Abstract
We report a novel, high throughput magnetic-tweezers based 3D microchannel electroporation system capable of transfecting 40,000 cells/cm2 on a single-chip for gene therapy, regenerative medicine and intracellular detection of target mRNA for screening cellular heterogeneity. A single cell or an ordered array of individual cells are remotely guided by programmable magnetic fields to poration sites with high (> 90%) cell alignment efficiency to enable various transfection reagents to be delivered simultaneously into the cells. The present technique, in contrast to the conventional vacuum based approach, is significantly gentler on the cellular membrane yielding > 90% cell viability and, moreover, allows transfected cells to be transported for further analysis. Illustrating the versatility of the system, the GATA2 molecular beacon was delivered into leukemia cells to detect the regulation level of the GATA2 gene that is associated with the initiation of leukemia. The uniform delivery and a sharp contrast of fluorescence intensity between GATA2 positive and negative cells demonstrate key aspects of the platform for gene transfer, screening and detection of targeted intracellular markers in living cells.
Keywords: electroporation, magnetic tweezers, high throughput, gene delivery, micro-channel array
1. Introduction
While genetic engineering of cells, including cancerous cells or embryonic stem cells, offers enormous potential for regenerative medicine and for basic understanding of human biology, it continues to be limited by difficulties of common methods used to introduce exogenous DNA or RNA. Efficient and high throughput transfection of nucleic acids and plasmids into cells without causing damage or death would indeed be a valuable experimental tool to probe the effects of gene delivery in these cells for research and clinical applications. In this regard, electroporation has been widely adapted as a useful technique.[1-3] Compared to other physical methods of gene delivery such as micro-injection[4], gene gun[5,6], laser irradiation[7,8] and sonoporation[9], electroporation has become more valuable for both in vitro and in vivo applications due to its simplicity and potential to transfect large numbers of cells.[10,11] A number of electroporation systems have been developed and commercialized.[12-15] For example, bulk electroporation (BEP) is a techniques in which millions of cells are simultaneously shocked with a high voltage between two electrodes. A serious drawback of this approach, however, is that a large fraction of the cells are damaged due to the nonuniform and hazardous electric-fields that affect individual cells. Therefore, three critical aspects - transfection efficiency, gene delivery to targeted cells and cell viability - are not guaranteed [16-17] with the BEP approach. Microchannel electroporation (MEP) provides a means to overcome these drawbacks by offering a gentler environment where each cell is porated under more controlled conditions.[17-19] By confining individual cells at a microscale pore, the electric field strength across the pore increases by several orders over those achieved by BEP.[20-22] Thus not only are low voltages (< 10 V) sufficient for cell poration [17,18,23-25], but delivery into the cell is confined to regions determined by the size of the pore. Furthermore, MEP offers the potential for versatile lab-on-chip systems that integrate cell-manipulation and real-time detection followed by cell transfer, thereby paving the road for comprehensive analysis of cellular behaviors in response to environment, signal pathways, cell-cell interactions and cellular dynamics in the post-transfection stage. [26]
Currently most MEP designs, however, only facilitate single-cell electroporation,[18,19,23,24] which is inadequate for clinical applications that require high throughput. Amongst recent approaches [17,18,24,27-29], microfluidic electroporation devices often operate in a sequential manner, and thus could be less conducive to scale-up for clinical applications.[30] On the other hand, 3D microchannel electroporation (3D MEP) could achieve high throughput by handling thousands of cells on a planar (X,Y) membrane while the applied electric field and transfection are in the vertical (Z) direction. [24,31-33] However, a critical requirement that is currently lacking for 3D MEP is an efficient approach to manipulate and safely align a large number of individual cells with an array of micropores for high throughput transfection at a low voltage.
In this work we report on the application of a versatile 3D MEP - magnetic tweezers (MT) based system capable of realizing the three important aspects of (a) individual-cell based electroporation, (b) high throughput transfection, and (c) retention of cell viability. To efficiently place a cell at a single micropore, an array of thin Permalloy (NiFe) magnetic disks fabricated on a silicon wafer are utilized as an effective multiplexed magnetic tweezers. Magnetically labeled cells are remotely controlled by weak external magnetic fields which operate over the entire array enabling simultaneous manipulation of tens of thousands of cells. Additionally, the weak magnetic fields (< 150 G) do not generate heat nor adversely damage the cells, concerns that arise with manipulation associated with other techniques, including vacuum force [24, 31-33], which is difficult to optimize without serious cell membrane damage,[34] and optical tweezers, which is burdened by low throughput[35-37] and laser-induced Joule heating.[30]
The present magnetic tweezers-based approach illustrates parallel manipulation, localization, electroporation, and subsequent transport of the transfected cells. The versatility of the approach with its potential for pre-clinical studies and gene therapy is demonstrated with several distinct cell types and transfection reagents. A highlight is the delivery of the GATA2 molecular beacon (MB) for detection of GATA2 mRNA expression. The GATA2 family of transcription factors play important roles in proliferation and differentiation of hematopoietic stem cells (HSCs).[38] Among them GATA2 is highly expressed in HSCs and progenitors regulating hematopoitic development, and its disorder has been implicated in the onset of leukemia.[39] Detection of GATA2 is thus of great significance for the study of heterogeneities of HSCs. As a transcription factor, however, few accessible technologies to date can achieve intracellular detection for GATA2 within living cells. In our experiment, a GATA2 molecular beacon was delivered into both K562 cells and Jurkat cells to detect the regulation of the GATA2 gene. The transfected GATA2-MB specifically hybridizes to GATA2 mRNA in the cytosol and subsequently fluoresces by unzipping its hairpin stucture, thereby distinguishing cells with high GATA2 levels from those with low levels. K562 is a common myloid progenitor with high GATA2 expression, while Jurkat is a T cell mature leukemic cell line with low level of GATA2 mRNA.[40] The present study provides a platform to test the performance of the GATA2-MB on identifying intracellular GATA2 mRNA within a large, statistically meaningful, framework of individual cells. The 3D MEPMT system, in combination with molecular beacon-based probes, thus offers a high throughput fluorescence bio-sensing system for detecting explicit intracellular markers within individual living cells with the potential to provide insight into the expression of genes and their role in diseases initiated by specific cells.
2. Results and Discussion
2.1. 3D MEP Chip
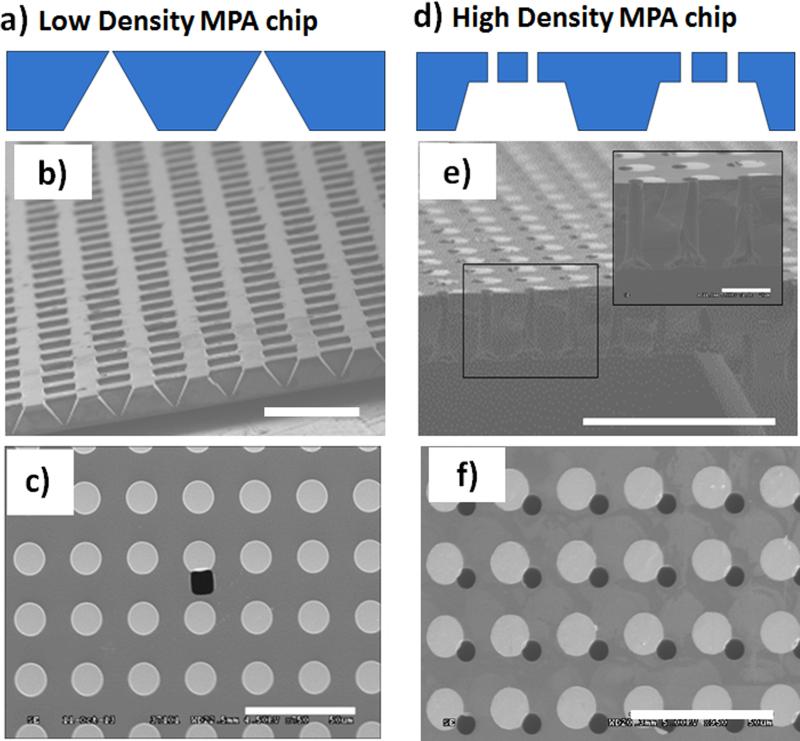
The 3D MEP chip is the primary element of this setup, serving as the platform for cell loading, alignment with micro-pores, and gene delivery by electroporation. Figure 1 shows two types of silicon MEP chips, the low pore density array chip, Fig. 1 (a-c), and the high pore density array chip, Fig. 1 (d-f), with well-ordered array patterns and uniform pore size, that were fabricated using cleanroom techniques.
Figure 1. SEM micrographs of the Low Pore Density (a-c) and High Pore Density (d-f) Micropore Array (MPA) Chips.
(a) The cross-sectional schematic of the low density MPA chip. (b) The pyramid-pit shaped chip was fabricated by anisotropic etch of silicon in KOH. scale bar = 500 μm. (c) The top surface of the chip shows a single micropore (black) and a magnetic disk array (white). Scale bar = 50μm. (d) The cross-sectional schematic of the high density MPA chip. (e) A terrace array with vertical microchannel array with high aspect ratio (pore size: 5 μm, pore length: 30 μm) were created using anisotropic wet etch and DRIE, respectively. Scale bar = 100 μm. Insert shows zoomed-in cross section of three individual micropores. Scale bar = 20 μm. (f) Each micropore is adjacent to an individual magnetic disk. Scale bar = 50 μm.
2.1.1. Low Pore Density Array Chip
A pyramid-shaped micropore array (MPA) chip was developed to test the performance of the MT on single-cell manipulation (Figure 1(a)). This low pore density chip was fabricated by anisotropic wet etch in potassium hydroxide (KOH). Masked by a metallic mask with a square-array feature (Details in Experimental Section), a pyramid-pit array was developed in the silicon wafer as shown in Figure 1(b). By controlling the etch-stop time, 5 μm diameter pores with 315 μm spacing were created on the other side of the chip. The Permalloy disk array for magnetic traps was fabricated on the flat surface, denoted the top surface (Figure 1(c)), with an architecture that enables single cells to be maneuvered to individual micropores. This particular low density chip has ~1000 micropores/cm2, capable of transfecting ~1000 cells under ideal trapping efficiency. Thus, though the pore density is relatively low, its transfection throughput is nevertheless much higher than those previously reported from microfluidics-based MEP. [18-24,30]
2.1.2. High Pore Density Array Chip
The ultimate goal is to provide a high-throughput MEP platform to simultaneously handle 104 ~ 106 cells on a single chip. For this purpose, a high density pore array was fabricated using both wet etching and Deep Reactive-Ion Etching (DRIE) (Fabrication protocol shown in Figure S1). Figure 1(d) provides the cross-sectional schematic of the high density MPA chip. Using DRIE (Bosch Process) offers uniform (< 10 % variations) and high aspect ratio (up to 30: 1) microchannels, while increasing the difficulties in fabrication compared to the low density MPA chip. The process benefitted from the combined advantages of wet etch (low-cost, rapid processing) and DRIE (uniform, high pore density) processes. A terrace-shaped trench array was first etched from one side with KOH using an etching mask with a large micro-square array pattern (each square 500 - 800 μm in width). DRIE was then used to etch a pore array (each pore 5 μm diameter) from the reverse side. The length of the vertical micropores (30 μm) was determined by the thickness of the silicon wafer and the depth of the micro-trench (Figure 1(e)). The portion of the pore connecting to the bottom of the micro-trench (termed “through-pore”) is the region of interest for the experiment (Figure S3). At the top surface, a well-aligned pore array is patterned such that each micropore is adjacent to one magnetic disk (Figure 1(f)). In contrast to the low pore density chip, the high throughput chip has ~ 40,000 through-pores/cm2, and thus closely aligns with transfection requirements that are suitable for clinical use.
2.2. 3D Microchannel Electroporation-Magnetic Tweezers System
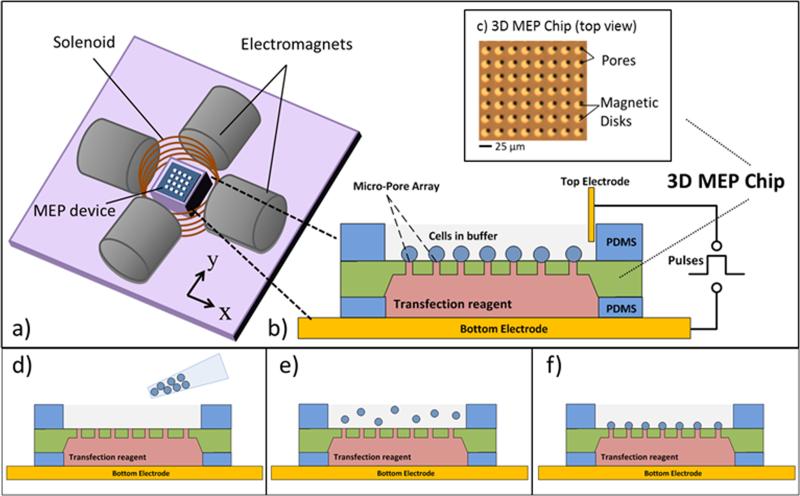
The 3D MEP-MT system is illustrated in Figure 2. The magnetic tweezers setup[41] consists of four orthogonal electromagnets (for X, Y fields) and a solenoid (for Z fields) (Figure 2(a)) surrounding the 3D MEP platform and providing nearly uniform magnetic fields needed for the high throughput cell manipulation. As illustrated in Figure 2(b), a planar electrode (glass slide coated with 100 nm Au) is used as a substrate and the bottom electrode for the electroporation. A PDMS (polydimethylsiloxane) spacer on the surface holds a solution of the molecules to be delivered. On top of the PDMS spacer is the 3D MEP chip, shown from top-down view in Figure 2(c). As mentioned in Section 2.1, two types of MPA chips were fabricated for different experiments. The low density MPA chip was suitable for testing single-cell on-chip manipulation, including trapping, alignment, and movement after electroporation. The high density MPA chip, on the other hand, enabled monitoring of the feasibility of high-throughput cell alignment and transfection. On both chips, by controlling the external magnetic fields, individual labeled cells are localized at the magnetic disk array aligned to the micropore array. Once the cells are aligned with the micropores, the top electrode is placed into the buffer solution and a low voltage (4 ~ 10 V) applied between the top and bottom electrodes. Due to the geometry of the system, the resulting potential drop is mainly across the micropore and cell. It is noted that viral particles have been previously captured and aligned in nanopores using chemical and centrifugation apporaches.[42] For tracking single-cell or cell-array manipulation on the chip, an upright microscope (Leica Microsystems DM2500 MH) was used as shown in Figure S2(a).
Figure 2. The 3D MEP- MT system setup.
(a) Electromagnet setup consists of 4 orthogonal electromagnets that create the X- and Y- (in-plane) components of the external magnetic field and a solenoid that creates Z- (out-of-plane) component. (b) The 3D MEP device sits at the center of the magnets. A gold substrate serves as the bottom electrode. A PDMS spacer holds the transfection reagents in solution. The 3D MEP wafer sits above the spacer with etched pores and magnetic disks. Another PDMS spacer sits on top to hold cells in solution, and a platinum electrode is located above. (c). Micrograph of a 3D MEP wafer, showing Permalloy disks aligned with 5 micron diameter pores. Cell seeding is performed by simple pipetting (d) of cells in PBS buffer solution into upper chamber and allowing them to settle (e) with magnetic fields turned on such that they fall into an aligned pattern with the pores (f).
2.3. Magnetic Tweezers
To manipulate the individual cells in parallel, an array of permalloy (Ni0.81Fe0.19) disks, 15 μm diameter and 50 nm thick, was patterned on the 3D MEP chip. Weak (<150 Gauss) external in-plane magnetic fields HXY = HX + HY are created by four orthogonal electromagnets (Magnetech OP-2025) and the out-of-plane field HZ is produced by a surrounding solenoid. Each field source (HX, HY, and HZ) is controlled by a separate current channel. In the presence of the external field HEXT = HXY + HZ, magnetic micro-beads residing on the device align in the direction of HEXT. The permalloy magnetic disks magnetize in the direction of HXY, creating either attractive traps (magnetic potential energy wells) or repulsive centers whose strengths are tuned with HZ.[41] Each disk captures a single magnetically labelled cell. Simple programmable routines that rotate HXY and reverse HZ are used to transport the labelled cells to electroporation sites, where they are then transfected. The transfected cells are subsequently maneuvered across the platform. Parallel transfection and manipulation enables easy scale up of the device (currently capable of 40,000 cells/cm2) to address millions of cells by merely increasing the number of micropores and associated magnetic disks that comprise the chip array.
2.4. Electric Field and Transmembrane Potential in 3D MEP
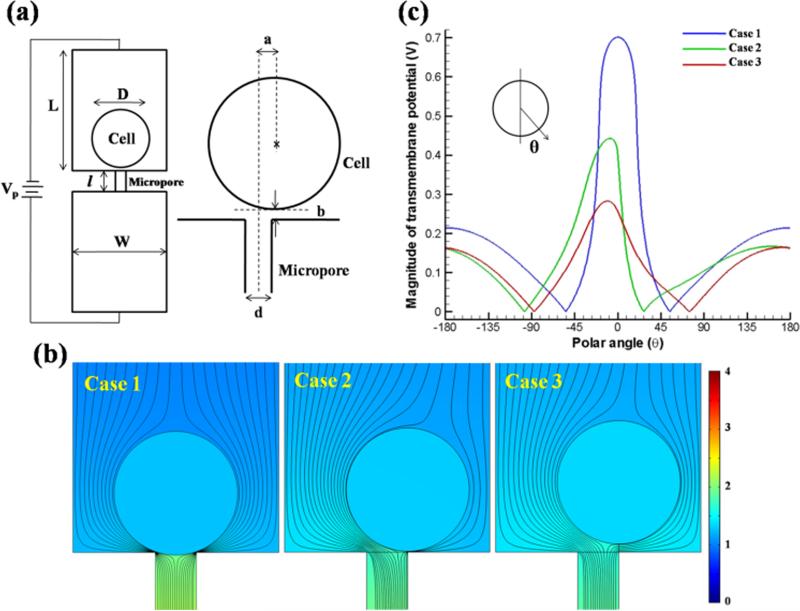
Micro-electroporation offers an advantage over BEP in being capable of permeabilizing the cell membrane using low voltages (< 10 V), resulting in high cell viability. This low cell mortality is facilitated by the localization of a single cell on an individual micro-pore. The transmembrane potential and electric fields corresponding to different cell-pore distances in the MEP-MT system were calculated as shown inFigure 3(a). To match with experiments, a cell (D = 15 μm) was placed on top of a micropore (d = 5 μm, l = 30 μm) with an applied voltage (Vp = 4 V) across the setup (L = 200 μm, W = 25 μm). To investigate the importance of cell alignment and attachment to the micropore, three different cell locations were assigned: Case 1 (a = 0 and b = 0.1 μm), Case 2 (a = 2.5 and b = 0.1 μm), and Case 3 (a = 2.5 and b = 1 μm), representing the situations of (1) aligned cell attached to the micropore, (2) cell misaligned but attached to substrate, and (3) cell misaligned and not attached to the substrate. Due to the limitation of creating the computational meshes, a tiny gap (b = 0.1 μm) exists between the cell surface and substrate at their contact points for Case 1 and 2. This tiny gap may mimic the real situation since the cell and substrate surface are not perfectly smooth. A three-layer cell model, in which the domain is divided into an external medium, cell membrane, and cell cytoplasm, was adopted.[43,44] The electric field was calculated by solving the Laplace equation using COMSOL®:
| (1) |
in which σ is the electrical conductivity and V the electrical potential. The electric field was determined by E = −∇V. In this three-layer model, the electrical conductivities of buffer, cytoplasm, and membrane were set to 0.8, 0.2, and 5×10−7 Sm−1, respectively. Figure 3(b) shows the cell locations and simulated electric fields around the cell. The field for Case 1 is symmetric and is focused by the tiny gap (0.1 μm) between the cell and micropore. In Cases 2 and 3, the electric field is distorted due to misalignment between cell and micropore. Based on the calculated electric potential distribution, the transmembrane potential (TMP) and its variation along the polar angle are summarized in Figure 3(c). Due to the focused electric field, the cell side facing the micropore has a higher TMP than that of the opposite side. As the cell moves away from the micropore (from Case 1 to Case 3), the TMP peak decreases. In Case 3, the micropore focusing effect is sharply reduced and the TMP profile approaches that of bulk electroporation. Clearly, due to the alignment and attachment to the surface, the cell under Case 1 experiences the highest TMP where the peak is about 0.7 V. In general, the threshold TMP for electroporation is in the range of 0.2 ~ 1 V.[45] Therefore, Case 1 has the highest likelihood and largest area of poration among the three situations considered. Furthermore, even if the cells of Case 2 and 3 were porated, misalignment and detachment from the surface would allow the charged transfection reagents to pass around the cell. Thus, the highest transfection efficiency and uniformity will be realized for a cell represented by Case 1, illustrating the importance of incorporating the magnetic tweezers into the MEP platform.
Figure 3. Simulation of low-voltage cell electroporation on 3D MEP.
(a) Simulation parameters for one set of cell electroporation. D: cell diameter, L: reservoir depth, W: cell-to-cell distance, l: micropore depth, d: micropore diameter, Vp: applied voltage, a: distance between cell center and micropore central line, and b: the smallest distance between cell edge and substrate surface. (b) Simulated electrical potential contours and electrical field streamlines around the cell for Case 1 to 3. (c) Simulated transmembrane potential magnitudes for Case 1 to 3. A polar angle θ is defined with respect to the negative Y-axis from the center of the cell, as depicted in the inset of (c).
2.5. Magnetic Tweezers (MT) Assisted On-chip Single-cell Manipulation
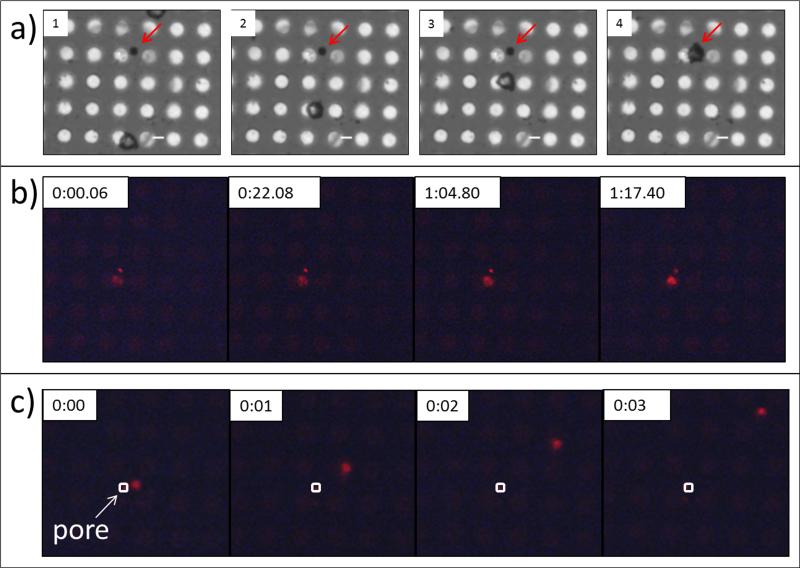
The low density pore array device demonstrated the complete control over cell manipulation and precise localization in a single-cell transfection experiment (Figure 4). Here, pre-programmed magnetic fields are first used to guide a magnetically labeled white blood cell directly onto the pore (Figure 4(a)). PI dye was delivered into the cells by low voltage electroporation (10 V, 10 ms, 5 pulses) in just over 1 minute (Figure 4(b)). Post transfection, the cell is rapidly removed from the pore via protocols associated with the magnetic tweezers (Figure 4(c)).[41] These experiments demonstrate the robust capability of the MT to not only optimally align cells on the pores, but also to manipulate them to or from any location on the array before and after transfection, indicating the versatility of the MT-based system for integration with post-transfection analysis.
Figure 4. On-chip single cell manipulation by MT.
(a) Programmed routines are utilized to manipulate a single magnetically labeled white blood cell toward a micropore (indicated by red arrow) and localize it on the pore (frame 4). (b) 10 V pulses are delivered, resulting in rapid PI dye uptake. (c) Post-transfection, the cell is rapidly removed from the pore (indicated by white square).
2.6. High-Throughput Cell Alignment and 3D MEP
2.6.1. Alignment Efficiency and Transfection Efficiency
The cell alignment for high-throughput 3D MEP is completely automated. Electromagnets apply a constant external magnetic field directed toward the pores (e.g. if pores are located on the +X side of disks, HXY = 50 Oe along +X direction, and HZ = 50 Oe in +Z direction). As the labeled cells settle to the bottom of the platform, they are gently attracted by the magnetic fields of the disks to locations directly above the pores. This alignment is critical, since when the voltage pulse is delivered it will evenly transfect all cells simultaneously. Besides being automated, this cell alignment technique takes only a few minutes, as shown in the sequential frames of Figure 5(a).
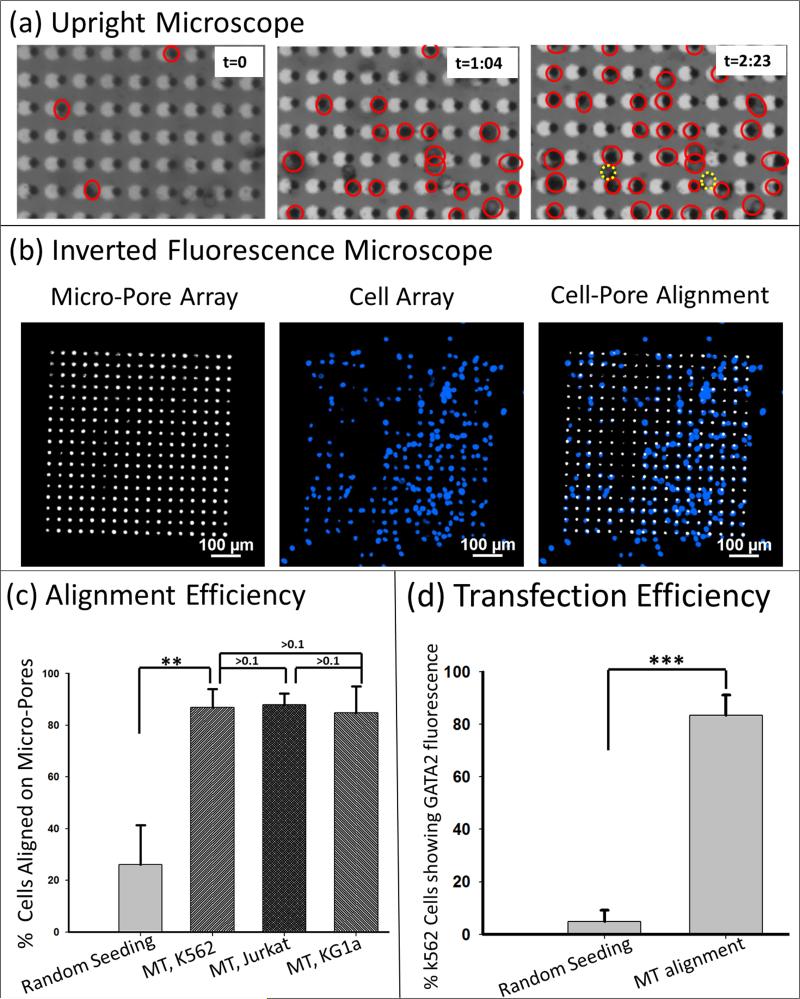
Figure 5. High throughput cell trapping and transfection efficiency by 3D MEP.
(a) Sequential frames taken during cell seeding show alignment of the cell array on pores (aligned cells circled in red) in one region of the chip with a constant external magnetic field. Misaligned cells are circled in dashed yellow lines in the third panel. (b) Resulting cell alignment on a 17 by 17 array of pores with phase contrast indicating the location of through-pores, Hoechst staining indicating the location of each cell, and a merged image demonstrating the cell-micropore alignment. (c) efficiencies on aligning K562, Jurkat and KG1a cells to the micropore by MT, compared to that from K562 random seeding (cell number, n=300). (d) the efficiency of K562 transfection with GATA2 MB after MT assisted cell trapping, in comparison with that after random cell seeding (n= 500). ***: p < 0.005; **: p < 0.01.
Figure 5(b) illustrates the cell array pattern resulting from the high throughput alignment. Cell nuclei are stained with Hoechst for visualizing cell location. Quantifications in Figure 5(c) demonstrates the MT assisted alignment efficiency (single-pore completely covered by a single-cell) as compared to that from random seeding (without magnetic fields). When cells randomly precipitate down to the chip, the chance of alignment to the micropores is low (~25%). However with inclusion of the magnetic tweezers, the efficiencies are dramatically enhanced and are at 87% for K562 cells, 88% for Jurkat cells, and 85% for KG1a cells. It is noteworthy that this MT alignment requires a simple magnetic labelling via a cellular surface receptor and weak external fields provided by low-cost electromagnets. All four types of cells used for this study were readily labelled with anti-CD45 magnetic beads; various kits are also commercially available to label different cell types.
As evident from Figure 5(d), MT-based cell alignment and localization directly impacts the subsequent transfection. As shown in the figure, ~83.4 % transfection efficiency was achieved by alignment and transfection of K562 cells with GATA2 MB. This is significantly higher than the mere ~ 4.9% transfection efficiency of the control group (p-value < 0.005; alignment and GATA2 fluorescence in Figure S4)), in which cells were randomly seeded and electroporated under the same conditions of 4 V, 50 ms pulse, 10 pulses. This result verifies findings from the trans-membrane potential simulation, and underscores the importance of the MT in increasing the efficiency of cellular poration by aligning cells to the micropores. Details of K562 cells transfection with GATA2 MB are given in Section 2.6.4.
2.6.2. High Throughput ODN-FAM delivery into cells
To test the high-throughput gene delivery capabilities of the 3D MEP-MT system, ODN-FAM (oligonucleotide with carboxyfluorescein fluorescent dye) was delivered into an array of KG1a cells (Figure 6(a)). Efficient alignment of the cells with the array of micropores is confirmed with phase contrast imaging and Hoechst staining. The cells were transfected with low voltage (4 V), and the green cellular fluorescence reported from FAM was visualized after pulse generation, indicating the intracellular delivery. These preliminary experiments demonstrate the high throughput functionality of the chip for gene delivery.
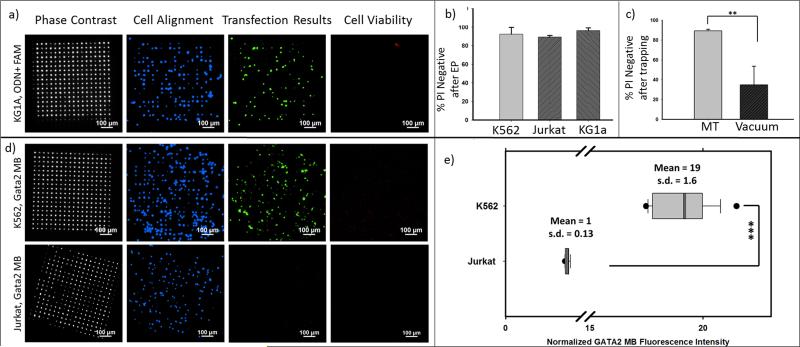
Figure 6. High throughput cell transfection with FAM-ODN and GATA2 molecular beacon on 3D MEP.
Phase contrast (showing locations of through-pores), Hoechst staining (showing cell location by staining cell nuclei), green fluorescence (showing fluorescence of ODN or GATA2 MB), and PI staining (showing cell mortality/viability) are illustrated for (a) ODN + FAM delivery into KG1a cells. (b) Quantitative analysis of PI negative fluorescence, showing cell viability for K562 (92% viable), Jurkat (89% viable), and KG1a (96% viable) cells. Performed after electroporation. (cell number n=1000) (c) Comparison of PI negative fluorescence, showing cell viability, for Jurkat cells aligned by magnetic tweezers (90% viable) and Jurkat cells aligned by vacuum (6~8 Psi, 34% viable). (n=600) (d)The same panels described in (a) are shown for GATA2 MB delivery and fluorescence in K562 cells (GATA2 positive) and Jurkat cells (GATA2 negative). (e) A normalized comparison of fluorescent intensity for GATA2 MB in K562 cells and Jurkat cells. (n=400, the average fluorescence intensity of Jurkat is normalized to 1). ***: p < 0.005; **: p < 0.01.
2.6.3. Cell Viability
For each high-throughput cell transfection experiment, cell viability was evaluated approximately 75 minutes after electroporation with propidium iodide (PI) dye. PI uptake will not occur in a cell with an intact membrane, but will rapidly enter a damaged cell membrane, intercalate into cellular nucleic acids and fluoresce brightly in the red. The images (4th panels of Figure 6(a) and (c)) show no obvious PI fluorescence confirming viable post-transfected cells with intact membranes. Quantitative results in Figure 6(b) confirm viability of 92% for K562 cells, 89% for Jurkat cells, and 96% for KG1a cells. These experimental results validate that magnetic labelling and the low voltage pulses do not damage the cell membranes.
Experiments were also carried out using a conventional vacuum approach for trapping cells on the same 3D MEP chip in order to estimate membrane damage and cell viability following transfection under this mode of localization.[46] Since it is difficult to optimize the vacuum pressure, as shown in Figure S5, achieving a high trapping efficiency was not possible without damaging the cell membrane. For instance, Ref.[25] reported a pressure difference of 0.5 kPa (=0.07 psi), while Ref.[33] used 34 kPa (=4.5 psi) to trap cells. A finalized and optimized value for achieving high trapping efficiency as well as high viability were however not reported.[25,33] Ref.[23] does not report a specific force or pressure difference.[23] In the present work, if the applied vacuum force was too weak, e.g. 4 - 6 psi, the hydrodynamics were insufficient to draw cells down to the micropore regions. Natural precipitation of cells eventually results in a random distribution without a clear array pattern. When the vacuum was increased from 6 to 8 psi, however, 90% of the cells were damaged. Although adjusting the vacuum to the 6 ~ 8 psi range yielded a 90% alignment efficiency in several attempts, the associated cell viability (Figure 6(e)) clearly illustrates the MT to be far more effective in keeping cell membranes intact (~90% cell viability) compared to the vacuum approach (~34% cell viability) for similar alignment efficiencies. Since the effective range of the MT traps is short, the cells experience the magnetic trap force only after they gently precipitate down to the immediate vicinity of the micropore. In contrast, the vacuum-generated forces are long ranged and would drive cells with finite speeds until they are abruptly brought to rest by the chip. The resulting momentum change upon reaching the edge of the micropore usually causes cell deformation and/or membrane rupture. In fact this method has been previously used to rupture cell membranes for gene delivery.[34]
2.6.4. Delivery of GATA2 MB for Detection of GATA2 mRNA Expression in Leukemia Cells
To demonstrate the value of the system for pre-clinical studies, the GATA2 molecular beacon (MB) was delivered into leukemia cells to detect the regulation level of intracellular GATA2. Cells were trapped at the pores via the MT, resulting in an ordered cell array as shown in the Hoechst panels of Figure 6(b). Electroporation pulses were applied immediately following cell trapping to transfect the GATA2 MB (green fluorescence panels of Figure 6(b)). K562 is a human immortalised myelogenous leukemia cell line and has been shown in a previous report to be GATA2high. Jurkat, a T lymphocyte mature leukemic cell line, has been shown to be GATA2low.[39,40] As a result, a remarkable expression of green fluorescence 1 hour after GATA2 MB delivery into the K562 cells is observed, indicating the up-regulation of GATA2 gene in this cell type. In contrast, the lack of expression of GATA2 in the negative control Jurkat cells was evident by the negligible GATA2 MB fluorescence even 1 hour post-delivery. Comparison of the relative normalized fluorescence intensity between K562 and Jurkat cells, (Figure 6(e)) shows that the GATA2 MB fluorescence from K562 is ~19 times higher than that from Jurkat (p-value < 0.005). The small variation in fluorescence intensity (s.d. = 1.6) reveals that uniform amounts of MB are delivered into cells with the 3D MEP-MT device. Note that the fluorescence intensity from Jurkat cells is weak but uniform (s.d. = 0.13), forming a sharp contrast to the strong positive signals from K562. These reults confirm that on-chip discrimination of GATA2 positive cells from negative cells at the single-cell level within a large population is possible through this approach. Importantly, it presents a simple and efficient method to detect target mRNA using intracellular probes within living cells.
3. Conclusion
In this paper, a 3D MEP-MT system capable of rapid, effective cell alignment resulting in high-throughput electroporation has been introduced. The transfection achieved through low voltages, as well as the weak magnetic fields that remotely control cell alignment and movement do not harm the cells. This platform provides a promising high throughput gene transfection device with the added advantages of high cell viability and uniform transfection at the single-cell level. We also demonstrate use of the system with high throughput on-chip screening of heterogeneous leukemic cells based on fluorescence signals from the GATA2 molecular beacon. This unified MEPMT device can be broadly integrated into experiments aimed at delivering and tracking intracellular molecular probes for gene detection as well as to understand basic processes associated with gene therapy outcomes and regenerative progressions in large populations of individual cells.
4. Experimental Section
4.1. 3D MEP Fabrication
4.1.1 Low Density Micropore Array (MPA) Chip
The following steps were involved in the fabrication of the chip. A Si wafer ((100) orientation, double side polished) was thinned to ~200 μm and a metal mask (Cr/Au, 30 nm / 100 nm, E-gun evaporation, Denton DV 520A) was deposited on both sides. A micro-square array (250 μm in width, 315 μm center-to-center distance) was patterned on the mask layer by photolithography (S1813, Shipley, 1.4 μm), followed by mask layer etch in which chromium (CR-7S) and gold (GE-8111) etchants were used to transfer the micro-square array to the etch mask. The masked silicon wafer was placed in a KOH wet-etching tank (45%, 80 ) for several hours. Due to the anisotropic etch properties of silicon for each crystal plane, an array of inverted pyramid pits, with an angle of 54.7° to (100) plane were created. By controlling the stop-time, the opposite side of the Si wafer was also thinned down until a small micropore through-hole (5 μm in diameter) appeared at the bottom of each pyramid-pit. The mask layer was removed by using both CR-7S and GE-8111. Following the MPA chip fabrication, the circular disk array (15 μm in diameter, 25 μm center-to-center distance) was fabricated with photolithography (LOR2A and S1813, Shipley). A 50 nm nickel-iron film was sputtered (AJO Orion) on the chip, followed by lift-off in which a well-ordered magnetic disk array was formed on the chip. Micropores adjacent to the magnetic disks are used for single-cell manipulation. A SiO2 dielectric film (~200 nm) was deposited on the chip by PECVD (Plasma Therm 770 SLR) for electrical isolation during electroporation.
4.1.2 High Density MPA Chip
The steps involved in the fabrication of the high density MPA chip are shown in Figure S1. The first four steps were similar to the protocol of the low pore density chip (Section 4.1.1), except a large terrace-shaped micro-trench array, instead of the pyramid pit array, were developed by use of a wider micro-square array (500 ~ 800 μm) with a metal mask. A micropore array (5 μm diameter, 25 μm pore-to-pore distance) was patterned on the flat surface of the silicon wafer using photolithography (SPR200-7, 6 μm thick). DRIE was used to etch the micropore array through the bottom of the terrace array to create the through-hole micropore array. Bosch process (SF6: 12s / 100 sccm gas flow / 700 W ICP power / 40 W RF power/ 30 mT APC pressure; C4F8: 7s / 100 sccm gas flow / 700 W ICP power / 10 W RF power / 30 mT APC pressure), with etch rate ~3 μm / min, was applied for DRIE (Oxford Plasma Lab 100). The magnetic disk array was created with the same protocol as that described in the low density MPA chip. Alignment was required to position each micropore next to the adjacent magnetic disk. Photolithography and optical alignment were conducted on the EVG 620 aligner.
4.2. Cells and Biomolecules
Four types of cells were used. Primary human white blood cells were purified from whole blood from the American Red Cross (Columbus, OH). KG1a (an acute myeloid leukemia cell line), K562 (human immortalised myelogenous leukemia) and Jurkat (human T lymphocyte cell lines) were obtained from ATCC. These three cell lines were cultured in RPMI 1640 (catalog No. 11875-093), with the addition of 10% (v/v) fetus bovine serum (FBS, heat-inactivated, catalog No. 26010). Propidium Iodide (PI, cat. No. P3566, Invitrogen, excitation / emitting wavelength, 535 / 617 nm) was delivered into cells aligned on the micropores. FAM-labeled oligonucleotides (FAM-ODN, alpha DNA company, cat No. 427520, excitation / emitting wavelength, 492 / 517 nm) and GATA2 molecular beacon (GATA2 MB, 50 μM/ ml, Sigma-Aldrich, excitation / emitting wavelength, 495 / 515 nm ) were delivered into cells to evaluatethe performance of the 3D MEP-MT systems. An inverted microscope (Nikon Elipse Ti) was used to check the fluorescent signal after electroporation. The nuclei of cells were stained with Hoechst (Sigma-Aldrich, catalog No. 654434, excitation / emitting wavelength, 350 / 461 nm) to indicate the cell location on the chip. The through-hole micropore array was visualized by phase contrast. Propidium iodide (PI) was also used to measure cell viability with the staining applied ~75 min after electroporation with transfection reagents other than PI.
For statistical analysis, two-sided student T-test was used to determine the significance for data with Gaussian distribution and equal variances. Groups with p values < 0.05 were considered statistically significant.
4.3. Labeling Cells with Magnetic Micro-beads
All cells were linked with superparamagnetic beads to enable remote control of the cells by weak external magnetic fields. Dextran-coated magnetic micro-beads (1 μm diameter, StemCell Technologies, Cat. No. 19250) were attached to cells via CD45 antigen, which is expressed on all cells used in this study. To attach these beads, depletion cocktail (EasySepTM Human CD45 Depletion Kit, Stem Cell Technologies, Cat. No. 18259) containing tetrameric antibody complexes targeting CD45 antigens was mixed with cells in phosphate buffered saline (PBS) and allowed to incubate at room temperature with regular mixing. Dextran coated magnetic micro-beads were then added and once again incubated with mixing. One wash step with a magnet removes any unlabelled cells.
4.4. Chip Treatment for Cell Manipulation
To protect the magnetic traps and to realize electrical isolation, the chip was coated in a thin (~200 nm) layer of SiO2 immediately after fabrication using plasma enhanced chemical vapor deposition (Plasma Therm 790 PECVD). To prevent cell adhesion during transfection, the chip surface was modified with polyethylene glycol-silane (PEG-silane, MW 2,000, Laysan Bio, Inc.) prior to each experiment. To do so, it was first treated with oxygen plasma (PTS Oxygen Plasma System), soaked in 1 mM PEG in ethyl alcohol at room temperature for 30 minutes, followed by a 30-minute bake at 110 °C. This treatment coats the surface with a layer of PEG molecules, which inhibit nonspecific protein adsorption.
4.5. Vacuum-based Cell Trapping on 3D MPA
Vacuum based cell trapping was carried out to compare the trapping efficiency and cell viability with that of the MT method. The method adopted here follows procedures reported in previous literature.[46] The vacuum was generated in a filter bottle with a porous cork and a nozzle linked to a vacuum pump (Figure S2(b)). The 3D MEP chip was placed on the cork while the vacuum pressure was adjusted by a control valve and read out by a pressure gauge. For cell trapping, a droplet of cell buffer with a defined concentration of cells was placed on the chip. Due to the pressure difference across the chip, the cell buffer entered through the MPA, which hydrodynamically drove cells to the micropores.
Supplementary Material
Acknowledgements
The authors are grateful to Dr. Jeffrey Chalmers, Jenny Park, Yongqi Wu, and Xi Zhao for providing white blood cells and KG1a cells. The authors thank the National Science Foundation (EEC-0914790), the Army Research Office (W911NF-10-1-0353), and the National Institutes of Health (RO1-CA135332) for supporting this study.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Lingqian Chang, NSEC Center for Affordable Nanoengineering of Polymeric Biomedical Devices, Ohio State University, Columbus, OH 43209, USA; Biomedical Engineering Department, Ohio State University, Columbus, OH 43209, USA.
Marci Howdyshell, NSEC Center for Affordable Nanoengineering of Polymeric Biomedical Devices, Ohio State University, Columbus, OH 43209, USA; Department of Physics, Ohio State University, Columbus, OH 43209, USA.
Wei-Ching Liao, NSEC Center for Affordable Nanoengineering of Polymeric Biomedical Devices, Ohio State University, Columbus, OH 43209, USA.
Chi-Ling Chiang, NSEC Center for Affordable Nanoengineering of Polymeric Biomedical Devices, Ohio State University, Columbus, OH 43209, USA; Division of Hematology, The Comprehensive Cancer Center, Ohio State University, Columbus, OH, 43209, USA.
Daniel Gallego-Perez, NSEC Center for Affordable Nanoengineering of Polymeric Biomedical Devices, Ohio State University, Columbus, OH 43209, USA.
Zhaogang Yang, NSEC Center for Affordable Nanoengineering of Polymeric Biomedical Devices, Ohio State University, Columbus, OH 43209, USA.
Wu Lu, NSEC Center for Affordable Nanoengineering of Polymeric Biomedical Devices, Ohio State University, Columbus, OH 43209, USA; Electrical and Computer Engineering Department, Ohio State University, Columbus, OH 43209, USA.
John C. Byrd, Comprehensive Cancer Center, The Ohio State University, Columbus, OH, 43209, USA. Division of Hematology, The Comprehensive Cancer Center, Ohio State University, Columbus, OH, 43209, USA
Natarajan Muthusamy, Division of Hematology, The Comprehensive Cancer Center, Ohio State University, Columbus, OH, 43209, USA.
L. James. Lee, NSEC Center for Affordable Nanoengineering of Polymeric Biomedical Devices, Ohio State University, Columbus, OH 43209, USA Chemical and Biomolecular Engineering Department, Ohio State University, Columbus, OH 43209, USA.
Ratnasingham Sooryakumar, NSEC Center for Affordable Nanoengineering of Polymeric Biomedical Devices, Ohio State University, Columbus, OH 43209, USA; Department of Physics, Ohio State University, Columbus, OH 43209, USA.
References
- 1.Mehier-Humbert S, Guy RH. Adv. Drug Delivery Rev. 2005;57:733–753. doi: 10.1016/j.addr.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 2.Aihara H, Miyazaki J. Nat. Biotechnol. 1998;16:867–870. doi: 10.1038/nbt0998-867. [DOI] [PubMed] [Google Scholar]
- 3.Stephens DJ, Pepperkok R. P.N.A.S. 2001;98:4295–4298. doi: 10.1073/pnas.081065198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diacumakos EG. Methods in cell biology. Academic Press; NY, USA: 1973. [Google Scholar]
- 5.Klein TM, Wolf ED, Wu R, Sanford JC. Nature. 1987;327:70–73. [Google Scholar]
- 6.Yang NS, Burkholder J, Roberts B, Martinell B, McCabe D. P.N.A.S. 1990;87:9568–9572. doi: 10.1073/pnas.87.24.9568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shirahata Y, Ohkohchi N, Itagak H, Satomi S. J. Investig. Med. 2001;49:184–190. doi: 10.2310/6650.2001.34045. [DOI] [PubMed] [Google Scholar]
- 8.Tirlapur UK, Konig K. Nature. 2002;418:290–291. doi: 10.1038/418290a. [DOI] [PubMed] [Google Scholar]
- 9.Kim HJ, Greenleaf JF, Kinnick RR, Bronk JT, Bolander ME. Hum. Gene Ther. 1996;7:1339–1346. doi: 10.1089/hum.1996.7.11-1339. [DOI] [PubMed] [Google Scholar]
- 10.Hoover F, Magne KJ. Anal.Biochem. 2000;285:175–178. doi: 10.1006/abio.2000.4730. [DOI] [PubMed] [Google Scholar]
- 11.Gehl J. Acta. Physiol. Scand. 2003;177:437–447. doi: 10.1046/j.1365-201X.2003.01093.x. [DOI] [PubMed] [Google Scholar]
- 12.BIO-RAD company [04, 2014];Gene pulser MXcellTM electroporation system. http://www.biorad.com/en-us/category/electroporation.
- 13.Life-technology [04, 2014];Neon transfection. https://www.lifetechnologies.com/us/en/home/life-science/cell-culture/transfection/transfection---selection-misc/neon-transfection-system/neon-transfection-system-information.html.
- 14.Lonza company [04, 2014];Nucleofector electroporation system. http://www.lonza.com/products-services/bio-research/transfection/nucleofector-technology.aspx.
- 15.Havard Apparatus [04, 2014];BTX single waveform electroporation systems. http://www.btxonline.com/
- 16.Fox MB, Esveld DC, Valero A, Luttge R, Mastwijk HC, Bartels PV, van den Berg A, Boom RM. Anal.Bioanal.Chem. 2006;385:474–485. doi: 10.1007/s00216-006-0327-3. [DOI] [PubMed] [Google Scholar]
- 17.Wang S, Lee LJ. Biomicrofluidics. DOI: 10.1063/1.4774071. [Google Scholar]
- 18.Valero A, Post JN, van Nieuwkasteele JW, ter Braak PM, Kruijer W, van den Berg A. Lab on Chip. 2008;8:62–67. doi: 10.1039/b713420g. [DOI] [PubMed] [Google Scholar]
- 19.Khine M, Lau A, Ionescu-Zanetti C, Seo J, Lee LP. Lab on Chip. 2005;5:38–43. doi: 10.1039/b408352k. [DOI] [PubMed] [Google Scholar]
- 20.Zheng Q, Chang DC. BBA Gene Structure and Expression. 1991;1088:104–110. doi: 10.1016/0167-4781(91)90158-i. [DOI] [PubMed] [Google Scholar]
- 21.Boukany PE, Morss A, Liao W, Henslee B, Zhang X, Yu B, Wang X, Wu Y, Jung H, Li L, Gao K, Hu X, Zhao X, Hemminger O, Lu W, Lafyatis GP, Lee LJ. Nat. Nano. 2011;6:747–754. doi: 10.1038/nnano.2011.164. [DOI] [PubMed] [Google Scholar]
- 22.Gao K, Li L, He L, Hinkle K, Wu Y, Ma J, Chang L, Zhao X, Gallego-Perez D, Eckardt S, Mclaughlin J, Liu B, Farson DF, Lee LJ. Small. 2014;10:1015–1023. doi: 10.1002/smll.201300116. [DOI] [PubMed] [Google Scholar]
- 23.Huang Y, Rubinsky B. Biomed. Microdev. 1999;2:145–150. [Google Scholar]
- 24.Khine M, Zanetti CI, Blatz A, Wang LP, Lee LP. Lab on Chip. 2007;7:457–462. doi: 10.1039/b614356c. [DOI] [PubMed] [Google Scholar]
- 25.Kurosawa O, Oana H, Matsuoka S, Noma A, Kotera H, Washizu M. Meas. Sci. Technol. 2006;17:3127–3133. [Google Scholar]
- 26.Przybyla L, Voldman J. Annu. Rev. Anal. Chem. 2012;5:293–315. doi: 10.1146/annurev-anchem-062011-143122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bao N, Le TT, Cheng JX, Lu C. Integr. Biol. 2010;2:113–120. doi: 10.1039/b919820b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang HY, Lu C. Anal.Chem. 2006;78:5158–5164. doi: 10.1021/ac060733n. [DOI] [PubMed] [Google Scholar]
- 29.Huang Y, Rubinsky B. Sensor. Actuat. A. 2003;104:205–212. [Google Scholar]
- 30.Lindstrom S, Andersson-Svahn H. Lab on Chip. 2010;10:3363–3372. doi: 10.1039/c0lc00150c. [DOI] [PubMed] [Google Scholar]
- 31.Fei Z, Hu X, Choi H, Wang S, Farson D, Lee LJ. Anal. Chem. 2010;82:353–358. doi: 10.1021/ac902041h. [DOI] [PubMed] [Google Scholar]
- 32.Huang Y, Rubinsky B. Sensor. Actuat. A. 2001;89:242–249. 25. [Google Scholar]
- 33.Fei Z, Wang S, Xie Y, Henslee BE, Koh CG, Lee LJ. Anal. Chem. 2007;79:5719–5722. doi: 10.1021/ac070482y. [DOI] [PubMed] [Google Scholar]
- 34.Sharei A, Zoldan J, Adamo A, Sim WY, Cho N, Jackson E, Mao S, Schneider S, Han MJ, Lytton-Jean A, Basto PA, Jhunjhunwala S, Lee J, Heller DA, Kang JW, Hartoularos GC, Kim KS, Anderson DG, Langer R, Jensen KF. P.N.A.S. 2013;110:2082–2087. doi: 10.1073/pnas.1218705110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grier DG. Nature. 2003;424:810–816. doi: 10.1038/nature01935. [DOI] [PubMed] [Google Scholar]
- 36.Yang AHJ, Moore SD, Schmidt BS, Klug M, Lipson K, Erickson D. Nature. 2009;457:71–75. doi: 10.1038/nature07593. [DOI] [PubMed] [Google Scholar]
- 37.Soni GV, Jonsson MP, Dekker C. Small. 2013;9:679–684. doi: 10.1002/smll.201201875. [DOI] [PubMed] [Google Scholar]
- 38.Nagai T, Harigae H, Ishihara H, Motohashi H, Minegishi N, Tsuchiya S, Hayashi N, Gu L, Andres B, Engel JD. Blood. 1994;84:1074–1084. [PubMed] [Google Scholar]
- 39.Ikonomi P, Rivera CE, Riordan M, Washington G, Schechter AN, Noguchi CT. Exp. Hematol. 2000;28:1423–1431. doi: 10.1016/s0301-472x(00)00553-1. [DOI] [PubMed] [Google Scholar]
- 40.Klein E, Ben-Bassat H, Neumann H, Ralph P, Zeuthen J, Polliack A, Vánky F. Int. J. Cancer. 1976;18:421–431. doi: 10.1002/ijc.2910180405. [DOI] [PubMed] [Google Scholar]
- 41.Henighan T, Chen A, Vieira G, Hauser AJ, Yang FY, Chalmers JJ, Sooryakumar R. Biophys. J. 2010;98:412–417. doi: 10.1016/j.bpj.2009.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moon JM, Akin D, Xuan Y, Ye PD, Guo P, Bashir R. Biomed. Microdev. 2009;11:135–142. doi: 10.1007/s10544-008-9217-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stewart DA, Gowrishankar TR, Smith KC, Weaver JC. IEEE Trans.Biomed. Eng. 2005;52:1643. doi: 10.1109/TBME.2005.856030. [DOI] [PubMed] [Google Scholar]
- 44.Zu Y, Huang S, Liao WC, Lu Y, Wang S. J. Biomed. Nanotech. 2014;10:982. doi: 10.1166/jbn.2014.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang MY, Orwar O, Olofsson J, Weber J. Anal. Bioanal. Chem. 2010;397:3235. doi: 10.1007/s00216-010-3744-2. [DOI] [PubMed] [Google Scholar]
- 46.Ferrell N, Gallego-Perez D, Higuita-Castro N, Butler RT, Reen RK, Gooch KJ, Hansford DJ. Anal.Chem. 2010;82:2380. doi: 10.1021/ac902596b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.