Abstract
Single cell explorations offer a unique window to inspect molecules and events relevant to mechanisms and heterogeneity constituting the central dogma of biology. A large number of nucleic acids, proteins, metabolites and small molecules are involved in determining and fine-tuning the state and function of a single cell at a given time point. Advanced optical platforms and nanotools provide tremendous opportunities to probe intracellular components with single-molecule accuracy, as well as promising tools to adjust single cell activity. In order to obtain quantitative information (e.g. molecular quantity, kinetics and stoichiometry) within an intact cell, achieving the observation with comparable spatiotemporal resolution is a challenge. For single cell studies both the method of detection and the biocompatibility are critical factors as they determine the feasibility, especially when considering live cell analysis. Although a considerable proportion of single cell methodologies depend on specialized expertise and expensive instruments, it is our expectation that the information content and implication will outweigh the costs given the impact on life science enabled by single cell analysis.
INTRODUCTION
“The central dogma of molecular biology deals with the detailed residue-by-residue transfer of sequential information,” in a seminal work by Francis Crick in 1960s.1 This was the first time that the major components of a single cell (i.e. DNA, RNA and protein) were proposed to be functionally connected in complicated biological systems. Metazoan species, especially human beings, are highly heterogeneous and dynamic, containing billions of variedly differentiated cells that play fundamental roles in biological processes. Even cells within the same tissue or region, at a given time point might have distinct morphological properties and functional states that could result from just a minute discrepancy in their surrounding physicochemical environment, signaling communication, epigenetic regulation, cell cycle status or others. The population- and end-point-based measurement can no longer meet the requirements in present biomedical research. Owing to innovations in optics and nanoscale materials, the spatiotemporal resolution of our observation has made “seeing inside cells” a reality. Therefore over the past few decades, our understanding of DNA replication, RNA transcription, protein translation and related intracellular events has been expanded and deepened at an unprecedented pace.2, 3 Elucidation of intracellular interactions and dynamics will aid in a better understanding of the structure and function of tissues and organs. One can anticipate that single cell studies will greatly reshape the architecture base of our biological knowledge to impact future practices in healthcare, ecology and environmental science.
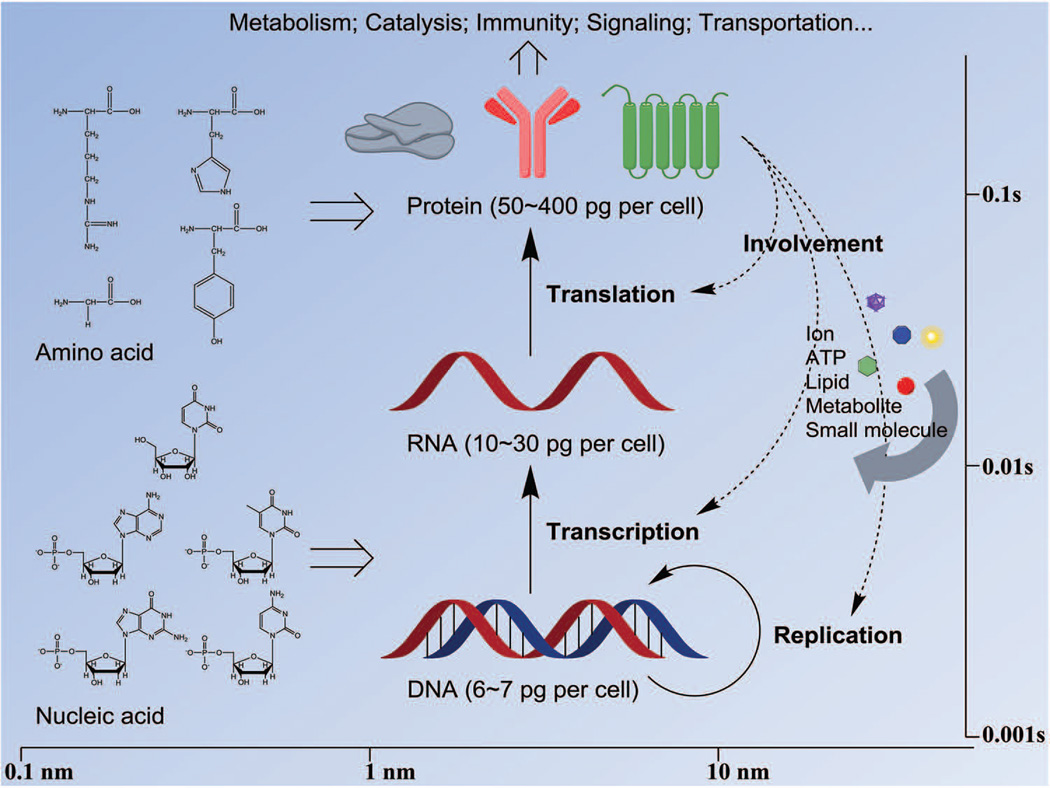
Since cell nucleus, cytoplasm, organelles and membranes have distinct biochemical and biophysical properties, studying the molecules and activities in subcellular compartments requires different sets of tools because of the complexity involved in probing and extracting the desired information. Given the spatiotemporal scale of molecular events taking place in living cells (Figure 1), an ideal platform for detection should, in essence provide information at the spatial resolution of nanometer and at the temporal resolution of millisecond simultaneously. For some highly dynamic processes, such as molecular rotation and enzyme catalysis, the temporal scale may extend down to the microsecond-level or below. The variation in transcription and translation of different genes may bring about another dimension in the challenges posed for quantitative single cell analysis. For a typical human cell, 3 billion base pairs of DNA are contained in its nucleus, which encode approximate 21,000 genes. For a differentiated cell to carry out its normal function, on an average 360,000 mRNA molecules from about 12,000 different transcript types could exist. In HeLa cells, around 2.3 billion protein molecules reside in each single cell, giving rise to a concentration of 1 million proteins per femtoliter.4 It also must be taken into consideration that the copy numbers of different transcripts and proteins vary radically: from tens of copies (e.g. HER2 mRNA in MCF-7 cells5) to thousands of copies (e.g. some housekeeping genes). This numeric cascade along with the transmission of genetic information from DNA to RNA to protein makes it a challenging proposition to detect, analyze and quantify the products even for a single gene at all the three levels.
FIGURE 1.
The spatiotemporal scales expected for single cell studies.
In this review we focus on two major aspects – optical approaches for single cell analysis and nanoprobes that enable single-molecule sensing. In this context, quantitative detection of nucleic acids, proteins, metabolites and small molecules at the single cell level is elaborated with representative applications and examples. In the final part, future prospects and developmental challenges are also briefly discussed.
NANOMATERIALS IN SINGLE CELL SENSING
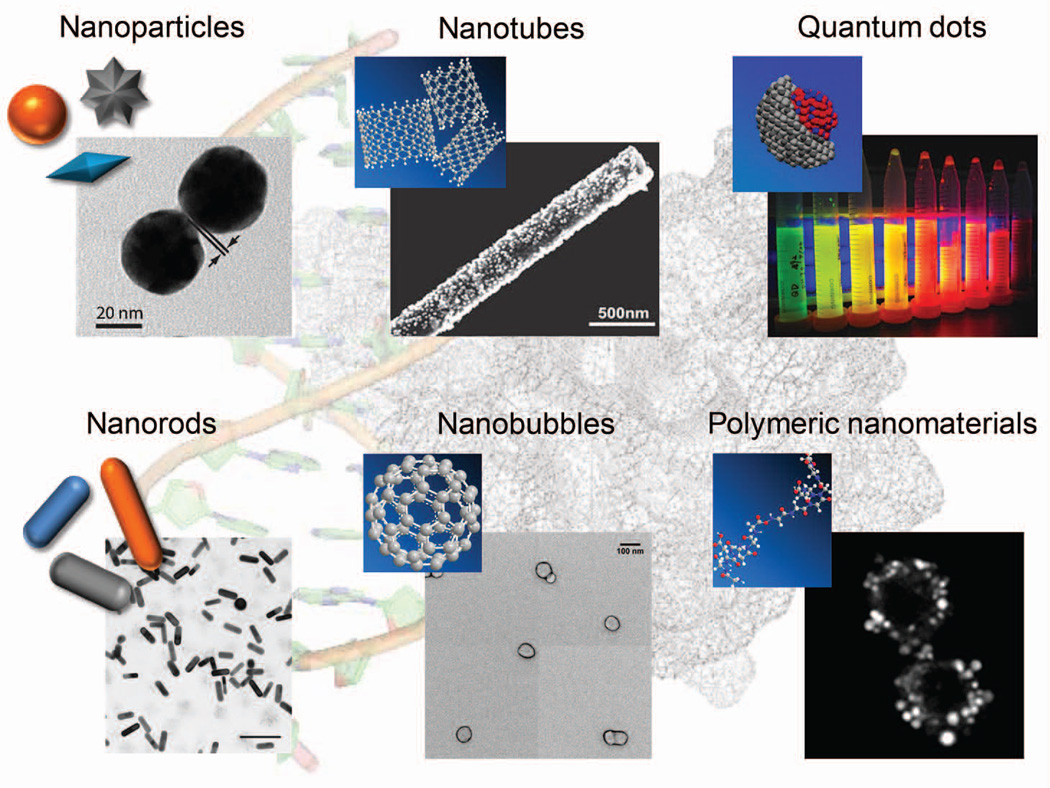
In this section we discuss the different aspects of nanomaterials that are instrumental in single-molecule sensing. Tunable metallic nanomaterials (e.g. gold, silver and iron), carbon-derived nanomaterials (e.g. graphene and nanotube) and polymer-based nanomaterials (e.g. nanobubble and origami) play increasingly pivotal roles in basic science research and engineering (Figure 2). They can be efficiently introduced into single cells to perform diverse functions with minimum perturbations.
FIGURE 2.
Nanomaterials, ranging from 1 nm to ~100 nm, are broadly applied in single cell studies.
Nanomaterials provide excellent stability and signal-to-noise ratio (SNR) and can serve as ideal reporters of sparse information from single cells. Of a myriad of nanomaterials, gold nanoparticles (GNPs) are the most extensively used contrast agents whose size and shape can be easily controlled. Due to localized surface plasmon resonance, GNPs have characteristic light extinction (absorption and scattering) patterns. Upon illumination, they exhibit different colors depending upon the morphology and inter-particle distance, and hence can serve as ideal single-molecule labeling materials for colorimetric assays and dark-field microscopy.6–8 When used as contrast agents in hyperspectral dark-field microscopy, the scattering of GNPs on a pixel-by-pixel basis can be extracted as a spectrum. Then by filtering the spectra, the spectral characteristics of interest can be reconstructed background-free and shown as images (Figure 6b).9–11 This appealing potential enables us to investigate the dynamic localization of nanomaterials inside single cells and also serve as a powerful tool for uncovering biological mechanisms not possible by conventional methods. Interestingly, with intense luminescence under red laser excitation, aspect ratio tunable gold nanorods (GNRs) have also been applied in single-molecule tracking (Figure 7c).12, 13 In addition to serving as a contrast agent, the intense surface plasmon field makes GNPs an ideal substrate for amplifying the spontaneous but weak Raman scattering signals. GNPs with different attributes have been successfully implemented for sensing trace amount of target molecules under a diversity of conditions.9, 14, 15
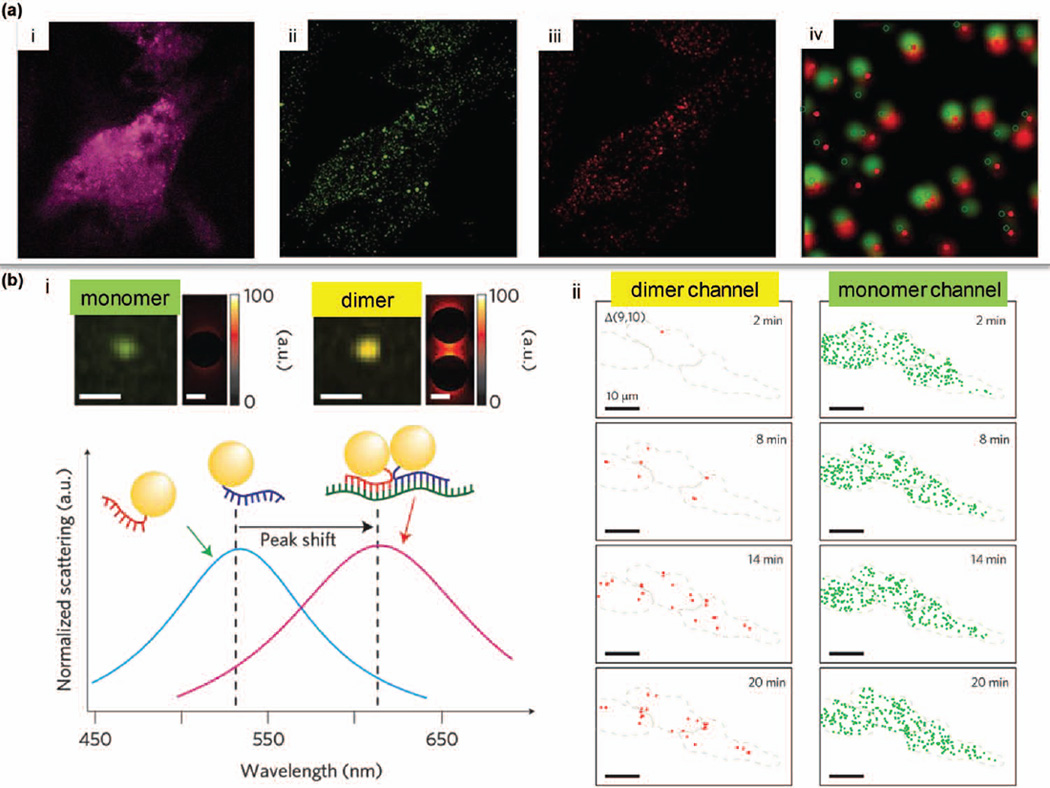
FIGURE 6.
Single cell studies at the RNA level. (a) HT1080-GFP-96mer cells were incubated with ratiometric bimolecular beacons (RBMBs) targeting GFP mRNAs. Pink signal (in i) is from the reference dye. Green fluorescence is from the reporter dye (in ii). Single-molecule FISH was performed (red in iii) to validate the targeting efficiency of RBMBs (overlay in iv). (Reprinted with permission from Ref 96. Copyright 2013 Oxford University Press) (b) Alternative splicing of BRCA1 was assessed by hyperspectral dark-field microscopy. GNP dimers exhibit a red-shifted color compared to monomers (in i). GNP-based probes flanking distant mRNA regions enable the identification of variants with spliced-out exons (in ii). (Reprinted with permission from Ref 10. Copyright 2014 Nature Publishing Group)
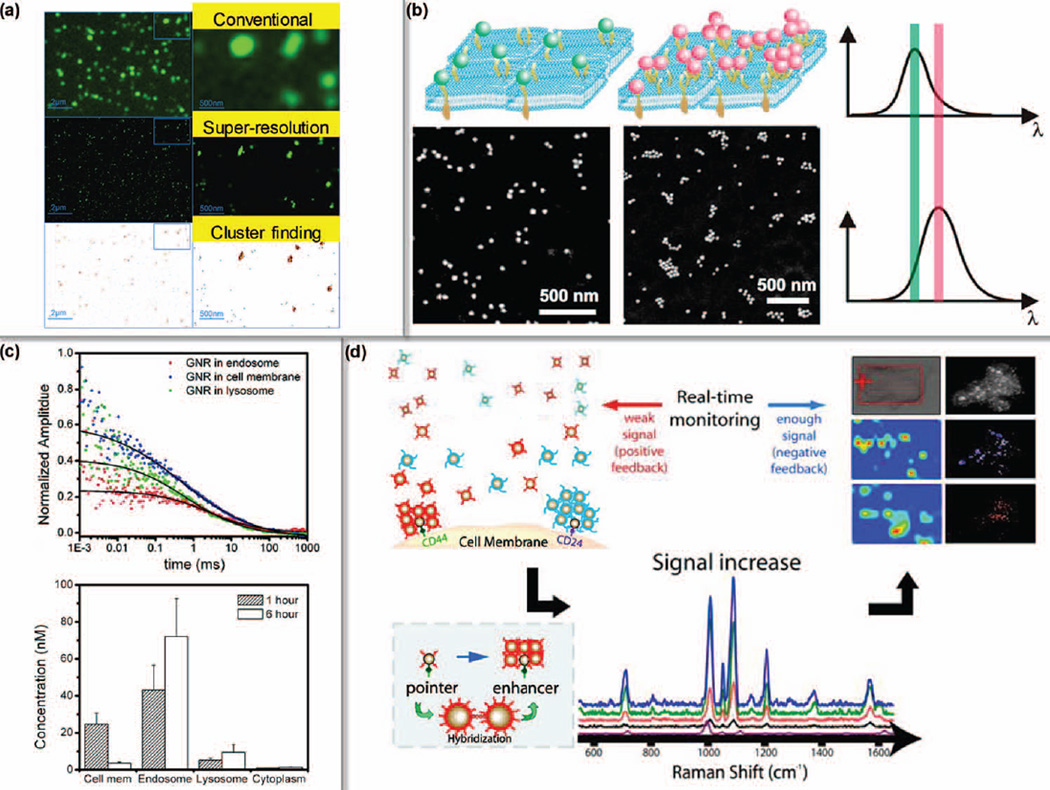
FIGURE 7.
Multimodal characterization of breast cancer surface markers. (a) HER2 clusters on SKBR3 cell membrane can be resolved by super-resolution microscopy. (Reprinted with permission from Ref 101. Copyright 2010 John Wiley & Sons, Inc.) (b) The density of HER2 clusters can also be evaluated with GNP labels by scattering microscopy. The density positively correlates with the plasmonic peak (λmax). (Reprinted with permission from Ref 102. Copyright 2012 American Chemical Society) (c) HER2 mediated cellular uptake was tracked by H-GNRs and FCS. (Reprinted with permission from Ref 12. Copyright 2009 American Chemical Society) (d) The ratio of CD44/CD24 was quantified by SERS to identify breast CSCs. (Reprinted with permission from Ref 9. Copyright 2011 American Chemical Society)
Another class of nanomaterials constitute the semiconductor nanocrystals, namely quantum dots (QDs), which have attracted extensive attention due to their outstanding quantum efficiency and tunability in emitted light. QDs are often synthesized with binary compounds, and the diameter of formed crystal structure is smaller than the exciton Bohr radius, which exhibits unique electronic characteristics – quantum confinement. The corresponding band gap energy of QDs, predominantly determined by size, is inversely proportional to the absorbance and emission. This physical property yields QDs in a range of colors from violet to infrared. Compared to conventional fluorophores, QDs are tens of times brighter and hundreds of times more stable, rendering them ideal for long-term single-molecule imaging and tracking.16–18 Recently, photoblinking QDs have been shown to be promising materials for localization-based super-resolution microscopy.19, 20 Furthermore, by tuning QDs to near-infrared region, probing single cells under in vivo conditions is now possible.21 A major concern in the extensive use of QDs is their underlying toxicity since the first generation of QDs often contains heavy metal elements (e.g. cadmium) requiring a long time for clearance from the body. Encouragingly, “nontoxic” QDs enabled by optimized synthesis and polymer coating are rapidly arising.22, 23
Metallic nanomaterials in addition to its signal generation properties can also be used as a quencher, for example, quenching fluorescence when used in molecular beacon (MB). Based on a distance-dependent mechanism, noble metal nanoparticles have been found to be powerful quenchers of fluorophores when held at a proximal distance.24, 25 This property provides us ample opportunities in single-molecule probing if an intracellular component needs to be selectively tagged. In a typical MB probe, the two termini are respectively conjugated with a nanoquencher and a fluorescent reporter.26 In the absence of target, the probe itself would form a loose stem-loop structure that brings together the nanoquencher and the fluorescent reporter. Therefore the non-specific emission can be potently inhibited. In the presence of target, the loop would be stretched upon hybridization, then generating a detectable signal. Besides gold and silver nanomaterials, graphene, graphene oxide and single-walled carbon nanotube have recently been tested to be more effective quenchers.27–29 Using this class of nanomaterials, the traditional MB approach has been significantly improved to detect mRNAs in live cells at a higher degree of specificity.30
With better instrumentation for fabrication, not only can nanomaterials be tuned to different conformations, but a variety of sophisticated nanodevices can be constructed, such as one-dimension nanoneedles for penetrating cell membranes in real-time measurements (Figure 9a).31, 32 These nanodevices can be further adapted to different platforms where the signals from ongoing processes and relevant molecules can be converted to quantifiable statistics. Nanoneedle-based biosensors have already been used for quantifying mRNA molecules, chemical carcinogens, apoptotic enzymes and reactive oxygen species (ROS).33–36 The advantage of nanodevices primarily comes from their extraordinary biocompatibility and superb surface-area-to-volume ratio. However, to ensure the durability and reproducibility of most nanodevices is a challenging task.
FIGURE 9.
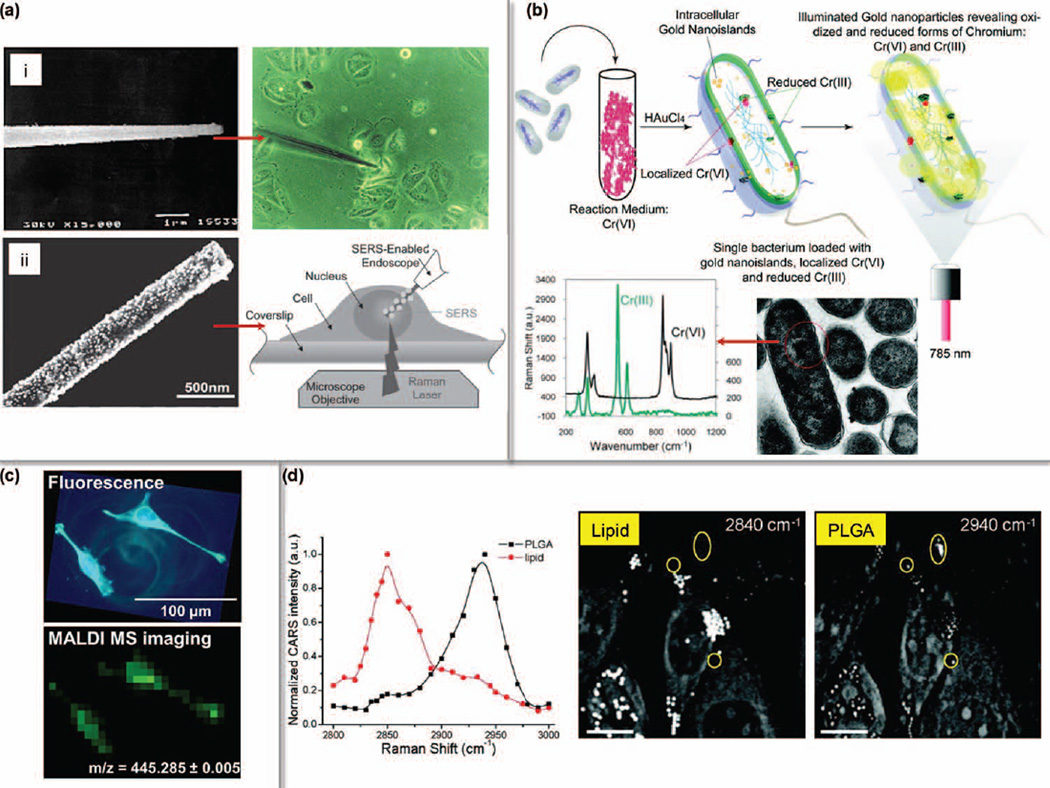
Label-free intracellular fingerprinting. (a) One-dimensional nanoneedle sensors for single cell probing. (i, Reprinted with permission from Ref 34. Copyright 2000 Nature Publishing Group) GNP-coated nanoneedle was used for SERS. (ii, Reprinted with permission from Ref 125. Copyright 2010 John Wiley & Sons, Inc.) (b) SERS mapping of trivalent and hexavalent Chromium was facilitated by intracellular growth of gold nanoislands. (Reprinted with permission from Ref 123. Copyright 2011 American Chemical Society) (c) 3, 3′-dihexyloxacarbocyanine iodide (DIOC6(3)) was imaged by MALDI-MSI at 7-µm resolution. (Reprinted with permission from Ref 48. Copyright 2012 American Chemical Society) (d) CARS for probing intracellular lipid contents and PLGA polymers. (Reprinted with permission from Ref 130. Copyright 2009 American Chemical Society)
Since the nanomaterials and nanodevices are exogenous to cells, biocompatibility and cytotoxicity are some of the crucial concerns for single cell analysis, especially when probing living cells. Introduction of any external materials into living cells would have unpredictable impact since the cellular physiology is subject to various stresses. A paradox is thus emerging in the development of nanomaterials: for single cell studies an ideal probe should be as small as possible; but for a number of nanomaterials, the smaller they are, the greater their surface-area-to-volume ratio, leading to a higher chemical activity which could generate excessive ROS to damage cells.37, 38 Additionally, quite a few nanomaterials are hydrophobic, preventing their uptake by cells. Hence, in order to lower any possible toxic impact and increase water solubility, the surface of nanomaterials is usually modified by polymers (e.g. polyethylene glycol) or biomimetic decoys.39, 40 Nevertheless, the fast growing field of nanotechnology is expected to refuel single cell-based research with thrilling precisions and new discoveries. The enriching toolbox of nanomaterials will not only empower us to see the microscopic world more clearly but also provide the foundation for single cell manipulation. In the near future, we expect nanotools to play a key role in the detection and correction of a single ill cell in vivo with single-molecule accuracy.
ADVANCED OPTICAL PLATFORMS FOR SINGLE CELL STUDIES
Around 340 years have passed since Robert Hooke and Van Leeuwenhoek popularized the optical microscope to observe cells and micro-organisms. Today optical microscopy with fitted modules or labeling materials is still the most direct way to visualize intracellular contents and events. Microscopic platforms such as electron microscope (EM) and atomic force microscope (AFM) are able to achieve sub-nanometer resolution however, when dissecting single cells or subcellular ultrastructures, specialized sample preparation/fixation techniques are required. In comparison, the capacity to capture dynamic processes and multiple components is an exceptional strength that optical microscopes possess. In the next several subsections, we will briefly discuss the newly developed optical approaches for single cell research.
Label-free detection
Undoubtedly, although we have a multitude of nanomaterial choices to label intracellular entities, non-invasive detection is attractive because it provides the ability to probe the intact structure and delicate metabolism of cells. Among the existing methods, scattered light-based spectroscopy and microscopy are outstanding in evaluating subcellular contents and events. Currently, one major approach that takes advantage of scattering signals is chemical fingerprinting by Raman spectroscopy. As a group of label-free tools, Raman spectroscopy and microscopy are able to generate characteristic molecular vibrational fingerprints corresponding to energy levels. For instance, the C-H stretch bond enriched in lipids can be used to map cholesterol storage and adipocyte under a variety of physiological and pathological conditions at the single cell level.41 Theoretically, because the inelastic scattering, spontaneous Raman signal has a much lower intensity compared to Rayleigh scattering (elastic scattering), a high laser excitation and long integration time might be necessary for measurement.42 With the emergence of novel techniques including the surface-enhanced Raman spectroscopy (SERS),43 tip-enhanced Raman spectroscopy (TERS),44 coherent anti-stokes Raman spectroscopy (CARS)45 and stimulated Raman spectroscopy (SRS),46 fast label-free profiling of a single live cell will become feasible.
Since quite a few intrinsic biomolecules and metabolites, such as tryptophan, phenylalanine, NAD(P)H and flavin, can emit autofluorescence upon appropriate excitations, they also attract considerable attention in probing single cells label-free.47 Another elegant technique worth mentioning is the mass spectroscopy imaging (MSI), although this may not be defined exclusively as optical. In the matrix-assisted laser desorption/ionization MSI (MALDI-MSI), thousands of fingerprints of intracellular biomolecules, in terms of mass-to-charge ratio (m/z), can be obtained in a label-free manner (Figure 9c). A prominent advantage for MSI is its capability to simultaneously collect all the characteristics of proteins, lipids, metabolites and chemicals in one single image with any desired combinations. MSI holds great promise for cell classification, cancer diagnosis, and metabolomics analysis.48–51 With further improvement of its spatiotemporal resolution, ambient MSI will be a valuable addition to the arsenal for single cell studies.
Despite their advantages in probing cells without extra labeling, several common limitations for label-free detection should be considered before application. First, in spite of the exceptional detection sensitivity, the chemical bond-dependence of the Raman signal can hardly be attributed to a unique source since most chemical bonds are shared by a large number of biomolecules. In light of this, it is necessary to integrate multiplexed characteristics of the target to improve the detection specificity, which is also true for MSI. Another limitation is the spatial resolution. Due to the limitation in specificity, at present label-free detection lags behind fluorescent microscopy to obtain high-quality images. To our excitement, by re-aligning the pattern of optical illumination, a label-free method has been shown to achieve the sub-diffraction resolution in far-field imaging, which holds tremendous promise for future single cell studies.52 Other concerns include limited penetration depth, potential photodamage and highly specialized instrumentation, which await future improvement.
Super-resolution microscopy for diffraction-unlimited interrogation
The resolution barrier imposed by the intrinsic diffraction limit of optical microscopy (discovered by Ernst Abbe) obscures the inspection of subcellular components and activities occurring below ~200 nm. Near-field scanning optical microscope (NSOM)53 and total internal reflection fluorescence microscope (TIRFM) were established to break the diffraction limit, but their instrumental setup constrains the focus to near-surface subjects. Over the last decade, with the development in optics and nanomaterials, several revolutionary methods have successfully achieved the diffraction-unlimited scales by far-field optical imaging.54–56
In general, the mechanisms of mainstream super-resolution microscopies can be divided into two major groups: patterned illumination-based imaging, such as stimulated emission depletion (STED) microscopy and structured-illumination microscopy (SIM); single-molecule localization-based imaging, such as stochastic optical reconstruction microscopy (STORM) and photoactivated localization microscopy (PALM). Detailed theories and instrumentations of these techniques are illustrated in other in-depth reviews.57, 58 Admittedly, to quantitatively describe the molecular events in a single live cell the spatial resolution needs to improve. The temporal resolution is another imperative hurdle that remains to be overcome. To some extent, a better spatial resolution, necessitating more emitted and collected photons from each pixel/voxel, is often at the cost of imaging speed. As illustrated above, the real-time events related to the genomic and proteomic kinetics mostly happen at the millisecond level, while at least hundreds of milliseconds are needed to obtain one super-resolution image for a mammalian cell in the range between 10–100 µm.59 The trade-off between spatial and temporal resolutions needs to be well-balanced. For example, the interferometric PALM (iPALM) has an outstanding spatial resolution of around 10 nm, but it requires 20,000–100,000 frames (> 1 min) to reconstruct one image, thus making it quite powerful in resolving static finer structures of the cellular components rather than dynamic monitoring.60 Key parameters related to the spatiotemporal resolutions of recently commercialized super-resolution microscopes are listed in Table 1.
TABLE 1.
Trade-off between spatial and temporal resolutions
| Achieved | Key limitation | In the market* | ||
|---|---|---|---|---|
| Spatial resolution | Temporal resolution | |||
| STED61, 62 | 20 nm (lateral) 100 nm (axial) |
0.04 s (2.5 × 1.8 µm) | Point-scanning mode | Leica |
| STORM63, 64 | 10 nm (lateral) 20 nm (axial) |
0.5 s (31 × 31 µm) | Photoswitchable probe | Nikon, Leica |
| SIM65, 66 | 40 nm (lateral) 100 nm (axial) |
0.3 s (32 × 32 µm) | Reconstruction algorithm | Nikon, Zeiss |
till manuscript preparation
To further improve the temporal resolution of high-resolution imaging, in addition to developing better fluorophores with higher quantum efficiency, the response frequency and photon collection efficiency of detectors are among the primary components that can be further improved despite their significant advances. Electron-multiplying CCD (EMCCD) and intensified CCD (ICCD) are some of the devices presently used to amplify the signal gain generated from photoelectrons thousands of times, hence conducive to fast-speed super-resolution imaging.64, 65 Given the advances, the prospects for the next generation of super-resolution microscopy are anticipated to revolutionize our understanding of the dynamic cell structures and behaviors.
Single-molecule systems for quantifying dynamics
The ability of single-molecule techniques to provide real-time information on intracellular dynamics is an unparalleled advantage. Methods such as Fluorescence Recovery After Photobleaching (FRAP), Fluorescence Correlation Spectroscopy (FCS) and Fluorescence Lifetime Imaging Microscopy (FLIM) are several of the key techniques that enable single-molecule sensitivity. The principle of FRAP is based on temporarily bleaching the local fluorophores by high-energy illumination followed by recording the in situ recovery rate of fluorescence intensity, from which molecular diffusion can be quantified.67 FRAP is quite useful for biological studies related to cell membrane diffusion and protein interactions occurring at the timescale of seconds (Figure 3).68, 69 However, a considerable number of molecular dynamics in live cells takes place within the time domain ranging from microseconds to milliseconds, which is beyond the scope of normal FRAP detection. FCS and fluorescence cross-correlation spectroscopy (FCCS), are also popular approaches (Figure 4) with potential to monitor the dynamic motion of biomolecules at single-molecule resolution.70–73 In FCS a femtoliter volume of the sample is probed to provide microsecond temporal resolution using single photon avalanche photodiode detectors (SPAD). The fluorescence fluctuation profile thus measured could be used to track the dynamic nature of proteins and other molecules with autocorrelation (in FCS) or cross-correlation (in FCCS) functions. By fitting with appropriate mathematical models, a number of parameters related to the number of molecules, hydrodynamic radius and kinetics can be determined. Extracted from the same fluorescence fluctuation profile, molecular stoichiometry can be further determined based on single-molecule brightness using Photon Counting Histogram (PCH).74, 75 Similarly, dark-field microscopy equipped with the analytical ability to monitor and quantify single-molecule dynamics without fluorescent labels can also be used for motion tracking (Figure 8c).76
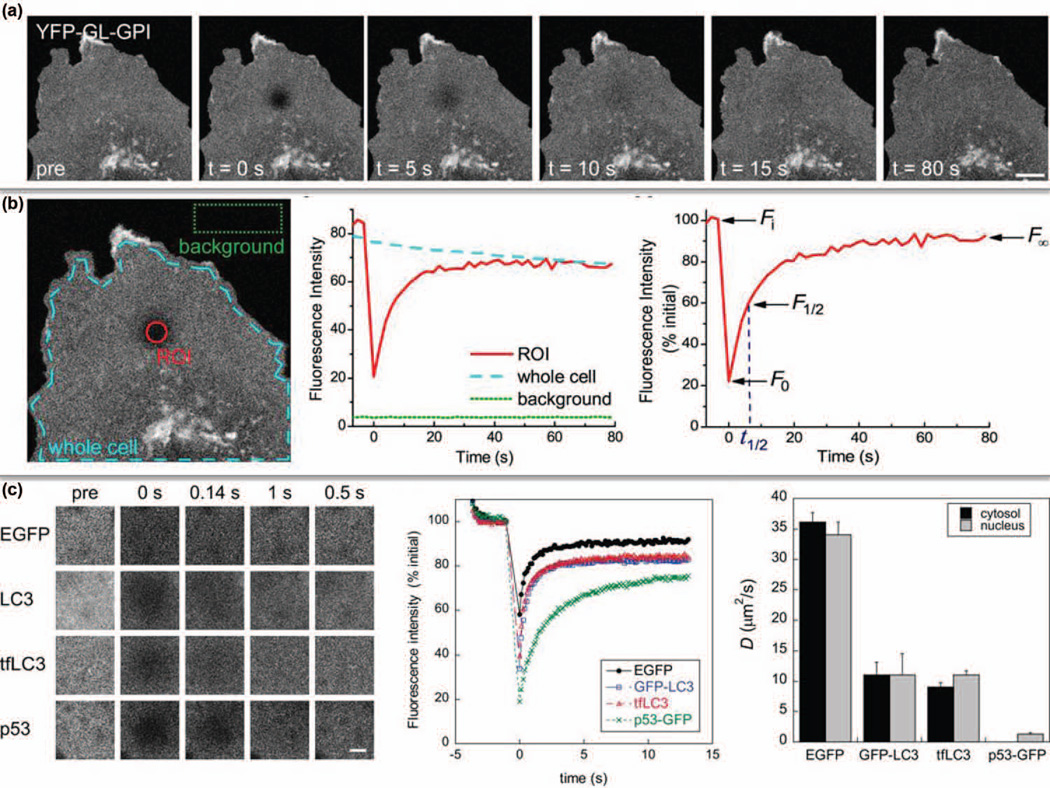
FIGURE 3.
FRAP for measuring molecular dynamics inside live single cells. (a) Recombinant YFP-GL-GPI plasmid was transfected into COS-7 cells. After photobleaching, the fluorescence recovery profile was recorded for 80 s. (b) Recovery curves of three regions are presented. A standard FRAP curve contains several critical points (right panel): initial intensity pre-bleach (Fi), starting point for post-bleach (F0), half maximal fluorescence recovery (F1/2), and ultimate recovered fluorescence (F∞). (c) Effective diffusion coefficients of EGFP, LC3, tfLC3 and p53 in live COS-7 cells were obtained. (Reprinted with permission from Ref 69. Copyright 2012 John Wiley & Sons, Inc.)
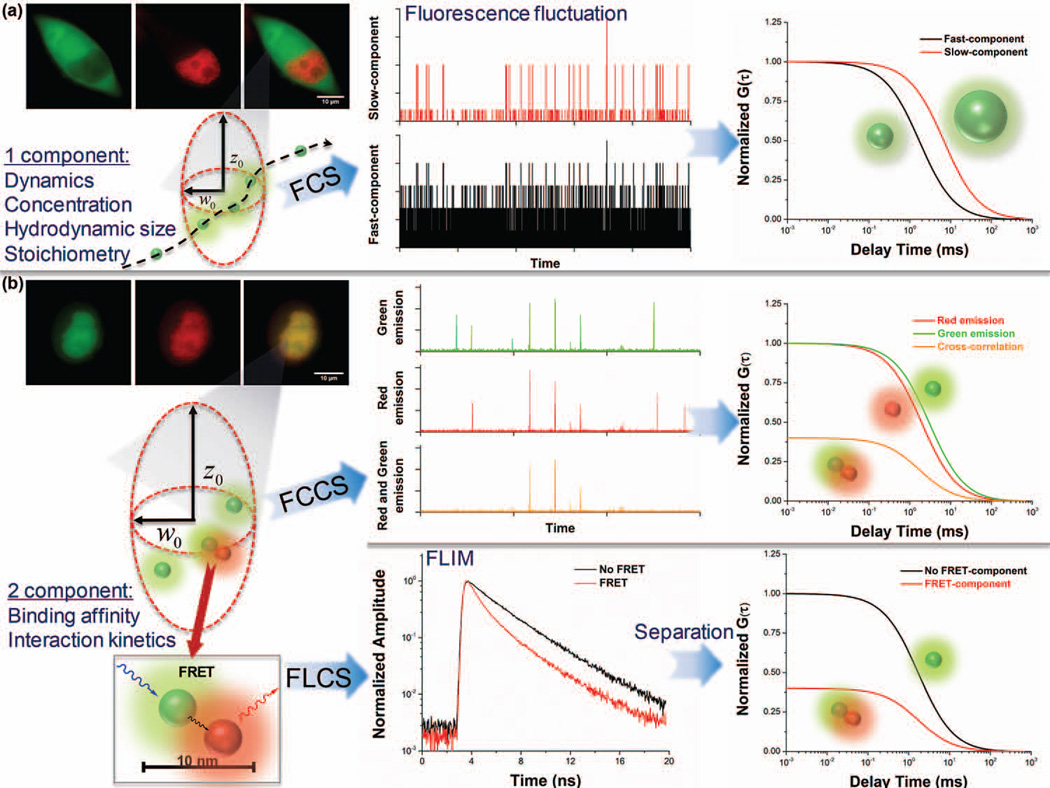
FIGURE 4.
FCS techniques for single cell analysis. (a) For fluorescent molecules without interaction, one-component FCS can be used to determine molecular dynamics, number, size, and stoichiometry. (b) For molecules with higher degrees of interaction, two-component FCCS or FLCS (in combination with FRET) can be applied. In FCCS, the association (cross-correlation) is determined when signals from different molecules are simultaneously detected within a diffraction limited spot. In FLCS, molecules with different fluorescence lifetimes can be separated, for example, distinguishing the FRET molecules from non-FRET ones.
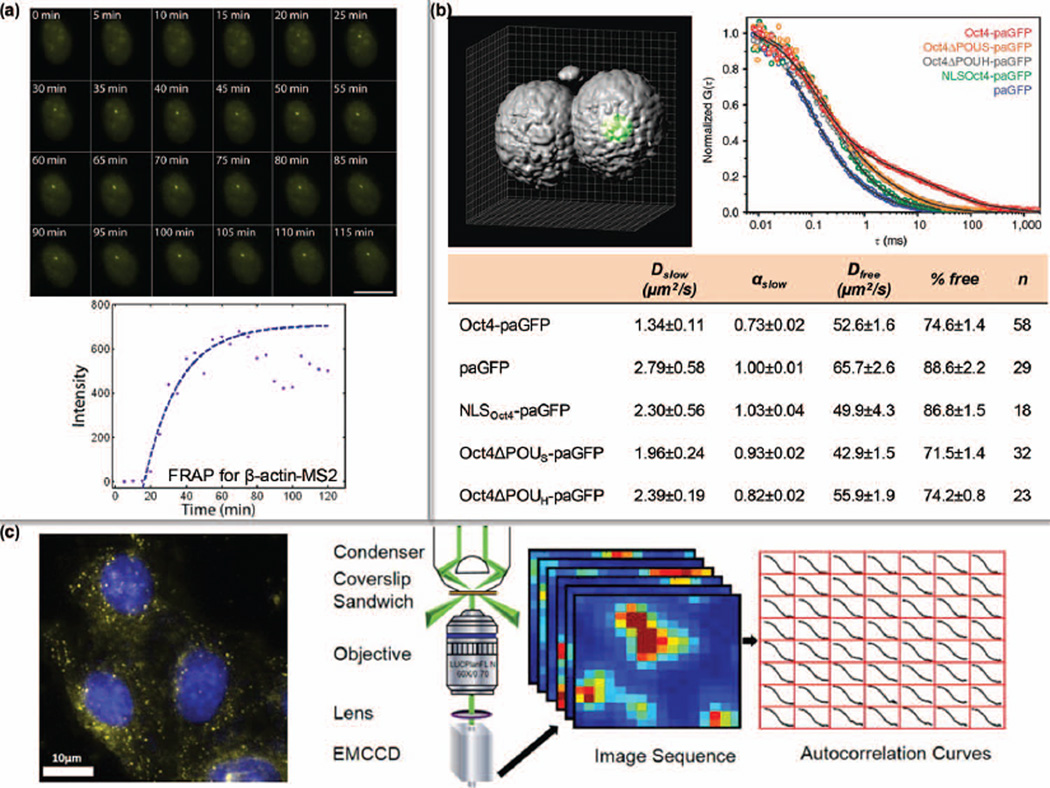
FIGURE 8.
Single-molecule dynamics inside cells. (a) The rate of mRNA transcription was quantified by MS2-labeling and FRAP. (Reprinted with permission from Ref 110. Copyright 2010 The Company of Biologists Ltd.) (b) The diffusion property of Oct4 variants in mouse embryos was profiled by FCS. D: diffusion coefficient; α: degree of anomalous diffusion; % free: percentage of the free component. (Reprinted with permission from Ref 114. Copyright 2013 Nature Publishing Group) (c) Dark-field illumination-based scattering correlation spectroscopy (DFSCS) enables the monitoring of intracellular dynamics of GNPs. (Reprinted with permission from Ref 76. Copyright 2014 American Chemical Society)
Depending upon the high temporal resolution and ultrashort pulsed laser of advanced single-molecule detection systems, fluorescence lifetime-based evaluations are becoming popular. Fluorescence lifetime describes the average time the fluorophore stays at the excited state prior to emitting the first photon, and is found to be independent on molecular concentrations but sensitive to physicochemical factors of the surrounding environment.77 Fluorescence lifetime has been used to assess the real-time status of intracellular microenvironment (e.g. pH and metabolites).78, 79 Some FLIM applications integrates Förster resonance energy transfer (FRET) to probe molecular interactions, where FRET is defined as the non-radiative energy transfer between appropriate fluorescent donor and acceptor through dipole-dipole coupling within 10 nm. FLIM-based FRET (FLIM-FRET) depends on the reduction in the fluorescence lifetime of donors upon energy transfer, and is a robust way to obtain quantitative information on spatial interactions.80–82 FCS, FLIM and FRET measurements constitute the core of single-molecule tools to probe intracellular events, and have given rise to a series of advanced derivatives such as fluorescence lifetime cross-correlation spectroscopy (FLCS).71, 83,84 In the future one can expect lable-free techniques to monitor biomolecule dynamics.
For both super-resolution microscopes and single-molecule systems, an unavoidable concern is phototoxicity, especially in FRAP experiments. A photon flux as low as 0.2 W/cm2 for imaging green fluorescent proteins (GFP) can irreversibly change the genetic activity, cell metabolism and finer structures.85 Hence, pulsed laser (e.g. in FCS and FLIM) and multi-photon excitation (e.g. in super-resolution microscopy) are suggested to be optimal for optical interrogation. The use of pulsed lasers in single cell studies, in contrast to continuous laser, permits a much lower excitation rate to be incident on samples so as to reduce the probability of photodamage and photobleaching. However, even though single-molecule tools such as FCS can be applied with ultrashort pulsed laser, the point-based measurement requires a considerable number of collection points and time from each cell, which might be detrimental to cell physiology. When multi-photon excitation is used, very limited cross-section (σ, in the order of 10−50 cm4s/photon for two-photon absorption) can be obtained to greatly reduce the unnecessary stimulation of the out-of-focus area.77 Taking into account the pros and cons of optical approaches, it is thus critical to choose the best fitted methodology and platform to probe different intracellular contents.
SELECTED APPLICATIONS IN SINGLE CELL PROBING
Characterization of DNA and chromatin contents
DNA is the basic building block that encodes genetic information in a species-conserved manner. The genetic information encrypted in DNA can be transmitted in two directions: the information is horizontally passed on to daughter cells through replication; or the template is hierarchically transmitted to form downstream proteins through transcription and translation. Nearly all of the physiological and pathological conditions are determined by these activities occurring in each single cell. Hence, quantitative assessment of DNA and chromatin related contents would pave the way for elucidating higher-order life phenomena.
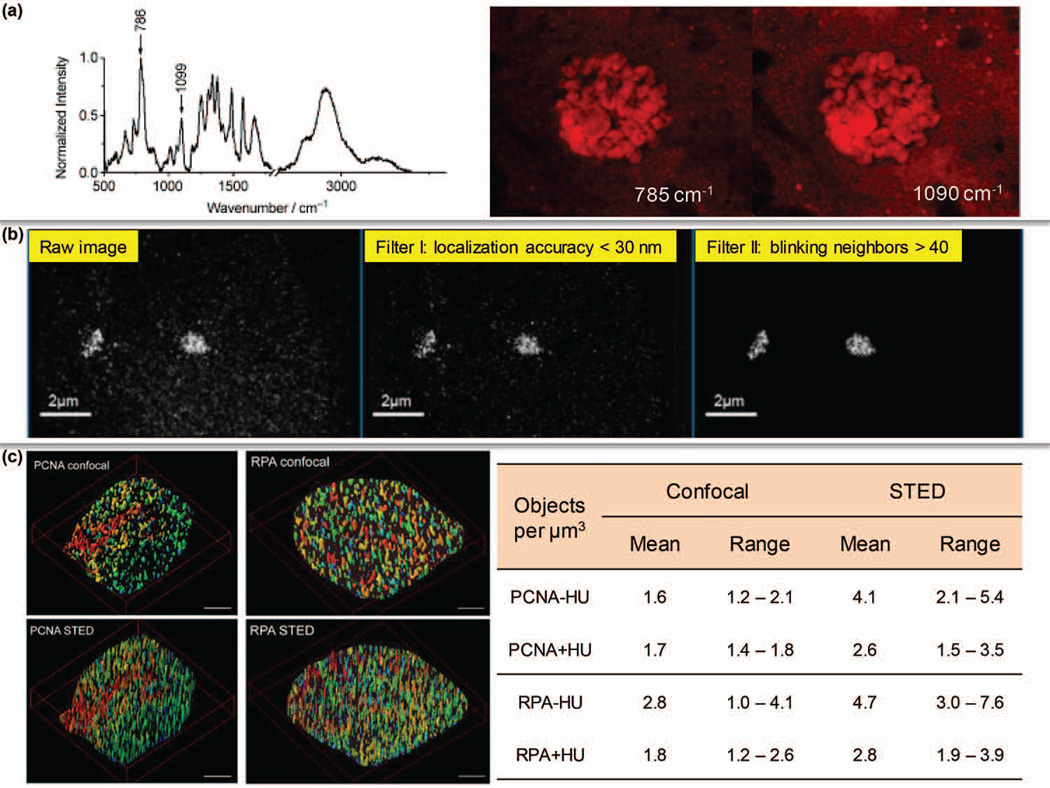
Currently, most of the single cell DNA related studies are driven by gene-specific detection rather than global quantification since it is uncommon for ensemble DNA contents to significantly alter even under pathological conditions. However, some attempts were made to assess the amount of DNA inside single cells with label-free imaging. By identifying the Raman peaks around 785 cm−1 and 1090 cm−1 as fingerprints, DNA has been quantitatively mapped in live cells by SRS to differentiate cell phases (Figure 5a).86
FIGURE 5.
Single cell studies at the DNA and chromatin levels. (a) Label-free SRS imaging of nucleic acids in live cells. (Reprinted with permission from Ref 86. Copyright 2012 John Wiley & Sons, Inc.) (b) Spectral Precision Distance/Position Determination Microscopy (SPDM) was applied to map the nanostructure of centromere 9 with an accuracy of 10–20 nm. (Reprinted with permission from Ref 87. Copyright 2010 MDPI AG, Basel, Switzerland) (c) Super-resolution imaging of DNA replication factories based on PCNA and RPA. HU: hydroxyurea treatment. (Reprinted with permission from Ref 92. Copyright 2009 BioMed Central Ltd)
The diffraction-unlimited precision of super-resolution microscopy also allows us to quantify chromatin contents. After the whole human genome and functional elements are mapped, the next challenge is to characterize the genomic landscape of natural chromatin where DNA is intertwined with histones and other factors. Applying fluorescence hybridization and super-resolution localization microscopy, a labeled locus can be imaged at 10–20 nm accuracy (e.g. centromeres as shown in Figure 5b).87, 88 Further, considerable efforts have also been devoted to visualizing the structure of chromatin or certain regions in single nucleus with quantifiable information.89–91 It is well-known that DNA replication takes place within specific machineries with the involvement of two major proteins – proliferating cell nuclear antigen (PCNA) and replication protein A (RPA). By labeling PCNA and RPA, STED microscopy successfully revealed that a single DNA replication factory is approximately 150 nm in size and up to 1,400 such factories could exist in an early S-phase nucleus (Figure 5c).92
Based on these achievements, it is anticipated that the dynamic process of DNA replication can be soon deciphered in live cells. Future efforts will also focus on evaluating 3D interaction of chromatin elements, drug targeting effects and real-time in situ gene regulation. In pursuit of these aims, the availability of single cell-sequencing will substantially complement the observations from phenotypic analysis.
Single mRNA counting
The number of transcripts serves as a direct indicator of the expression of specific genes. By labeling different mRNAs with fluorophores in a “barcode” pattern and applying STORM microscopy, 32 different mRNAs were simultaneously quantified in a single Saccharomyes cerevisiae cell.93 To date nanomaterial-based MB has experienced drastic development and is extensively applied for quantitative mRNA imaging in single cells with superior sensitivity and specificity (Figure 6a).94–97 An interesting combination was to couple a nanoneedle with MB probes to count single copies of mRNAs inside living HeLa cells;33 Other than hybridization methods, mRNA can also be labeled via MS2 system, a recognition mechanism derived from bacteriophage MS2. Upon inserting the MS2 sequence into the gene of interest, its transcribed mRNA would contain a unique stem-loop structure that can be recognized by MS2 coat protein (MCP). By combining MCP with FCS, the β-actin mRNA was quantified at the nanomolar level in single cells.98
In human cells, the number of genes is much lower than the number of identified mRNAs and proteins because of alternative splicing. However, very limited tools are available to evaluate the dynamic quantity and distribution of spliced transcripts in a single cell. Recently, utilizing the coupling between nanoplasmonic dimers of GNPs, three splice variants of the breast cancer susceptibility gene, BRCA1, were counted at single-copy resolution in living cells by hyperspectral dark-field imaging (Figure 6b).10 A key limitation of hyperspectral dark-field microscopy is in the speed since it relies on spectroscopy-based collection. Instrumentation that can fast select spectral windows is expected to increase the data acquisition rate. Future developments of counting single mRNAs could lie in multiplex detection of different mRNAs, identification of genetic mutation at the transcript level, and coordination with single protein counting to unlock the translation machinery.
Labeling-dependent protein profiling
It is estimated that more than 100,000 different types of proteins exist in the human body. Except water, half of the human body is composed of proteins, and nearly all the cellular activities are carried out with the involvement of proteins. Correctly forming a protein molecule is the final step in the vertical transmission of genetic information. Techniques for determining the protein content and distribution inside a single cell would greatly advance modern biomedicine.
Quantification of proteins has been demonstrated due to the excellent resolution and sensitivity of advanced optical microscopies.99, 100 For instance, the plasma membrane located HER2 tyrosine kinase protein (human epidermal growth factor receptor 2), a diagnostic biomarker of breast cancers, was found to form clusters with a mean size of 67 nm by super-resolution microscopy (Figure 7a),101 which was also characterized by scattering spectroscopy with a GNPs coupling-based plasmon resonance strategy (Figure 7b).102 Such cross-validation greatly enhances the credibility of single cell studies, and might lead to a better stratification of patients in clinics. A detailed protocol for quantifying the number, size and density of protein clusters within subcellular compartments based on PALM was recently documented as well.103 Using gold nanorods conjugated with monoclonal HER2 antibody-Herceptin® (H-GNRs), the HER2 mediated intracellular transition was tracked in single SKBR3 cells by FCS. At different time points, the concentration of H-GNRs in different cell organelles was precisely determined (Figure 7c).12 In another study, employing FLCS to simultaneously monitor the quantities of epidermal growth factor receptor and antagonist antibody, their real-time association was quantitatively evaluated in HEK293 cells with a single blue laser excitation.71
In oncology, cancer stem cells (CSCs) are thought to play a crucial role in the initiation and recurrence of cancers. An innovative methodology comprising of SERS and hyperspectral dark-field microscopy was developed to quantify the surface CD44/CD24 ratio in living single cells by growing DNA-GNP-based network on the cell membrane, facilitating the identification of breast CSCs (Figure 7d).9
Although a range of high-resolution tools to analyze proteins in single cells exist, most of research findings are still from fixed cells. Live single cell analysis will provide valuable insights to better interpret protein quantity and distribution in different cell types.
Dissecting intracellular dynamics
Dynamics of transcription
Several quantitative models have been established to characterize transcription via single-molecule tools and nano-manipulation, but mostly in vitro.104–106 Considering the multi-component involvement and intricate regulation of transcription, information obtained from live cells will become the wave of the future and might even be the norm. For instance, it has been reported that the transcription dynamics involves the clustering of RNA polymerase II (RNAP II); with time-correlated PALM (tcPALM) the real-time clustering rate of RNAP II was characterized and the average lifetime for a transient cluster was determined to be 5.1 ± 0.4 seconds, while external stimuli could have a significant impact on the clustering during genetic regulation.107
In eukaryotic cells, transcription is controlled by transcription factors (TFs), a group of proteins capable of binding to specific DNA sequences and recruiting RNAP. Hence, the process of TFs assembling at the target sites becomes a rate-limit step in transcription. In a recent study, single-molecule tools were implemented in embryonic stem cells to investigate the intracellular kinetics of Sox2.108 A “trial-and-error” model was proposed and it was determined that Sox2 would experience 84–97 events of free diffusion (3.3–3.7 second), interspersed with transient collision (0.75–0.9 second), before identifying and anchoring onto the target sequence (12.0–14.6 second). A follow-up study also uncovered the distinct searching modes of c-Myc and P-TEFb.109 Once RNAP is recruited by TFs, the next step is mRNA elongation. By engineering β-actin gene with MS2 system, the progression rate for RNAP II was determined to be 3.3 kb/minute in human O2OS cells by FRAP (Figure 8a).110 With novel nanoscopic tools, critical dynamic properties implicated in single cell transcription could be further discovered.
Dynamics of nuclear proteins
Over the past few years, single-molecule techniques have considerably contributed to the elucidation of dynamics and heterogeneity of nuclear proteins, which can be well-exemplified by the evaluation of interaction between chromatin and DNA methyltransferase 1 (DNMT1) in single nucleus. DNMT1 is the major enzyme to catalyze the maintenance DNA methylation that is critical for genomic activity and stability. Utilizing FRAP the loading dynamics of DNMT1 onto chromatin was profiled and found to be associated with the DNA replication machinery.111, 112 In addition, by applying 3D SIM, the coupling model of DNMT1 on chromatin was refined and two alternatives were proposed to access hemi-methylated DNA: a PCNA-binding domain-dependent interaction in early S-phase (residence time ≤ 10 seconds), and a targeting sequence domain-dependent interaction (residence time ~ 22 seconds), which provides a new perspective in viewing the function of nuclear proteins under physiological circumstances.113 For proteins of millisecond mobility, FCS has been used to quantify their dynamics and unravel the behavioral modes.114, 115 As shown in Figure 8b, by constructing different fusion variants, the interplay between DNA and Oct4 was comprehensively investigated in two-cell mouse embryos.
Dynamics of molecular interactions
Oftentimes proteins do not stay in the monomeric state. It is thus of interest to assess their associations, which necessitates the development of methods to determine protein-DNA interactions and kinetics of oligomerization. For biomolecules labeled with fluorophores, PCH can be applied to define the stoichiometry of homogeneous oligomers. For example, upon measuring the brightness of monomeric GFP as control, it was found that toll-like receptor 9 with a GFP tag (TLR9-GFP) binds to DNA with a 2:1 stoichiometric ratio.116 On the other hand, FCCS and FRET are powerful tools to quantitatively resolve a heterogeneous complex, but with respective strengths: FCCS can determine the binding affinity while FRET is superior in the assessment of inter-molecule distance. Based on these technical features, the mitogen activated protein kinase (MAPK) cascade in live single yeast was mapped in relation to membrane association and regulation by the Ste5 scaffold protein.117 It is known that activation of MAPK pathway will initiate DNA replication and cell proliferation. Considering the fact that chromatin contains a plethora of epigenetic modifications on histones, to conserve this set of epigenetic information the newly synthesized chromatin has to establish the same pattern as its parental template. From FLIM-FRET it was confirmed that PCNA can interact with histone acetyltransferases SAS-I and Rtt109p (distance ~6–6.2 nm) during S-phase to maintain the epigenetic memory.118 Collectively, the explosion of advances in optics and nanoscale interrogation have empowered biology not only to scrutinize the intracellular world that was long-unknown, but also to acquire the dynamic connections with unprecedented accuracy.
Label-free fingerprinting metabolites, peptides and small molecules
Given the complexity of a single cell, a number of various biomolecules are involved and participate in diverse functions, serving as structural scaffolds, signaling messengers, redox substrates, enzyme co-factors, energy carriers, regulatory components and indicative biomarkers. Conventional approaches can only resolve a fraction of the whole puzzle due to the limited means of labeling. In comparison, the enriched tools for label-free molecular fingerprinting can provide a convenient measure of cell identity and metabolism.
Metabolites and small molecules are excellent targets for label-free single cell probing. Single cell SERS detection can probe abundant chemical bonds if a biocompatible Raman-enhancing substrate is introduced.119, 120 Several strategies are developed to tackle this. The first is to grow the cells directly on a SERS substrate. Using this strategy, cancer cells in 2D and 3D configuration at different stages of tumorigenesis could be readily classified with almost 100% accuracy.121 The second is to deliver nanomaterials, such as GNPs, to desired subcellular compartments. GNPs functionalized with nuclear localization signaling peptide can efficiently penetrate nuclear membranes for in situ biochemical sensing.122 A more intriguing approach constituting the intracellular growth of gold nanoislands to map the reduction of toxic chromate have also been illustrated and the detection of trivalent and hexavalent Chromium was possible by SERS in singe living cells (Figure 9b).123 The fourth is to compose specialized nanodevices, for example, the use of nanoneedles for SERS detection.124 With the carbon nanotube-based nanoneedle coated with 20-nm GNPs, 1 pM of glycine can be detected inside cells.125
Until today a dominant portion of single cell studies using CARS and SRS have focused on evaluating lipids since their enriched C-H bond produces a superb Raman contrast around 2850 cm−1.126 Upon identifying and quantifying this vibrational stretch in single cells, understanding lipid synthesis and movement,127 neuronal demyelination and remyelination,128 reagents targeting lipid metabolism and cholesterol storage129 have been explored. CARS was also used to monitor the cellular uptake of poly(lactic-co-glycolic acid) (PLGA) nanoparticle that is a degradable drug carrier and has a characteristic CH3 signal at 2940 cm−1 (Figure 9d).130
Besides Raman scattering, autofluorescence emitted by intrinsic molecules of cells also contains useful information. By classifying flavin species into three major groups (flavin mononucleotide (FMN), free flavin adenine dinucleotide (FAD) and bound FAD) based on different fluorescence lifetimes under a blue excitation, an integrated lifetime measured by FLIM system can be mathematically deconvoluted to differentiate malignant breast cancer cells from non-malignant cells.78
As mentioned above, MSI is an effective label-free technique for peptide mapping, and particularly helpful in cell classification. A characteristic m/z value of the target generates the fingerprint for MSI to identify cancer cells, for example, an m/z value of 9,744 for proteasome activator subunit 1 (PSME1, overexpressed in ovarian tumors)131 and an m/z value of 8,404 for cysteine-rich intestinal protein 1 (CRIP1, overexpressed in HER2-positive breast cancers).132 Besides those two, MSI is employed in a variety of other malignancies,133, 134 contributing to early diagnosis and prognosis monitoring. Moreover, due to its multiplex capability and the data obtained, MSI appears to be a potent supplement in drug discovery and single cell metabolomics.135–137
The number of nanotools and methods for label-free assays is rapidly increasing. However, only a small fraction of these methods are applicable to probe at the single cell scale, while a majority of the approaches are performed with cell lysates or bulky samples under limited conditions. Customizing platforms for single-molecule sensing in intact cells is still very challenging. Another immediate goal is to establish platform-oriented fingerprint libraries of specific molecules for high-throughput screening.
CONCLUSION
During the historical transition from superstitious astrology and alchemy to modern physics and chemistry, quantitative understanding of mechanical forces, electricity, light, chemical elements and reaction kinetics have resulted in revolutionary advances. Today, to understand a single cell in modern biology, quantitative measurements of various biomolecules and intracellular events hold the key. In conventional molecular biology, methods such as DNA sequencing, RNA polymerase chain reaction and protein western blot rely on the ensemble observations from a population of cells, overlooking the cellular heterogeneity and intermediate dynamics. Therefore, dissecting those components at the subcellular level provides a new dimension in interpreting life and diseases.
Admittedly, a number of facts with regard to optical methods and nanomaterials need further optimization. First, to achieve high-throughput readout from most single cell studies is a significant challenge since the trade-off between speed and precision limits the number of assessed cells. Combining single cell tools with flow cytometry could be a possible solution. Second, although we have obtained a large number of data from cultured cells, it is more crucial to understand the behavior of a cell within its natural growth environment. Thus 3D ex vivo models could be used as an intermediate step. Third, nowadays fully biocompatible nanomaterials are still scarce. Therefore biomolecule-based nanomaterials, such as peptide nanosensor,79 xeno-nucleic acid138 and DNA origami,139 might be future options. Last but not the least, to correlate the micro-scale genotype with the macro-scale phenotype requires a combinatorial elucidation. With the groundbreaking development in single cell-sequencing techniques, the hope of obtaining informative DNA and RNA sequences from single cells is becoming a reality.140–142 Further efforts in understanding the function of genes and their regulatory contribution in a loci-specific manner can also be expected.
The biggest challenge brings about the best solutions as well. Single cell study is still not commonplace in biology and only a fraction of the possibilities, if any have been explored. Opportunities abound in enhancing our knowledge of embryogenesis, development, aging, chronic disease (e.g. diabetes) and cancer – solution to these questions exclusively hinges upon how much we know about the function of a single cell ~1 picoliter in volume where an unknown number of reactions (possibly, millions) occur in the timescale ranging from microseconds or lower to hours/days.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge funding from the National Science Foundation (Award# 1249315), National Institute of Health (Award# CA157395-01), and the Center for Food Safety Engineering (USDA-ARS-Purdue University) grant.
REFERENCES
- 1.Crick F. Central dogma of molecular biology. Nature. 1970;227:561–563. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- 2.Bustamante C, Cheng W, Meija YX. Revisiting the Central Dogma One Molecule at a Time. Cell. 2011;144:480–497. doi: 10.1016/j.cell.2011.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li GW, Xie XS. Central dogma at the single-molecule level in living cells. Nature. 2011;475:308–315. doi: 10.1038/nature10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Milo R. What is the total number of protein molecules per cell volume? A call to rethink some published values. Bioessays. 2013;35:1050–1055. doi: 10.1002/bies.201300066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orjalo A, Jr, Johansson HE, Ruth JL. Stellaris™ fluorescence in situhybridization (FISH) probes: a powerful tool for mRNA detection. Nature Methods. 2011:1. [Google Scholar]
- 6.Ungureanu F, Wasserberg D, Yang N, Verdoold R, Kooyman RPH. Immunosensing by colorimetric darkfield microscopy of individual gold nanoparticle-conjugates. Sensors and Actuators B-Chemical. 2010;150:529–536. [Google Scholar]
- 7.Verdoold R, Gill R, Ungureanu F, Molenaar R, Kooyman RPN. Femtomolar DNA detection by parallel colorimetric darkfield microscopy of functionalized gold nanoparticles. Biosensors & Bioelectronics. 2011;27:77–81. doi: 10.1016/j.bios.2011.06.019. [DOI] [PubMed] [Google Scholar]
- 8.Rosman C, Pierrat S, Henkel A, Tarantola M, Schneider D, Sunnick E, Janshoff A, Sonnichsen C. A New Approach to Assess Gold Nanoparticle Uptake by Mammalian Cells: Combining Optical Dark-Field and Transmission Electron Microscopy. Small. 2012;8:3683–3690. doi: 10.1002/smll.201200853. [DOI] [PubMed] [Google Scholar]
- 9.Lee K, Drachev VP, Irudayaraj J. DNA-gold nanoparticle reversible networks grown on cell surface marker sites: application in diagnostics. ACS Nano. 2011;5:2109–2117. doi: 10.1021/nn1030862. [DOI] [PubMed] [Google Scholar]
- 10.Lee K, Cui Y, Lee LP, Irudayaraj J. Quantitative imaging of single mRNA splice variants in living cells. Nat Nanotechnol. 2014;9:474–480. doi: 10.1038/nnano.2014.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fairbairn N, Christofidou A, Kanaras AG, Newman TA, Muskens OL. Hyperspectral darkfield microscopy of single hollow gold nanoparticles for biomedical applications. Physical Chemistry Chemical Physics. 2013;15:4163–4168. doi: 10.1039/c2cp43162a. [DOI] [PubMed] [Google Scholar]
- 12.Chen JJ, Irudayaraj J. Quantitative Investigation of Compartmentalized Dynamics of ErbB2 Targeting Gold Nanorods in Live Cells by Single Molecule Spectroscopy. Acs Nano. 2009;3:4071–4079. doi: 10.1021/nn900743v. [DOI] [PubMed] [Google Scholar]
- 13.Wang C, Chen J, Talavage T, Irudayaraj J. Gold nanorod/Fe3O4 nanoparticle "nano-pearl-necklaces" for simultaneous targeting, dual-mode imaging, and photothermal ablation of cancer cells. Angew Chem Int Ed Engl. 2009;48:2759–2763. doi: 10.1002/anie.200805282. [DOI] [PubMed] [Google Scholar]
- 14.Yu C, Nakshatri H, Irudayaraj J. Identity profiling of cell surface markers by multiplex gold nanorod probes. Nano Lett. 2007;7:2300–2306. doi: 10.1021/nl070894m. [DOI] [PubMed] [Google Scholar]
- 15.Ando J, Fujita K, Smith NI, Kawata S. Dynamic SERS Imaging of Cellular Transport Pathways with Endocytosed Gold Nanoparticles. Nano Letters. 2011;11:5344–5348. doi: 10.1021/nl202877r. [DOI] [PubMed] [Google Scholar]
- 16.Chan WC, Nie S. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 17.Dahan M, Levi S, Luccardini C, Rostaing P, Riveau B, Triller A. Diffusion dynamics of glycine receptors revealed by single-quantum dot tracking. Science. 2003;302:442–445. doi: 10.1126/science.1088525. [DOI] [PubMed] [Google Scholar]
- 18.Ballou B, Lagerholm BC, Ernst LA, Bruchez MP, Waggoner AS. Noninvasive imaging of quantum dots in mice. Bioconjug Chem. 2004;15:79–86. doi: 10.1021/bc034153y. [DOI] [PubMed] [Google Scholar]
- 19.Watanabe TM, Fukui S, Jin T, Fujii F, Yanagida T. Real-Time Nanoscopy by Using Blinking Enhanced Quantum Dots. Biophysical Journal. 2010;99:L50–L52. doi: 10.1016/j.bpj.2010.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Fruhwirth G, Cai E, Ng T, Selvin PR. 3D Super-Resolution Imaging with Blinking Quantum Dots. Nano Letters. 2013;13:5233–5241. doi: 10.1021/nl4026665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim S, Lim YT, Soltesz EG, De Grand AM, Lee J, Nakayama A, Parker JA, Mihaljevic T, Laurence RG, Dor DM, et al. Near-infrared fluorescent type II quantum dots for sentinel lymph node mapping. Nat Biotechnol. 2004;22:93–97. doi: 10.1038/nbt920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pelley JL, Daar AS, Saner MA. State of academic knowledge on toxicity and biological fate of quantum dots. Toxicol Sci. 2009;112:276–296. doi: 10.1093/toxsci/kfp188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anc MJ, Pickett NL, Gresty NC, Harris JA, Mishra KC. Progress in Non-Cd Quantum Dot Development for Lighting Applications. Ecs Journal of Solid State Science and Technology. 2013;2:R3071–R3082. [Google Scholar]
- 24.Dulkeith E, Morteani AC, Niedereichholz T, Klar TA, Feldmann J, Levi SA, van Veggel FC, Reinhoudt DN, Moller M, Gittins DI. Fluorescence quenching of dye molecules near gold nanoparticles: radiative and nonradiative effects. Phys Rev Lett. 2002;89:203002. doi: 10.1103/PhysRevLett.89.203002. [DOI] [PubMed] [Google Scholar]
- 25.Dulkeith E, Ringler M, Klar TA, Feldmann J, Javier AM, Parak WJ. Gold nanoparticles quench fluorescence by phase induced radiative rate suppression. Nano Letters. 2005;5:585–589. doi: 10.1021/nl0480969. [DOI] [PubMed] [Google Scholar]
- 26.Goel G, Kumar A, Puniya AK, Chen W, Singh K. Molecular beacon: a multitask probe. J Appl Microbiol. 2005;99:435–442. doi: 10.1111/j.1365-2672.2005.02663.x. [DOI] [PubMed] [Google Scholar]
- 27.Kasry A, Ardakani AA, Tulevski GS, Menges B, Copel M, Vyklicky L. Highly Efficient Fluorescence Quenching with Graphene. Journal of Physical Chemistry C. 2012;116:2858–2862. [Google Scholar]
- 28.Wang Y, Li ZH, Weber TJ, Hu DH, Lin CT, Li JH, Lin YH. In Situ Live Cell Sensing of Multiple Nucleotides Exploiting DNA/RNA Aptamers and Graphene Oxide Nanosheets. Analytical Chemistry. 2013;85:6775–6782. doi: 10.1021/ac400858g. [DOI] [PubMed] [Google Scholar]
- 29.Yang RH, Jin JY, Chen Y, Shao N, Kang HZ, Xiao Z, Tang ZW, Wu YR, Zhu Z, Tan WH. Carbon nanotube-quenched fluorescent oligonucleotides: Probes that fluoresce upon hybridization. Journal of the American Chemical Society. 2008;130:8351–8358. doi: 10.1021/ja800604z. [DOI] [PubMed] [Google Scholar]
- 30.Huang K, Marti AA. Recent trends in molecular beacon design and applications. Anal Bioanal Chem. 2012;402:3091–3102. doi: 10.1007/s00216-011-5570-6. [DOI] [PubMed] [Google Scholar]
- 31.Vo-Dinh T, Zhang Y. Single-cell monitoring using fiberoptic nanosensors. Wiley Interdisciplinary Reviews-Nanomedicine and Nanobiotechnology. 2011;3:79–85. doi: 10.1002/wnan.112. [DOI] [PubMed] [Google Scholar]
- 32.Gao Y, Longenbach T, Vitol EA, Orynbayeva Z, Friedman G, Gogotsi Y. One-dimensional nanoprobes for single-cell studies. Nanomedicine (Lond) 2014;9:153–168. doi: 10.2217/nnm.13.196. [DOI] [PubMed] [Google Scholar]
- 33.Kihara T, Yoshida N, Kitagawa T, Nakamura C, Nakamura N, Miyake J. Development of a novel method to detect intrinsic mRNA in a living cell by using a molecular beacon-immobilized nanoneedle. Biosensors & Bioelectronics. 2010;26:1449–1454. doi: 10.1016/j.bios.2010.07.079. [DOI] [PubMed] [Google Scholar]
- 34.Vo-Dinh T, Alarie JP, Cullum BM, Griffin GD. Antibody-based nanoprobe for measurement of a fluorescent analyte in a single cell. Nat Biotechnol. 2000;18:764–767. doi: 10.1038/77337. [DOI] [PubMed] [Google Scholar]
- 35.Kasili PM, Song JM, Vo-Dinh T. Optical sensor for the detection of caspase-9 activity in a single cell. Journal of the American Chemical Society. 2004;126:2799–2806. doi: 10.1021/ja037388t. [DOI] [PubMed] [Google Scholar]
- 36.Zheng XT, Hu WH, Wang HX, Yang HB, Zhou W, Li CM. Bifunctional electro-optical nanoprobe to real-time detect local biochemical processes in single cells. Biosensors & Bioelectronics. 2011;26:4484–4490. doi: 10.1016/j.bios.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 37.Nel A, Xia T, Madler L, Li N. Toxic potential of materials at the nanolevel. Science. 2006;311:622–627. doi: 10.1126/science.1114397. [DOI] [PubMed] [Google Scholar]
- 38.Fischer HC, Chan WCW. Nanotoxicity: the growing need for in vivo study. Current Opinion in Biotechnology. 2007;18:565–571. doi: 10.1016/j.copbio.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 39.Hezinger AF, Tessmar J, Gopferich A. Polymer coating of quantum dots--a powerful tool toward diagnostics and sensorics. Eur J Pharm Biopharm. 2008;68:138–152. doi: 10.1016/j.ejpb.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 40.Zhang F, Lees E, Amin F, Gil PR, Yang F, Mulvaney P, Parak WJ. Polymer-Coated Nanoparticles: A Universal Tool for Biolabelling Experiments. Small. 2011;7:3113–3127. doi: 10.1002/smll.201100608. [DOI] [PubMed] [Google Scholar]
- 41.Yu Y, Ramachandran PV, Wang MC. Shedding new light on lipid functions with CARS and SRS microscopy. Biochim Biophys Acta. 2014 doi: 10.1016/j.bbalip.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Manen HJ, Kraan YM, Roos D, Otto C. Single-cell Raman and fluorescence microscopy reveal the association of lipid bodies with phagosomes in leukocytes. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:10159–10164. doi: 10.1073/pnas.0502746102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang YL, Irudayaraj J. Surface-enhanced Raman spectroscopy at single-molecule scale and its implications in biology. Philosophical Transactions of the Royal Society B-Biological Sciences. 2013;368 doi: 10.1098/rstb.2012.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deckert-Gaudig T, Deckert V. Nanoscale structural analysis using tip-enhanced Raman spectroscopy. Current Opinion in Chemical Biology. 2011;15:719–724. doi: 10.1016/j.cbpa.2011.06.020. [DOI] [PubMed] [Google Scholar]
- 45.Evans CL, Xie XS. Coherent Anti-Stokes Raman Scattering Microscopy: Chemical Imaging for Biology and Medicine. Annual Review of Analytical Chemistry. 2008;1:883–909. doi: 10.1146/annurev.anchem.1.031207.112754. [DOI] [PubMed] [Google Scholar]
- 46.Min W, Freudiger CW, Lu SJ, Xie XS. Coherent Nonlinear Optical Imaging: Beyond Fluorescence Microscopy. Annual Review of Physical Chemistry, Vol 62. 2011;62:507–530. doi: 10.1146/annurev.physchem.012809.103512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Monici M. Cell and tissue autofluorescence research and diagnostic applications. Biotechnol Annu Rev. 2005;11:227–256. doi: 10.1016/S1387-2656(05)11007-2. [DOI] [PubMed] [Google Scholar]
- 48.Schober Y, Guenther S, Spengler B, Rompp A. Single cell matrix-assisted laser desorption/ionization mass spectrometry imaging. Anal Chem. 2012;84:6293–6297. doi: 10.1021/ac301337h. [DOI] [PubMed] [Google Scholar]
- 49.Passarelli MK, Ewing AG. Single-cell imaging mass spectrometry. Curr Opin Chem Biol. 2013;17:854–859. doi: 10.1016/j.cbpa.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Passarelli MK, Ewing AG, Winograd N. Single-cell lipidomics: characterizing and imaging lipids on the surface of individual Aplysia californica neurons with cluster secondary ion mass spectrometry. Anal Chem. 2013;85:2231–2238. doi: 10.1021/ac303038j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Penner-Hahn JE. Technologies for detecting metals in single cells. Met Ions Life Sci. 2013;12:15–40. doi: 10.1007/978-94-007-5561-1_2. [DOI] [PubMed] [Google Scholar]
- 52.Wang P, Slipchenko MN, Mitchell J, Yang C, Potma EO, Xu X, Cheng JX. Far-field Imaging of Non-fluorescent Species with Sub-diffraction Resolution. Nat Photonics. 2013;7:449–453. doi: 10.1038/nphoton.2013.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Lange F, Cambi A, Huijbens R, de Bakker B, Rensen W, Garcia-Parajo M, van Hulst N, Figdor CG. Cell biology beyond the diffraction limit: near-field scanning optical microscopy. Journal of Cell Science. 2001;114:4153–4160. doi: 10.1242/jcs.114.23.4153. [DOI] [PubMed] [Google Scholar]
- 54.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 55.Hell SW. Far-field optical nanoscopy. Science. 2007;316:1153–1158. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- 56.Huang B, Bates M, Zhuang X. Super-resolution fluorescence microscopy. Annu Rev Biochem. 2009;78:993–1016. doi: 10.1146/annurev.biochem.77.061906.092014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang B, Babcock H, Zhuang X. Breaking the diffraction barrier: super-resolution imaging of cells. Cell. 2010;143:1047–1058. doi: 10.1016/j.cell.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Requejo-Isidro J. Fluorescence nanoscopy. Methods and applications. J Chem Biol. 2013;6:97–120. doi: 10.1007/s12154-013-0096-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reece JB. Campbell biology : concepts and connections. (Eighth edition) [Google Scholar]
- 60.Shtengel G, Galbraith JA, Galbraith CG, Lippincott-Schwartz J, Gillette JM, Manley S, Sougrat R, Waterman CM, Kanchanawong P, Davidson MW, et al. Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proc Natl Acad Sci U S A. 2009;106:3125–3130. doi: 10.1073/pnas.0813131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Westphal V, Rizzoli SO, Lauterbach MA, Kamin D, Jahn R, Hell SW. Video-rate far-field optical nanoscopy dissects synaptic vesicle movement. Science. 2008;320:246–249. doi: 10.1126/science.1154228. [DOI] [PubMed] [Google Scholar]
- 62.Wildanger D, Medda R, Kastrup L, Hell SW. A compact STED microscope providing 3D nanoscale resolution. J Microsc. 2009;236:35–43. doi: 10.1111/j.1365-2818.2009.03188.x. [DOI] [PubMed] [Google Scholar]
- 63.Xu K, Babcock HP, Zhuang X. Dual-objective STORM reveals three-dimensional filament organization in the actin cytoskeleton. Nat Methods. 2012;9:185–188. doi: 10.1038/nmeth.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jones SA, Shim SH, He J, Zhuang X. Fast, three-dimensional super-resolution imaging of live cells. Nat Methods. 2011;8:499–508. doi: 10.1038/nmeth.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rego EH, Shao L, Macklin JJ, Winoto L, Johansson GA, Kamps-Hughes N, Davidson MW, Gustafsson MG. Nonlinear structured-illumination microscopy with a photoswitchable protein reveals cellular structures at 50-nm resolution. Proc Natl Acad Sci U S A. 2012;109:E135–E143. doi: 10.1073/pnas.1107547108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shao L, Isaac B, Uzawa S, Agard DA, Sedat JW, Gustafsson MG. I5S: wide-field light microscopy with 100-nm-scale resolution in three dimensions. Biophys J. 2008;94:4971–4983. doi: 10.1529/biophysj.107.120352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Axelrod D, Koppel DE, Schlessinger J, Elson E, Webb WW. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys J. 1976;16:1055–1069. doi: 10.1016/S0006-3495(76)85755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sprague BL, Pego RL, Stavreva DA, McNally JG. Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys J. 2004;86:3473–3495. doi: 10.1529/biophysj.103.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Day CA, Kraft LJ, Kang M, Kenworthy AK. Analysis of protein and lipid dynamics using confocal fluorescence recovery after photobleaching (FRAP) Curr Protoc Cytom. 2012;Chapter 2(Unit2):19. doi: 10.1002/0471142956.cy0219s62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dong C, Chowdhury B, Irudayaraj J. Probing site-exclusive binding of aqueous QDs and their organelle-dependent dynamics in live cells by single molecule spectroscopy. Analyst. 2013;138:2871–2876. doi: 10.1039/c3an36906d. [DOI] [PubMed] [Google Scholar]
- 71.Chen J, Irudayaraj J. Fluorescence lifetime cross correlation spectroscopy resolves EGFR and antagonist interaction in live cells. Anal Chem. 2010;82:6415–6421. doi: 10.1021/ac101236t. [DOI] [PubMed] [Google Scholar]
- 72.Wang Y, Chen J, Irudayaraj J. Nuclear targeting dynamics of gold nanoclusters for enhanced therapy of HER2+ breast cancer. ACS Nano. 2011;5:9718–9725. doi: 10.1021/nn2032177. [DOI] [PubMed] [Google Scholar]
- 73.Pack CG, Yukii H, Toh EA, Kudo T, Tsuchiya H, Kaiho A, Sakata E, Murata S, Yokosawa H, Sako Y, et al. Quantitative live-cell imaging reveals spatio-temporal dynamics and cytoplasmic assembly of the 26S proteasome. Nat Commun. 2014;5:3396. doi: 10.1038/ncomms4396. [DOI] [PubMed] [Google Scholar]
- 74.Chen Y, Muller JD, So PT, Gratton E. The photon counting histogram in fluorescence fluctuation spectroscopy. Biophys J. 1999;77:553–567. doi: 10.1016/S0006-3495(99)76912-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen Y, Muller JD, Ruan Q, Gratton E. Molecular brightness characterization of EGFP in vivo by fluorescence fluctuation spectroscopy. Biophys J. 2002;82:133–144. doi: 10.1016/S0006-3495(02)75380-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu H, Dong CQ, Ren JC. Tempo-Spatially Resolved Scattering Correlation Spectroscopy under Dark-Field Illumination and Its Application to Investigate Dynamic Behaviors of Gold Nanoparticles in Live Cells. Journal of the American Chemical Society. 2014;136:2775–2785. doi: 10.1021/ja410284j. [DOI] [PubMed] [Google Scholar]
- 77.Lakowicz JR. Principles of fluorescence spectroscopy. 3rd ed. New York: Springer; 2006. p. 954. [Google Scholar]
- 78.Damayanti NP, Craig AP, Irudayaraj J. A hybrid FLIM-elastic net platform for label free profiling of breast cancer. Analyst. 2013;138:7127–7134. doi: 10.1039/c3an01097j. [DOI] [PubMed] [Google Scholar]
- 79.Damayanti NP, Parker LL, Irudayaraj JM. Fluorescence lifetime imaging of biosensor peptide phosphorylation in single live cells. Angew Chem Int Ed Engl. 2013;52:3931–3934. doi: 10.1002/anie.201209303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vidi PA, Chen J, Irudayaraj JM, Watts VJ. Adenosine A(2A) receptors assemble into higher-order oligomers at the plasma membrane. FEBS Lett. 2008;582:3985–3990. doi: 10.1016/j.febslet.2008.09.062. [DOI] [PubMed] [Google Scholar]
- 81.Chen J, Miller A, Kirchmaier AL, Irudayaraj JM. Single-molecule tools elucidate H2A.Z nucleosome composition. J Cell Sci. 2012;125:2954–2964. doi: 10.1242/jcs.101592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vidi PA, Liu J, Salles D, Jayaraman S, Dorfman G, Gray M, Abad P, Moghe PV, Irudayaraj JM, Wiesmuller L, et al. NuMA promotes homologous recombination repair by regulating the accumulation of the ISWI ATPase SNF2h at DNA breaks. Nucleic Acids Res. 2014;42:6365–6379. doi: 10.1093/nar/gku296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sahoo H, Schwille P. FRET and FCS-Friends or Foes? Chemphyschem. 2011;12:532–541. doi: 10.1002/cphc.201000776. [DOI] [PubMed] [Google Scholar]
- 84.Kapusta P, Wahl M, Benda A, Hof M, Enderlein J. Fluorescence lifetime correlation spectroscopy. Journal of Fluorescence. 2007;17:43–48. doi: 10.1007/s10895-006-0145-1. [DOI] [PubMed] [Google Scholar]
- 85.Magidson V, Khodjakov A. Circumventing Photodamage in Live-Cell Microscopy. Digital Microscopy. (4th Edition) 2013;114:545–560. doi: 10.1016/B978-0-12-407761-4.00023-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang X, Roeffaers MBJ, Basu S, Daniele JR, Fu D, Freudiger CW, Holtom GR, Xie XS. Label-Free Live-Cell Imaging of Nucleic Acids Using Stimulated Raman Scattering Microscopy. Chemphyschem. 2012;13:1054–1059. doi: 10.1002/cphc.201100890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Muller P, Schmitt E, Jacob A, Hoheisel J, Kaufmann R, Cremer C, Hausmann M. COMBO-FISH Enables High Precision Localization Microscopy as a Prerequisite for Nanostructure Analysis of Genome Loci. International Journal of Molecular Sciences. 2010;11:4094–4105. doi: 10.3390/ijms11104094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Weiland Y, Lemmer P, Cremer C. Combining FISH with localisation microscopy: Super-resolution imaging of nuclear genome nanostructures. Chromosome Research. 2011;19:5–23. doi: 10.1007/s10577-010-9171-6. [DOI] [PubMed] [Google Scholar]
- 89.Wombacher R, Heidbreder M, van de Linde S, Sheetz MP, Heilemann M, Cornish VW, Sauer M. Live-cell super-resolution imaging with trimethoprim conjugates. Nat Methods. 2010;7:717–719. doi: 10.1038/nmeth.1489. [DOI] [PubMed] [Google Scholar]
- 90.van de Corput MP, de Boer E, Knoch TA, van Cappellen WA, Quintanilla A, Ferrand L, Grosveld FG. Super-resolution imaging reveals three-dimensional folding dynamics of the beta-globin locus upon gene activation. J Cell Sci. 2012;125:4630–4639. doi: 10.1242/jcs.108522. [DOI] [PubMed] [Google Scholar]
- 91.Recamier V, Izeddin I, Bosanac L, Dahan M, Proux F, Darzacq X. Single cell correlation fractal dimension of chromatin: A framework to interpret 3D single molecule super-resolution. Nucleus. 2014;5 doi: 10.4161/nucl.28227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cseresnyes Z, Schwarz U, Green CM. Analysis of replication factories in human cells by super-resolution light microscopy. BMC Cell Biol. 2009;10:88. doi: 10.1186/1471-2121-10-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lubeck E, Cai L. Single-cell systems biology by super-resolution imaging and combinatorial labeling. Nat Methods. 2012;9:743–748. doi: 10.1038/nmeth.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kam Y, Rubinstein A, Nissan A, Halle D, Yavin E. Detection of Endogenous K-ras mRNA in Living Cells at a Single Base Resolution by a PNA Molecular Beacon. Molecular Pharmaceutics. 2012;9:685–693. doi: 10.1021/mp200505k. [DOI] [PubMed] [Google Scholar]
- 95.Catrina IE, Marras SA, Bratu DP. Tiny molecular beacons: LNA/2'-O-methyl RNA chimeric probes for imaging dynamic mRNA processes in living cells. ACS Chem Biol. 2012;7:1586–1595. doi: 10.1021/cb300178a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang XM, Song Y, Shah AY, Lekova V, Raj A, Huang L, Behlke MA, Tsourkas A. Quantitative assessment of ratiometric bimolecular beacons as a tool for imaging single engineered RNA transcripts and measuring gene expression in living cells. Nucleic Acids Research. 2013;41 doi: 10.1093/nar/gkt561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang XM, Zajac AL, Huang LY, Behlke MA, Tsourkas A. Imaging the Directed Transport of Single Engineered RNA Transcripts in Real-Time Using Ratiometric Bimolecular Beacons. Plos One. 2014;9 doi: 10.1371/journal.pone.0085813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wu B, Chao JA, Singer RH. Fluorescence Fluctuation Spectroscopy Enables Quantitative Imaging of Single mRNAs in Living Cells. Biophysical Journal. 2012;102:2936–2944. doi: 10.1016/j.bpj.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dani A, Huang B, Bergan J, Dulac C, Zhuang X. Superresolution imaging of chemical synapses in the brain. Neuron. 2010;68:843–856. doi: 10.1016/j.neuron.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lee SH, Shin JY, Lee A, Bustamante C. Counting single photoactivatable fluorescent molecules by photoactivated localization microscopy (PALM) Proc Natl Acad Sci U S A. 2012;109:17436–17441. doi: 10.1073/pnas.1215175109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kaufmann R, Muller P, Hildenbrand G, Hausmann M, Cremer C. Analysis of Her2/neu membrane protein clusters in different types of breast cancer cells using localization microscopy. J Microsc. 2011;242:46–54. doi: 10.1111/j.1365-2818.2010.03436.x. [DOI] [PubMed] [Google Scholar]
- 102.Wang J, Yu X, Boriskina SV, Reinhard BM. Quantification of differential ErbB1 and ErbB2 cell surface expression and spatial nanoclustering through plasmon coupling. Nano Lett. 2012;12:3231–3237. doi: 10.1021/nl3012227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sengupta P, Jovanovic-Talisman T, Lippincott-Schwartz J. Quantifying spatial organization in point-localization superresolution images using pair correlation analysis. Nature Protocols. 2013;8:345–354. doi: 10.1038/nprot.2013.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tang GQ, Roy R, Bandwar RP, Ha T, Patel SS. Real-time observation of the transition from transcription initiation to elongation of the RNA polymerase. Proc Natl Acad Sci U S A. 2009;106:22175–22180. doi: 10.1073/pnas.0906979106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Friedman LJ, Mumm JP, Gelles J. RNA polymerase approaches its promoter without long-range sliding along DNA. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:9740–9745. doi: 10.1073/pnas.1300221110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang Z, Revyakin A, Grimm JB, Lavis LD, Tjian R. Single-molecule tracking of the transcription cycle by sub-second RNA detection. Elife. 2014;3:e01775. doi: 10.7554/eLife.01775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cisse II, Izeddin I, Causse SZ, Boudarene L, Senecal A, Muresan L, Dugast-Darzacq C, Hajj B, Dahan M, Darzacq X. Real-time dynamics of RNA polymerase II clustering in live human cells. Science. 2013;341:664–667. doi: 10.1126/science.1239053. [DOI] [PubMed] [Google Scholar]
- 108.Chen J, Zhang Z, Li L, Chen BC, Revyakin A, Hajj B, Legant W, Dahan M, Lionnet T, Betzig E, et al. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell. 2014;156:1274–1285. doi: 10.1016/j.cell.2014.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Izeddin I, Recamier V, Bosanac L, Cisse II, Boudarene L, Dugast-Darzacq C, Proux F, Benichou O, Voituriez R, Bensaude O, et al. Single-molecule tracking in live cells reveals distinct target-search strategies of transcription factors in the nucleus. Elife. 2014:e02230. doi: 10.7554/eLife.02230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ben-Ari Y, Brody Y, Kinor N, Mor A, Tsukamoto T, Spector DL, Singer RH, Shav-Tal Y. The life of an mRNA in space and time. J Cell Sci. 2010;123:1761–1774. doi: 10.1242/jcs.062638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Easwaran HP, Schermelleh L, Leonhardt H, Cardoso MC. Replication-independent chromatin loading of Dnmt1 during G2 and M phases. Embo Reports. 2004;5:1181–1186. doi: 10.1038/sj.embor.7400295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schermelleh L, Haernmer A, Spada F, Rosing N, Meilinger D, Rothbauer U, Cardoso MC, Leonhardt H. Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation. Nucleic Acids Research. 2007;35:4301–4312. doi: 10.1093/nar/gkm432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schneider K, Fuchs C, Dobay A, Rottach A, Qin WH, Wolf P, Alvarez-Castro JM, Nalaskowski MM, Kremmer E, Schmid V, et al. Dissection of cell cycle-dependent dynamics of Dnmt1 by FRAP and diffusion-coupled modeling. Nucleic Acids Research. 2013;41:4860–4876. doi: 10.1093/nar/gkt191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kaur G, Costa MW, Nefzger CM, Silva J, Fierro-Gonzalez JC, Polo JM, Bell TD, Plachta N. Probing transcription factor diffusion dynamics in the living mammalian embryo with photoactivatable fluorescence correlation spectroscopy. Nat Commun. 2013;4:1637. doi: 10.1038/ncomms2657. [DOI] [PubMed] [Google Scholar]
- 115.Cui Y, Cho IH, Chowdhury B, Irudayaraj J. Real-time dynamics of methyl-CpG-binding domain protein 3 and its role in DNA demethylation by fluorescence correlation spectroscopy. Epigenetics. 2013;8:1089–1100. doi: 10.4161/epi.25958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen J, Nag S, Vidi PA, Irudayaraj J. Single molecule in vivo analysis of toll-like receptor 9 and CpG DNA interaction. PLoS One. 2011;6:e17991. doi: 10.1371/journal.pone.0017991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Slaughter BD, Schwartz JW, Li R. Mapping dynamic protein interactions in MAP kinase signaling using live-cell fluorescence fluctuation spectroscopy and imaging. Proc Natl Acad Sci U S A. 2007;104:20320–20325. doi: 10.1073/pnas.0710336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Miller A, Chen J, Takasuka TE, Jacobi JL, Kaufman PD, Irudayaraj JM, Kirchmaier AL. Proliferating cell nuclear antigen (PCNA) is required for cell cycle-regulated silent chromatin on replicated and nonreplicated genes. J Biol Chem. 2010;285:35142–35154. doi: 10.1074/jbc.M110.166918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shamsaie A, Jonczyk M, Sturgis J, Robinson JP, Irudayaraj J. Intracellularly grown gold nanoparticles as potential surface-enhanced Raman scattering probes. Journal of Biomedical Optics. 2007;12 doi: 10.1117/1.2717549. [DOI] [PubMed] [Google Scholar]
- 120.Palonpon AF, Ando J, Yamakoshi H, Dodo K, Sodeoka M, Kawata S, Fujita K. Raman and SERS microscopy for molecular imaging of live cells. Nat Protoc. 2013;8:677–692. doi: 10.1038/nprot.2013.030. [DOI] [PubMed] [Google Scholar]
- 121.Damayanti NP, Fang Y, Parikh MR, Craig AP, Kirshner J, Irudayaraj J. Differentiation of cancer cells in two-dimensional and three-dimensional breast cancer models by Raman spectroscopy. J Biomed Opt. 2013;18:117008. doi: 10.1117/1.JBO.18.11.117008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Xie W, Wang L, Zhang Y, Su L, Shen A, Tan J, Hu J. Nuclear targeted nanoprobe for single living cell detection by surface-enhanced Raman scattering. Bioconjug Chem. 2009;20:768–773. doi: 10.1021/bc800469g. [DOI] [PubMed] [Google Scholar]
- 123.Ravindranath SP, Henne KL, Thompson DK, Irudayaraj J. Raman chemical imaging of chromate reduction sites in a single bacterium using intracellularly grown gold nanoislands. ACS Nano. 2011;5:4729–4736. doi: 10.1021/nn201105r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vitol EA, Orynbayeva Z, Bouchard MJ, Azizkhan-Clifford J, Friedman G, Gogotsi Y. In Situ Intracellular Spectroscopy with Surface Enhanced Raman Spectroscopy (SERS)-Enabled Nanopipettes. Acs Nano. 2009;3:3529–3536. doi: 10.1021/nn9010768. [DOI] [PubMed] [Google Scholar]
- 125.Niu JJ, Schrlau MG, Friedman G, Gogotsi Y. Carbon Nanotube-Tipped Endoscope for In Situ Intracellular Surface-Enhanced Raman Spectroscopy. Small. 2011;7:540–545. doi: 10.1002/smll.201001757. [DOI] [PubMed] [Google Scholar]
- 126.Pezacki JP, Blake JA, Danielson DC, Kennedy DC, Lyn RK, Singaravelu R. Chemical contrast for imaging living systems: molecular vibrations drive CARS microscopy. Nature Chemical Biology. 2011;7:137–145. doi: 10.1038/nchembio.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wilfling F, Wang HJ, Haas JT, Krahmer N, Gould TJ, Uchida A, Cheng JX, Graham M, Christiano R, Frohlich F, et al. Triacylglycerol Synthesis Enzymes Mediate Lipid Droplet Growth by Relocalizing from the ER to Lipid Droplets. Developmental Cell. 2013;24:384–399. doi: 10.1016/j.devcel.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shi YZ, Kim S, Huff TB, Borgens RB, Park K, Shi RY, Cheng JX. Effective repair of traumatically injured spinal cord by nanoscale block copolymer micelles. Nature Nanotechnology. 2010;5:80–87. doi: 10.1038/nnano.2009.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang MC, Min W, Freudiger CW, Ruvkun G, Xie XS. RNAi screening for fat regulatory genes with SRS microscopy. Nat Methods. 2011;8:135–138. doi: 10.1038/nmeth.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Xu PS, Gullotti E, Tong L, Highley CB, Errabelli DR, Hasan T, Cheng JX, Kohane DS, Yeo Y. Intracellular Drug Delivery by Poly(lactic-co-glycolic acid) Nanoparticles, Revisited. Molecular Pharmaceutics. 2009;6:190–201. doi: 10.1021/mp800137z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lemaire R, Menguellet SA, Stauber J, Marchaudon V, Lucot JP, Collinet P, Farine MO, Vinatier D, Day R, Ducoroy P, et al. Specific MALDI imaging and profiling for biomarker hunting and validation: fragment of the 11S proteasome activator complex, Reg alpha fragment, is a new potential ovary cancer biomarker. J Proteome Res. 2007;6:4127–4134. doi: 10.1021/pr0702722. [DOI] [PubMed] [Google Scholar]
- 132.Rauser S, Marquardt C, Balluff B, Deininger SO, Albers C, Belau E, Hartmer R, Suckau D, Specht K, Ebert MP, et al. Classification of HER2 receptor status in breast cancer tissues by MALDI imaging mass spectrometry. J Proteome Res. 2010;9:1854–1863. doi: 10.1021/pr901008d. [DOI] [PubMed] [Google Scholar]
- 133.Schwamborn K, Caprioli RM. Molecular imaging by mass spectrometry--looking beyond classical histology. Nat Rev Cancer. 2010;10:639–646. doi: 10.1038/nrc2917. [DOI] [PubMed] [Google Scholar]
- 134.Boggio KJ, Obasuyi E, Sugino K, Nelson SB, Agar NY, Agar JN. Recent advances in single-cell MALDI mass spectrometry imaging and potential clinical impact. Expert Rev Proteomics. 2011;8:591–604. doi: 10.1586/epr.11.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Miura D, Fujimura Y, Yamato M, Hyodo F, Utsumi H, Tachibana H, Wariishi H. Ultrahighly sensitive in situ metabolomic imaging for visualizing spatiotemporal metabolic behaviors. Anal Chem. 2010;82:9789–9796. doi: 10.1021/ac101998z. [DOI] [PubMed] [Google Scholar]
- 136.Marko-Varga G, Fehniger TE, Rezeli M, Dome B, Laurell T, Vegvari A. Drug localization in different lung cancer phenotypes by MALDI mass spectrometry imaging. J Proteomics. 2011;74:982–992. doi: 10.1016/j.jprot.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 137.Chaurand P, Cornett DS, Angel PM, Caprioli RM. From whole-body sections down to cellular level, multiscale imaging of phospholipids by MALDI mass spectrometry. Mol Cell Proteomics. 2011;10:O110 004259. doi: 10.1074/mcp.O110.004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang Q, Chen L, Long YT, Tian H, Wu JC. Molecular Beacons of Xeno-Nucleic Acid for Detecting Nucleic Acid. Theranostics. 2013;3:395–408. doi: 10.7150/thno.5935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chen H, Weng TW, Riccitelli M, Cui Y, Irudayaraj J, Choi JH. Understanding the Mechanical Properties of DNA Origami Tiles and Controlling the Kinetics of their Folding and Unfolding Reconfiguration. J Am Chem Soc. 2014 doi: 10.1021/ja500612d. [DOI] [PubMed] [Google Scholar]
- 140.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Xu X, Hou Y, Yin X, Bao L, Tang A, Song L, Li F, Tsang S, Wu K, Wu H, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148:886–895. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Eberwine J, Sul JY, Bartfai T, Kim J. The promise of single-cell sequencing. Nat Methods. 2014;11:25–27. doi: 10.1038/nmeth.2769. [DOI] [PubMed] [Google Scholar]