synapsin I and GFAP can be used to target expression to neurons or astrocytes respectively. However, options for promoter-defined expression are limited by the 4.7kb payload of AAV vectors and expression levels vary with promoter activity. To circumvent these shortcomings, AAV vectors have been developed that contain the transgene in a flip-excision (FLEX’d) configuration, wherein the cre-mediated recombination FLips the gene to the forward direction and EXcises loxP sites to make this change permanent. When these viruses are used to infect a cre-driver mouse, cre-mediated recombination leads to permanent expression of the transgene (typically under the control of a constitutively highly active promoter such as CMV) in the subset of cells expressing cre recombinase. Similarly AAV-cre vectors can be used to deliver cre recombinase to reporter animals to achieve sparse and/or brain-region specific expression of a floxed gene. FLEX’d virus can be injected at a desired site to get expression in a genetically defined cell type that project to the site of injection (e.g. by infecting the left dorsal striatum of D1-cre animals with AAV-FLEX-GCaMP, it is possible to target neurons of the basal ganglia that express D2 and project axons to the striatum45).
Logistically, AAV vectors have several advantages as well. When a new fluorescent-protein is developed and the plasmid is made available, a virus can be made to deliver the gene in a matter of weeks (often even before publication of the new gene), compared with typically ~1 year for transgenic mouse development. Core facilities such as those at the University of North Carolina (http://genetherapy.unc.edu/services.htm) and University of Pennsylvania (http://www.med.upenn.edu/gtp/vectorcore) maintain stocks of high titer AAV vectors for the most popular optogenetic constructs (indicators and activators) used in the brain and generate custom vectors for delivery of any DNA sequence that will fit in the AAV backbone.
Pseudo-rabies virus for retrograde synaptic labelling
Recently the retrogradely infectious rabies virus has been modified and used to trace small neural circuits with fluorescent proteins46,47. In this approach, a single (or small number of) “starter” cell is transduced with plasmids for the rabies glycoprotein and the avian viral receptor TVA, which is not endogenously found in mammals. The modified rabies virus used is pseudotyped to target only TVA-expressing cells and is glycoprotein deficient, meaning it cannot continue to spread after leaving the glycoprotein-producing starter cell. Thus, the rabies virus genome, which may include a variety of optogenetic constructs48, is delivered to cells providing monosynaptic input to the starter cell. While the cytotoxicity of rabies virus is substantially greater than that of AAV, it enables the study of synaptically defined neuronal microcircuits and will undoubtedly prove to be a valuable research tool.
High spatiotemporal resolution and multimodal imaging
A critical component for imaging activity in neural tissue is choosing a microscopy technique that matches the requirements for a given experimental paradigm. Confocal, multi-photon, and single-photon widefield imaging all have distinct advantages depending on requirements for sampling rate, spatial resolution, imaging depth, and sensitivity.
Confocal microscopy
Confocal microscopy is a very mature technology49, capable of achieving excellent axial (z-plane) resolution, to image fine structures even in densely expressing tissue. Fast laser scanning techniques such as the Nipkow spinning disk50 or swept-field scan51 produce frame rates of >100Hz. As with any visible wavelength single-photon imaging used in neural tissue, scattering prevents imaging deeper in tissue than ~50μm and out-of-focus excitation leads to increased photobleaching and phototoxicity rates in the sample.
Multi-photon imaging
With two-photon imaging, excitation light has double the wavelength (half the energy/photon) required to excite the fluorophore. Thus, two photons must strike the fluorescent molecule nearly simultaneously to generate emitted light. There is only a significant probability of this happening where photons are very dense: at the apex of the focal cone. Since out-of-focus fluorescence is eliminated and longer wavelength excitation light penetrates brain tissue with much less scattering, it is practical to image up to ~1mm deep in vivo 52,53. Parallel beam scanning approaches similar to those used in confocal imaging have been applied to multi-photon microscopy to achieve high frame rates54, however, the power requirements of two-photon excitation substantially limit the imaging depth for split-beam scanning. Reduced scan paths have also been used to sample at high rates fluorescence from distant cells (at the cost of not scanning the entire sample). For example, spiral scanning has been used to sparsely sample (at least one pixel) in 90% of neurons in a 250x250x200μm volume of brain tissue55 at rates of 10Hz. Similarly, acouto-optic deflectors (AODs) are being used to rapidly sample >500 cells in a 400x400x500μm volume56. Since AODs cause significant spatial and temporal dispersion, their use requires significant dispersion compensation hardware57. Targeted path scanning (TPS) uses a manually directed approach to sample neurons with a specified number of pixels at path-dependent speeds (e.g. 30 cells at >30Hz, Figure 1C,58). TPS has the advantage of working on a standard galvanometer-based two-photon microscope hardware (requiring only a software modification), but is speed-limited by the inertia of the scanning mirrors.
Single-photon imaging of sparsely expressing tissue
Confocal and multiphoton microscopy enable imaging of fluorescent probes with subcellular resolution by rejecting out-of-focus fluorescence or selectively exciting molecules in the focal plane respectively. Another way to eliminate out-of-focus fluorescence is to use a system with sparse expression of the fluorophore. This can provide cellular and even sub-cellular resolution using simple, inexpensive wide-field single-photon imaging. A particularly useful new class of cameras, based on the recently developed scientific CMOS (sCMOS) image sensors, make it possible to image these sparsely expressing samples with low noise (<1e−), high resolution (4MP), and high bit-depth (16-bits) at rates of 100Hz. As is often the case with cutting edge camera technologies, the data flow from these cameras pushes the limits of current computing speeds, producing a staggering (as of this writing) 50GB/min.
In vivo imaging
The capacity of multiphoton imaging to image deep in tissue has made chronic in vivo imaging through implanted cranial windows a reality59. This approach has been used in epilepsy research to look for changes in dendritic spine morphology induced by seizures60. Although well-established protocols making use of stage-mounted spherical treadmills have made it routine to image calcium in awake, behaving (albeit head-fixed) animals61, functional imaging during seizure remains challenging. Miniaturized, head-mounted two-photon62 and single photon17 microscopes are currently being developed and may prove to be critical enabling technologies for obtaining stable recordings from unrestrained seizing animals.
Given the scale of the challenge of understanding brain function (and dysfunction), new tools will be needed to record from neural circuits. Here we have summarized some of the latest tools being used to measure neural activity in epilepsy research. Current technologies enable the simultaneous recording from hundreds to thousands of neurons. Although ongoing development of probes, gene delivery pathways, and imaging techniques surely will continue to advance our ability to record activity in increasingly larger populations, the current state of the art has begun to provide fundamentally novel insight into how cells of the nervous system work together to produce normal and pathological brain rhythms.
In conclusion, understanding functional and structural resting state and epileptiform dynamics at the molecular, cellular, and network levels will contribute an integral missing part to our understanding of epilepsy and other neurological network disorders. Continuing advancements in multimodal imaging techniques will continue to enhance our ability to reveal hierarchical complexity and spatiotemporal dynamics with a simultaneous resolution spanning from single molecule to an entire microcircuit, and ultimately the whole brain. Given the rapid rate of development in imaging techniques, this frontier now does not seem far beyond our reach.
Acknowledgments
Dr. Lillis received support from the National Institutes of Health grants R01NS074772 and R01NS034700.
Dr. Maheshwari received funding from the Caroline Wiess Law Fund for Molecular Medicine.
Footnotes
Confirmation of ethical publication:
We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
Disclosure
None of the authors has any conflict of interest to disclose.
References
- 1.Claes L, Del-Favero J, Ceulemans B, et al. De Novo Mutations in the Sodium-Channel Gene SCN1A Cause Severe Myoclonic Epilepsy of Infancy. Am J Hum Genet. 2001;68:1327–32. doi: 10.1086/320609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Leary T, Williams AH, Caplan JS, et al. Correlations in ion channel expression emerge from homeostatic tuning rules. Proc Natl Acad Sci. 2013;110:E2645–E2654. doi: 10.1073/pnas.1309966110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hüttmann K, Sadgrove M, Wallraff A, et al. Seizures preferentially stimulate proliferation of radial glia-like astrocytes in the adult dentate gyrus: functional and immunocytochemical analysis. Eur J Neurosci. 2003;18:2769–78. doi: 10.1111/j.1460-9568.2003.03002.x. [DOI] [PubMed] [Google Scholar]
- 4.Avignone E, Ulmann L, Levavasseur F, et al. Status Epilepticus Induces a Particular Microglial Activation State Characterized by Enhanced Purinergic Signaling. J Neurosci. 2008;28:9133–44. doi: 10.1523/JNEUROSCI.1820-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Danzer SC, He X, Loepke AW, et al. Structural plasticity of dentate granule cell mossy fibers during the development of limbic epilepsy. Hippocampus. 2010;20:113–24. doi: 10.1002/hipo.20589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–7. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 7.Ando R, Hama H, Yamamoto-Hino M, et al. An optical marker based on the UV-induced green-tored photoconversion of a fluorescent protein. Proc Natl Acad Sci U S A. 2002;99:12651–6. doi: 10.1073/pnas.202320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ando R, Mizuno H, Miyawaki A. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science. 2004;306:1370–3. doi: 10.1126/science.1102506. [DOI] [PubMed] [Google Scholar]
- 9.Patterson GH. Highlights of the optical highlighter fluorescent proteins: HIGHLIGHTS OF THE OPTICAL HIGHLIGHTER FLUORESCENT PROTEINS. J Microsc. 2011;243:1–7. doi: 10.1111/j.1365-2818.2011.03505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Betzig E, Patterson GH, Sougrat R, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–5. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 11.Hofmann M, Eggeling C, Jakobs S, et al. Breaking the diffraction barrier in fluorescence microscopy at low light intensities by using reversibly photoswitchable proteins. Proc Natl Acad Sci U S A. 2005;102:17565–9. doi: 10.1073/pnas.0506010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyawaki A, Llopis J, Heim R, et al. Fluorescent indicators for Ca2+based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–7. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 13.Thestrup T, Litzlbauer J, Bartholomäus I, et al. Optimized ratiometric calcium sensors for functional in vivo imaging of neurons and T lymphocytes. Nat Methods. 2014;11:175–82. doi: 10.1038/nmeth.2773. [DOI] [PubMed] [Google Scholar]
- 14.Chen T-W, Wardill TJ, Sun Y, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grewe BF, Langer D, Kasper H, et al. High-speed in vivo calcium imaging reveals neuronal network activity with near-millisecond precision. Nat Methods. 2010;7:399–405. doi: 10.1038/nmeth.1453. [DOI] [PubMed] [Google Scholar]
- 16.Mank M, Santos AF, Direnberger S, et al. A genetically encoded calcium indicator for chronic in vivo two-photon imaging. Nat Methods. 2008;5:805–11. doi: 10.1038/nmeth.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghosh KK, Burns LD, Cocker ED, et al. Miniaturized integration of a fluorescence microscope. Nat Methods. 2011;8:871–8. doi: 10.1038/nmeth.1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akerboom J, Chen T-W, Wardill TJ, et al. Optimization of a GCaMP Calcium Indicator for Neural Activity Imaging. J Neurosci. 2012;32:13819–40. doi: 10.1523/JNEUROSCI.2601-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian L, Hires SA, Mao T, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods. 2009;6:875–81. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maheshwari A, Nahm WK, Noebels JL. Paradoxical proepileptic response to NMDA receptor blockade linked to cortical interneuron defect in stargazer mice. Front Cell Neurosci. 2013;7:156. doi: 10.3389/fncel.2013.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang D, Zhang Z, Chanda B, et al. Improved Probes for Hybrid Voltage Sensor Imaging. Biophys J. 2010;99:2355–65. doi: 10.1016/j.bpj.2010.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao G, Platisa J, Pieribone VA, et al. Genetically Targeted Optical Electrophysiology in Intact Neural Circuits. Cell. 2013;154:904–13. doi: 10.1016/j.cell.2013.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.St-Pierre F, Marshall JD, Yang Y, et al. High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nat Neurosci. doi: 10.1038/nn.3709. [Internet]. Epub 2014 Apr 23; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hochbaum DR, Zhao Y, Farhi SL, et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat Methods. 2014;11:825–33. doi: 10.1038/nmeth.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zou P, Zhao Y, Douglass AD, et al. Bright and fast multicoloured voltage reporters via electrochromic FRET. Nat Commun. doi: 10.1038/ncomms5625. [Internet]. Epub 2014 Dec 29; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coulter DA, Yue C, Ang CW, et al. Hippocampal microcircuit dynamics probed using optical imaging approaches. J Physiol. 2011;589:1893–903. doi: 10.1113/jphysiol.2010.202184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hazra A, Rosenbaum R, Bodmann B, et al. β-Adrenergic modulation of spontaneous spatiotemporal activity patterns and synchrony in hyperexcitable hippocampal circuits. J Neurophysiol. 2012;108:658–71. doi: 10.1152/jn.00708.2011. [DOI] [PubMed] [Google Scholar]
- 28.Gu F, Hazra A, Aulakh A, et al. Purinergic control of hippocampal circuit hyperexcitability in Dravet syndrome. Epilepsia. 2014;55:245–55. doi: 10.1111/epi.12487. [DOI] [PubMed] [Google Scholar]
- 29.Kuner T, Augustine GJ. A Genetically Encoded Ratiometric Indicator for Chloride: Capturing Chloride Transients in Cultured Hippocampal Neurons. Neuron. 2000;27:447–59. doi: 10.1016/s0896-6273(00)00056-8. [DOI] [PubMed] [Google Scholar]
- 30.Lillis KP, Kramer MA, Mertz J, et al. Pyramidal cells accumulate chloride at seizure onset. Neurobiol Dis. 2012;47:358–66. doi: 10.1016/j.nbd.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamy CM, Chatton J-Y. Optical probing of sodium dynamics in neurons and astrocytes. NeuroImage. 2011;58:572–8. doi: 10.1016/j.neuroimage.2011.06.074. [DOI] [PubMed] [Google Scholar]
- 32.Chance B, Williams GR. A method for the localization of sites for oxidative phosphorylation. Nature. 1955;176:250–4. doi: 10.1038/176250a0. [DOI] [PubMed] [Google Scholar]
- 33.Kann O, Kovács R, Njunting M, et al. Metabolic dysfunction during neuronal activation in the ex vivo hippocampus from chronic epileptic rats and humans. Brain J Neurol. 2005;128:2396–407. doi: 10.1093/brain/awh568. [DOI] [PubMed] [Google Scholar]
- 34.Dulla C, Tani H, Okumoto S, et al. Imaging of Glutamate in Brain Slices Using FRET Sensors. J Neurosci Methods. 2008;168:306–19. doi: 10.1016/j.jneumeth.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hires SA, Zhu Y, Tsien RY. Optical measurement of synaptic glutamate spillover and reuptake by linker optimized glutamate-sensitive fluorescent reporters. Proc Natl Acad Sci U S A. 2008;105:4411–6. doi: 10.1073/pnas.0712008105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marvin JS, Borghuis BG, Tian L, et al. An optimized fluorescent probe for visualizing glutamate neurotransmission. Nat Methods. 2013;10:162–70. doi: 10.1038/nmeth.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dulla CG, Tani H, Brill J, et al. Glutamate biosensor imaging reveals dysregulation of glutamatergic pathways in a model of developmental cortical malformation. Neurobiol Dis. 2013;49:232–46. doi: 10.1016/j.nbd.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miesenböck G, De Angelis DA, Rothman JE. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 1998;394:192–5. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- 39.Upreti C, Otero R, Partida C, et al. Altered neurotransmitter release, vesicle recycling and presynaptic structure in the pilocarpine model of temporal lobe epilepsy. Brain J Neurol. 2012;135:869–85. doi: 10.1093/brain/awr341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sauer B. Functional expression of the cre-lox site-specific recombination system in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:2087–96. doi: 10.1128/mcb.7.6.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng Z, Zhang N, Wei W, et al. A Reorganized GABAergic Circuit in a Model of Epilepsy: Evidence from Optogenetic Labeling and Stimulation of Somatostatin Interneurons. J Neurosci. 2013;33:14392–405. doi: 10.1523/JNEUROSCI.2045-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu H, Cavendish JZ, Agmon A. Not all that glitters is gold: off-target recombination in the somatostatin-IRES-Cre mouse line labels a subset of fast-spiking interneurons. Front Neural Circuits. doi: 10.3389/fncir.2013.00195. [Internet]. Epub 2014 May 28; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Howard DB, Powers K, Wang Y, et al. Tropism and toxicity of adeno-associated viral vector serotypes 1,2,5,6,7,8,9 in rat neurons and glia in vitro. Virology. 2008;372:24–34. doi: 10.1016/j.virol.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aschauer DF, Kreuz S, Rumpel S. Analysis of Transduction Efficiency, Tropism and Axonal Transport of AAV Serotypes 1, 2, 5, 6, 8 and 9 in the Mouse Brain. PLoS One. doi: 10.1371/journal.pone.0076310. [Internet]. Epub 2013 Nov 29; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cui G, Jun SB, Jin X, et al. Concurrent activation of striatal direct and indirect pathways during action initiation. Nature. 2013;494:238–42. doi: 10.1038/nature11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wickersham IR, Finke S, Conzelmann K-K, et al. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat Methods. 2007;4:47–9. doi: 10.1038/NMETH999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wickersham IR, Lyon DC, Barnard RJO, et al. Monosynaptic Restriction of Transsynaptic Tracing from Single, Genetically Targeted Neurons. Neuron. 2007;53:639–47. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Osakada F, Mori T, Cetin AH, et al. New Rabies Virus Variants for Monitoring and Manipulating Activity and Gene Expression in Defined Neural Circuits. Neuron. 2011;71:617–31. doi: 10.1016/j.neuron.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Minsky M. US3013467 A. Microscopy apparatus. 1961
- 50.Xiao GQ, Corle TR, Kino GS. Real-time confocal scanning optical microscope. Appl Phys Lett. 1988;53:716–8. doi: 10.1364/AO.29.003769. [DOI] [PubMed] [Google Scholar]
- 51.Vogt WI, Szulczewski MJ, Wolf DJ. US6856457 B2. Single and multi-aperture, translationally-coupled confocal microscope. 2005
- 52.Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–6. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 53.Svoboda K, Yasuda R. Principles of Two-Photon Excitation Microscopy and Its Applications to Neuroscience. Neuron. 2006;50:823–39. doi: 10.1016/j.neuron.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 54.Bewersdorf J, Pick R, Hell SW. Multifocal multiphoton microscopy. Opt Lett. 1998;23:655–7. doi: 10.1364/ol.23.000655. [DOI] [PubMed] [Google Scholar]
- 55.Göbel W, Kampa BM, Helmchen F. Imaging cellular network dynamics in three dimensions using fast 3D laser scanning. Nat Methods. 2007;4:73–9. doi: 10.1038/nmeth989. [DOI] [PubMed] [Google Scholar]
- 56.Katona G, Szalay G, Maák P, et al. Fast two-photon in vivo imaging with three-dimensional random-access scanning in large tissue volumes. Nat Methods. 2012;9:201–8. doi: 10.1038/nmeth.1851. [DOI] [PubMed] [Google Scholar]
- 57.Iyer V, Losavio BE, Saggau P. Compensation of spatial and temporal dispersion for acousto-optic multiphoton laser-scanning microscopy. J Biomed Opt. 2003;8:460–71. doi: 10.1117/1.1580827. [DOI] [PubMed] [Google Scholar]
- 58.Lillis KP, Eng A, White JA, et al. Two-photon imaging of spatially extended neuronal network dynamics with high temporal resolution. J Neurosci Methods. 2008;172:178–84. doi: 10.1016/j.jneumeth.2008.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trachtenberg JT, Chen BE, Knott GW, et al. Long-term in vivo imaging of experience-dependent synaptic plasticity in adult cortex. Nature. 2002;420:788–94. doi: 10.1038/nature01273. [DOI] [PubMed] [Google Scholar]
- 60.Rensing N, Ouyang Y, Yang X-F, et al. In vivo imaging of dendritic spines during electrographic seizures. Ann Neurol. 2005;58:888–98. doi: 10.1002/ana.20658. [DOI] [PubMed] [Google Scholar]
- 61.Dombeck DA, Khabbaz AN, Collman F, et al. Imaging large scale neural activity with cellular resolution in awake mobile mice. Neuron. 2007;56:43–57. doi: 10.1016/j.neuron.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Helmchen F, Fee MS, Tank DW, et al. A Miniature Head-Mounted Two-Photon Microscope: High-Resolution Brain Imaging in Freely Moving Animals. Neuron. 2001;31:903–12. doi: 10.1016/s0896-6273(01)00421-4. [DOI] [PubMed] [Google Scholar]
- 63.Grimley JS, Li L, Wang W, et al. Visualization of Synaptic Inhibition with an Optogenetic Sensor Developed by Cell-Free Protein Engineering Automation. J Neurosci. 2013;33:16297–309. doi: 10.1523/JNEUROSCI.4616-11.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harootunian AT, Kao JP, Eckert BK, et al. Fluorescence ratio imaging of cytosolic free Na+ in individual fibroblasts and lymphocytes. J Biol Chem. 1989;264:19458–67. [PubMed] [Google Scholar]