Abstract
Lower grade (WHO II-III) gliomas vary widely in clinical behavior and are classified as astrocytic, oligodendroglial, or mixed forms. Anaplasia depends greatly on mitotic activity, with CDKN2A loss considered the most common mechanism for cell cycle dysregulation. We investigated whether loss of the CDKN2A gene is associated with overall survival across pathologically and genetically defined glioma subtypes. After adjusting for IDH mutation, sex, and age, CDKN2A deletion was strongly associated with poorer overall survival in astrocytomas but not in oligodendrogliomas or oligoastrocytomas. Molecular classification of astrocytomas by IDH mutation, TP53 mutation, and/or ATRX loss of expression revealed that CDKN2A loss in IDH/TP53 mutated tumors was strongly associated with worse overall survival. CDKN2A loss in IDH mutated tumors with ATRX loss was only weakly associated with worse overall survival. These findings suggest that CDKN2A testing may provide further clinical aid in lower-grade glioma sub-stratification beyond IDH mutation and 1p19q codeletion status, particularly in IDH/TP53 mutated astrocytomas.
Keywords: Astrocytoma, ATRX, Biomarker, CDKN2A, IDH, Infiltrating glioma, Oligodendroglioma, p16, TP53
INTRODUCTION
Infiltrating gliomas comprise approximately 60% of primary malignant intracranial tumors (1) and have a devastating course given their widespread invasiveness, tendency toward biological progression, and resistance to available adjuvant therapies. The World Health Organization (WHO) classifies infiltrating gliomas as diffuse (WHO grade II) astrocytoma (A-II), oligodendroglioma (O-II), or oligoastrocytoma (OA-II), anaplastic (WHO grade III) astrocytoma (A-III), oligodendroglioma (O-III), or oligoastrocytoma (OA-III), and glioblastoma ([GBM]; WHO grade IV) (2). While diagnostic reproducibility among neuropathologists is high for GBM, there is far lower concordance for cell type determination in other subtypes and in the distinction between WHO grades II and III. The latter difficulty stems in part from different grading criteria for astrocytic and oligodendroglial tumors, vagaries in WHO grading criteria, and inter-observer variability in the detection of mitotic figures and microvascular proliferation. Additionally, whereas molecular criteria have been published recently to distinguish diffuse astrocytomas from oligodendrogliomas in adult patients more objectively, it remains unclear whether previously devised grading criteria based solely on morphology can be appropriately applied to newly defined glioma subtypes, which are based mostly on molecular definitions (3). As such, the development of clinically useful “grading” biomarkers for molecularly defined glioma subsets is sorely needed to improve diagnostic reproducibility.
The large majority of grade II and III gliomas, as well as so-called secondary GBM that derive from lower grade gliomas, harbor mutations in an IDH gene, with IDH1 being most common (4–7). Mutations in TP53 and ATRX also occur frequently and are present in approximately 60% to 70% of tumors of astrocytic differentiation (2). In a recent study of lower-grade gliomas, ATRX mutation was closely associated with TP53 mutation and was restricted to IDH mutated tumors (8). A consequence of ATRX mutation is activation of the alternative lengthening of telomeres pathway and telomerase-independent immortalization of tumor cells (9, 10). Conversely, approximately 70% of oligodendrogliomas harbor 1p19q codeletion and lack TP53 and ATRX alterations (2, 8, 11–14). Approximately 80% of oligodendrogliomas have mutations in the TERT gene promoter, which reactivates telomerase activity (15). Mixed oligoastrocytomas remain poorly defined molecularly, with the vast majority showing classic molecular features of either astrocytoma or oligodendroglioma (16).
In addition to the alterations described above, homozygous and hemizygous losses involving 9p21 have been observed at high frequency in infiltrating gliomas, with homozygous loss being most common (17–20). One of the consequences of 9p21 deletion is loss of the cyclin-dependent kinase inhibitor CDKN2A gene, which results in cellular proliferation and dysregulation of pro-apoptotic pathways (21). Cell cycle progression from G1 to S phase relies on complex formation between cyclin dependent-kinases CDK4 or CDK6 and D-type cyclins, which subsequently leads to phosphorylation of the retinoblastoma (RB1) protein, release of the elongation factor (EF2) transcriptional factor, and activation of genes involved in G1 to S transition (22, 23). Alterations in this pathway appear to be rare in grade II gliomas but are frequent in higher-grade (III and IV) tumors, suggesting a key role for the CDKN2A–CDK4-RB pathway in malignant progression (24–26). Since the main criteria for anaplastic designation in gliomas are either the presence of mitoses (astrocytomas) or high mitotic activity (oligodendroglial tumors), loss of the CDKN2A gene or p16 protein (the CDKN2A product) appears an ideal candidate for distinguishing the molecular phenotypes of WHO grade II and III gliomas.
Several studies have reported worse prognosis for CDKN2A loss in gliomas (27–33). However, these studies were conducted before the realization that IDH mutation is an independent favorable prognostic factor in adult type diffuse gliomas (6, 34, 35). Therefore, it is unclear whether CDKN2A loss remains a statistically significant prognostic factor after correcting for the effect of IDH mutation. Furthermore, prior studies have not explored the effect of CDKN2A loss on the newly defined ‘molecular’ grade II-III astrocytomas, i.e. IDH mutated, TP53 mutated, often with associated loss of ATRX protein expression, or ‘molecular’ grade II-III oligodendrogliomas, i.e. that are IDH mutated and 1p19q codeleted. Our study addresses these questions by testing the hypothesis that CDKN2A loss determined by fluorescence in situ hybridization (FISH) and/or loss of p16 protein expression determined by immunohistochemistry (IHC) can enhance prognostication of overall survival in molecularly-characterized lower-grade (WHO II-III) adult type diffuse gliomas. If so, these molecular biomarkers could potentially enhance future WHO grading criteria and improve inter-observer concordance rates among pathologists.
MATERIALS AND METHODS
Study Participants and Selection Criteria
Cases were selected from among participants of the San Francisco Bay Area Adult Glioma Study (AGS), which was conducted at the University of California, San Francisco, as previously described (36, 37). Briefly, the AGS enrolled patients who were newly diagnosed with a histologically confirmed glioma at age ≥18 years between 1991 and 2012. All participants gave informed consent, and the study was conducted under protocols approved by the UCSF Institutional Review Board.
The following criteria were used to select cases from the AGS for this analysis: Only cases classified as grade II or III astrocytoma, oligodendroglioma, or oligoastrocytoma during the study’s neuropathology review were included. In addition, only cases with sufficient available tissue for the planned assays were included. Because these subjects were also a subset of another study relating tumor markers to inherited single nucleotide polymorphism data, subjects were of European ancestry and had constitutive DNA available. Cases were prioritized with respect to available mutational status for isocitrate dehydrogenase (IDH1 or IDH2) obtained by DNA sequencing as previously described (37, 38). Tumor TP53 mutation was also available for many of the subjects and assayed as described previously (39).
FISH Analysis
We assessed CDKN2A copy number alterations by FISH using commercially available probes (LSI CDKN2A (9p21) orange and CEP9 green Spectrum; Abbott Laboratories, North Chicago, IL). In brief, green and orange fluorescent signals were enumerated under an Olympus BX60 fluorescence microscope with appropriate filters (Olympus, Melville, NY). One hundred non-overlapping nuclei were assessed for numbers of green and red signals, and cases were considered deleted regardless of whether the pattern suggested hemizygous or homozygous loss. An interpretation of deletion was made when the orange to green ratio was <0.8. Partial hybridization failure was ruled out by evaluating orange and green signals within vascular endothelial cells as an internal control. Failed or weak hybridizations were repeated; uninterpretable cases were considered “non-informative.” Interpretable results were available for 253 cases (93.7%).
FISH images were captured using a black and white, high-resolution COHU CCD camera, Z-stack motor and a CytoVision basic workstation (Applied Imaging, Santa Clara, CA) with sequential DAPI (1 level), FITC (5 levels), and rhodamine (5 levels) filter settings. The resulting images were reconstituted with blue, green, and orange pseudocolors using CytoVision software. 1p19q deletion status was assayed according to published methods (40, 41). Tumors classified histologically as astrocytoma were not assayed for 1p19q deletion.
Immunohistochemistry
Immunohistochemistry for ATRX and p16 was performed at the Brain Tumor Research Center and Clinical Immunohistochemistry Laboratories, University of California San Francisco, utilizing an automated staining processor (Ventana, Tucson, AZ). ATRX loss of expression was assessed using the rabbit polyclonal antibody (HPA001906; 1:100, Sigma, St. Louis, MO). Only cases with retained staining of endothelial cells (internal positive control) were considered interpretable. For p16 expression, the Mtm Lab kit (Ventana) and the JC8 mouse antibody (1:100, Santa Cruz Biotechnology, Dallas, TX) were used, with the majority of cases evaluated using the Mtm Lab kit. Quantitative analysis of the staining pattern for 10 cases using both antibodies demonstrated a high correlation (r2 = 0.8; p = 0.0004). Nuclear staining was required for a positive p16 result.
Image acquisition for p16 quantification was done using an Olympus BX-41 microscope, 20X objective, and Olympus DP72 digital camera. Areas of maximal staining were photographed for assessing labeling index. JPEG images were imported into Image J (http://imagej.nih.gov/ij/), and the ImmunoRatio plug-in (http://153.1.200.58/sites/default/files/software/immunoratio-plugin/index.html) was used to determine labeling index. Approximately 1000 cells were required for an interpretable result.
Statistics and Survival Analysis
Comparisons among categorical variables were made using chi-square and Fisher exact tests. Cox proportional hazards regression was employed to estimate overall survival. Models built based on histologic subtype (astrocytoma, oligodendroglioma, oligoastrocytoma) or pathologic diagnosis (A-II, A-III, O-II, O-III, AO-II, AO-III) were adjusted for sex, age, and IDH status while models built based on molecular parameters (IDH, TP53, ATRX, 1p19q status) were adjusted only for sex and age. Molecular subtypes of astrocytoma were defined based on the presence of IDH/TP53 mutations (IDH/TP53 group), combined IDH/TP53 mutations with ATRX loss (IDH/TP53/ATRX group), or IDH mutation and ATRX loss (IDH/ATRX group). Molecular oligodendroglioma was defined as IDH mutated, 1p19q codeleted tumors. A logistic analysis was used to evaluate the association between CDKN2A loss by FISH and p16 labeling index by IHC, stratified by histologic subtype or pathologic diagnosis. The area under the curve was obtained from the receiver operating characteristic curve (ROC), which provides a graphical representation of the relationship between false-positive and true-positive rates (true-positive as defined by FISH). The optimal labeling index for p16 was extracted from the ROC table and used to dichotomize the continuous p16 labeling index data into "deleted" and "intact" groups. Results with a p value <0.05 were deemed significant.
RESULTS
Patient Characteristics
The clinical and pathological characteristics of the 270 study subjects are summarized in Table 1. The percentages of astrocytomas and oligodendrogliomas were similar (41.9% and 38.5%, respectively); tumors with mixed histology comprised 19.6% of cases. The median follow-up time was 10.6 years (mean: 10.5 years; range 0.2–22.3 years), with 105 deaths (38.9%) and 165 patients (61.1%) alive at last follow-up.
Table 1.
Distribution of histologic subtype, pathologic diagnosis, demographic, and clinical characteristics of glioma patients from the UCSF Adult Glioma Study.
| N | Percent | Mean age (y) | SE (y) | Median age (y) | |
|---|---|---|---|---|---|
| All Cases | 270 | 100.0% | 42.2 | 0.7 | 41.0 |
| Histology | |||||
| Astrocytoma | 113 | 41.9% | 42.4 | 1.3 | 41.0 |
| Oligodendroglioma | 104 | 38.5% | 44.5 | 1.0 | 44.5 |
| Oligoastrocytoma | 53 | 19.6% | 36.9 | 1.3 | 36.0 |
| Pathology | |||||
| Diffuse astrocytoma (A-II) | 49 | 18.2% | 41.4 | 1.9 | 40.0 |
| Anaplastic astrocytoma (A-III) | 64 | 23.7% | 43.2 | 1.8 | 41.5 |
| Oligodendroglioma (O-II) | 80 | 29.6% | 43.9 | 1.2 | 44.0 |
| Anaplastic oligodendroglioma (O-III) | 24 | 8.9% | 46.8 | 2.0 | 51.0 |
| Oligoastrocytoma (OA-II) | 42 | 15.6% | 37.7 | 1.6 | 36.0 |
| Anaplastic oligoastrocytoma (OA-III) | 11 | 4.1% | 34.1 | 2.4 | 32.0 |
| Gender | |||||
| Male | 147 | 54.4% | |||
| Female | 123 | 45.6% | |||
| Surgery | |||||
| Biopsy | 20 | 7.4% | |||
| Resection | 250 | 92.6% | |||
| Adjunct therapy | |||||
| Chemotherapy | |||||
| No | 107 | 40.7% | |||
| Yes | 154 | 58.6% | |||
| Unknown | 2 | 0.8% | |||
| Temozolomide | 129 | 47.8% | |||
| Radiotherapy | |||||
| No | 127 | 47.0% | |||
| Yes | 143 | 53.0% | |||
| Outcome | |||||
| Alive | 165 | 61.1% | |||
| Dead | 105 | 38.9% |
CDKN2A Loss Occurred More Frequently in Astrocytoma than Other Histologic Subtypes and Was Present in Relatively Similar Percentages in A-II and A-III
Examples of CDKN2A results by FISH are illustrated in Figure 1. The frequency of loss for each group is summarized in Table 2 and illustrated in Supplementary Figure 1. Astrocytomas demonstrated the highest frequency of CDKN2A loss (astrocytoma: 52/108 cases (55.3%); oligodendroglioma: 23/96 cases (24.5%); oligoastrocytoma: 19/49 cases (20.2%); p = 0.006, chisquare). No significant difference was seen in the relative percentage of deleted cases in A-II vs. A-III tumors (A-II: 21/47 cases (44.7%); A-III: 31/61 cases (50.8%); p = 0.6, Fisher exact test).
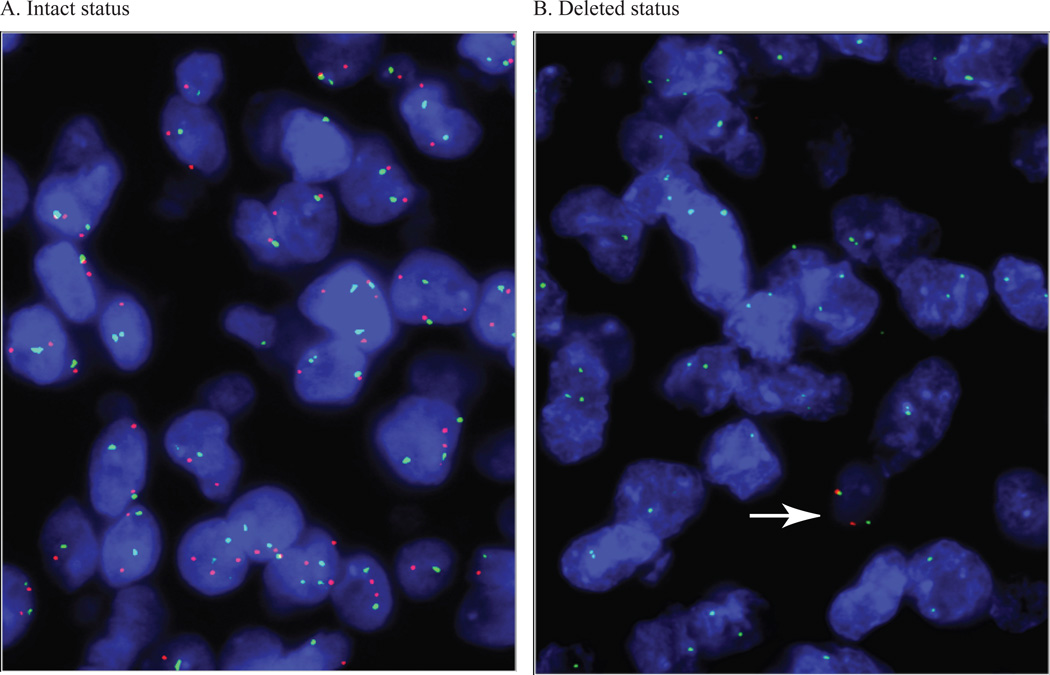
Figure 1. Fluorescence in situ hybridization for CDKN2A.
(A, B) Results of fluorescence in situ hybridization demonstrating intact (A: CEP9/CDKN2A ratio near one) and deleted (B) CDKN2A status. In the latter case, a non-neoplastic cell containing 2 green CEP9 signals and 2 red CDKN2A signals is seen (white arrow), whereas all the tumor cells have only CEP9 signals.
Table 2.
Distribution of CDKN2A, IDH, ATRX, TP53 and 1p19q status by pathologic diagnosis in tumors from glioma patients in the UCSF Adult Glioma Study
| All cases | Diffuse astrocytoma (A-II) |
Anaplastic astrocytoma (A-III) |
Oligodendroglioma (O-II) |
Anaplastic oligodendroglioma (O-III) |
Oligoastrocytoma (OA-II) |
Anaplastic oligoastrocytoma (OA-III) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Percent | N | Percent | N | Percent | N | Percent | N | Percent | N | Percent | N | Percent | |
| CDKN2A | ||||||||||||||
| Intact | 159 | 63% | 26 | 55% | 30 | 49% | 57 | 77% | 16 | 73% | 25 | 63% | 5 | 56% |
| Deleted | 94 | 37% | 21 | 45% | 31 | 51% | 17 | 23% | 6 | 27% | 15 | 38% | 4 | 44% |
| Total | 253 | 47 | 61 | 74 | 22 | 40 | 9 | |||||||
| IDH | ||||||||||||||
| Wild type | 65 | 24% | 10 | 21% | 26 | 41% | 9 | 11% | 8 | 33% | 8 | 19% | 4 | 36% |
| Mutated | 202 | 76% | 37 | 79% | 37 | 59% | 71 | 89% | 16 | 67% | 34 | 81% | 7 | 64% |
| Total | 267 | 47 | 63 | 80 | 24 | 42 | 11 | |||||||
| ATRX | ||||||||||||||
| Intact | 156 | 60% | 22 | 47% | 24 | 39% | 68 | 86% | 20 | 87% | 18 | 44% | 4 | 36% |
| Lost | 106 | 40% | 25 | 53% | 37 | 61% | 11 | 14% | 3 | 13% | 23 | 56% | 7 | 64% |
| Total | 262 | 47 | 61 | 79 | 23 | 41 | 11 | |||||||
| TP53 | ||||||||||||||
| Wild type | 107 | 63% | 9 | 31% | 24 | 48% | 42 | 89% | 14 | 82% | 13 | 65% | 5 | 63% |
| Mutated | 64 | 37% | 20 | 69% | 26 | 52% | 5 | 11% | 3 | 18% | 7 | 35% | 3 | 38% |
| Total | 171 | 29 | 50 | 47 | 17 | 20 | 8 | |||||||
| 1p19q | ||||||||||||||
| Intact | 65 | 42% | NA* | NA* | 19 | 25% | 5 | 23% | 29 | 74% | 8 | 89% | ||
| Codeleted | 89 | 58% | 58 | 75% | 17 | 77% | 10 | 26% | 1 | 11% | ||||
| Total | 154 | 77 | 22 | 39 | 9 | |||||||||
NA, 1p19q data not available for tumors classified histologically as astrocytoma.
The results for p16 expression by IHC are summarized in Supplementary Table 1. The p16 labeling index was highly variable across cases within a given histopathologic group.
CDKN2A Loss Was Associated with Poorer Survival in Astrocytomas But Not Oligodendrogliomas or Oligoastrocytomas
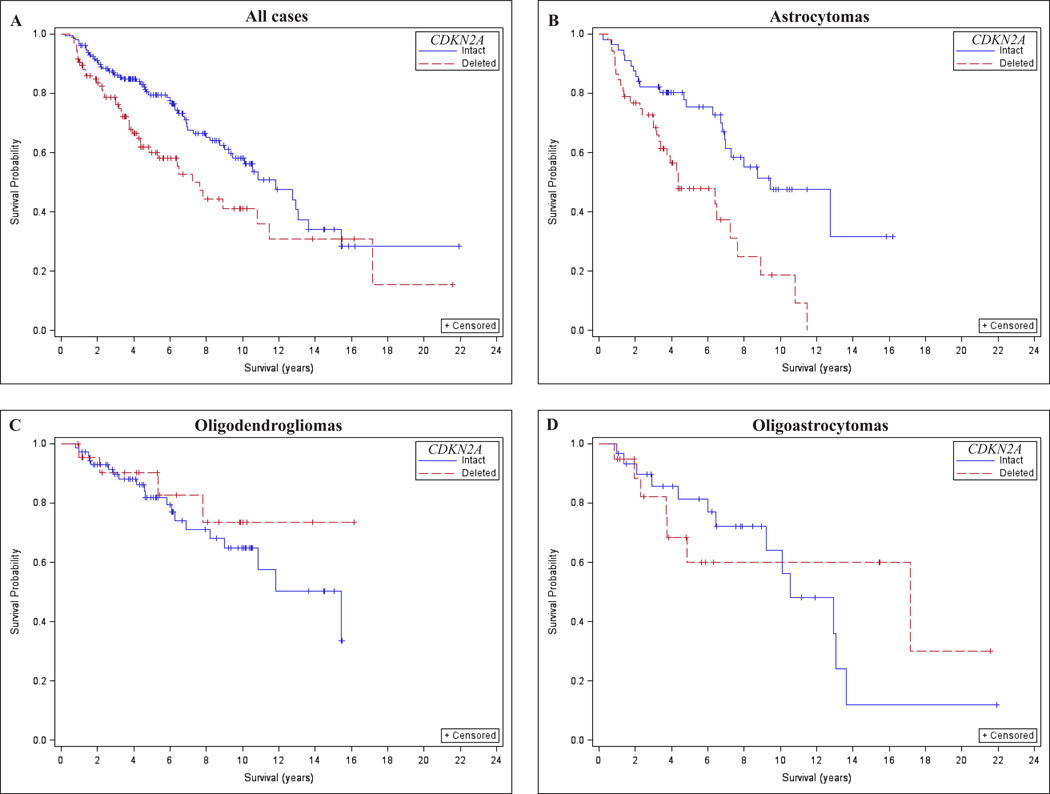
As hypothesized, CDKN2A loss determined by FISH was associated with significantly worse overall survival in grade II and III gliomas after adjusting for age, sex, and IDH mutation (Table 3; hazard ratio [HR] = 1.6, 95% confidence interval [CI] = 1.0–2.4, p = 0.03). This result was mostly related to a strong adverse effect of CDKN2A loss among astrocytoma patients; HR = 2.0, 95% CI = 1.1–3.5 (Table 3; Fig. 2A–D). Results were similar for grade II and III astrocytomas (Table 3). There were non-significant inverse associations of CDKN2A loss and overall survival among patients with oligodendroglioma or oligoastrocytoma (Table 3). Thus, CDKN2A loss was associated with poor survival among patients with astrocytoma, but this effect was not observed among patients with oligodendroglioma or oligoastrocytoma.
Table 3.
Association of CDKN2A loss by fluorescence in situ hybridization with overall survival stratified by histologic subtype, pathologic diagnosis, and molecular subtypes using Cox proportional hazards models in glioma patients from the UCSF Adult Glioma Study
| CDKN2A intact | CDKN2A deleted | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | N deceased |
Mean overall survival (years) |
Median overall survival (years) |
N | N deceased |
Mean overall survival (years) |
Median overall survival (years) |
HR | 95% CI | p-value | HR | 95% CI | p-value | |
| Unadjusted | Adjusted for age and sex | |||||||||||||
| All cases* | 159 | 56 | 10.3 | 11.8 | 94 | 42 | 8.8 | 7.2 | 1.7 | 1.1–2.5 | 0.01a | 1.6 | 1.0–2.4 | 0.03 |
| Astrocytoma* | 56 | 23 | 8.7 | 9.5 | 52 | 31 | 5.5 | 4.4 | 2.6 | 1.5–4.6 | 0.0008a | 2.0 | 1.1–3.5 | 0.02 |
| Oligodendroglioma* | 73 | 20 | 11.1 | 15.4 | 19 | 4 | 7.0 | 0.7 | 0.2–1.7 | 0.4 | 0.7 | 0.2–2.00 | 0.5 | |
| Oligoastrocytoma* | 30 | 13 | 9.8 | 10.6 | 23 | 7 | 11.5 | 17.2 | 0.9 | 0.3–2.2 | 0.7 | 0.8 | 0.3–2.4 | 0.7 |
| A-II* | 26 | 9 | 10.2 | 12.8 | 21 | 9 | 6.6 | 6.4 | 3.9 | 1.4–11.2 | 0.01a | 2.7 | 0.8–8.8 | 0.1 |
| A-III* | 30 | 14 | 5.0 | 6.9 | 31 | 22 | 4.7 | 3.3 | 2.1 | 1.1–4.2 | 0.03a | 1.8 | 0.8–3.6 | 0.1 |
| O-II* | 57 | 14 | 9.4 | 11.8 | 17 | 2 | 7.6 | NA | 0.4 | 0.07–1.6 | 0.3 | 0.4 | 0.07–1.5 | 0.2 |
| O-III* | 16 | 6 | 11.2 | 15.4 | 6 | 2 | 1.9 | NA | 1.4 | 0.2–6.6 | 0.7 | 1.6 | 0.2–9.9 | 0.6 |
| OA-II* | 25 | 12 | 9.6 | 10.6 | 15 | 5 | 12.3 | 17.2 | 0.7 | 0.2–2.0 | 0.5 | 1.0 | 0.3–3.1 | 0.9 |
| OA-III* | 5 | 1 | 1.0 | NA | 4 | 2 | 3.4 | 3.7 | 2.8 | 0.3–60.0 | 0.4 | 0.2 | 0.0004–21.2 | 0.6 |
| IDH/TP53 mutated, ATRX lost | 21 | 10 | 10.2 | 12.9 | 21 | 14 | 6.4 | 4.8 | 3.3 | 1.4–8.8 | 0.008a | 3.4 | 1.4–8.4 | 0.008a |
| IDH/TP53 mutated | 28 | 12 | 10.6 | 12.8 | 22 | 15 | 6.3 | 4.8 | 4.3 | 1.8–10.1 | 0.001a | 4.4 | 1.8–10.3 | 0.0008a |
| IDH mutated, ATRX lost | 49 | 19 | 9.6 | 9.2 | 39 | 18 | 8.7 | 7.8 | 1.6 | 0.8–3.0 | 0.2 | 1.6 | 0.8–3.0 | 0.2 |
| IDH mutated, 1p19q codeleted** | 56 | 16 | 9.2 | 10.6 | 16 | 0 | NA | NA | NA | NA | NA | NA | NA | NA |
HR, Hazard ratio; CI, Confidence interval; NA, Median survival is not available due to lack of events.
Hazard ratios for histologic groups and pathologic diagnosis were adjusted for IDH mutation status in addition to age and sex.
These tumors were also ATRX intact.
A-II, Diffuse astrocytoma; A-III, Anaplastic astrocytoma; O-II, Oligodendroglioma, O-III, Anaplastic oligodendroglioma, OA-II, Oligoastrocytoma; OA-III, Anaplastic oligoastrocytoma.
statistical significance at the 0.05 level.
Figure 2.
Effect of CDKN2A loss on overall survival of histologically classified lower grade (World Health Organization II-III) gliomas. (A–D) CDKN2A loss was associated with worse overall survival in lower grade gliomas not stratified by histology (A) and in astrocytomas (B), but not in oligodendrogliomas (C) or oligoastrocytomas (D).
Logistic analysis demonstrated a poor association between CDKN2A deletion by FISH and p16 expression by IHC (Supplementary Table 2). Furthermore, the loss of p16 expression by IHC was weakly associated with poor overall survival in lower-grade (II-III) gliomas (Supplementary Table 3).
CDKN2A Loss Was Strongly Associated With Poor Survival in Certain Molecular Subsets of Astrocytoma
As an alternative to using the histologic classification, we evaluated the effect of CDKN2A loss on the overall survival of certain molecular subsets of grade II-III astrocytoma, defined as IDH mutated tumors with both TP53 mutation and ATRX loss (termed IDH/TP53/ATRX group), or either combined with TP53 mutation (termed IDH/TP53 group) or ATRX loss (termed IDH/ATRX group) (Table 2; Supplementary Fig. 1). The co-occurrence of IDH, TP53, and ATRX alterations is provided in Table 4. Among 123 IDH mutated tumors with known TP53 and ATRX status, 42 cases (34.1%) harbored both mutation in TP53 and loss of ATRX expression, 21 harbored only ATRX loss (17.1%), and 9 harbored only TP53 mutation (7.3%) (Table 4). Fifty-one cases (41.5%) showed no alteration in either TP53 or ATRX, of which 38 (74.5%) were 1p19q codeleted “molecular oligodendroglioma.” Overall, a strong association was noted for the co-occurrence of TP53 mutation and ATRX loss (p < 0.0001, Fisher exact test). Similarly, a strong association was found for the co-occurrence of IDH mutation and ATRX loss both in IDH mutated cases with 1p19q intact status (p = 0.0004, Fisher exact test; Table 4) and IDH mutated cases not evaluated for 1p19q since they were classic astrocytomas on histopathology (p < 0.0001; Fisher exact test; Table 4); the latter was similar to observations made in IDH/TP53 mutated cases. These data support a strong association for the co-occurrence of IDH mutation with TP53 mutation and/or ATRX loss in grade II-III astrocytomas.
Table 4.
Distribution of IDH/TP53 mutation status and ATRX loss of expression in tumors of patients from the UCSF Adult Glioma Study
| IDH mutated cases with both ATRX and TP53 data available (n = 123) | ||||
| ATRX | TP53 | N | Percent | Fisher Exact test p-value |
| Intact | Wild type | 51 | 41.5% | <0.0001* |
| Intact | Mutated | 9 | 7.3% | |
| Lost | Wild type | 21 | 17.1% | |
| Lost | Mutated | 42 | 34.1% | |
| Total | 123 | |||
| Cases with both IDH and ATRX data available (N=259) | ||||
| IDH | ATRX | N | Percent | Fisher’s Exact test p-value |
| Wild type | Intact | 47 | 18.1% | 0.0017* |
| Wild type | Lost | 14 | 5.4% | |
| Mutated | Intact | 108 | 41.7% | |
| Mutated | Lost | 90 | 34.7% | |
| Total | 259 | |||
| Cases with both IDH and ATRX data available (stratified by 1p19q status) 1p19q codeleted (N=87) | ||||
| IDH | ATRX | N | Percent | Fisher’s Exact test p-value |
| Wild type | Intact | 5 | 5.7% | 0.3 |
| Wild type | Lost | 1 | 1.1% | |
| Mutated | Intact | 78 | 89.7% | |
| Mutated | Lost | 3 | 3.4% | |
| Total | 87 | |||
| 1p19q intact (N=63) | ||||
| IDH | ATRX | N | Percent | Fisher’s Exact test p-value |
| Wild type | Intact | 16 | 25.4% | 0.0004* |
| Wild type | Lost | 7 | 11.1% | |
| Mutated | Intact | 9 | 14.3% | |
| Mutated | Lost | 31 | 49.2% | |
| Total | 63 | |||
| 1p19q not assayed (tumors histologically classified as astrocytoma; N=101) | ||||
| IDH | ATRX | N | Percent | Fisher’s Exact test p-value |
| Wild type | Intact | 26 | 25.7% | <0.0001* |
| Wild type | Lost | 6 | 5.9% | |
| Mutated | Intact | 17 | 16.8% | |
| Mutated | Lost | 52 | 51.5% | |
| Total | 101 | |||
Statistical significance at the 0.05 level.
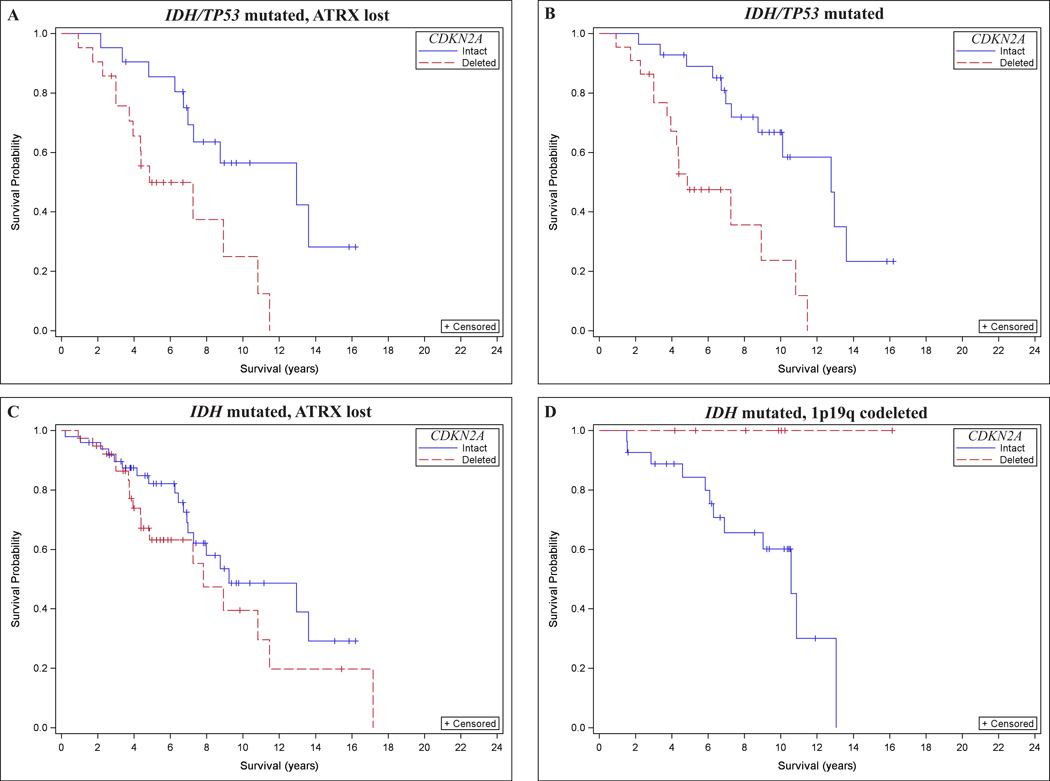
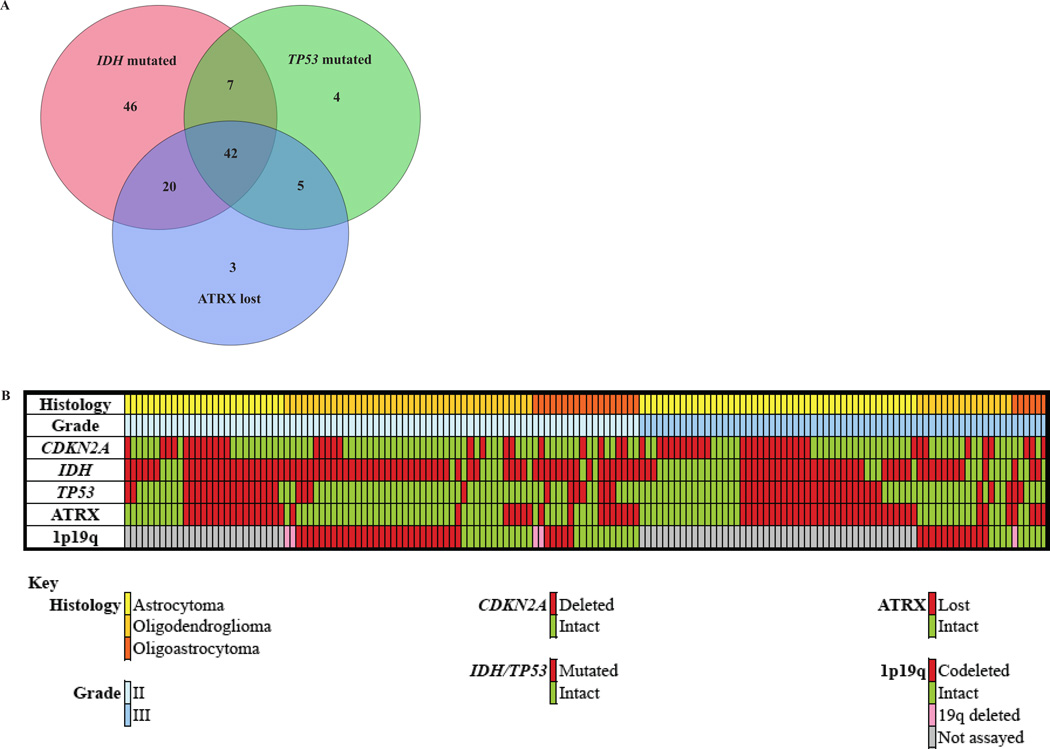
In the IDH/TP53/ATRX group, CDKN2A loss was associated with worse overall survival after adjusting for age and sex (Fig. 3A; HR = 3.4, 95% CI = 1.4–8.4, p = 0.008; Table 3). CDKN2A loss was strongly associated with poor survival in IDH/TP53 mutated tumors (Fig. 3B; HR = 4.4, 95% CI = 1.8–10.3, p = 0.0008; Table 3), but this association did not reach statistical significance in the IDH/ATRX group (Fig. 3C; HR = 1.6, 95% CI = 0.8–3.0, p = 0.2; Table 3). The 2 groups overlapped partially, such that of the 115 IDH mutated cases with known status for CDKN2A, TP53, and ATRX, 42 cases (36.5%) showed ATRX loss and TP53 mutation, 7 cases (6.1%) were ATRX intact and TP53 mutated, and 20 cases (17.4%) showed ATRX loss without TP53 mutation (Fig. 4). The IDH/TP53 and IDH/ATRX groups contained a similar proportion of grade II and grade III tumors. Furthermore, the subjects in each group were of nearly identical age at the time of diagnosis (IDH/TP53: 36.5 ± 1.3 years; IDH/ATRX: 36.4 ± 1.2 years, mean ± SE). Thus, in both histologically and molecularly defined astrocytoma (in particular, in astrocytomas defined by the presence of IDH and TP53 mutations), CDKN2A loss was strongly associated with poor survival after adjusting for age and sex.
Figure 3.
Effect of CDKN2A loss on overall survival in subsets of molecularly defined astrocytomas and oligodendrogliomas. (A–C) CDKN2A loss by fluorescence in situ hybridization (FISH) was associated with worse overall survival in IDH/TP53 mutated tumors with ATRX loss (IDH/TP53/ATRX group) (A) and IDH/TP53 mutated tumors (IDH/TP53 group) (B) but not in IDH mutated tumors with ATRX loss (IDH/ATRX group) (C). (D) Patients with molecularly defined oligodendrogliomas in the CDKN2A deleted group appeared to have improved survival; however, the statistical significance of the association between CDKN2A loss and overall survival could not be accurately assessed because none of the subjects in the CDKN2A deleted group died during the follow-up period.
Figure 4.
Overlap between IDH/TP53 and IDH/ATRX groups. (A) The astrocytoma groups defined by IDH/TP53 mutation (IDH/TP53 group) or IDH mutated tumors with ATRX loss (IDH/ATRX group) showed partial overlap such that among 115 IDH mutated cases with known status for CDKN2A IDH TP53, and ATRX, 42 cases (36.5%) showed ATRX loss and TP53 mutation, 7 cases (6.1%) were TP53 mutated and ATRX intact; 20 cases (17.4%) were ATRX intact and TP53 wild type. (B) OncoPrint of molecular parameters grouped by histology and grade.
With respect to molecularly defined oligodendrogliomas, the effect of CDKN2A loss on IDH mutated, 1p19q codeleted tumors was examined (Fig. 3D). This group consisted of 16 patients with no deaths (Table 3); therefore, a comparison using Cox proportional hazards would not be reliable and an estimate of the association between CDKN2A loss and survival in molecular oligodendroglioma could not be determined.
DISCUSSION
This study demonstrates an independent adverse effect of CDKN2A loss on the overall survival of patients with lower grade (WHO II-III) astrocytomas after accounting for IDH mutation, age, and sex. Tumors for 270 patients were classified and graded according to WHO criteria by expert neuropathologists, and molecular data for CDKN2A, IDH, TP53, ATRX, and 1p19q were included. While most patients (61.1%) were alive at censoring, the survival analysis yielded unequivocal results with good concordance between the histologic and molecular analysis, in particular for the molecular subset of astrocytoma defined by IDH/TP53 mutation. Unfortunately, a conclusion could not be reached for molecular oligodendrogliomas because of the absence of deaths in the CDKN2A deleted group. This analysis will require a follow-up interval longer than the median 10.9 years obtained for the molecular oligodendroglioma subgroup included here.
In previous studies, loss of CDKN2A was associated with worse survival in both astrocytomas and oligodendrogliomas (27–33). This study shows that CDKN2A loss is associated with worse survival in astrocytomas, but this effect was not observed in oligodendrogliomas. The disagreement between the findings here and those of prior studies may stem from the powerful effect of IDH mutation on survival, which was not considered previously because the role of IDH mutation in gliomas was not yet known. However, we cannot exclude other factors such as limited statistical power in the oligodendroglioma subgroup since there were few deaths among these patients in our cohort.
Most cases of CDKN2A loss in infiltrating gliomas are homozygous deletions in the 9p21 region (17–20). Such genomic alterations would be predicted to result in loss of p16 protein expression. Surprisingly, the expression of p16 by IHC correlated poorly with CDKN2A deletion by FISH. Several factors could explain this discrepancy. For example, while hemizygous losses are relatively straightforward to detect by FISH, the loss of p16 expression might be minimal and result in no significant detectable change in p16 expression by IHC. Indeed, in relating our findings to those of the The Cancer Genome Atlas (TCGA) lower-grade glioma data set, (publically available at http://www.cbioportal.org/), a significant decrease in CDKN2A mRNA expression was observed only for homozygous deletion of the CDKN2A gene but not hemizygous deletion (Supplementary Fig. 2). Alternatively, CDKN2A gene expression may be lost through promoter hypermethylation (42, 43) or point mutations (26), which are seen in a small subset of cases but are undetectable by FISH. If both copies of CDKN2A were inactivated by either mechanism, p16 expression by IHC would be expected to decrease while the status of CDKN2A by FISH would be “intact.” Regardless of the reason(s) for the discrepancy, it should be emphasized that the strong association between CDKN2A loss by FISH and poor survival in astrocytoma, as opposed to the weak association between p16 loss of expression by IHC and poor survival in astrocytoma, argues strongly in favor of using FISH to evaluate the status of CDKN2A when prognosticating astrocytomas.
The analysis of molecular subsets of astrocytomas revealed a strong association between CDKN2A loss and poor survival in patients harboring IDH/TP53 mutations, but a weaker effect was observed in the subset of molecular astrocytomas defined by IDH mutation and ATRX loss of expression. The concordance between ATRX loss and TP53 mutation in our study (76%) was similar to that reported in previous studies on ATRX (range: 71%-78%) (8, 10, 44). Though a small fraction of cases with missense mutations (~10%) may be missed by IHC, the majority of mutations in ATRX result in loss of protein expression (9, 44). Prior data strongly suggest that biological differences may exist between the IDH/TP53 and IDH/ATRX groups (9, 44–46). For example, other pathways of gliomagenesis, including telomerase-dependent mechanisms (47, 48), could predominate in IDH/TP53 mutated tumors with intact ATRX. Alternatively, mutations in other genes involved in the alternative lengthening of telomeres pathway could be present. Future studies focusing on these questions should help clarify this issue.
Whether the combination of IDH mutation and TP53 mutation or the combination of IDH mutation and ATRX loss of expression should be considered the “gold standard” for defining molecular astrocytomas in the clinical setting remains unresolved. The strong association between CDKN2A loss and poor survival in IDH/TP53 astrocytomas reported here relied on determination of CDKN2A status by FISH and TP53 mutation by sequencing. The poor correlation between CDKN2A status by FISH and p16 loss by IHC shows that the assay of choice can play a crucial role. Since molecular information on these markers will be an integral component of the standard integrated diagnoses in clinical reports as recently proposed by the WHO (3), our findings suggest that comparative studies using both sequencing and IHC methods should be conducted to better define the standard of care and avoid confusion.
In conclusion, our study demonstrates that CDKN2A loss is highly prognostic in IDH/TP53 mutated astrocytomas. Testing for CDKN2A loss could be a potentially useful “grading” biomarker for this adult glioma subtype.
Supplementary Material
ACKNOWLEDGMENTS
Sources of support: Work at University of California, San Francisco was supported by the National Institutes of Health (grant numbers R01CA52689, P50CA097257, R01CA126831, R01CA139020 and R25CA112355), as well as the National Brain Tumor Foundation, the Stanley D. Lewis and Virginia S. Lewis Endowed Chair in Brain Tumor Research, the Robert Magnin Newman Endowed Chair in Neuro-oncology, and by donations from families and friends of John Berardi, Helen Glaser, Elvera Olsen, Raymond E. Cooper, and William Martinusen. This publication was also supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through UCSF-CTSI Grant Number UL1 RR024131.
We thank Cynthia Cowdrey, King Chiu, Yunita Lim, Sunny Evans, Tanya Zaitsev, and Yuri Murphy for assistance with tissue histology and immunohistochemistry. We thank Dr. Kenneth Aldape and Tarik Tihan for assisting in the neuropathology review of cases. Contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. The ideas and opinions expressed herein are those of the author(s) and endorsement by the State of California Department of Public Health, the National Cancer Institute, and the Centers for Disease Control and Prevention or their Contractors and Subcontractors is not intended nor should be inferred.
REFERENCES
- 1.Dolecek TA, Propp JM, Stroup NE, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neuro Oncol. 2012;(14) Suppl 5:v1–v49. doi: 10.1093/neuonc/nos218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis DN, Cavenee WK, Ohgaki H, et al. WHO classification of tumours of the central nervous system. Lyon. 2007 doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Louis DN, Perry A, Burger P, et al. International Society of Neuropathology-Haarlem Consensus Guidelines for Nervous System Tumor Classification and Grading. Brain Pathol. 2014;24:429–435. doi: 10.1111/bpa.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourne TD, Schiff D. Update on molecular findings, management and outcome in low-grade gliomas. Nat Rev Neurol. 2010;6:695–701. doi: 10.1038/nrneurol.2010.159. [DOI] [PubMed] [Google Scholar]
- 5.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hartmann C, Meyer J, Balss J, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118:469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 8.Kannan K, Inagaki A, Silber J, et al. Whole-exome sequencing identifies ATRX mutation as a key molecular determinant in lower-grade glioma. Oncotarget. 2012;3:1194–1203. doi: 10.18632/oncotarget.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heaphy CM, de Wilde RF, Jiao Y, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333:425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiao Y, Killela PJ, Reitman ZJ, et al. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3:709–722. doi: 10.18632/oncotarget.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okamoto Y, Di Patre PL, Burkhard C, et al. Population-based study on incidence, survival rates, and genetic alterations of low-grade diffuse astrocytomas and oligodendrogliomas. Acta Neuropathol. 2004;108:49–56. doi: 10.1007/s00401-004-0861-z. [DOI] [PubMed] [Google Scholar]
- 13.Watanabe T, Nakamura M, Kros JM, et al. Phenotype versus genotype correlation in oligodendrogliomas and low-grade diffuse astrocytomas. Acta Neuropathol. 2002;103:267–275. doi: 10.1007/s004010100464. [DOI] [PubMed] [Google Scholar]
- 14.Reifenberger G, Louis DN. Oligodendroglioma: toward molecular definitions in diagnostic neuro-oncology. J Neuropathol Exp Neurol. 2003;62:111–126. doi: 10.1093/jnen/62.2.111. [DOI] [PubMed] [Google Scholar]
- 15.Killela PJ, Reitman ZJ, Jiao Y, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110:6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sahm F, Reuss D, Koelsche C, et al. Farewell to oligoastrocytoma: in situ molecular genetics favor classification as either oligodendroglioma or astrocytoma. Acta Neuropathol. 2014;128:551–559. doi: 10.1007/s00401-014-1326-7. [DOI] [PubMed] [Google Scholar]
- 17.Ohgaki H, Kleihues P. Genetic alterations and signaling pathways in the evolution of gliomas. Cancer Science. 2009;100:2235–2241. doi: 10.1111/j.1349-7006.2009.01308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rasheed A, Herndon JE, Stenzel TT, et al. Molecular markers of prognosis in astrocytic tumors. Cancer. 2002;94:2688–2697. doi: 10.1002/cncr.10544. [DOI] [PubMed] [Google Scholar]
- 19.Perry A, Anderl K, Borell TJ, et al. Detection of p16, RB, CDK4, and p53 gene deletion and amplification by fluorescence in situ hybridization in 96 gliomas. Am J Clin Pathol. 1999;112:801–809. doi: 10.1093/ajcp/112.6.801. [DOI] [PubMed] [Google Scholar]
- 20.Maruno M, Yoshimine T, Muhammad AK, et al. Loss of heterozygosity of microsatellite loci on chromosome 9p in astrocytic tumors and its prognostic implications. J Neurooncol. 1996;30:19–24. doi: 10.1007/BF00177439. [DOI] [PubMed] [Google Scholar]
- 21.Ruas M, Peters G. The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim Biophys Acta. 1998;1378:F115–F177. doi: 10.1016/s0304-419x(98)00017-1. [DOI] [PubMed] [Google Scholar]
- 22.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 23.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 24.He J, Olson JJ, James CD. Lack of p16INK4 or retinoblastoma protein (pRb), or amplification-associated overexpression of cdk4 is observed in distinct subsets of malignant glial tumors and cell lines. Cancer Res. 1995;55:4833–4836. [PubMed] [Google Scholar]
- 25.Watanabe T, Yokoo H, Yokoo M, et al. Concurrent inactivation of RB1 and TP53 pathways in anaplastic oligodendrogliomas. J Neuropathol Exp Neurol. 2001;60:1181–1189. doi: 10.1093/jnen/60.12.1181. [DOI] [PubMed] [Google Scholar]
- 26.Ueki K, Ono Y, Henson JW, et al. CDKN2/p16 or RB alterations occur in the majority of glioblastomas and are inversely correlated. Cancer Res. 1996;56:150–153. [PubMed] [Google Scholar]
- 27.Jeon YK, Park K, Park CK, et al. Chromosome 1p and 19q status and p53 and p16 expression patterns as prognostic indicators of oligodendroglial tumors: a clinicopathological study using fluorescence in situ hybridization. Neuropathology. 2007;27:10–20. doi: 10.1111/j.1440-1789.2006.00735.x. [DOI] [PubMed] [Google Scholar]
- 28.Bigner SH, Matthews MR, Rasheed BK, et al. Molecular genetic aspects of oligodendrogliomas including analysis by comparative genomic hybridization. Am J Pathol. 1999;155:375–386. doi: 10.1016/S0002-9440(10)65134-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perry A, Nobori T, Ru N, et al. Detection of p16 gene deletions in gliomas: a comparison of fluorescence in situ hybridization (FISH) versus quantitative PCR. J Neuropathol Exp Neurol. 1997;56:999–1008. doi: 10.1097/00005072-199709000-00005. [DOI] [PubMed] [Google Scholar]
- 30.Fuller CE, Schmidt RE, Roth KA, et al. Clinical utility of fluorescence in situ hybridization (FISH) in morphologically ambiguous gliomas with hybrid oligodendroglial/astrocytic features. J Neuropathol Exp Neurol. 2003;62:1118–1128. doi: 10.1093/jnen/62.11.1118. [DOI] [PubMed] [Google Scholar]
- 31.Bortolotto S, Chiado-Piat L, Cavalla P, et al. CDKN2A/p16 inactivation in the prognosis of oligodendrogliomas. Int J Cancer. 2000;88:554–557. doi: 10.1002/1097-0215(20001115)88:4<554::aid-ijc6>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 32.Cairncross JG, Ueki K, Zlatescu MC, et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst. 1998;90:1473–1479. doi: 10.1093/jnci/90.19.1473. [DOI] [PubMed] [Google Scholar]
- 33.Miettinen H, Kononen J, Sallinen P, et al. CDKN2/p16 predicts survival in oligodendrogliomas: comparison with astrocytomas. J Neurooncol. 1999;41:205–211. doi: 10.1023/a:1006185220369. [DOI] [PubMed] [Google Scholar]
- 34.Wick W, Hartmann C, Engel C, et al. NOA-04 randomized phase III trial of sequential radiochemotherapy of anaplastic glioma with procarbazine, lomustine, and vincristine or temozolomide. J Clin Oncol. 2009;27:5874–5880. doi: 10.1200/JCO.2009.23.6497. [DOI] [PubMed] [Google Scholar]
- 35.Shibahara I, Sonoda Y, Kanamori M, et al. New insights into glioma classification based on isocitrate dehydrogenase 1 and 2 gene status. Brain Tumor Pathol. 2011;28:203–208. doi: 10.1007/s10014-011-0050-4. [DOI] [PubMed] [Google Scholar]
- 36.Walsh KM, Codd V, Smirnov IV, et al. Variants near TERT and TERC influencing telomere length are associated with high-grade glioma risk. Nat Genet. 2014;46:731–735. doi: 10.1038/ng.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rice T, Zheng S, Decker PA, et al. Inherited variant on chromosome 11q23 increases susceptibility to IDH-mutated but not IDH-normal gliomas regardless of grade or histology. Neuro-oncology. 2013;15:535–541. doi: 10.1093/neuonc/nos324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Christensen BC, Smith AA, Zheng S, et al. DNA methylation, isocitrate dehydrogenase mutation, and survival in glioma. J Natl Cancer Inst. 2011;103:143–153. doi: 10.1093/jnci/djq497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiencke JK, Aldape K, McMillan A, et al. Molecular features of adult glioma associated with patient race/ethnicity, age, and a polymorphism in O6-methylguanine-DNA-methyltransferase. Cancer epidemiology, biomarkers & prevention. 2005;14:1774–1783. doi: 10.1158/1055-9965.EPI-05-0089. [DOI] [PubMed] [Google Scholar]
- 40.Stacey SN, Sulem P, Jonasdottir A, et al. A germline variant in the TP53 polyadenylation signal confers cancer susceptibility. Nat Genet. 2011;43:1098–1103. doi: 10.1038/ng.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perry A, Fuller CE, Banerjee R, et al. Ancillary FISH analysis for 1p and 19q status: preliminary observations in 287 gliomas and oligodendroglioma mimics. Front Biosci. 2003;8:a1–a9. doi: 10.2741/896. [DOI] [PubMed] [Google Scholar]
- 42.Costello JF, Berger MS, Huang HS, et al. Silencing of p16/CDKN2 expression in human gliomas by methylation and chromatin condensation. Cancer research. 1996;56:2405–2410. [PubMed] [Google Scholar]
- 43.Alves MK, Faria MH, Neves Filho EH, et al. CDKN2A promoter hypermethylation in astrocytomas is associated with age and sex. International journal of surgery. 2013;11:549–553. doi: 10.1016/j.ijsu.2013.05.030. [DOI] [PubMed] [Google Scholar]
- 44.Liu XY, Gerges N, Korshunov A, et al. Frequent ATRX mutations and loss of expression in adult diffuse astrocytic tumors carrying IDH1/IDH2 and TP53 mutations. Acta neuropathologica. 2012;124:615–625. doi: 10.1007/s00401-012-1031-3. [DOI] [PubMed] [Google Scholar]
- 45.Jiao Y, Shi C, Edil BH, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwartzentruber J, Korshunov A, Liu XY, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 47.Shay JW, Wright WE. Role of telomeres and telomerase in cancer. Semin Cancer Biol. 2011;21:349–353. doi: 10.1016/j.semcancer.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walsh KM, Rice T, Decker PA, et al. Genetic variants in telomerase-related genes are associated with an older age at diagnosis in glioma patients: evidence for distinct pathways of gliomagenesis. Neuro Oncol. 2013;15:1041–1047. doi: 10.1093/neuonc/not051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.