Abstract
All organisms encode transfer RNAs (tRNAs) that are synthesized as precursor molecules bearing extra sequences at their 5′ and 3′ ends; some tRNAs also contain introns, which are removed by splicing. Despite commonality in what the ultimate goal is (i.e., producing a mature tRNA), mechanistically, tRNA splicing differs between Bacteria and Archaea or Eukarya. The number and position of tRNA introns varies between organisms and even between different tRNAs within the same organism, suggesting a degree of plasticity in both the evolution and persistence of modern tRNA splicing systems. Here we will review recent findings that not only highlight nuances in splicing pathways but also provide potential reasons for the maintenance of introns in tRNA. Recently, connections between defects in the components of the tRNA splicing machinery and medically relevant phenotypes in humans have been reported. These differences will be discussed in terms of the importance of splicing for tRNA function and in a broader context on how tRNA splicing defects can often have unpredictable consequences.
INTRODUCTION
Introns are everywhere! And because they interrupt functional regions in RNA, universally, their precise removal ensures the functionality of many essential transcripts. Thus, RNA splicing is at the very heart of cell viability. Despite their widespread occurrence in many coding and noncoding RNAs and their broad phylogenetic distribution, the strategies for intron removal vary between different RNA types, between different organisms and even within different RNAs within single organisms. To date, there are 4 major intron types classified based on their particular splicing mechanism. At one end of the scale we have spliceosomal introns, which are processed by dedicated ribonucleoprotein complexes, where the proteins provide a scaffold for their RNA components (the snRNPs).1–3 It is within these, sometimes intricate, multiprotein-RNA complexes that the task of splicing occurs in a highly orchestrated manner where the RNA components by themselves are not catalytic. A second group formed by the group II introns, shares structural and mechanistic features with the spliceosomal introns, with one major difference: these are auto-catalytic requiring in principle no proteins for their excision.4 Just as widely distributed in the three domains of life are the intervening sequences that disrupt transfer RNAs (tRNAs), which are removed by two distinct pathways: One, RNA mediated in the case of Group I introns found in tRNAs of bacteria5,6; the other, found in Archaea and Eukarya is strictly protein-enzyme catalyzed.7 It is this tRNA family of introns and their processing that will be the major focus of this review.
The number and position of tRNA introns varies depending on the organism.8 In Eukarya, intron-containing tRNAs can range from a single tRNA (tRNATyr) in trypanosomatids, to more that 60 in other eukaryotes such as Saccharomyces cerevisiae, plants or mammals.8 Yet in all cases there is a single intron per tRNA that is always found between position 37 of the anticodon loop (adjacent to the anticodon triplet) and position 38, thus effectively dividing the tRNA into nonfunctional halves. In Archaea, on the other hand, introns can be found at the canonical anticodon position as in Eukaryal tRNAs but also at other positions including the D-arm and the TΨC-arm.9 Although there are many arguments for how tRNA introns appeared (or for that matter disappear) from genomes, the positional variability described above directly reflects on changes in splicing enzyme specificity, which presumably is the result of co-evolution between the introns and the enzymes that remove them.
In terms of the actual mechanism of tRNA splicing, the bacterial group I introns require no protein help and both the cutting of the intron and the gluing together of the exon halves are performed concomitantly with intron removal in a now classical mechanism of double-transesterification that uses a guanosine, bound in trans, as a nucleophile.10 In Archaea and Eukarya, the situation is quite different, intron removal is orchestrated by several enzymatic activities: an endonuclease, a kinase, a phospodiesterase, a phosphotransferase, and a ligase.11 The general requirement for each activity depends on the organism. The evolution of one type of pathway or another is in fact determined by how, following endonuclease cleavage, the free exons are further processed. In the following pages, we will discuss the finer details of tRNA splicing in various systems and delve into the growing connection between tRNA splicing components and disease in humans. Lastly, we will summarize recent findings about the fate of introns and speculate on why they still exist today.
DETERMINANTS OF SPLICING
Unlike the self-splicing introns of Bacteria,6 Archaea and Eukarya have adopted a protein-catalyzed method to handle tRNA splicing, where, despite varying sequence and length, introns are efficiently cleaved by the tRNA splicing endonuclease; a reaction in which the intron only plays a minor role (Figure 1). This process depends little on conserved sequence-specific recognition and is primarily based on proximity and base pair interactions between the intron and the tRNA body to form the proper structure for cleavage.
FIGURE 1.
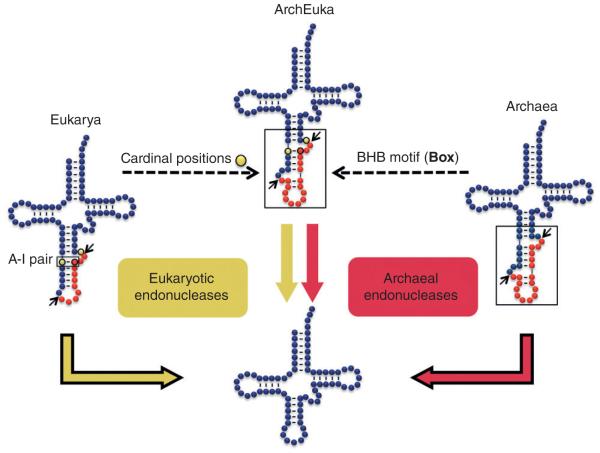
The substrates of tRNA-splicing endonucleases of Archaea and Eukarya. Dashed lines represent the splicing determinants taken from eukaryotic and archaeal tRNA to create tRNAArchEuka. tRNAArchEuka is composed of the body of a eukaryotic tRNA (top half) with the anticodon stem of an Archaeal tRNA (box). The box identifies the canonical bulge-helix-bulge (BHB) motif of Archaeal endonucleases, while the eukaryotic cardinal positions and the A–I interaction are denoted in yellow.
Early work began uncovering the basis for endonuclease recognition.12 It all started with the inference that since all tRNAs share a similar tertiary structure, the overall shape of the tRNA, and its conserved features, must then be a determining factor for splicing. In support of this idea, insertion of extra base pairs in the anticodon stem of tRNAs led to a predictable shift of the endonuclease cleavage sites. Inserting one base pair in the anticodon stem shifted the cleavage sites by two nucleotides. Similarly, insertion of two base pairs into the stem increased it by four nucleotides.12 This predictable alteration in intron recognition led to what became known as the ‘anchor and measure’ model.13 This implied that the endonuclease would recognize the mature tRNA body and then measure ‘downward’ a particular distance to identify the splice sites. The intron–exon boundaries would be determined by this specific set distance from the tRNA mature domain, allowing for a predictable change in intron cleavage corresponding to the alteration of base pairs in the anticodon stem.
Although the intron–exon boundaries are not specified at the sequence level, this is not to say that there is a complete lack of sequence specificity in splice-site selection.14 The splicing process does depend on a few key nucleotides located within the anticodon stem. These are known as cardinal positions 1 and 2 (CP1 and CP2, respectively) (Figure 1). Found predominantly two nucleotides upstream of the anticodon, CP1 is usually a pyrimidine, which forms a canonical base pair with a purine located three nucleotides upstream of the 3′ splice site within the intron.14 This vital interaction between the anticodon and intron is referred to as the A–I (for anticodon-intron) base pair and is required for splicing. The A-I base pair likely stabilizes the intron-mature tRNA structure and allows for proper positioning of the endonuclease. CP2 marks the 3′ intron–exon boundary, which is always found in a bulge formed within a single stranded region of the anticodon stem (position 38 of the mature anticodon loop and 3′ downstream nucleotides).
Aside from structural stabilization, the A-I base pair is a major determinant for correct 5′ and 3′ splice-site selection. In a series of elegant experiments, it was shown that an intron-containing circularly permuted tRNA, where the intron had a free end, was still efficiently processed by the endonuclease. This construct allowed the separation of the introns into two independent segments permitting surveying of the two splice sites separately. If the A-I pair is present, and the intron is at least three nucleotides in length, the endonuclease will cleave only at the 3′ splice site of the intron. In the absence of the A-I base pair, cleavage will take place only at the 5′-splice site as long as the intron is at least eight nucleotides in length.13 This highlighted the importance of the A-I base pair for splice-site recognition. Also, the presence of two distinct minimalist segments (or domains) of the intron became apparent. Domain I consisting of the three nucleotides from the A-I pair to the 3′ splice junction, and domain II being the eight nucleotides on the 5′ half of the intron, both needed for proper splicing. Therefore, without the A-I base pair, splicing of the intron cannot occur efficiently.13
Perhaps one of the most intriguing mechanisms to generate a canonical intron structure for splicing occurs in the tRNATyr of Trypanosoma brucei, and presumably in related trypanosomatids. T. brucei encodes a single intron-containing tRNA with an 11-nucleotide intron, which is among the shortest known. The intron sequence encoded in the genome does not conform to all conserved sequence requirements above. This problem is solved post-transcriptionally by the editing of the intron at 2 or 3 positions, which leads to regeneration of the proposed canonical mature tRNA-intron structure. Although the nature of the editing events is unusual (G to A and A to U changes) and does not belong to any of the known editing mechanisms, intron editing is required for splicing and thus for T. brucei viability.15 It suggests that shorter introns may require more strict structural constrains whereby U-G pairs in the A-I stem may not be sufficient to ensure cleavage.
Archaea differ mainly in structural requirements for cleavage. The Archaeal splicing substrate consists of a conserved bulge-helix-bulge (BHB) motif, where two bulges (3 nucleotides each) are separated by a four-nucleotide helix.16 Unlike Eukarya, the intron itself is the main splicing determinant while the body of the tRNA can withstand major alterations without affecting the splicing reaction (12). As a result Archaeal introns are found in different positions in the tRNA in addition to the canonical position found in Eukarya.9 As long as the BHB motif is formed, the Archaeal endonuclease will recognize and cleave the intron. Despite these disparities, Eukarya and Archaea are still similar in that they both contain the A-I base pair near the 3′ splice site,13 but unique to Archaea is the occurrence of a second A-I like base pair near their 5′ splice site created by the requirement for the BHB motif17(Figure 1).
PROTEIN MEDIATED CATALYSIS
Once ribozymes were discovered it was natural to think that the first step in tRNA splicing could be RNA catalyzed, in line with the prevailing thoughts at the time. However, tRNA splicing catalysis leaped forward with the purification and characterization of the yeast splicing endonuclease, an all protein enzyme.18 Initially, this enzyme was thought to consist of three subunits19 acting as a membrane-associated trimeric complex of low abundance in cells, but shortly after a fourth subunit was discovered.20 Interestingly, this ‘new’ 15 kDa protein had escaped identification by virtue that it migrated with the dye front in gels. It is now clear that the endonuclease in yeast, plants, and mammals is formed by four nonidentical subunits of approximately 15, 34, 44, and 54 kDa (Sen15p, Sen34p, Sen2p, and Sen54p) respectively.20,21 Evidence supporting the heterotetramerization of these subunits came from further purification of stoichiometric amounts of each protein in a complex, as well as the co-sedimentation of the complex in density gradients.20 Even before, however, there were indications that the Sen2 gene was involved in tRNA splicing, given that yeast mutants of Sen2 (for splicing endonuclease 2) accumulated a ‘2/3’ tRNA molecule which was only cleaved at the 3′ splice site, yielding a product formed by the 5′-exon still attached to the intron, indicating the importance of Sen2p for 5′ splice-site cleavage.21 The amino acid sequence of Sen2p contains a predicted transmembrane domain possibly linking the endonuclease to a membrane, partly explaining the membrane association seen during the purification procedure. The evidence that Sen2p is responsible only for 5′ splice-site cleavage then suggested that another subunit in the heterote-tramer would be responsible for cleavage at the 3′ site. Sequence comparative analysis of the subunits revealed strong similarity between Sen2p and Sen34p. Both were later found to be part of a family of homodimeric proteins responsible for tRNA splicing in Archaea, also suggesting their function in these organisms.17 Sen34 mutants also accumulate ‘2/3’ tRNA molecules, yet this time the intron remained attached to the 3′-exon supporting its role in providing the second active site needed for complete cleavage.21
Pinning a function to the remaining subunits was more difficult since they did not share sequence similarities to the other active site containing subunits. To establish their interaction, a yeast two-hybrid assay was implemented that showed the surprising associations between Sen2p-Sen54p and Sen15p-Sen34p to the exclusion of any other combinations.20 The implication from this modular association was that the two heterodimers could independently recognize each splice site, while cleavage was dependent on individual determinants (in an αβγδconfiguration). The work from several laboratories on the specificity of the endonuclease led to the current model whereby Sen2p and Sen34p act to cleave the intron and Sen54p acts as a molecular ruler measuring the distance from the mature body of the tRNA down the anticodon stem to determine site of cleavage.20 The experiments showing the importance of the A-I pair for splicing, then allowed for the extrapolation that Sen15 must be required to monitor the A-I base pair and further help position the endonuclease over the intron all while forming a complete heterotetramer.
Coincident with the characterization of the yeast endonuclease, the nature of the archaeal endonucleases of Haloferax volcanii and Methanococcus jannaschii were also described. Remarkably, different quaternary structures were shown for each, with the H. volcanii enzyme working as a homodimer (α2) and the M. jannaschii counterpart having a homotetramer (α4) configuration.17,22 Considering that both types of archaeal endonucleases must cleave the same BHB motif, each of M. jannaschii’s dimers must be akin to each individual monomer of the endonuclease of H. volcanii. In fact, closer inspection of the endonuclease of H. volcanii revealed an in frame gene duplication.23 Even though the enzyme acts as a homodimer, it can be viewed as a pseudodimer of sorts comprised of two pseudomonomers each connected by a small linking segment. This extra domain allows for the folding of the enzyme to resemble the homotetramer configuration and still permit for efficient cleavage of the BHB motif.
A fourth family of endonucleases was discovered in the Crenarchaeota and Nanoarchaeum equitans consisting of dimers of heterodimers (αβ)2 configuration.24 This requirement for an additional subunit is in contrast to the other Archaea, which employ homodimers or homotetramers. The change in subunit composition may coincide with the diversity of substrates observed between different Archaea.25 These organisms contain examples of tRNA introns lacking the correct nucleotide arrangement necessary for folding into two 3-nucleotide bulges separated by a 4-nucleotide helix. The dimer of heterodimers seems to be more accepting of structural divergence from the canonical BHB motif possibly providing a practical explanation for the existence of the β subunit.26
The ARMAN (Archaeal Richmond Mine Acidophilic Nano-organism), a group of recently discovered Archaea, harbor another unique endonuclease. Candidatus Micrarchaeum acidiphilum (ARMAN-2) and its related species ARMAN-1 contain an endonuclease of three subunits (ε)2.27 One subunit of 166 or 163 amino acids depending on the organism and two additional subunits of 127 amino acids and 97 amino acids. Phylogenetic analysis guided by structural alignment showed that the active site of the larger subunit is similar to the active site of the αsubunit of the (αβ)2 enzymes, while the 127 amino acid protein was related to the nonactive βsubunit of the (αβ)2 endonuclease. Sequence comparisons also suggested that the 127-amino acid protein contained the key active-site residues found in the active αsubunit, while the larger 166/163 amino acid proteins would not be functional in catalysis due to the loss of those key residues. This led to the idea that the 166/163 amino acid subunit was pseudocatalytic.27 Organisms with the (ε)2 endonuclease were similar to the (αβ)2 family in that they contained tRNAs with introns at noncanonical positions (i.e., other than in the anticodon loop). Their tRNAs are split into multiple pieces which then have to be joined together before cleavage could occur and in addition they contained imperfect BHB motifs.28 Despite this diversity of substrates, thus far organisms possessing noncanonical introns all can be grouped into the (αβ)2 or (ε)2 endonuclease families, based on the relaxation in their substrate recognition in comparison to the BHB motifs for the α2α4 enzymes.11
Setting their differences aside, the active sites of archaeal endonucleases do share similarity with eukaryotic splicing domains of Sen2p and Sen34p, in a region approximately 100 amino acids in length that encompasses the active site. In fact their similarity can be appreciated in their cleavage mechanism, which in all cases generates a 2′, 3′ cyclic phosphate intermediate at the 3′-end of the 5′-exon and a 5′-hydroxyl termini of the 3′ exon. The degree of active-site conservation was elegantly recapitulated by incorporating the different nuances in cleavage determinants into a single hybrid substrate, tRNAArcheuka, containing eukaryal splicing determinants from the body of the tRNA combined with the BHB motif recognized by archaeal endonucleases (Figure 1). The tRNAArcheuka is efficiently cleaved by both eukaryotic and archaeal endonucleases, making it a true universal substrate.29 In tRNAArcheuka, addition of two base pairs to the A-I stem does not shift the cleavage sites indicating that, in this structural context, the eukaryal endonuclease is able to utilize the archaeal BHB recognition motif to select the correct cleavage sites.29
HERE ENDO, THERE ENDO, EVERYWHERE ENDO
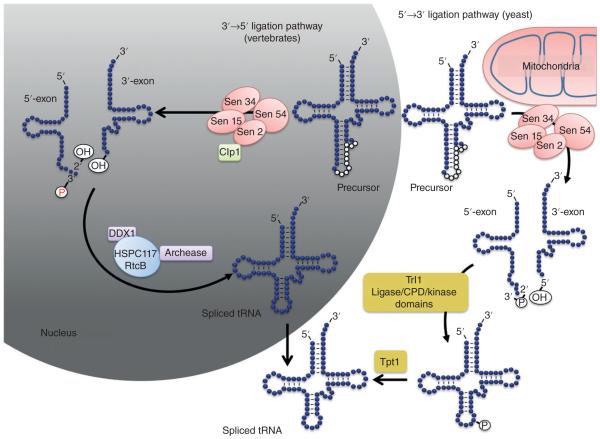
One intriguing facet of endonuclease studies is the seemingly drastic differences in the intracellular localization of splicing complex components in different eukaryotes. In mammals, all the tRNA splicing proteins localize to the nucleus, which makes practical sense since tRNAs are transcribed, end processed, and matured there.30 An added benefit of such localization is that it may provide a layer of control to ensure that only matured tRNAs reach the cytoplasm to engage in translation. Plants offer a more mixed picture and a developing story. The endonuclease of Arabidopsis thaliana is localized to both the nucleus and the mitochondria.31 The reasons for such a disparate cellular distribution have yet to be understood.
One of the most interesting cases occurs in S. cerevisiae, where the Sen subunits are associated with the outer mitochondrial membrane presumably facing the cytoplasm.32 This surprising observation is supported by several lines of evidence including fluorescent localization of GFP-tagged Sen subunits, localization of tRNA ligase to the cytoplasm and the accumulation of intron-containing tRNAs in the cytoplasm with a temperature sensitive endonuclease mutant.32 This localization does however raise interesting questions of the functional significance of moving the endonuclease away from the nucleus. The spatial separation could be involved in some form of regulation of tRNA biogenesis or possibly indicative of an alternative function served by these components.33 In line with this proposal, localization of the endonuclease to the nucleus of yeast fails to rescue growth defects associated with nonmitochondrial surface localization of splicing, again suggesting an essential function for the endonuclease complex in the cytoplasm that is unrelated to splicing.33
The tRNA splicing endonuclease varying subunit composition suggests alterations made over time to accommodate diversity of substrates; as introns evolved so did their processing endonucleases. Introns positioned in different locations of the tRNA, or ones of unique shape, required adaptation of endonuclease subunit configuration to allow for their removal. In cases, like Eukarya, where introns are situated in a canonical conserved position between nucleotides 37 and 38 of the anticodon loop, the endonuclease could possibly take a more specific role and mode of recognition. Despite the endonuclease becoming synonymous with tRNA splicing, several reactions have been described that involve excision of introns from other RNAs such as ribosomal RNAs in Archaea34 and the maturation of certain mRNAs in Humans.35 In addition, various Archaea use the splicing endonuclease to excise pre-rRNA spacers containing BHB motifs, which remove pre-16S and pre-23S rRNA from the operon primary transcripts.36 Lastly, the recent association of HsClp1, involved in 3′ mRNA maturation in humans, with tRNA splicing further links the endonuclease complex with components of seemingly unrelated cellular processes.37,38
SEALING THE DEAL
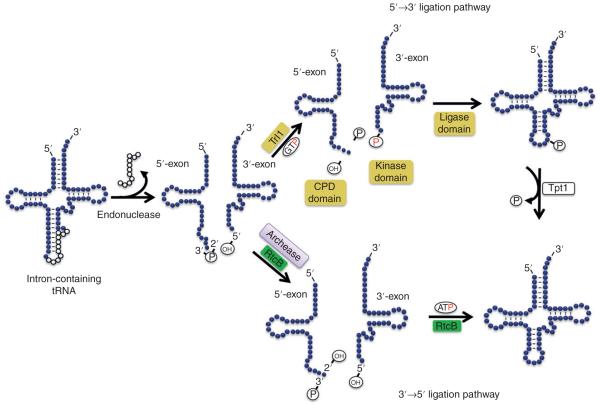
Following cleavage, the remaining exon halves are ligated to generate the full-length tRNA. While the intron removal mechanism is conserved among the three domains of life, the degree of conservation of the tRNA splicing ligases is still a matter of debate. There are two main tRNA ligation mechanisms described: 5′→3′ and 3′→5′ ligation pathways39 (Figure 2). The first uses a 5′-PO4 terminus at the 3′-exon and a 2′-PO4 and a 3′-OH at the 3′-end of the 5′-exon (5′→3′ pathway) and involves the Trl1 ligase.40 The second pathway involves ligation of the 3′-PO4, left at the 5′-exon after intron removal, and the 5′-OH at the 3′-exon (3′→5′ pathway) by the RtcB enzyme. Still, however, some organisms encode both types of ligases (Trl1 and RtcB) in their genomes (e.g., trypanosomatids),41 suggesting either additional functions for the extra enzymes or some sort of ligase back up mechanism.
FIGURE 2.
The two main tRNA-splicing pathways. The 5′→3′ ligation pathway (top) has been extensively studied in yeast. Following cleavage by the tRNA-splicing endonuclease, the resulting exon halves are processed by the CPD and kinase domains; the CPD domains generates a 3′ terminus 2′-phospate on the 5′ exon and the kinase domain phosporylates the 5′ end of the 3′ exon using GTP. The two exons are joined by the ligase domain of the same protein (Trl1) using ATP, the dangling 2′-phosphate is finally removed by Tpt1 which is a free-standing enzyme. All domains reside in a single Trl1 protein and are shown in yellow boxes. The 3′→5′ ligation pathway has been described in archaea and vertebrates. RtcB (green box) is the enzyme responsible for the opening of the 2′3′ cyclic phosphate (in some organisms but not in mammals) and for the joining of the halves. In humans RtcB named HSPC117 has been identified as part of the human ligase complex.
tRNA Ligases in Phages, Yeast, and Plants
tRNA splicing ligases were originally identified in yeast,40 where a 95 kDa protein (Trl1), encoded by the essential RLG1 gene, was purified and has been studied extensively.42 Trl1 is a multifunctional enzyme composed of three different domains: An N-terminal adenylyltransferase/ligase domain, a central polynucleotide kinase domain that resembles that of the phage T4 polynucleotide kinase, and a C-terminal cyclic phosphodiesterase domain. Although all domains are essential, they can function separately in vivo. Each domain is responsible for catalyzing distinct reactions that are absolutely required for tRNA splicing39 (Figure 2). Likewise, plants also encode similar enzymes (AtTRNL, in A. thaliana and tRNA ligase in Oryza sativa) with the same protein domains catalyzing the same reactions as the yeast tRNA ligase. These ligases are even able to complement a Δtrl1 yeast strain.43 Joining the two exons following cleavage involves three important steps: the ‘healing’ of the 5′-exon and 3′-exon termini and a final ‘sealing’ step.44 Intron removal by the tRNA endonuclease generates a 2′-3′-cyclic phosphate at the 3′-end of the 5′-exon and a 5′-OH at the 5′-end of the 3′-exon (Figure 2). The first step performed by Trl1 is the hydrolysis of the 2′-3′ cyclic phosphate of the 5′-exon by its CPD domain, leaving a 3′-OH, 2′-PO4 terminus. In the second step, the kinase domain phosphorylates the 5′-OH terminus of the 3′-exon, using GTP, not ATP, as the phosphate donor. In the last step, the ligase domain forms a ligase-(lysyl)-AMP intermediate, using ATP. AMP is then transferred to the 5′-PO4 RNA terminus and the ligase seals it to the 3′-OH of the other tRNA half, with AMP as the leaving group. This ligation reaction results in a full-length tRNA molecule, but still containing a 2′ residual phosphate, which will be removed in a subsequent step by a 2′-phosphotransferase (Tpt1) acting alone.45
The ligase domains of these enzymes are similar to that of the T4 bacteriophage ligase15 with one key difference, Trl1 and plant ligases absolutely require a dangling 2′-phosphate at the 3′-end of the 5′ exon. Nevertheless, many important features are shared between these ligases, including the conserved motifs that define each domain. These shared features suggested a common evolutionary pathway for these enzymes.5 Similarly, the putative trypanosomatid Trl1 also contains the l, Ia, IV, and V motifs present in the N-terminal ligase domain, the GxGKT and RxxxR motifs found in the kinase domain, and the HΦTΦ motifs of the CPD domains found in the yeast and plant Trl1.45
Another interesting aspect of yeast and plant tRNA ligases is their subcellular localization. In yeast, all necessary components for tRNA splicing are in the cytoplasm. On the other hand, Arabidopsis and Oryza tRNA ligases fused to GFP were detected in the chloroplast.39 Considering that chloroplasts genomes do not encode eukaryotic-type intron-containing tRNAs, the unusual organellar localization of the ligase suggests a secondary function for this enzyme. Notably, alternative functions for the tRNA splicing ligases have been reported, e.g., yeast Trl1 participates in the HAC1 mRNA splicing pathway, which encodes a transcriptional factor involved in the unfolded protein response (UPR).46 In this case, Trl1 is responsible for joining the two HAC1 mRNA halves created by Ire1p after intron removal, generating a final product with a 2′ residual phosphate that also has to be removed by Tpt1.46
tRNA Ligases in Bacteria, Archaea, and Vertebrates
In vertebrates, the in vivo tRNA ligation pathway has been the subject of much discussion. Free-standing cyclic phosphodiesterase (HsCNP) and phosphotransferase (HsTRPT1) activities were described in bovine and human brain cells.47,48 The mammalian CNP is able to heal the 3′ terminus of yeast tRNAs, in a genetic background of otherwise lethal mutations in the yeast Trl1 CPD domain, rescuing normal growth of the strains.48 Although an activity, similar to yeast Trl1, has been detected in crude extracts from HeLa cells, the enzyme has never been identified. Furthermore, no Trl1 homolog is evident from the human genome databases. This observation suggests that the yeast and plants tRNA ligases might be a more recent acquisition than the enzymes from Archaea. In line with this suggestion, RtcB homologs have been found in Bacteria, albeit for the purpose of tRNA repair in a function unrelated to splicing.49,50
The 3′→5′ ligation is regarded as the main tRNA ligation pathway used in vertebrates (Figure 2). In humans, the protein catalyzing this reaction was identified as HSPC117.51 HSPC117 homologs are found in many organisms including vertebrates, protozoa, and insects.39 In Archaea, HSPC117 homologs are known as RtcB, an RNA ligase that catalyzes a GTP and Mn(II)-dependent ligation reaction. Human RtcB (HSPC117) is part of a 200 kDa tRNA ligase complex which includes a DEAD-box helicase (DDX1).39 This complex has a nucleo-cytoplasmic distribution,52 which suggests functions in both compartments. Recent studies described archease, a 16 kDa protein distributed in all domains of life, as another component of the human tRNA ligase complex.53 Addition of Pyrococcus horikoshii archease to the RtcB reaction improved RNA-3′P formation, also increasing ligation efficiency54 (Figure 2). In Archaea and Bacteria archease and RtcB are found in an operon, again suggesting related functions. In humans, archease and DDX1 are important contributors for full activity of the tRNA ligase complex, facilitating the formation of the RtcB-guanylate intermediate necessary for the subsequent steps of RNA ligation.53 Using RNAi, archease and RtcB were depleted in HeLa cells and the cell extracts were used in tRNA maturation assays. The archease RNAi affected tRNA maturation in vitro to comparable levels as RtcB RNAi.53 Unlike mammalian RtcB, expression of E. coli RtcB in Trl1-depleted yeast is able to recover growth. Furthermore, E. coli RtcB is able to replace yeast Trl1 in joining the HAC1 mRNA exons in UPR events.55–57 In mammals a similar UPR pathway has been described, XBP1u mRNA is cleaved by IREα leading to the removal of a 26-nucleotide intron. While in yeast HAC1 mRNA splicing is performed by a multistep reaction catalyzed by Trl1; in mammals, the XBP1u exons are joined by a 3′→5′ ligation mechanism in a single step. The enzyme responsible for joining these exons remained unknown for many years. However, recent studies showed that human RtcB is the RNA ligase for the mammalian UPR pathway.57,58
While all the needed activities, with the exception of the phosphotransferase, reside in a single polypeptide in Trl1, these are found as independent enzymes in other organisms. In humans, a kinase (hClp1) initially described as a component of the cleavage factor I in mRNAs in S. cerevisiae and as a component of cleavage factor II acting at the 3-end of mRNAs in humans,59,60 was identified as a component of the tRNA splicing complex.35 Later it was shown that hClp1 is able to phosphorylate tRNA exons in vitro37 and also function in vivo.61 In fact, purification of Sen2 from human cell extracts showed that the fractions contained not only endonuclease but also the kinase activity, suggesting that the two act in a complex that localizes in the nucleus. Additionally, a knockdown of the hClp1 in HeLa cells resulted in low tRNA ligation activity.37 Hence, hClp1 has been regarded as the crucial kinase enzyme needed for the tRNA splicing pathway in mammals. Coincidentally, hClp1 has ATP/GTP binding sites resembling those found in phage T4 polynucleotide kinase, but In contrast to yeast Trl1, which uses GTP as a phosphate donor, hClp1 uses ATP to phosphorylate the 5′-terminus of the 3′-exon. What is odd at the moment is why is Clp1 required when endonuclease cleavage itself generates the phosphate needed for the 3′→5′ pathway. It may well be that there is needed redundancy in the system. In fact in mice, although CNP and the Tpt1 (the phosphotransferase for the 2-P) are encoded in the genome, neither is essential.62 More recently, it was shown that a marine chordate, Branchiostoma floridae (Bf), has the RNA ligase and the PNK/CPD activities in two different proteins. The Bf RNA ligase is able to seal two RNA substrates in which one has a 5′-phosphate at the 5′ terminus and the other, a 3′-OH containing 2′-phosphate at the 3′ terminus,63 accomplishing a 5′→3′ type ligation. These two Bf polypeptides were also able to complement a Δtrl1 yeast strain supporting their role in RNA ligation.
tRNA PHOSPHOTRANFERASE
In the 5′→3′ ligation pathway after the sealing of the two tRNA halves by the tRNA ligase, a remaining 2′-phosphate must be removed by a phosphotransferase enzyme45 (Figures 2 and 3). In yeast, the enzyme responsible for this final step, ScTpt1, catalyzes an NAD+-dependent phosphotransfer reaction generating ADP-ribose cyclic phosphate.64 Conditional mutants of the otherwise essential Tpt1 accumulate tRNA molecules with 2′-phosphates.64 Tpt1 localizes to the cytoplasm, however, it is also present in the nucleus, again suggesting a secondary function aside from tRNA splicing that may involve 2′-phosphate removal from some other substrate. In addition, a Δtpt1Δtrl1 yeast strain could be complemented by the expression of phage T4 ligase and kinase resulting in a mature tRNA lacking the dangling 2′-phosphate characteristic of the Trl1 reaction. Therefore, Tpt1 is essential for growth only in the presence of the Trl1 pathway. Moreover, these results show that Tpt1 has a single essential role in yeast, which is the removal of 2′-phosphate at the ligated tRNA exon junctions.45
FIGURE 3.
Subcellular localization of the tRNA-splicing machinery. In yeast (in pink), the endonuclease complex is localized on the outer membrane surface of mitochondria. The precursor tRNA is cleaved and the halves are processed and joined by Trl1 in the cytoplasm. The remaining 2′-phosphate is removed by Tpt1, which is also localized in the cytoplasm, generating the spliced tRNA. In humans, splicing is accomplished by a nuclear macromolecular complex formed by the endonuclease (pink), the Clp1 kinase (green), HSPC117/RtcB (light blue), and other proteins (purple).
Tpt1 homologs are found in all three domains of life. The ability to complement yeast has been demonstrated using Tpt1 orthologs from other fungi, Arabidopsis thaliana, mouse and human, suggesting a conserved function among these enzymes. Interestingly, TRPT1 knockout mice are viable, and no alterations are detected in tyrosine-rich protein production, which needs properly spliced tRNATyr. This evidence reinforces the existence of a tRNA ligation pathway in vertebrates that differs from yeast and does not involve Tpt1. TRPT1 knockout mice cells do not exhibit any noticeable defects in Xbp-1 translation, thus it does not seem to be involved in the UPR response.62
Bacteria, also encode the Tpt1 homolog KptA, an unexpected finding considering that in these organisms introns are of the self-splicing type.65 Nevertheless, KptA is able to complement a Δtpt1 yeast strain indicating that the reaction catalyzed by KptA may be the same performed by Tpt1. The real function of KptA is puzzling for many reasons. First, KptA shows similar kinetics parameters and substrate specificity to yeast Tpt1. Second, why do bacteria conserve a Tpt1 homolog if there is no evidence for a bacterial RNA ligase that generates 2′-phosphates? These observations suggest that Tpt1/TRPT1/KptA have distinct roles in each organism and many questions involving the function of these enzymes still need answering.65
WHEN tRNA SPLICING SUFFERS, CELLS SUFFER
Knowing that tRNA splicing is an essential event in cells, it is not surprising that abnormalities and diseases related to alterations in the tRNA splicing pathways have been reported.66 One important example is pontocerebellar hypoplasia (PCH), a neurodegenerative disorder that compromises the development, growth, and function of the brain stem and cerebellum.67 Microcephaly, mental and motor impairment have also been noted. Mutations in three of four subunits of the tRNA splicing endonuclease in PCH-patient subtypes 2 and 4 have been identified.67 Since the endonuclease is essential for mature tRNA production and, thus, for cell survival, it is unlikely that the endonuclease activity is completely impaired. It may be that the activity is only reduced, suggesting that tRNA splicing may not be the sole cause of all the alterations observed.
In Caenorhabditis elegans, an RNAi-based genetic screen yielded RTCB-1 as one of the most significant modifiers of α-syn protein misfolding. As a consequence of this, RTCB-1 depletion leads to age-dependent neuron degeneration. This connection is not likely due to tRNA splicing defects but rather to the aforementioned involvement of the tRNA splicing ligase in XBP-1 mRNA splicing. Still, once again this highlights the global connection between seemingly unrelated pathways converging in an unusual way to affect neuroprotection.68 Importantly just like in the mammalian system, mutations in C. elegans RTCB-1 should impact neuron function.
hClp1 mutations have been reported as the molecular cause of a new syndrome that affects the central/peripheral nervous system.38,69 In mice, mutations in Clp1 led to the accumulation of tRNATyr fragments that resulted from a compromised tRNA splicing process. High levels of small tRNA fragments led to p53 hyperphosphorylation mediated by oxidative-stress and thereafter p53-induced cell death.38 In humans, mutations in hClp1 have been reported as the cause of cerebellar development impairment and neurodegeneration in patients who were homozygous for a single-nucleotide substitution. The mutant hClp1 contains a mutation in the ATP-binding loop, a highly conserved region in kinases.38,70 Neurons from patients showed a reduced kinase activity in vitro and altered Clp1 nuclear localization leading to intron-containing pre-tRNA accumulation and mature tRNA depletion.38 A study performed concomitantly showed that mutant hClp1 is still able to phosphorylate RNA in vitro but with a reduced activity.69 Furthermore, the hClp1 mutant is not able to interact with the TSEN complex, resulting in reduced pre-tRNA cleavage and accumulation of intron-containing tRNAs.38 Although all the alterations in the tRNA splicing process are clear, the machinery composed by TSEN proteins and hClp1 is also able to act on other substrates such as mRNAs and siRNAs. This suggests that other pathways can be linked to the phenotypes observed in these studies, making the underlying reasons for such defects very complex.
THE PERSISTENCE AND FATE OF tRNA INTRONS
Much has been written about intron evolution and arguments have been put forth and against their early or late appearance. Still a lingering question is why are introns still maintained? It has become clear now that mRNA introns are not so ‘junky’ after all, as they are part and parcel of the newly discovered world of noncoding RNAs and their eccentric ways of controlling RNA metabolism and gene expression.
For tRNAs, the same question can be raised, but the answer may not be so clear. Although in some well-documented cases, the intron is necessary for formation of post-transcriptional modifications crucial to function; for example, the case of pseudouridine formation in yeast tRNATyr and similar findings in other organisms.71 Still for the most parts, whether the introns serve a purpose has not yet been adequately answered. It is provoking to think that the intron once removed may serve a secondary function such as guiding modification of other RNAs or serve as some new kind of microRNA in cells; such proposals would indeed help rationalize the persistence of tRNA introns. Recently, every intron-containing tRNATrp genes in yeast were painstakingly replaced by a mature copy,72 the results were quite revealing in that nothing happened (i.e., there was no observable phenotype). So at least in that particular case the released intron seems to serve no obvious secondary function. This is not surprising given that there is great deal of variability on what tRNA contains an intron in different organisms as mentioned before in this review. The one glaring surprise is the fact that tRNATyr contains an intron in all eukaryotes. This observation led to the suggestion that this may have been the original intron-containing species that drove the evolution of tRNA splicing as we know it today, yet from its conservation we learn little as to why is still there.
CONCLUSION
The general principle that derives from so many studies is that tRNA splicing can be very complex, requiring several enzymatic activities. That the process has to be precise is highlighted by the fact that defects in tRNA components tend to have dire consequences for cells, causing a growing list of cellular defects. Remarkably, while archaeal and eukaryal tRNA splicing requires so many activities, the same is simply accomplished by bacterial self-splicing introns without the obvious need for proteins, begging the question as to what forces favored one mechanism over the other. Perhaps other factors deeply rooted in secondary functions for introns are at play.
One thing is clear, if introns are going to provide alternative functions, then the fate of introns following cleavage should be at the heart of the argument. As shown recently, in yeast the enzymes used to ‘heal’ tRNA ends for their protection also ‘heal’ intron ends for rapid destruction.73 Whether introns are diced away rapidly in other systems remains to be seen but at least in patients with mutations in CLP1 free introns accumulate suggesting that the presence of free introns may indeed affect normal physiology.70 Regardless of what the answers are to these more philosophical questions, the field of tRNA splicing is full of nuances that have provided fertile grounds for investigation. We hope that in this review we have raised as many questions as currently there are answers and convincingly show that there is still much road to travel in the expanding field of tRNA splicing and its often unexpected implications for cell physiology and metabolism.
ACKNOWLEDGMENTS
This work was supported by a GM084065-07 grant to Juan D. Alfonzo and WHO-TDR, CNPq, and FAPERJ grants to Carla Polycarpo.
Footnotes
Conflict of interest: The authors have declared no conflicts of interest for this article.
Notes
The use of the term ‘healing and sealing’ is accredited to the Stewart Shuman Laboratory, which first coined the term. Please see Ref 42 below.
REFERENCES
- 1.Fischer U, Englbrecht C, Chari A. Biogenesis of spliceosomal small nuclear ribonucleoproteins. WIREs RNA. 2011;2:718–731. doi: 10.1002/wrna.87. [DOI] [PubMed] [Google Scholar]
- 2.Nilsen TW. The spliceosome: the most complex macromolecular machine in the cell? Bioessays. 2003;25:1147–1149. doi: 10.1002/bies.10394. [DOI] [PubMed] [Google Scholar]
- 3.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 4.Pyle AM. The tertiary structure of group II introns: implications for biological function and evolution. Crit Rev Biochem Mol Biol. 2010;45:215–232. doi: 10.3109/10409231003796523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuhsel MG, Strickland R, Palmer JD. An ancient group I intron shared by eubacteria and chloroplasts. Science. 1990;250:1570–1573. doi: 10.1126/science.2125748. [DOI] [PubMed] [Google Scholar]
- 6.Reinhold-Hurek B, Shub DA. Self-splicing introns in tRNA genes of widely divergent bacteria. Nature. 1992;357:173–176. doi: 10.1038/357173a0. [DOI] [PubMed] [Google Scholar]
- 7.Abelson J, Trotta CR, Li H. tRNA splicing. J Biol Chem. 1998;273:12685–12688. doi: 10.1074/jbc.273.21.12685. [DOI] [PubMed] [Google Scholar]
- 8.Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marck C, Grosjean H. Identification of BHB splicing motifs in intron-containing tRNAs from 18 archaea: evolutionary implications. RNA. 2003;9:1516–1531. doi: 10.1261/rna.5132503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 11.Yoshihisa T. Handling tRNA introns, archaeal way and eukaryotic way. Front Genet. 2014;5:213. doi: 10.3389/fgene.2014.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reyes VM, Abelson J. Substrate recognition and splice site determination in yeast tRNA splicing. Cell. 1988;55:719–730. doi: 10.1016/0092-8674(88)90230-9. [DOI] [PubMed] [Google Scholar]
- 13.Di Nicola NE, Fabbri S, Bufardeci E, Baldi MI, Gandini Attardi D, Mattoccia E, Tocchini-Valentini GP. The eucaryal tRNA splicing endonuclease recognizes a tripartite set of RNA elements. Cell. 1997;89:859–866. doi: 10.1016/s0092-8674(00)80271-8. [DOI] [PubMed] [Google Scholar]
- 14.Baldi MI, Mattoccia E, Bufardeci E, Fabbri S, Tocchini-Valentini GP. Participation of the intron in the reaction catalyzed by the Xenopus tRNA splicing endonuclease. Science. 1992;255:1404–1408. doi: 10.1126/science.1542788. [DOI] [PubMed] [Google Scholar]
- 15.Rubio MA, Paris Z, Gaston KW, Fleming IM, Sample P, Trotta CR, Alfonzo JD. Unusual noncanonical intron editing is important for tRNA splicing in Trypanosoma brucei. Mol Cell. 2013;52:184–192. doi: 10.1016/j.molcel.2013.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson LD, Daniels CJ. A tRNA(Trp) intron endonuclease from Halobacterium volcanii. Unique substrate recognition properties. J Biol Chem. 1988;263:17951–17959. [PubMed] [Google Scholar]
- 17.Kleman-Leyer K, Armbruster DW, Daniels CJ. Properties of H. volcanii tRNA intron endonuclease reveal a relationship between the archaeal and eucaryal tRNA intron processing systems. Cell. 1997;89:839–847. doi: 10.1016/s0092-8674(00)80269-x. [DOI] [PubMed] [Google Scholar]
- 18.Peebles CL, Gegenheimer P, Abelson J. Precise excision of intervening sequences from precursor tRNAs by a membrane-associated yeast endonuclease. Cell. 1983;32:525–536. doi: 10.1016/0092-8674(83)90472-5. [DOI] [PubMed] [Google Scholar]
- 19.Rauhut R, Green PR, Abelson J. Yeast tRNA-splicing endonuclease is a heterotrimeric enzyme. J Biol Chem. 1990;265:18180–18184. [PubMed] [Google Scholar]
- 20.Trotta CR, Miao F, Arn EA, Stevens SW, Ho CK, Rauhut R, Abelson JN. The yeast tRNA splicing endonuclease: a tetrameric enzyme with two active site subunits homologous to the archaeal tRNA endonucleases. Cell. 1997;89:849–858. doi: 10.1016/s0092-8674(00)80270-6. [DOI] [PubMed] [Google Scholar]
- 21.Ho CK, Rauhut R, Vijayraghavan U, Abelson J. Accumulation of pre-tRNA splicing ’2/3’ intermediates in a Saccharomyces cerevisiae mutant. EMBO J. 1990;9:1245–1252. doi: 10.1002/j.1460-2075.1990.tb08232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lykke-Andersen J, Garrett RA. RNA-protein interactions of an archaeal homotetrameric splicing endoribonuclease with an exceptional evolutionary history. EMBO J. 1997;16:6290–6300. doi: 10.1093/emboj/16.20.6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Trotta CR, Abelson J. Crystal structure and evolution of a transfer RNA splicing enzyme. Science. 1998;280:279–284. doi: 10.1126/science.280.5361.279. [DOI] [PubMed] [Google Scholar]
- 24.Calvin K, Hall MD, Xu F, Xue S, Li H. Structural characterization of the catalytic subunit of a novel RNA splicing endonuclease. J Mol Biol. 2005;353:952–960. doi: 10.1016/j.jmb.2005.09.035. [DOI] [PubMed] [Google Scholar]
- 25.Xue S, Calvin K, Li H. RNA recognition and cleavage by a splicing endonuclease. Science. 2006;312:906–910. doi: 10.1126/science.1126629. [DOI] [PubMed] [Google Scholar]
- 26.Calvin K, Li H. RNA-splicing endonuclease structure and function. Cell Mol Life Sci. 2008;65:1176–1185. doi: 10.1007/s00018-008-7393-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujishima K, Sugahara J, Miller CS, Baker BJ, Di Giulio M, Takesue K, Sato A, Tomita M, Banfield JF, Kanai A. A novel three-unit tRNA splicing endonuclease found in ultrasmall Archaea possesses broad substrate specificity. Nucleic Acids Res. 2011;39:9695–9704. doi: 10.1093/nar/gkr692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanai A. Welcome to the new tRNA world! Front Genet. 2014;5:336. doi: 10.3389/fgene.2014.00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fabbri S, Fruscoloni P, Bufardeci E, Di Nicola NE, Baldi MI, Attardi DG, Mattoccia E, Tocchini-Valentini GP. Conservation of substrate recognition mechanisms by tRNA splicing endonucleases. Science. 1998;280:284–286. doi: 10.1126/science.280.5361.284. [DOI] [PubMed] [Google Scholar]
- 30.De Robertis EM, Black P, Nishikura K. Intranuclear location of the tRNA splicing enzymes. Cell. 1981;23:89–93. doi: 10.1016/0092-8674(81)90273-7. [DOI] [PubMed] [Google Scholar]
- 31.Englert M, Latz A, Becker D, Gimple O, Beier H, Akama K. Plant pre-tRNA splicing enzymes are targeted to multiple cellular compartments. Biochimie. 2007;89:1351–1365. doi: 10.1016/j.biochi.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 32.Yoshihisa T, Yunoki-Esaki K, Ohshima C, Tanaka N, Endo T. Possibility of cytoplasmic pre-tRNA splicing: the yeast tRNA splicing endonuclease mainly localizes on the mitochondria. Mol Biol Cell. 2003;14:3266–3279. doi: 10.1091/mbc.E02-11-0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dhungel N, Hopper AK. Beyond tRNA cleavage: novel essential function for yeast tRNA splicing endonuclease unrelated to tRNA processing. Genes Dev. 2012;26:503–514. doi: 10.1101/gad.183004.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kjems J, Garrett RA. Novel splicing mechanism for the ribosomal RNA intron in the archaebacterium Desulfurococcus mobilis. Cell. 1988;54:693–703. doi: 10.1016/s0092-8674(88)80014-x. [DOI] [PubMed] [Google Scholar]
- 35.Paushkin SV, Patel M, Furia BS, Peltz SW, Trotta CR. Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3’ end formation. Cell. 2004;117:311–321. doi: 10.1016/s0092-8674(04)00342-3. [DOI] [PubMed] [Google Scholar]
- 36.Lykke-Andersen J, Aagaard C, Semionenkov M, Garrett RA. Archaeal introns: splicing, intercellular mobility and evolution. Trends Biochem Sci. 1997;22:326–331. doi: 10.1016/s0968-0004(97)01113-4. [DOI] [PubMed] [Google Scholar]
- 37.Weitzer S, Martinez J. The human RNA kinase hClp1 is active on 3′ transfer RNA exons and short interfering RNAs. Nature. 2007;447:222–226. doi: 10.1038/nature05777. [DOI] [PubMed] [Google Scholar]
- 38.Hanada T, Weitzer S, Mair B, Bernreuther C, Wainger BJ, Ichida J, Hanada R, Orthofer M, Cronin SJ, Komnenovic V, et al. CLP1 links tRNA metabolism to progressive motor-neuron loss. Nature. 2013;495:474–480. doi: 10.1038/nature11923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Popow J, Schleiffer A, Martinez J. Diversity and roles of (t)RNA ligases. Cell Mol Life Sci. 2012;69:2657–2670. doi: 10.1007/s00018-012-0944-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greer CL, Peebles CL, Gegenheimer P, Abelson J. Mechanism of action of a yeast RNA ligase in tRNA splicing. Cell. 1983;32:537–546. doi: 10.1016/0092-8674(83)90473-7. [DOI] [PubMed] [Google Scholar]
- 41.Wang LK, Shuman S. Structure-function analysis of yeast tRNA ligase. RNA. 2005;11:966–975. doi: 10.1261/rna.2170305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phizicky EM, Schwartz RC, Abelson J. Saccharomyces cerevisiae tRNA ligase. Purification of the protein and isolation of the structural gene. J Biol Chem. 1986;261:2978–2986. [PubMed] [Google Scholar]
- 43.Englert M, Beier H. Plant tRNA ligases are multifunctional enzymes that have diverged in sequence and substrate specificity from RNA ligases of other phylogenetic origins. Nucleic Acids Res. 2005;33:388–399. doi: 10.1093/nar/gki174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwer B, Sawaya R, Ho CK, Shuman S. Portability and fidelity of RNA-repair systems. Proc Natl Acad Sci U S A. 2004;101:2788–2793. doi: 10.1073/pnas.0305859101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Culver GM, McCraith SM, Consaul SA, Stanford DR, Phizicky EM. A 2′-phosphotransferase implicated in tRNA splicing is essential in Saccharomyces cerevisiae. J Biol Chem. 1997;272:13203–13210. doi: 10.1074/jbc.272.20.13203. [DOI] [PubMed] [Google Scholar]
- 46.Sidrauski C, Cox JS, Walter P. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell. 1996;87:405–413. doi: 10.1016/s0092-8674(00)81361-6. [DOI] [PubMed] [Google Scholar]
- 47.Hu QD, Lu H, Huo K, Ying K, Li J, Xie Y, Mao Y, Li YY. A human homolog of the yeast gene encoding tRNA 2′-phosphotransferase: cloning, characterization and complementation analysis. Cell Mol Life Sci. 2003;60:1725–1732. doi: 10.1007/s00018-003-3107-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwer B, Aronova A, Ramirez A, Braun P, Shuman S. Mammalian 2′,3′ cyclic nucleotide phosphodiesterase (CNP) can function as a tRNA splicing enzyme in vivo. RNA. 2008;14:204–210. doi: 10.1261/rna.858108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanaka N, Chakravarty AK, Maughan B, Shuman S. Novel mechanism of RNA repair by RtcB via sequential 2′,3′-cyclic phosphodiesterase and 3′-Phosphate/5′-hydroxyl ligation reactions. J Biol Chem. 2011;286:43134–43143. doi: 10.1074/jbc.M111.302133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanaka N, Shuman S. RtcB is the RNA ligase component of an Escherichia coli RNA repair operon. J Biol Chem. 2011;286:7727–7731. doi: 10.1074/jbc.C111.219022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Popow J, Englert M, Weitzer S, Schleiffer A, Mierzwa B, Mechtler K, Trowitzsch S, Will CL, Luhrmann R, Soll D, et al. HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science. 2011;331:760–764. doi: 10.1126/science.1197847. [DOI] [PubMed] [Google Scholar]
- 52.Perez-Gonzalez A, Pazo A, Navajas R, Ciordia S, Rodriguez-Frandsen A, Nieto A. hCLE/C14orf166 associates with DDX1-HSPC117-FAM98B in a novel transcription-dependent shuttling RNA-transporting complex. PLoS One. 2014;9:e90957. doi: 10.1371/journal.pone.0090957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Popow J, Jurkin J, Schleiffer A, Martinez J. Analysis of orthologous groups reveals archease and DDX1 as tRNA splicing factors. Nature. 2014;511:104–107. doi: 10.1038/nature13284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Desai KK, Cheng CL, Bingman CA, Phillips GN, Jr, Raines RT. A tRNA splicing operon: archease endows RtcB with dual GTP/ATP cofactor specificity and accelerates RNA ligation. Nucleic Acids Res. 2014;42:3931–3942. doi: 10.1093/nar/gkt1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kosmaczewski SG, Edwards TJ, Han SM, Eckwahl MJ, Meyer BI, Peach S, Hesselberth JR, Wolin SL, Hammarlund M. The RtcB RNA ligase is an essential component of the metazoan unfolded protein response. EMBO Rep. 2014;15:1278–1285. doi: 10.15252/embr.201439531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu Y, Liang FX, Wang X. A synthetic biology approach identifies the mammalian UPR RNA ligase RtcB. Mol Cell. 2014;55:758–770. doi: 10.1016/j.molcel.2014.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jurkin J, Henkel T, Nielsen AF, Minnich M, Popow J, Kaufmann T, Heindl K, Hoffmann T, Busslinger M, Martinez J. The mammalian tRNA ligase complex mediates splicing of XBP1 mRNA and controls antibody secretion in plasma cells. EMBO J. 2014;33:2922–2936. doi: 10.15252/embj.201490332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tanaka N, Meineke B, Shuman S. RtcB, a novel RNA ligase, can catalyze tRNA splicing and HAC1 mRNA splicing in vivo. J Biol Chem. 2011;286:30253–30257. doi: 10.1074/jbc.C111.274597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gross S, Moore C. Five subunits are required for reconstitution of the cleavage and polyadenylation activities of Saccharomyces cerevisiae cleavage factor I. Proc Natl Acad Sci U S A. 2001;98:6080–6085. doi: 10.1073/pnas.101046598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weitzer S, Martinez J. hClp1: a novel kinase revitalizes RNA metabolism. Cell Cycle. 2007;6:2133–2137. doi: 10.4161/cc.6.17.4642. [DOI] [PubMed] [Google Scholar]
- 61.Ramirez A, Shuman S, Schwer B. Human RNA 5′-kinase (hClp1) can function as a tRNA splicing enzyme in vivo. RNA. 2008;14:1737–1745. doi: 10.1261/rna.1142908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harding HP, Lackey JG, Hsu HC, Zhang Y, Deng J, Xu RM, Damha MJ, Ron D. An intact unfolded protein response in Trpt1 knockout mice reveals phylogenic divergence in pathways for RNA ligation. RNA. 2008;14:225–232. doi: 10.1261/rna.859908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Englert M, Sheppard K, Gundllapalli S, Beier H, Soll D. Branchiostoma floridae has separate healing and sealing enzymes for 5′-phosphate RNA ligation. Proc Natl Acad Sci U S A. 2010;107:16834–16839. doi: 10.1073/pnas.1011703107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Culver GM, McCraith SM, Zillmann M, Kierzek R, Michaud N, LaReau RD, Turner DH, Phizicky EM. An NAD derivative produced during transfer RNA splicing: ADP-ribose 1′′-2′′ cyclic phosphate. Science. 1993;261:206–208. doi: 10.1126/science.8392224. [DOI] [PubMed] [Google Scholar]
- 65.Steiger MA, Kierzek R, Turner DH, Phizicky EM. Substrate recognition by a yeast 2′-phosphotransferase involved in tRNA splicing and by its Escherichia coli homolog. Biochemistry. 2001;40:14098–14105. doi: 10.1021/bi011388t. [DOI] [PubMed] [Google Scholar]
- 66.Weitzer S, Hanada T, Penninger JM, Martinez J. CLP1 as a novel player in linking tRNA splicing to neurodegenerative disorders. WIREs RNA. 2014;6:47–63. doi: 10.1002/wrna.1255. [DOI] [PubMed] [Google Scholar]
- 67.Budde BS, Namavar Y, Barth PG, Poll-The BT, Nurnberg G, Becker C, van Ruissen F, Weterman MA, Fluiter K, te Beek ET, et al. tRNA splicing endonuclease mutations cause pontocerebellar hypoplasia. Nat Genet. 2008;40:1113–1118. doi: 10.1038/ng.204. [DOI] [PubMed] [Google Scholar]
- 68.Ray A, Zhang S, Rentas C, Caldwell KA, Caldwell GA. RTCB-1 mediates neuroprotection via XBP-1 mRNA splicing in the unfolded protein response pathway. J Neurosci. 2014;34:16076–16085. doi: 10.1523/JNEUROSCI.1945-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Karaca E, Weitzer S, Pehlivan D, Shiraishi H, Gogakos T, Hanada T, Jhangiani SN, Wiszniewski W, Withers M, Campbell IM, et al. Human CLP1 mutations alter tRNA biogenesis, affecting both peripheral and central nervous system function. Cell. 2014;157:636–650. doi: 10.1016/j.cell.2014.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schaffer AE, Eggens VR, Caglayan AO, Reuter MS, Scott E, Coufal NG, Silhavy JL, Xue Y, Kayserili H, Yasuno K, et al. CLP1 founder mutation links tRNA splicing and maturation to cerebellar development and neurodegeneration. Cell. 2014;157:651–663. doi: 10.1016/j.cell.2014.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johnson PF, Abelson J. The yeast tRNATyr gene intron is essential for correct modification of its tRNA product. Nature. 1983;302:681–687. doi: 10.1038/302681a0. [DOI] [PubMed] [Google Scholar]
- 72.Mori S, Kajita T, Endo T, Yoshihisa T. The intron of tRNA-TrpCCA is dispensable for growth and translation of Saccharomyces cerevisiae. RNA. 2011;17:1760–1769. doi: 10.1261/rna.2851411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu J, Hopper AK. Healing for destruction: tRNA intron degradation in yeast is a two-step cytoplasmic process catalyzed by tRNA ligase Rlg1 and 5′-to-3′ exonuclease Xrn1. Genes Dev. 2014;28:1556–1561. doi: 10.1101/gad.244673.114. [DOI] [PMC free article] [PubMed] [Google Scholar]