Abstract
The successful extraction of metabolites is a critical step in metabolite profiling. By optimizing metabolite extraction, the range and quantitative capacity of metabolomics studies can be improved. We considered eight separate extraction protocols for the preparation of a metabolite extract from cultured mammalian cells. Parameters considered included temperature, pH, and cell washing before extraction. The effects on metabolite recovery were studied using a high resolution liquid chromatography mass spectrometry (LC-HRMS) platform that measures metabolites of diverse chemical classes including among others amino acids, lipids, and sugar derivatives. The temperature considered during the extraction or the presence of formic acid, a commonly used additive, was shown to have minimal effects on the measured ion intensities of metabolites. However, washing of samples before metabolite extraction whether with water or PBS (both commonly considered practices) exhibited dramatic effects on measured intensities of both intra- and extra-cellular metabolites. Together these findings present a systematic assessment of extraction conditions for metabolite profiling.
Keywords: metabolomics, extraction
1 Introduction
Metabolomics is an increasingly appreciated tool for probing the function of biological systems in the study of human biology and disease [1]. Metabolomics has shed light on the changes in metabolism that accompany diseases and drug treatment [2], identified changes in metabolism of human diseases such as cancer [3, 4], and provide vital biological insights through the study of a wide variety of organisms [5, 6].
Carrying out a high throughput metabolomics study, i.e. quantitating a wide variety of metabolites, requires a platform such as high resolution liquid chromatography-mass spectrometry (LC-HRMS) [7]. LC-HRMS is able to quantitate a large number of targeted metabolites with reproducibility often leading to a typical coefficient of variation of less than 20%. Untargeted metabolomics can also be carried out on LC-HRMS, allowing measurement of unknown metabolites. A variety of extracts can be prepared from yeast [8], plants [9, 10], mammalian cells [11, 12], mammalian serum [13, 14] and mammalian tissues and fluids [15].
Different metabolite extraction methods result in different recoveries of metabolites [16]. Simple modifications, such as changing the temperature or composition of the extraction solvent will have effects on the metabolites that can be detected and studied [17], while special extraction methods are required to study specific metabolites, such as lipids and metabolites involved in carbon metabolism [16, 18]. Hence, the protocol for metabolite extraction plays an extremely important role in determining the scope of metabolites that can be studied. Better understanding of extraction parameters will allow optimization of metabolite extraction protocols, leading to more robust metabolomics studies. Given the importance of sample preparation there is an unmet need to systematically examine the effects of critical extraction parameters on the quantitation of a wide variety of metabolites that this study attempts to address.
This paper examines the effects of three extraction parameters and combinations thereof on the detection and quantitation of known metabolites. The three different extraction parameters studied are the addition of acid to methanol extraction solvent, temperature and washing of the sample before extraction of metabolites. These parameters are thought to be important to metabolite extraction, and are not as well studied or understood compared to other parameters [19]. Optimization studies of metabolite extraction tend to focus on the chemical composition of the extraction solvent or on data analysis [20–22]. A recent review reported that temperature and pH are important factors in quenching biological activity prior to metabolite extraction, while washing is a common practice thought to remove metabolites found outside the cell [23]. Washing cells is reported to result in decreased recovery of metabolites found inside cells [24], and improve signal-to-noise ratio [19]. Washing also requires additional experimental manoeuvring that could lead to larger variability. As each of these extraction parameters is important in metabolite extraction, studying the extent to which they affect the detection and quantitation of common metabolite classes, including amino acids, free nucleotides, lipids and active cellular metabolites, will allow better understanding and design of metabolite extraction for metabolomics studies.
In our study, a cold methanol extraction solvent is used to simultaneously quench and extract metabolites. This extraction method is quick and recovers a large variety of metabolites [17, 25]. The broad range of metabolites recovered using a cold methanol extraction solvent allows the study of systematic variation brought about by extraction parameters on a broad spectrum of metabolites. In addition, studying an established set of targeted metabolites allows us to relate metabolite recovery with chemical properties of metabolites. A previously reported LC-HRMS platform was considered [26]. The biological system considered are colorectal cancer cells grown in adherent culture. This serves as a common model for studying mammalian cell biology and an identical number of cells was used under each extraction condition.
In comparing the different extraction protocols, a direct comparison across different conditions was carried out. Given the broad nature of classes of metabolites that cold methanol extraction can recover and quantitate, the effects of these extraction parameters may be applicable to other extraction methods. In addition, these extraction parameters may be used to improve quantitation of metabolites in cell culture metabolomics studies by providing insight into the metabolite extraction process. This will allow better design of metabolite extraction protocols, leading to optimization of metabolite extraction for future metabolomics studies.
2 Materials and Methods
2.1 Materials
The colorectal cancer cell line HCT116 was provided as a generous gift from Dr. Lewis Cantley’s laboratory. RPMI 1640 medium was purchased from Cellgro. Fetal Bovine Serum (FBS), penicillin, and streptomycin were purchased from Hyclone Laboratories. Optima-grade ammonium acetate, ammonium hydroxide, acetonitrile, methanol and water were purchased from Fisher Scientific. All solvents used were LC-MS grade and purchased from Fisher Scientific.
2.2 Cell culture and sample preparation
HCT116 cells were grown in RPMI 1640 supplemented with 10% heat inactivated Fetal Bovine Serum and 100,000 units/L penicillin and 100mg/L streptomycin. Cells were incubated at 37 °C. HCT116 cells were seeded in 6-well plates at a density of 500,000 cells per well. After overnight incubation at 37°C, media were removed, cells were washed with PBS (2ml) and fresh RPMI medium was added.
2.3 Metabolite extraction from cells
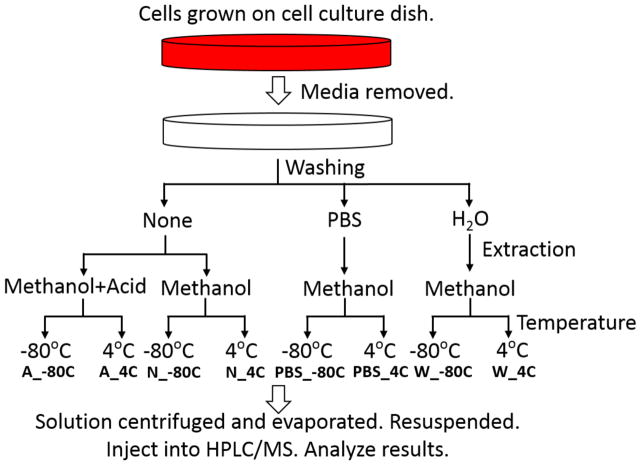
Metabolites were extracted according to the workflow presented in Figure 1. Plates with HCT116 cells were taken out of the 37°C incubator and immediately placed on dry ice (−80°C) or ice (4°C). 20μl of the growth media in the plates was collected, and the rest was removed. Then, half the plates were washed either with 2ml of PBS, or with 2ml of millipore H2O, while the other half was not washed. Subsequently, 1 ml extraction solvent (either 80% MeOH/H2O or 80%MeOH/H2O with 5% formic acid added), pre-cooled in −80°C freezer for at least 1 hour, was added to the cell culture dishes. The dishes on dry ice (−80°C) were transferred to the −80°C freezer. The plates were left for 15 min and then cells were scraped into the extraction solvent on dry ice (−80°C) or ice (4°C). The solution was transferred to eppendorf tubes, and centrifuged with at 20 000 g and 4°C for 10 min. Every extraction condition was prepared in three biological replicates. The supernatant is then split to two new eppendorf tubes, and dried in a SpeedVac. After drying, one tube of each sample was stored in the −80°C freezer as a backup, while the other one was re-constituted in 15μl water and then diluted with 15μl 50% MeOH/ACN on ice. Finally, 5μl was applied to the LC-MS.
Figure 1.
Schematic diagram of eight metabolite extraction conditions with differences in washing of samples, extraction solvent and temperature of extraction.
2.4 Metabolite Detection from Growth Media
To the collected media, 30μl of ice cold H2O was added together with 200μl ice cold methanol (Fisher, optima LC/MS grade). After vigorous vortexing, the solution was then centrifuged with 20 000g at 4°C for 10min and supernatant was dried in a SpeedVac. Preparation of cell extracts was described in the previous chapter. The medium extract was then re-constituted in 30μl water and diluted with 30μl 50% MeOH/ACN. 5μl of the final solution was applied to the LC-MS.
2.5 Liquid Chromatography
For metabolite separation and detection, the liquid chromatography system, Ultimate 3000 UHPLC, coupled to a Q Exactive mass spectrometer was used. An Xbridge amide column (100 x 2.1 mm i.d., 3.5 μm; Waters) is employed for compound separation at room temperature. The mobile phase A is 20 mM ammonium acetate and 15 mM ammonium hydroxide in water with 3% acetonitrile, pH 9.0 and mobile phase B was 100% acetonitrile. Linear gradient as follows: 0 min, 85% B; 1.5 min, 85% B, 5.5 min, 35% B; 10min, 35% B, 10.5 min, 35% B, 14.5 min, 35% B, 15 min, 85% B, and 20 min, 85% B. The flow rate was 0.15 ml/min from 0 to 10 min and 15 to 20 min, and 0.3 ml/min from 10.5 to 14.5 min.
2.6 Mass Spectrometry
The Q Exactive MS (Thermo Scientific) is equipped with a heated electrospray ionization probe (HESI), and the relevant parameters are as listed: evaporation temperature, 120 °C; sheath gas, 30; auxiliary gas, 10; sweep gas, 3; spray voltage, 3.6 kV for positive mode and 2.5 kV for negative mode. Capillary temperature was set at 320°C, and S-lens was 55. A full scan range from 60 to 900 (m/z) was used. The resolution was set at 70 000. The maximum injection time was 200 ms. Automated gain control (AGC) was targeted at 3,000,000 ions.
2.7 Peak Extraction and Statistical Analysis
Raw data collected from the LC-QE-MS were processed on Thermo Scientific, Sieve 2.0. Peak alignment and detection were performed according to manufacturer protocols. A frameseed including 166 metabolites were for analysis with data collected in positive mode while a frameseed of 119 metabolites was used for negative mode, where m/z width is set at 10ppm. A MS intensity (integrated peak area) of 103 was set as the noise level, and metabolites with signals lower than 103 treated as below the detection limit. Calculations were performed in R computing language (www.r-project.org). Unsupervised hierarchical clustering (Spearmann rank) and heat map were made using the Gene-E package for high-dimensional data visualization (Broad Institute, http://www.broadinstitute.org/cancer/software/GENE-E/index.html).
3 Results
To study the effects of the extraction process on metabolite quantitation, three different parameters were varied: temperature, addition of acid to extraction solvent and washing of cells before solvent extraction (Figure 1). In total, eight different extraction conditions were examined for colorectal cancer cell line HCT116: methanol containing formic acid added to the cells without washing at −80°C (A_−80°C), methanol extraction containing formic acid added to the cells without washing at 4°C (A_4°C), methanol extraction without washing at −80°C (N_−80°C), methanol without washing at 4°C (N_4°C), washing with PBS prior to methanol extraction at −80°C (PBS_−80°C), washing with PBS prior to methanol extraction at 4°C (PBS_4°C), washing with water prior to methanol extraction at −80°C (W_−80°C) and washing with water prior to methanol extraction at 4°C (W_4°C).
3.1 Resulting Metabolite Detection and Global Profile
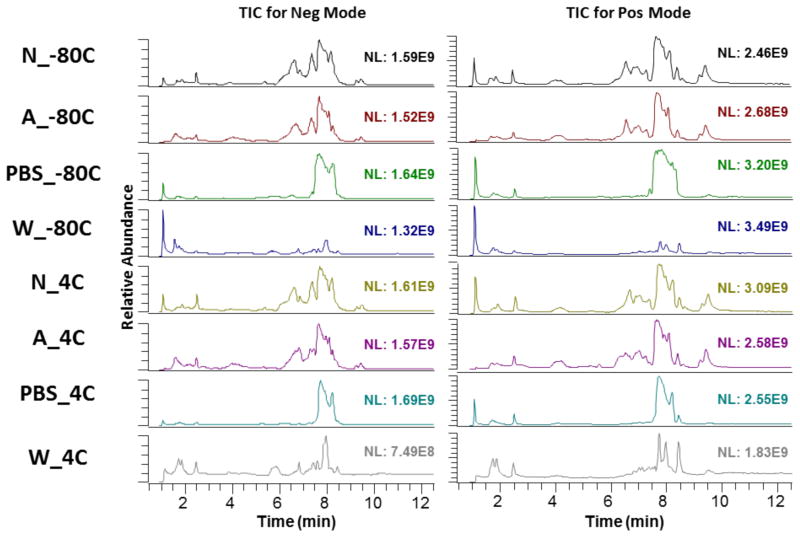
The effect of the different extraction conditions on the total spectrum of metabolites can be observed from their total ion chromatogram (TIC) (Figure 2). The TIC for methanol solvent without washing is similar to the TIC for methanol solvent with formic acid added without washing, with no observable differences when extraction is done at −80°C or at 4°C. This indicates a close similarity between the extraction conditions of A_−80°C, A_4°C, N_−80°C, N_4°C. Differences arise when comparing the TIC for washing with PBS and washing with water, at both −80°C and at 4°C, with the methanol solvent without washing condition. The peaks from around the 7 minute mark are absent when samples are washed with PBS, while the peaks at from the 6 to 8 minute mark are absent when samples are washed with water. This indicates that washing of the samples before solvent extraction of metabolites greatly affects the ion matrix detected by mass spectrometry.
Figure 2.
Total Ion Chromatogram (TIC) for each of eight different extraction methods detected in negative and positive mode.
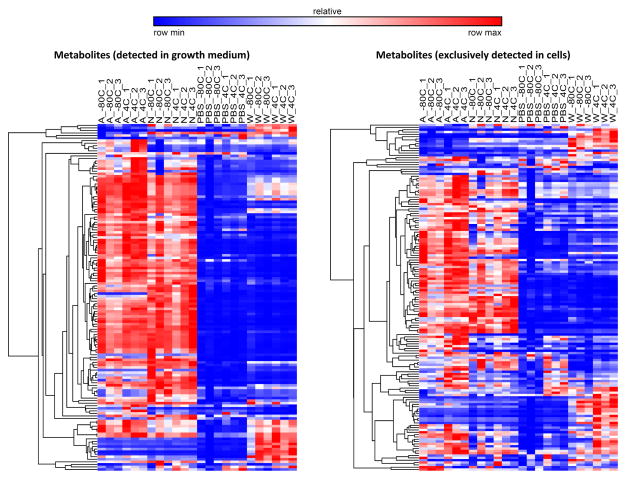
To understand the effects of the different extraction conditions on the matrix of metabolites that are detected and quantified, a further examination on the recovery of known metabolites was done (Supplementary Table 1). 136 metabolites were detected in both the growth medium of the cells and in the cells (Figure 3). These metabolites are likely to be excreted by cells and can be loosely classified as extracellular metabolites for the purposes of this study. 149 metabolites were found and detected in the cells but not in the growth media, and these metabolites are termed intracellular metabolites (Figure 3). An unsupervised hierarchical clustering of the mass spectrometry intensity levels for each metabolite under each of the eight conditions shows that washing with either PBS or water decreases the signal intensity for detected metabolites, regardless of whether these metabolites are extracellular or intracellular. Despite this general decrease, there are some exceptions where washing with water results in increased MS signal intensities. Some nucleotides, such as UDP, CMP and ADP and metabolites in the citric acid (TCA) cycle, such as fumarate, malate and succinate are detected at higher MS signal intensities when cells were washed with water. While washing seems to decrease the signal intensity, the addition of formic acid to the methanol extraction solvent seems to produce similar patterns of signal intensities for metabolites detected.
Figure 3.
Heat maps, generated by hierarchical pearson clustering, for 136 metabolites detected in both growth medium and in cells and for 149 metabolites that are detected in cells only.
3.2 Effects of Extraction Parameters
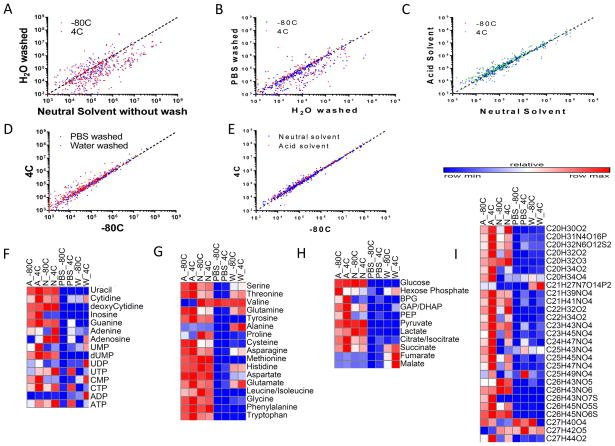
Pairwise comparisons of the MS signal intensities from the different extraction conditions was done to better understand the effects of each extraction parameter on metabolite quantitation. Comparing the MS signal intensities of washing with water and without washing (Figure 4A) shows that a higher MS signal intensity is obtained when cells are not pre-washed before metabolite extraction with methanol solvent. A comparison between washing with PBS and washing with water shows that washing with PBS decreases the MS signal intensities as compared to washing with water, with more metabolites being detected at a higher signal when sample is washed with water (Figure 4B). The addition of formic acid to the methanol solvent seems not to greatly disturb the intensities of metabolites, as observed in Figure 4C. In fact, more metabolites are detected at a higher intensity when formic acid is added to the methanol extraction solvent. Pairwise comparisons of the third extraction parameter, temperature, show that washing samples with PBS at 4°C returns a higher signal intensity than at −80°C, while washing with water results in a smaller difference between signal intensity at 4°C and at −80°C (Figure 4D). Pairwise comparison of the unwashed methods using methanol solvent or methanol solvent with formic acid show that there is an even distribution between metabolites that favour 4°C or −80°C (Figure 4E).
Figure 4.
A Comparison of washing with water against without washing B Comparison of washing with water against washing with PBS C Comparison of methanol solvent extraction against methanol solvent with formic acid extraction D Comparison of 4°C against −80°C extraction for washing with PBS and washing with water E Comparison of 4°C against −80°C for methanol solvent extraction and methanol solvent with formic acid extraction F Heat Map for Nucleotide metabolites G Heat Map for Free amino acids H Heat map for metabolites involved in glycolysis and the citric acid cycle I Heat map for lipid-like metabolites.
Chemical Abbreviations: UMP: uridine monophosphate. dUMP: deoxyuridine monophosphate. UDP: uridine diphosphate. UTP: uridine triphosphate. CMP: cytidine monophosphate. CTP: cytidine triphosphate. ADP: adenosine diphosphate. ATP: adenosine triphosphate. BPG: 2,3-bisphosphoglycerate. GAP: glyceraldehyde 3-phosphate. DHAP: dihydroxyacetone phosphate.
The different extraction methods have an effect on some commonly studied metabolites. Common patterns of signal intensities detected are seen across nucleotides (Figure 4F), free amino acids (Figure 4G), metabolites involved in glycolysis and TCA cycle (Figure 4H) and lipid-like molecules (Figure 4I). Signal intensities are decreased when samples were washed with PBS or with water, with notable exceptions (e.g. alanine, succinate, fumarate and malate). Using PBS as a washing solvent suppresses most metabolite signals except for ATP, UTP and CTP. This may be due to PBS stabilizing the high energy phosphate bonds in these molecules. Extraction carried at 4°C improves signals of nucleotides and lipids like metabolites (Figure 4F and 4I) while no dramatic effects are observed on amino acids or glycolysis and TCA cycle intermediates. Addition of formic acid to methanol solvent seems to increase the intensity for most lipid like compounds, with more compounds showing an increased intensity (red), especially when extraction was done at 4°C. This suggests that extraction of metabolites without any washing steps tends to return the highest signal intensities for these commonly studied classes of metabolites.
3.3 Variation across extraction conditions
The different extraction conditions used introduces variation in metabolite signals. Some metabolites are more sensitive to changes in extraction parameters, such as diphosphate and inosine. A list of metabolites that have the greatest variation in signal intensities is shown in Table 1. These metabolites have a maximum fold change in metabolite intensity that is larger than 100 when comparing the different extraction methods. Most of these metabolites are extracellular metabolites, with some exceptions such as sn-glycero-3-phosphocholine and diphosphate. This variation in metabolite signals is more pronounced when we consider which extraction condition returns the highest signal intensity or the lowest signal intensity (Table 2). Among the eight extraction conditions, methanol containing formic acid at 4°C gives the highest signal intensity for the most number of metabolites (136 metabolites) while extraction with PBS washing at −80°C gives the lowest signal intensity for the most number of metabolites (151 metabolites). The higher signal intensity for a greater number of metabolites allows for greater sensitivity in measuring a broader range of metabolites. This suggests that the method using methanol containing formic acid at 4°C is most suitable for unbiased metabolomics experiments.
Table 1.
List of metabolites that have large variation of MS signal across eight different extraction methods.
| Compound Name | Elemental Composition |
A_−80C | A_4C | B_−80C | B_4C_1 | PBS_−80C_1 | PBS_4C_1 | W_−80C_1 | W_4C_1 | Maxchange | CV (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Diphosphate* | H4O7P2 | 5.510E+06 | 5.970E+06 | 2.080E+06 | 2.371E+06 | 9.524E+05 | 4.492E+06 | 2.270E+04 | 9.133E+03 | 653.7 | 89.1 |
| 5-Amino-1-(5-Phospho-D-ribosyl)imidazole-4-carboxamide | C9H15N4O8P | 9.441E+05 | 1.177E+06 | 6.916E+05 | 8.762E+05 | 5.904E+04 | 6.340E+04 | 2.694E+03 | 3.396E+03 | 436.7 | 103.5 |
| sn-Glycero-3-phosphocholine* | C8H20NO6P | 9.193E+05 | 9.554E+05 | 4.558E+05 | 4.534E+05 | 4.337E+03 | 9.553E+03 | 1.541E+06 | 1.309E+06 | 355.3 | 80.7 |
| S-(2-Methylpropanoyl)-dihydrolipoamide | C12H23NO2S2 | 5.580E+05 | 5.421E+05 | 2.233E+05 | 2.331E+05 | 2.153E+03 | 1.331E+04 | 7.533E+04 | 6.916E+04 | 259.2 | 104.5 |
| dUMP | C9H13N2O8P | 1.884E+05 | 2.527E+05 | 2.317E+05 | 2.059E+05 | 1.000E+03 | 1.000E+03 | 2.276E+03 | 1.358E+03 | 252.7 | 106.9 |
| 3-dehydro-L-gulonate/D-glucuronate/L-iduronate | C6H10O7 | 1.373E+05 | 1.534E+05 | 9.957E+04 | 1.103E+05 | 1.000E+03 | 1.075E+03 | 1.607E+04 | 1.676E+04 | 153.4 | 96.4 |
| 2-oxoglutarate | C5H6O5 | 5.907E+04 | 6.052E+04 | 3.544E+04 | 3.933E+04 | 1.000E+03 | 4.277E+03 | 1.069E+05 | 1.419E+05 | 141.9 | 86.3 |
| Phosphoenol pyruvate | C3H5O6P | 8.209E+04 | 1.308E+05 | 3.165E+04 | 4.787E+04 | 1.000E+03 | 1.775E+04 | 2.535E+03 | 3.523E+03 | 130.8 | 116.3 |
| inosine | C10H12N4O5 | 5.510E+06 | 5.970E+06 | 2.080E+06 | 2.371E+06 | 9.524E+05 | 4.492E+06 | 2.270E+04 | 9.133E+03 | 653.7 | 148.8 |
| ADP | C10H15N5O10P2 | 9.441E+05 | 1.177E+06 | 6.916E+05 | 8.762E+05 | 5.904E+04 | 6.340E+04 | 2.694E+03 | 3.396E+03 | 436.7 | 96.4 |
Table 2.
Number of metabolites with highest or lowest MS signal intensity across all eight extraction methods in each extraction method.
| Extraction Method | # of Metabolites Highest Signal | # of Metabolites Lowest Signal |
|---|---|---|
| Acid solvent, −80C | 10 | 9 |
| (Neg:6, Pos:4) | (Neg:3, Pos:6) | |
| Acid solvent, 4C | 136 | 11 |
| (Neg:81, Pos:55) | (Neg:6, Pos:5) | |
| Neutral solvent, −80C | 52 | 16 |
| (Neg:30, Pos:22) | (Neg:10, Pos:6) | |
| Neutral solvent, 4C | 32 | 5 |
| (Neg:19, Pos:13) | (Neg:4, Pos:1) | |
| PBS wash | 0 | 151 |
| Neutral solvent, −80C | (Neg:0, Pos:0) | (Neg:89, Pos:62) |
| PBS wash | 10 | 44 |
| Neutral solvent, 4C | (Neg:8, Pos:2) | (Neg:28, Pos:16) |
| H2O wash | 7 | 34 |
| Neutral solvent, −80C | (Neg:3, Pos:4) | (Neg:16, Pos:18) |
| H2O wash | 38 | 40 |
| Neutral solvent, 4C | (Neg:19, Pos:19) | (Neg:28, Pos:12) |
4 Discussion
The range of metabolites that can be detected and quantitated by LC-HRMS is dependent on the metabolites that can be extracted from the biological samples. There is a great amount of variation in terms of metabolite detection and quantitation among the eight different extraction methods in this study, with each extraction parameter having different effects on the MS signal intensity.
Among the three extraction parameters, temperature in the range we have studied seems to have negligible effects on the measurement of most metabolites. Extraction between 4°C and −80°C exert little differences in the signal intensity of metabolites detected. This is contrary to the findings presented in previous studies, where temperatures of 0°C and −75°C was found to significantly affect metabolite levels for 16 out of 27 metabolites measured [19]. However, we examined a much larger range of metabolites of 285 metabolites. The lack of difference between temperatures could be due to the fast nature of metabolite quenching with the methanol extraction, whereby the quickness of the metabolite extraction minimizes metabolite degradation at temperature as high as 4°C.
Unlike the extraction parameter of temperature, the addition of formic acid, although not contributing to major overall effects on the global profile, seems to improve the signal intensity of metabolites for some chemical classes. Higher MS signal intensities were measured for a majority of amino acids and lipid-like metabolites with the addition of formic acid at 4°C. Some nucleotides (e.g. guanine, inosine, cytidine) and metabolites from glycolysis and TCA cycle (e.g. citrate/isocitrate and succinate) were also measured at higher signal intensities. This finding is in agreement with results reported that an acidic pH increases recovery of unknown metabolites, and increases the range of the metabolome recovered [27]. The increased recovery of metabolites due to an acidic pH might be due to the stabilization of charged compounds during the extraction process due to the increased acidity of the extraction solvent.
The last extraction parameter, washing of samples with either water or PBS before extraction of metabolites with the extraction solvent, seems to have the greatest effect on metabolite intensities among the three parameters studied. Washing decreased the signal intensities of metabolites for both metabolites found in the growth media and for metabolites found inside cells. The dramatic reduction of signal intensities is due several factors including the leakage of metabolites into water washing solution, and the ion suppression brought about by the introduction and saturation of ions in the PBS washing solution [23]. This is contrary to the common expectation that washing of samples allows better quantitation of intracellular metabolites by removing extracellular metabolites with minimal effects on intracellular metabolites [19, 20]. Hence, when quantifying intracellular metabolism, washing should be avoided as it severely reduces the ability of the LC-HRMS platform to measure and quantitate a wide variety of metabolites.
5 Concluding Remarks
Metabolite extraction parameters can play a significant role in determining the detection and quantitation of metabolites. In this study, we have shown that temperature (between −80°C and 4°C ) does not change metabolite quantitation while other parameters such as addition of acid to methanol extraction solvent and washing of samples before extraction play a big role in increasing or reducing the metabolite signals detected by mass spectrometry. As a general metabolomics extraction protocol, extraction with cold methanol without any washing is satisfactory, even though for few targeted metabolites or special application where intra and extra metabolites should be distinguished, an optimized extraction protocol should be chosen. This insights will allow us to improve our current extraction methods and provide a more robust and optimized method for metabolomics studies.
Supplementary Material
Acknowledgments
Work was supported by R00 CA168997 and RO1 AI110613 from the National Institutes of Health. We gratefully acknowledge members of the Locasale lab for their support and helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Patti GJ, Yanes O, Siuzdak G. Innovation: Metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012;13(4):263–9. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rabinowitz JD, et al. Metabolomics in drug target discovery. Cold Spring Harb Symp Quant Biol. 2011;76:235–46. doi: 10.1101/sqb.2011.76.010694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Witherspoon M, et al. Unbiased metabolite profiling indicates that a diminished thymidine pool is the underlying mechanism of colon cancer chemoprevention by alpha-difluoromethylornithine. Cancer Discov. 2013;3(9):1072–81. doi: 10.1158/2159-8290.CD-12-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sreekumar A, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457(7231):910–4. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 5.Dettmer K, Aronov PA, Hammock BD. Mass spectrometry-based metabolomics. Mass Spectrom Rev. 2007;26(1):51–78. doi: 10.1002/mas.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weckwerth W. Metabolomics in systems biology. Annual Review of Plant Biology. 2003;54:669–689. doi: 10.1146/annurev.arplant.54.031902.135014. [DOI] [PubMed] [Google Scholar]
- 7.Zhou B, et al. LC-MS-based metabolomics. Mol Biosyst. 2012;8(2):470–81. doi: 10.1039/c1mb05350g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Canelas AB, et al. Quantitative Evaluation of Intracellular Metabolite Extraction Techniques for Yeast Metabolomics. Analytical Chemistry. 2009;81(17):7379–7389. doi: 10.1021/ac900999t. [DOI] [PubMed] [Google Scholar]
- 9.t’Kindt R, et al. Joint GC-MS and LC-MS platforms for comprehensive plant metabolomics: repeatability and sample pre-treatment. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877(29):3572–80. doi: 10.1016/j.jchromb.2009.08.041. [DOI] [PubMed] [Google Scholar]
- 10.Theodoridis G, et al. LC-MS based global metabolite profiling of grapes: solvent extraction protocol optimisation. Metabolomics. 2012;8(2):175–185. [Google Scholar]
- 11.Sellick CA, et al. Evaluation of extraction processes for intracellular metabolite profiling of mammalian cells: matching extraction approaches to cell type and metabolite targets. Metabolomics. 2010;6(3):427–438. [Google Scholar]
- 12.Dietmair S, et al. Towards quantitative metabolomics of mammalian cells: Development of a metabolite extraction protocol. Analytical Biochemistry. 2010;404(2):155–164. doi: 10.1016/j.ab.2010.04.031. [DOI] [PubMed] [Google Scholar]
- 13.Wang XD, Wang YC. On-line extraction coupled with liquid chromatography tandem mass spectrometry for quantitation of pravastatin and its metabolite in human serum. Biomedical Chromatography. 2008;22(7):719–726. doi: 10.1002/bmc.989. [DOI] [PubMed] [Google Scholar]
- 14.Want EJ, et al. Solvent-dependent metabolite distribution, clustering, and protein extraction for serum profiling with mass spectrometry. Anal Chem. 2006;78(3):743–52. doi: 10.1021/ac051312t. [DOI] [PubMed] [Google Scholar]
- 15.Romisch-Margl W, et al. Procedure for tissue sample preparation and metabolite extraction for high-throughput targeted metabolomics. Metabolomics. 2012;8(1):133–142. [Google Scholar]
- 16.Ivanisevic J, et al. Toward ‘omic scale metabolite profiling: a dual separation-mass spectrometry approach for coverage of lipid and central carbon metabolism. Anal Chem. 2013;85(14):6876–84. doi: 10.1021/ac401140h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dettmer K, et al. Metabolite extraction from adherently growing mammalian cells for metabolomics studies: optimization of harvesting and extraction protocols. Analytical and Bioanalytical Chemistry. 2011;399(3):1127–1139. doi: 10.1007/s00216-010-4425-x. [DOI] [PubMed] [Google Scholar]
- 18.Kamphorst JJ, et al. Liquid chromatography-high resolution mass spectrometry analysis of fatty acid metabolism. Anal Chem. 2011;83(23):9114–22. doi: 10.1021/ac202220b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lorenz MA, Burant CF, Kennedy RT. Reducing time and increasing sensitivity in sample preparation for adherent mammalian cell metabolomics. Anal Chem. 2011;83(9):3406–14. doi: 10.1021/ac103313x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noack S, Wiechert W. Quantitative metabolomics: a phantom? Trends Biotechnol. 2014;32(5):238–44. doi: 10.1016/j.tibtech.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 21.Bi H, et al. Optimization of harvesting, extraction, and analytical protocols for UPLC-ESI-MS-based metabolomic analysis of adherent mammalian cancer cells. Anal Bioanal Chem. 2013;405(15):5279–89. doi: 10.1007/s00216-013-6927-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paglia G, et al. Monitoring metabolites consumption and secretion in cultured cells using ultra-performance liquid chromatography quadrupole-time of flight mass spectrometry (UPLC-Q-ToF-MS) Anal Bioanal Chem. 2012;402(3):1183–98. doi: 10.1007/s00216-011-5556-4. [DOI] [PubMed] [Google Scholar]
- 23.Leon Z, et al. Mammalian cell metabolomics: experimental design and sample preparation. Electrophoresis. 2013;34(19):2762–75. doi: 10.1002/elps.201200605. [DOI] [PubMed] [Google Scholar]
- 24.Munger J, et al. Dynamics of the cellular metabolome during human cytomegalovirus infection. PLoS Pathog. 2006;2(12):e132. doi: 10.1371/journal.ppat.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maharjan RP, Ferenci T. Global metabolite analysis: the influence of extraction methodology on metabolome profiles of Escherichia coli. Anal Biochem. 2003;313(1):145–54. doi: 10.1016/s0003-2697(02)00536-5. [DOI] [PubMed] [Google Scholar]
- 26.Liu X, Ser Z, Locasale JW. Development and quantitative evaluation of a high-resolution metabolomics technology. Anal Chem. 2014;86(4):2175–84. doi: 10.1021/ac403845u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sana TR, Waddell K, Fischer SM. A sample extraction and chromatographic strategy for increasing LC/MS detection coverage of the erythrocyte metabolome. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;871(2):314–21. doi: 10.1016/j.jchromb.2008.04.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.