Abstract
Autism spectrum disorders (ASD) are characterized by both phenotypic and genetic heterogeneity. Our analysis of functional networks perturbed in ASD suggests that both truncating and non-truncating de novo mutations contribute to autism, although there is a strong bias against truncating mutations in early embryonic development. We find that functional mutations are preferentially observed in genes likely to be haploinsufficient. Multiple cell types and brain areas are affected, but the impact of ASD mutations appears to be strongest in the cortical neurons and the medium spiny neurons of the striatum, implicating corticostriatal brain circuits. In females, truncating ASD mutations on average impact genes with 50–100% higher brain expression levels compared to males. Our study also suggests that truncating de novo mutations play a smaller role in the etiology of high-functioning ASD cases. Overall, we find that stronger functional insults usually lead to more severe intellectual, social and behavioral ASD phenotypes.
INTRODUCTION
Autism spectrum disorders (ASD) are associated with a wide range of cognitive and behavioral abnormalities1, 2. It is estimated that many hundreds of genes may ultimately contribute to autism and related phenotypes2, 3. Functionally important de novo mutations associated with ASD are individually rare, but their collective contribution to sporadic ASD cases is likely to be substantial due to a large number of target genes3-10. Several recent studies identified a large collection of de novo mutations associated with ASD, including copy number variations (CNVs)6 and single nucleotide variations (SNVs)3, 8, 11. These studies demonstrated that truncating SNVs (such as nonsense, splice site, and frameshift mutations) and large CNVs are likely to play a causal role in ASD.
To explore these underlying biological pathways, we previously developed a computational approach (NETBAG+) that searches for cohesive biological networks using a diverse collection of disease-associated genetic variants12, 13. Using network-based approaches we and others have recently demonstrated that genetic variations associated with ASD and other psychiatric disorders converge on several biological networks involved in brain development and synaptic function12-15. In parallel with the identification of disease-associated genetic variations, complementary datasets of brain-related functional and phenotypic resources are rapidly being accumulated. These include a comprehensive database of gene expression data across different cell types, distinct anatomical brain regions, and developmental stages16, 17. In addition, resources such as the Simons Simplex Collection (SSC)18 have assembled a large compendium of ASD-related phenotypic data, including intelligence and social phenotypic scores. In the present study we focus our analyses on a set of genes implicated by our network-based computational approach and also on all de novo truncating mutations from several recent studies. These two approaches provide complementary genes sets with a significant fraction of causal ASD mutations. We investigate the temporal, spatial, and cell-specific expression profiles of implicated genes. We also explore how expression, network, and functional properties of autism-associated genes affect ASD phenotypes.
RESULTS
Functional gene networks affected by de novo mutations
To elucidate functional networks perturbed in ASD, we applied NETBAG+ to a set of genes affected by de novo CNVs and SNVs observed in autistic patients from the Simons Simplex Collection (SSC) 3, 6-8. Of note, all the mutations used as the input for our analyses were obtained using genome-wide methodologies and are therefore not biased by any pre-existing hypotheses of ASD etiology. The combined input data contained a total of 991 unique genes from 624 independent genomic loci, including 580 unique genes with de novo SNVs, and 434 genes within de novo CNVs; we note that the number of genomic loci used in this study is considerably larger than the 47 loci considered in our previous analysis of de novo CNV events in autism15. We used NETBAG+ to identify a subset of the input genes that are strongly connected in the underlying phenotypic network (see Methods). The NETBAG+ search revealed a functional network containing 159 genes (P = 0.036, Fig. 1), of which 131 genes were affected by de novo SNVs (Fig. 1, circles), and 31 by de novo CNVs (squares). The network’s significance was estimated using random input sets that matched the real data in terms of protein length and network connectivity. Notably, no significant networks were detected using genes associated with the 368 non-synonymous de novo mutations identified in siblings.
Figure 1.
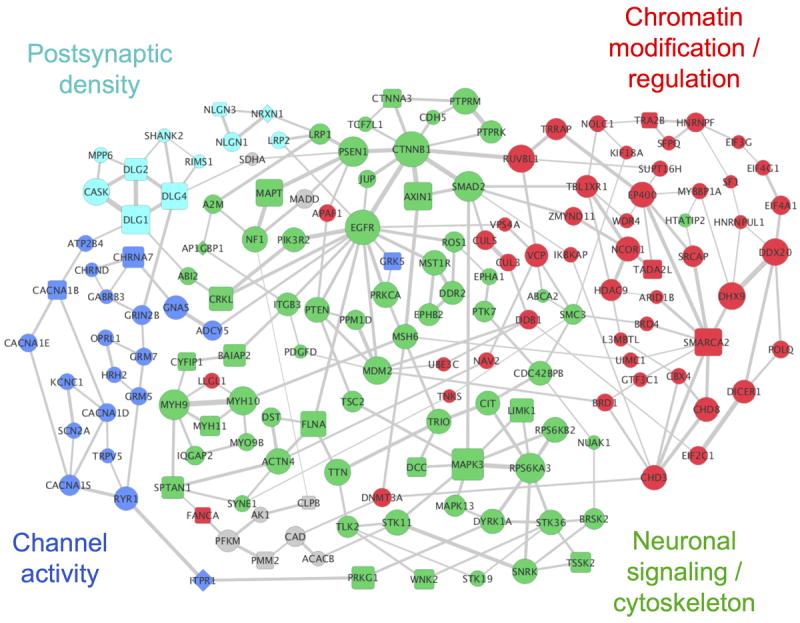
The network implicated by NETBAG+ based on ASD-associated de novo SNVs and CNVs from recent studies (network is comprised of 159 genes; P = 0.036). Node sizes are proportional to the contributions of each gene to the overall network score, and edge widths are proportional to the likelihood that the corresponding gene pair contributes to the same genetic phenotype (see Methods). For clarity, only the two strongest edges for each gene are shown. Node shapes indicate types of the corresponding mutations: circles represent genes from SNVs, squares represent genes from CNVs, and diamonds represent genes affected by both mutation types. The network was divided into cohesive functional clusters (indicated by node colors) using hierarchical clustering; general functions of these clusters determined using DAVID are shown in the figure (see Supplementary Table S3 for a complete list of GO terms associated with each cluster). Grey nodes represent genes that are not members of the network clusters.
To explore the biological functions associated with the functional network, we used DAVID19 to identify Gene Ontology (GO) terms that are significantly enriched among network gene annotations (Table 1). This analysis identified a diverse set of functions associated with the implicated network, including synaptic functions, chromatin modifications, and calcium channel activity. To better understand the functional relationships between genes in the network, we performed hierarchical clustering of the network genes using the strength of interactions in the phenotypic network as a metric. This analysis identified four major clusters in the network, each of which was associated with distinct and non-overlapping biological functions (Supplementary Table S3; node colors in Fig. 1 indicate cluster assignment).
Table 1. Gene Ontology (GO) terms associated with the implicated network.
| GO ID | Ontology | Term | P-value |
|---|---|---|---|
| GO:0045202 | CC | Synapse | 3×10−7 |
| GO:0016568 | BP | chromatin modification | 6×10−6 |
| GO:0051015 | MF | actin filament binding | 7×10−6 |
| GO:0005262 | MF | calcium channel activity | 1×10−5 |
| GO:0031252 | CC | cell leading edge | 2×10−5 |
| GO:0004386 | MF | helicase activity | 3×10−5 |
| GO:0015629 | CC | actin cytoskeleton | 6×10−5 |
| GO:0030036 | BP | actin cytoskeleton organization | 7×10−5 |
| GO:0016887 | MF | ATPase activity | 7×10−5 |
| GO:0005913 | CC | cell-cell adherens junction | 8×10−5 |
| GO:0045211 | CC | postsynaptic membrane | 8×10−5 |
| GO:0044456 | CC | synapse part | 0.0001 |
| GO:0007611 | BP | learning or memory | 0.0001 |
| GO:0030029 | BP | actin filament-based process | 0.0001 |
| GO:0005938 | CC | cell cortex | 0.0001 |
| GO:0005911 | CC | cell-cell junction | 0.0002 |
| GO:0014069 | CC | postsynaptic density | 0.002 |
| GO:0043005 | CC | neuron projection | 0.004 |
| GO:0030425 | CC | Dendrite | 0.004 |
Gene Ontology (GO) terms enriched in the implicated network (Fig. 1), as identified by DAVID (david.abcc.ncifcrf.gov). P-values shown in the table were corrected for multiple hypotheses testing using the Benjamini-Hochberg procedure in DAVID. The ontology column indicates GO domain: BP for biological process, MF for molecular function, and CC for cellular component. Non-specific terms, i.e. terms associated with more than 400 human genes, are not shown. For a list of GO terms associated with each network cluster see Supplementary Table S3. The enrichment calculations were performed using the default background set of all human genes; similar enrichment results were also obtained using brain-expressed genes as background (see Supplementary Table S4).
One cluster in our network (Fig. 1, cyan) contains genes responsible for synapse formation and function. This cluster contains neurexin (NRXN) and neuroligin (NRLG), and important components (SHANK2 and DLG2/DLG4) of the postsynaptic density (PSD) of excitatory synapses. A related cluster (blue) includes a diverse set of ion channels and receptors. This cluster contains genes (CACNA1B/D/E/S) for subunits of several voltage-dependent calcium channels, which play an important role in learning and memory20. The largest cluster (green) of the implicated network contains genes primarily associated with neuronal signaling and migration (NF1, DCC, EPHA1/B2), intracellular signaling (MAPK3, EGFR, PTEN, MDM2, CTNNB1, PRKCA, LIMK1), and the development of neuronal projections and actin cytoskeleton (CYFIP1, TRIO, SPTAN1, FLNA, ACTN4). As we have previously demonstrated, ASD-associated mutations often perturb multiple signaling pathways that converge on the regulation of actin cytoskeleton and other structural processes that are required for neuron migration, cell-cell adhesion, and the development of neuronal projections12, 21, 22. Many genes in the clusters described above encode proteins that participate in various processes related to the growth and function of dendritic spines, which have been implicated in our previous analyses of genetic insults in ASD and schizophrenia12, 13. The final cluster (red) is primarily related to functions associated with chromatin modifications, chromatin remodeling, and transcriptional regulation. Notably, there is growing evidence that chromatin regulatory mechanisms crucially affect various stages of neural development, neuroplasticity, and learning23. This cluster also contains genes involved in other processes that have recently been implicated in ASD: RNA interference (DICER1, AGO1/EIF2C1), translation (EIF4A1, EIF4G1, EIF3G) and splicing (SFPQ, TRA2B)24.
Association of the implicated genes with specific functional subsets
In addition to the molecular and biological GO categories discussed above, it is interesting to investigate an overlap between the network and specific gene subsets that may highlight the phenotypic and functional properties of the network genes. Recent exome sequencing studies revealed recurrent truncating mutations in several genes (ADNP, ANK2, ARID1B, CHD8, CUL3, DYRK1A, GRIN2B, KATNAL2, POGZ, SCN2A, and TBR1)3, 8, 11, 34. Although only 131/580 (~23%) of all genes harboring missense SNVs form the implicated network, six out of the eleven genes with recurrent truncating mutations are in the network (Fisher’s exact one-tail test P = 0.02). This suggests that there is a significant enrichment of causal genes in the implicated network. Because ASD de novo mutations are predominantly heterozygous, it is likely that true causal mutations would preferentially target haploinsufficient genes. Indeed, using haploinsufficiency probabilities from a recent study25, we found that genes in the implicated network are significantly more likely to be haploinsufficient than genes not selected by NETBAG+ (median probability for network and non-network genes are 0.57 and 0.32, respectively; Wilcoxon rank-sum two-tail test P < 10−14); a similar result is also observed for genes with recurrent truncating mutations (median probability for genes with recurrent truncating mutations and unaffected sibling SNV genes are 0.69 and 0.39, respectively; Wilcoxon rank-sum two-tail P = 0.016). We also considered the Genomic Evolutionary Rate Profiling (GERP) score to characterize the severity of SNV mutations. Interestingly, the average GERP score of network genes is significantly higher than the average GERP score of genes not selected into the network (average network and non-network GERP scores are 4.0 and 3.3, respectively; Wilcoxon rank-sum one-tail P = 0.001). As higher GERP scores correspond to more damaging mutations, this analysis demonstrates that NETBAG predominantly selects genes harboring SNV mutations with potentially higher functional impact.
Notably, a recent exome study showed a significant overlap between genes harboring truncating ASD de novo SNVs and targets of the fragile X mental retardation protein (FMRP) 3, 26. FMRP is a RNA-binding protein, essential for a wide array of cognitive functions including synaptic plasticity and learning27, 28. Failure to properly express FMRP can cause fragile X syndrome, a genetic disorder resulting in a spectrum of cognitive disabilities and often accompanied by autistic symptoms. Following the analysis by Iossifov et al. of truncating de novo SNVs, we calculated the expected number of FMRP targets for ASD network genes by inferring gene mutabilities from a large exome sequencing study29 (see Methods); the expected number of FMRP targets was compared to the observed values in various ASD gene sets (Table 2). This analysis showed that, similar to truncating SNVs genes, genes in the implicated network are significantly enriched for FMRP targets (truncating SNV gene enrichment 1:2.13, two-tail binomial test P = 3×10−4; network gene enrichment 1:2.78, P = 3×10−11). The enrichment remains significant when considering only network genes harboring non-truncating SNVs (enrichment = 1:2.67, P = 4×10−9). Significant enrichment is also observed for FMRP targets from another recent study (Supplementary Table S5)30. In contrast, there is no significant enrichment for proband SNV genes not selected to the network (P = 0.2) and de novo SNV genes from unaffected siblings (P = 0.15). Furthermore, enrichment for FMRP targets remains significant when we separately consider each of the four functional clusters of the implicated network (Table 2). Overall, these analyses confirm that FMRP targets play an important role in the autism etiology and suggest a causal role for a significant fraction of genes with non-truncating de-novo SNVs.
Table 2. Overlap between ASD gene sets and FMRP targets.
| Gene set | Number of genes | Expected : observed | P-value |
|---|---|---|---|
| Network genes | 159 | 17.3 : 48 (1 : 2.78) | 3×10−11 |
| Truncating SNV genes | 108 | 11.7 : 25 (1 : 2.13) | 0.0003 |
| Network non-truncating-SNV genes | 138 | 15.0 : 40 (1 : 2.67) | 4×10−9 |
| Neuronal signaling/cytoskeleton cluster genes | 69 | 7.49 : 15 (1 : 2.00) | 0.01 |
| Chromatin modification/regulation cluster genes | 50 | 5.43 : 17 (1 : 3.13) | 1×10−5 |
| Postsynaptic density cluster genes | 11 | 1.19 : 5 (1 : 4.19) | 0.004 |
| Channel activity cluster genes | 21 | 2.28 : 9 (1 : 3.95) | 0.0002 |
| Non-network SNV genes | 449 | 48.8 : 40 (1 : 0.82) | 0.2 |
| Sibling SNV genes | 355 | 38.6 : 47 (1 : 1.22) | 0.15 |
The expected and observed number of fragile X mental retardation protein (FMRP) targets26 among different sets of implicated genes. The expected numbers of FMRP targets were obtained based on the proportion of rare synonymous variants observed in a recent large-scale survey of human genetic variation29. The observed and expected overlaps based on another recent study of FMRP targets by Ascano et al.30 are given in Supplementary Table S5. The significances of the overlaps were established using the two-tail binomial test.
Genes forming the postsynaptic density (PSD) are also likely to play an important role in ASD and other psychiatric disorders12, 31. The PSD is localized at the postsynaptic membrane of excitatory synapses and is crucial to synaptic communications and plasticity32-34. There is a significant enrichment of PSD-associated genes in the implicated network (enrichment 1:3.4, two-tail binomial test P = 7×10−9, Supplementary Table S5) and marginal enrichment for truncating de novo SNVs in probands (enrichment 1:1.9, P = 0.05). No enrichment is observed for genes harboring SNVs in unaffected siblings (enrichment 1:1.05, P = 0.8). For genes not selected to the network, PSD genes are significantly under-represented (enrichment 1:0.58, P=0.04).
Temporal and spatial brain expression patterns of implicated genes
Gene expression patterns provide important clues for understanding physiological and developmental contexts of gene function. Therefore, we next investigated brain expression of the ASD-implicated genes using the Human Brain Transcriptome database (HBT, hbatlas.org)16. Based on postmortem transcriptional analysis of tissue samples from healthy individuals, the HBT database provides a comprehensive map of mRNA expression in multiple regions of the human brain and across developmental stages. The average expression levels across developmental periods are shown in Fig. 2a for various gene sets that form the implicated network as well as for genes with truncating de novo SNVs. The brain expression of network genes (Fig. 2a, red) is significantly higher (Wilcoxon rank-sum two-tail test P < 10−15) than the expression of SNV-containing genes that were not selected to the network (purple) or all human genes in the HBT database (dashed line). This result confirms that the implicated network is significantly enriched in brain-related genes. Notably, high brain expression of network genes is not simply a consequence of their length or connectivity in the phenotypic network; randomly selected human genes with equivalent length or network connectivity are expressed in the brain at significantly lower levels (P < 10−4) across all developmental periods (Supplementary Fig. S2).
Figure 2.
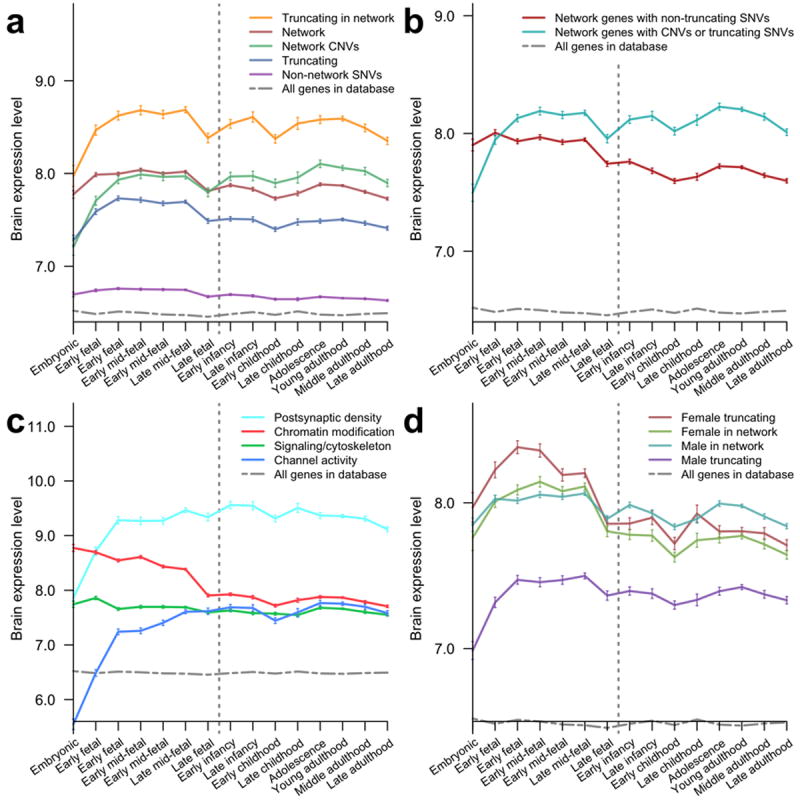
Temporal gene expression profiles in the human brain across developmental stages for implicated gene sets. Expression data were obtained from the HBT database; average expression levels at each developmental stage were calculated using all genes in a given set and error bars represent SEM. Vertical dashed lines indicate birth. (a) Expression profiles for truncating SNV genes in the network (orange), all network genes (red), network genes from CNVs (green), all truncating SNV genes (blue), and non-network SNV genes (purple). (b) Expression profiles for network genes with truncating (cyan) and non-truncating (red) mutations observed in probands. (c) Expression profiles for functional clusters (Fig. 1) of the implicated network: postsynaptic density genes (cyan), chromatin modification/regulation genes (red), signaling/cytoskeleton genes (green), and channel activity genes (blue). (d) Expression profiles for network and truncating female/male SNV genes: female truncating SNV genes (red), female network genes (green), male network genes (blue), and male truncating SNV genes (purple). Human expression data were obtained from the HBT database. Vertical dashed lines separate prenatal and postnatal developmental stages. Error bars represent SEM.
Expression of the network genes (Fig. 2a, red) is on average highest during the early fetal to early mid-fetal periods (8 to 19 post-conception weeks), possibly due to higher activity of ASD-related genes within brain cells or changes in brain cellular composition. To quantify the prenatal expression bias, we calculated the difference between the expression during prenatal developmental periods and during postnatal periods for every gene harboring de novo mutations. We used a Wilcoxon rank-sum test (PWT) to estimate the significance of the expression difference between sets of biological samples, and a permutation-based test (PPT) to estimate the probability of observing a greater bias in random probe sets of equal size (see Methods). The prenatal bias was highly significant for network genes (bias 0.16, one-tail test PWT < 10−15/PPT < 10−4) and also for genes with truncating SNVs (bias 0.18, PWT < 10−15/PPT < 10−4). However, we did not observe the high prenatal expression of ASD network genes in the embryonic period. This effect is particularly dramatic for genes with truncating de novo SNVs in the network (Fig. 2a, orange) and for genes from de novo CNVs in the network (Fig. 2a, green) in which embryonic expression is substantially lower compared to later developmental periods (two-tail test PWT = 6×10−7/PPT < 10−4 and PWT = 10−14/PPT < 10−4 respectively); Fig. 2b shows the average expression profiles for network genes with truncating (CNVs and truncating SNVs) and non-truncating mutations. Reassuringly, the bias against mutations in the embryonic period is absent for non-synonymous de novo SNV genes not selected to the network (PWT = 0.4/PPT = 0.3) and for genes with de novo SNVs in siblings (PWT = 0.2/PPT = 0.2). The observed bias against de novo truncating SNVs and CNVs during the earliest phase of embryonic development suggests strong selection against harmful mutations as they likely lead to more severe developmental consequences compared to mutations typically associated with autism.
We also examined the temporal expression profiles of each of the four clusters forming the implicated network (Fig. 2c). The cluster that is primarily associated with the postsynaptic density (Fig. 2c, cyan) has the highest overall expression in the brain. Both the PSD cluster and the cluster associated with various channel activities (blue) show a distinct rise during early fetal development, consistent with the start of synaptogenesis at the early mid-fetal developmental stage. In contrast, the cluster associated with chromatin modification and regulation (red) shows high embryonic expression, consistent with a developmental peak of neuronal proliferation and differentiation, and then gradually decreases to a postnatal plateau. Sustained expression of chromatin modification genes after neurodevelopmental stages may be attributed to their involvement in synaptic plasticity35. Finally, the cluster associated with neuronal signaling and cytoskeleton (green) is relatively constant across all developmental stages. This is consistent with important roles that signaling and structural genes play across all stages: from neural differentiation and migration to neuron projection development, and from synaptogenesis to learning and memory. Importantly, while the average expression profiles of clusters are shown in Fig. 2c, there is significant variability in the expression of individual genes (Supplementary Fig. S3). This variability is likely to be at least partially explained by different dosage requirements of individual genes. Normalized expression trajectories (see Supplementary Fig. S3) show that the temporal profiles of individual genes are generally consistent with the average expression profiles of corresponding functional clusters.
A high male to female incidence ratio, estimated at more than 4:1 for high-functioning individuals, is one of the most consistent and dramatic findings in ASD36, 37. Evidence suggests that this bias may be due to a female protective effect that requires a higher threshold of genetic insults to trigger ASD in females compared to males38. Consistent with this hypothesis is the observation that females in the SSC collection have a greater burden of truncating mutations than males (Fisher’s exact one-tail test P = 0.07) and this gender dimorphism is even stronger for the genes in the implicated network (30% of SNV mutations in females are truncating compared to 13% in males; Fisher’s exact one-tail test P = 0.03). Notably, the average brain expression is significantly higher for genes harboring truncating de novo mutations in females than in males (Fig. 2d; 7.98 the average log2 expression for genes in females compared to 7.40 for males; one-tail test PWT < 10−15/PPT = 2×10−3). Thus, the expression data also suggest that relatively stronger genetic perturbations, i.e. truncating mutations in genes with higher brain expression, are preferentially associated with ASD in females. Importantly, the observed expression patterns cannot be explained by normal expression differences between males and females. For genes with truncating SNV mutations in females, the average brain expression levels in females and males are 8.02 and 7.95 (< 1% difference), respectively; for genes with truncating SNVs in males, the average brain expression levels in females and males are 7.42 and 7.39 (< 0.5% difference), respectively. We also note that the relative difference in brain expression for genes harboring mutations in females versus males is larger for genes harboring truncating SNVs than for implicated network genes. This result is likely a consequence of the severe impact of truncating mutations on protein function, which makes the expression of corresponding genes an important factor in explaining the variability of ASD phenotypes across patients.
To investigate whether ASD-associated mutations preferentially affects specific brain regions, we analyzed spatial expression data available in the HBT database. For each brain region, we compared the average expression levels of network genes to the expression levels of genes with SNVs in unaffected sibling. Compared to controls, network genes show significantly higher expression across all brain regions (Supplementary Table S7), a result consistent with the wide spectrum of phenotypic abnormalities observed in ASD. Furthermore, we investigated the prenatal expression bias for network and truncating genes and compared these to the control bias for genes with SNVs in siblings (Table 3). This analysis demonstrated that the prenatal versus postnatal bias was generally similar and statistically significant for all brain regions (Supplementary Table S8). We note however that the amygdala, which is known to play a crucial role in processing emotional stimuli39, shows a numerically, though not significantly, larger prenatal bias compared to other regions. But overall, the lack of regional specificity underscores the pleiotropic nature of genes and functional processes involved in ASD.
Table 3.
Prenatal versus postnatal brain expression biases across brain regions.
| Network genes | Truncating SNVs | Sibling SNVs | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Brain region | Bias | SEM | P-value | Bias | SEM | P-value | Bias | SEM | P-value |
| Amygdala | 0.25 | 0.08 | 1×10−12 | 0.26 | 0.10 | 2×10−6 | 0.02 | 0.05 | 1.0 |
| Cerebellum | 0.23 | 0.08 | 3×10−9 | 0.17 | 0.10 | 4×10−3 | 0.07 | 0.05 | 0.1 |
| Frontal | 0.12 | 0.08 | 1×10−16 | 0.17 | 0.09 | 4×10−10 | 0.03 | 0.05 | 1.0 |
| Hippocampus | 0.15 | 0.07 | 1×10−5 | 0.18 | 0.09 | 4×10−4 | 0.02 | 0.05 | 1.0 |
| Occipital | 0.17 | 0.08 | 3×10−7 | 0.15 | 0.10 | 3×10−2 | 0.04 | 0.05 | 1.0 |
| Parietal | 0.16 | 0.08 | 3×10−11 | 0.17 | 0.10 | 3×10−4 | 0.05 | 0.05 | 0.7 |
| Striatum/ganglia | 0.24 | 0.08 | 2×10−12 | 0.22 | 0.10 | 3×10−5 | 0.04 | 0.05 | 1.0 |
| Temporal | 0.16 | 0.07 | 2×10−17 | 0.19 | 0.09 | 2×10−8 | 0.05 | 0.05 | 1.0 |
| Thalamus | 0.14 | 0.08 | 1×10−4 | 0.22 | 0.09 | 1×10−5 | 0.03 | 0.05 | 1.0 |
The biases were calculated using human expression data obtained from the HBT database. To quantify the expression bias for each brain region, we calculated the difference between the average log2 prenatal expression and the average log2 postnatal expression, such that positive values in the table indicate higher expression levels in the prenatal periods. The significances of the expression biases were evaluated using the Wilcoxon rank-sum one-tail test and corrected using the Bonferroni procedure. Bias SEM was calculated separately for each brain region.
The human brain contains a variety of neuronal and non-neuronal cell types, and it is interesting to investigate possible biases of implicated genes towards specific cell types. To explore this question we used an independent dataset of cell-specific gene expression generated using translating ribosome affinity purification (TRAP)17; the dataset contains gene expression profiles for 25 distinct cell types from the mouse central nervous system (CNS). To assess cell-specific expression biases, we compared, for each cell type, the expression of mouse orthologs for the implicated network genes with the expression of orthologs for genes harboring mutations in unaffected siblings. This analysis revealed that multiple neuronal and non-neuronal cell types are likely affected by de novo ASD mutation in the network genes (Fig. 3 and Supplementary Table S8). The diversity of affected cells can be explained, at least partially, by shared usage of common signaling, structural, and neural pathways across diverse cell types. While multiple cell types are affected, particularly strong and significant expression biases for implicated network genes are observed in cortical neurons, especially layer 5 pyramidal projection neurons, cerebellar granule cells, and medium spiny neurons of the striatum (Fig. 3, red). Notably, these cell types are also independently implicated by considering expression biases for 11 genes with recurrent truncating de novo mutations (Fig. 3, blue). On the other hand, some other cell types, such as motor neurons and astroglial cells, are markedly less affected. Notably, deep layer cortical glutamatergic projection neurons were also identified recently as a point of spatio-temporal convergence in co-expression networks built around high confidence ASD genes40.
Figure 3.
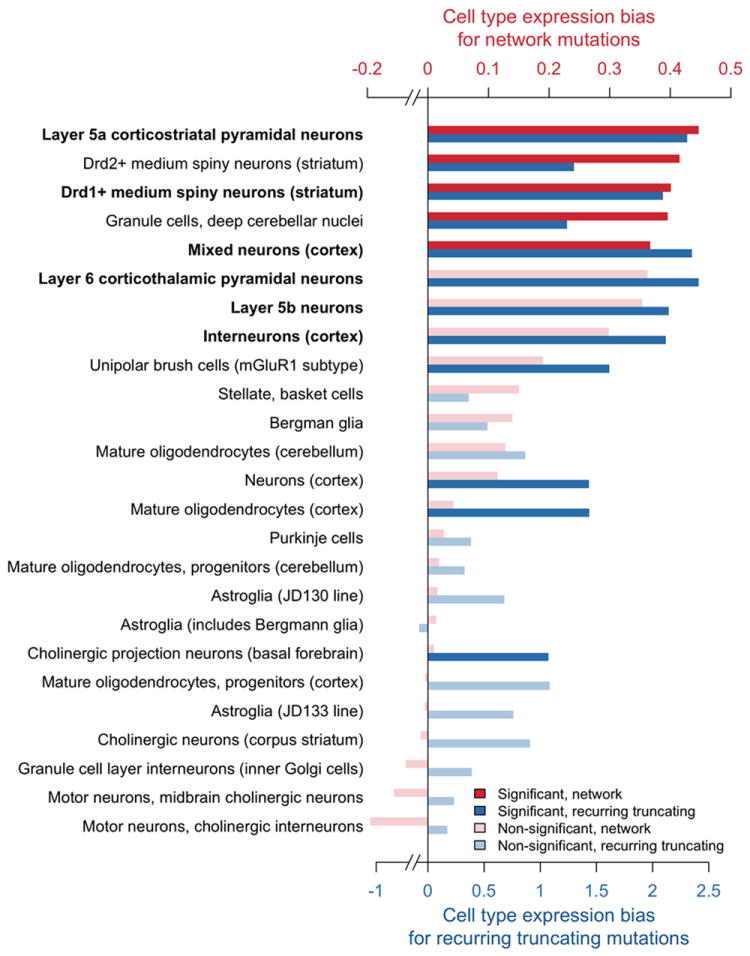
Cell type expression biases for network mutations and recurrent truncating mutations. Probands versus unaffected siblings expression biases were computed across 25 cell types of the central nervous system for implicated network genes (shown in red) and for 11 genes with recurrent truncating SNVs (blue). The biases were calculated using Mus. musculus expression data from the study by Doyle et al.17 To quantify the expression biases, we calculated for each cell type the difference between the average log2 expression of mouse orthologs for implicated human genes and the average log2 expression of mouse ortholog for human genes with de novo SNVs in unaffected siblings. Cell types in the figure are ordered by the magnitude of the cell type expression bias for network genes (red). The significance of the expression biases was evaluated using the Wilcoxon rank-sum one-tail test and corrected for multiple hypothesis testing using the Benjamini-Hochberg procedure with FDR=10%; significant cell types are shown in dark red/blue, and non-significant in light red/blue. P-values obtained from the two independent approaches — one based on network genes (red) and the other based on genes affected by recurrent truncating mutations (blue) — were combined using Fisher’s and Stouffer’s meta-analysis methods. The combined P-values were corrected using the Bonferroni method, and the cell types passing the significance cutoff for both meta-analyses are shown in bold. All P-values associated with the figure are presented in Supplementary Table S8.
Functional properties of implicated genes and disease phenotypes
Because ASD manifest significant pathophysiological differences across probands, it is important to understand how functional properties of implicated genes and associated mutations affect phenotypic characteristics of the disease. To investigate the effect of truncating de novo mutations on IQ, we calculated — separately for CNVs and truncating SNVs — the number of truncating mutations per individual across the IQ spectrum (Fig. 4a). Interestingly, for high-functioning ASD probands, the fractions of truncating mutations (CNVs and truncating SNVs) in all protein-altering events decreases and becomes similar to the average fractions in unaffected siblings. The combined fraction of truncating mutations (CNVs and truncating SNVs) for probands with IQ less than 100 is about two times higher compared to probands with IQ greater than or equal to 100 (0.13 for IQ<100, 0.067 for IQ≥100; Fisher’s exact two-tail test P = 0.001). In contrast, similar analyses for non-truncating mutations (synonymous and non-truncating SNVs) in probands show that the average number of non-truncating mutations per individual is relatively constant across IQs (Fig. 4b), without a prominent decrease for high-functioning ASD probands (0.43 for IQ < 100, 0.46 for IQ ≥ 100; Fisher’s exact two-tail test P = 0.7). Relative to synonymous mutations, non-truncating non-synonymous mutations are significantly enriched in probands with IQs above 70 compared to those below 70: 160 non-truncating and 72 synonymous mutations (2.2 ratio) are observed in probands with IQ ≤ 70, while 330 non-truncating and 110 synonymous mutations (3.3 ratio) in probands with IQ > 70 (Fisher’s exact one-tail test P = 0.04). Overall, these analyses suggest that truncating mutations are likely to play a less prominent role in high-functioning ASD cases. Notably, a recent study by Samocha et al.41 also demonstrated no excess of de novo loss-of-function mutations for high-functioning ASD probands.
Figure 4.
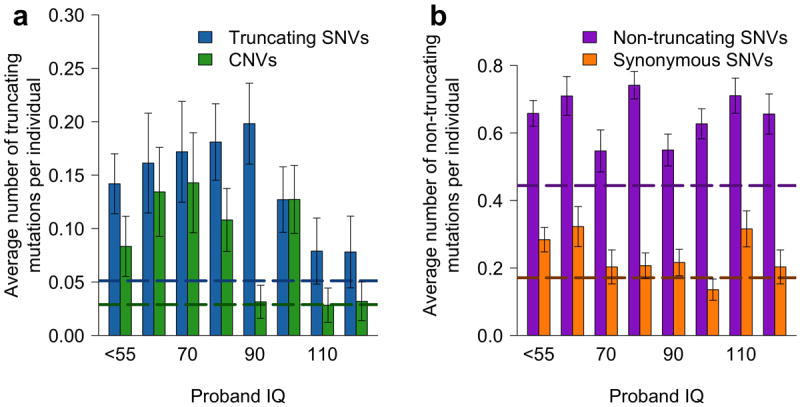
Average numbers of de novo mutations per individual for probands with different IQs. (a) The average number of truncating SNVs (blue) and CNVs (green) per individual. (b) The average number of non-truncating SNVs (purple) and synonymous SNVs (orange) per individual. Horizontal dashed lines represent the corresponding average numbers of mutations per individual for all unaffected siblings. Error bars represent SEM.
Notably, there is a significant difference in IQs between probands with network genes affected by CNV deletions and duplications. The average IQs for probands with CNV deletions and duplications are 64.8 and 83.9, respectively (Wilcoxon rank-sum one-tail test P = 6×10−3). This result suggests a significantly higher functional impact of an increase in gene dosage compared to a decrease.
Genes implicated in ASD exhibit diverse patterns of brain expression (Fig. 2c). Therefore, we asked whether the overall level of gene expression in the brain is associated, on average, with different phenotypic outcomes. For this analysis we considered the full-scale IQ and Autism Diagnostic Interview-Revised (ADIR) social interaction and repetitive behavior scores. The ADIR scores are based on structured interviews with proband parents and reflect patterns in reciprocal social interactions (ADIR-S) and repetitive/restrictive behaviors (ADIR-R)42. To explore the relationship between the average expression level of affected genes and corresponding phenotypes, we divided the ASD cases into low- and high-scoring phenotypic subsets relative to the corresponding median phenotype scores. We then calculated the average expression levels across these subsets for genes forming the implicated network and for all genes with truncating de novo SNVs (Fig. 5). This analysis reveals that affected genes associated with lower IQ scores or higher ADIR scores (both indicating more severe phenotypes) usually have significantly higher brain expression: network genes (solid lines in Fig. 5) one-tail test PWT < 10−15/PPT = 0.01 for IQ, PWT < 10−15/PPT = 0.3 for ADIR-S, and PWT < 10−15/PPT = 0.024 for ADIR-R; genes with truncating mutations (dashed lines) one-tail test PWT < 10−15/PPT < 10−4 for IQ, PWT < 10−15/PPT = 1.5×10−3 for ADIR-S, and PWT < 10−15/PPT = 0.08 for ADIR-R.
Figure 5.
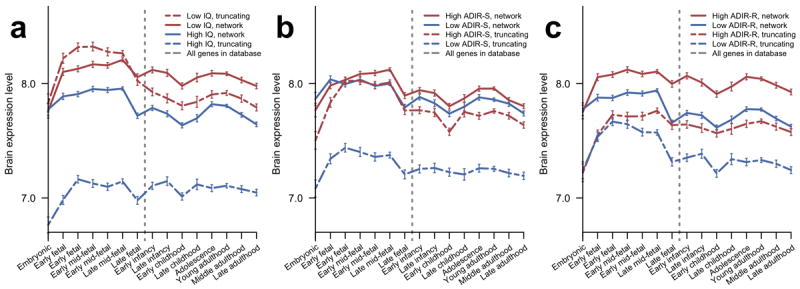
Temporal gene expression profiles in the human brain across developmental stages for genes affected in subsets of probands with different phenotypic scores. Expression data were obtained from the HBT database; average expression levels at each developmental stage were calculated using all genes in a given set and error bars represent SEM. In each figure, probands were divided into two groups (high and low) relative to the corresponding median phenotypic scores. Expression profiles for network genes are displayed as solid lines and profiles for truncating SNVs as dashed lines. Profiles for genes affected in probands with more severe phenotypes are shown in red and less severe phenotypes in blue. Vertical dashed lines indicate birth. (a) Profiles for probands with high/low IQ. (b) Profiles for probands with low/high ADIR-S scores. (c) Profiles for probands with low/high ADIR-R scores.
DISCUSSION
Our study suggests that the pathophysiological heterogeneity of ASD is matched by the diversity of genetic and functional insults associated with the disorder. We find that affected genes have diverse developmental expression profiles and therefore likely play important roles in multiple stages of neurogenesis, neuron mobility, synaptogenesis, and brain development. Although implicated genes and processes are active across multiple cells, some cell types, such as cortical neurons and medium spiny neuron of the striatum, seem to be more strongly impacted. Importantly, layer 5 cortical pyramidal neurons often project to the striatum, and the corresponding corticostriatal circuits are known to mediate diverse emotional, motor, habit forming, and motivational behaviors that are often perturbed in ASD43-46. Consequently, our unbiased genetic analysis implicates specific functional neural circuits that may mediate stereotypical and repetitive behaviors in ASD.
Although previous studies primarily emphasized the role of truncating de novo mutations in ASD3, 7, 8, our analysis suggests that a substantial fraction of non-truncating de novo missense mutations observed in probands also contributes to the disorder. Notably, we find that functional mutations are preferentially observed in haploinsufficient genes, which confirms that dosage effects play an important role in the disease mechanisms. We find that functional characteristics of affected genes — such as brain expression levels -- are likely to influence the observed phenotypic consequences of de novo ASD mutations. Stronger functional insults lead, on average, to more severe ASD phenotypes. Therefore, the distinction between intellectual disability and autism may lie primarily in the degree of overall functional impact rather than specific genes and pathways affected in the two disorders.
The presented analysis of brain expression provides further evidence for the hypothesis that stronger functional impacts, such as perturbation of genes with higher brain expression, are associated with female autistic phenotypes. Stronger functional perturbations in females compared to males were previously demonstrated based on the analysis of autism CNVs sizes6, 47 and gene network properties13. Thus, multiple independent sources of evidence suggest a protective effect in females, although the mechanisms of this effect remain to be elucidated.
We also find, in agreement with recently published studies41, 48, that truncating mutations play a relatively smaller role in high-functioning ASD cases. Because truncating mutations are usually associated with a loss-of-function, this result suggests that high-functioning autism phenotypes are less likely to be mediated by a loss of normal gene function in the brain. Functional gain and other types of genetic variations, such as non-truncating de novo mutations, common polymorphisms, or mutations in non-coding regulatory regions may predominantly contribute to high-functioning ASD cases.
Taken together, our analyses suggest that various functional properties of mutations and target genes may be useful in predicting ASD phenotypic severity. In future studies it will be important to investigate the extent to which individual ASD patients can be stratified based on affected pathways, biological functions, and cell-types. Such patient stratification — now a common practice in cancer — may lead to individualized diagnostic and prognostic predictions, and, ultimately, to targeted ASD therapies.
METHODS
ASD associated de novo variants
Variants were obtained from several recent studies of families in the Simons Simplex Collection and included de novo copy number variants (CNVs)6 and de novo single nucleotide variants (SNVs)3, 7, 8. Overlapping CNVs were combined into a single event and CNVs larger than 5 Mb were ignored. When a gene affected by a SNV was also contained within a CNV event, the SNV gene was considered individually and the remaining genes in the CNV were considered as a separate event. Combining overlaps and removing duplicate genes resulted in 991 genes at 624 distinct genomic loci that were used as input in our analysis. Eleven probands considered in our study had multiple SNV mutations; in various tests comparing mutations and corresponding functional properties, these probands were counted multiple times, once for each SNV.
Phenotypic network and the NETBAG+ algorithm
The NETBAG+ approach relies on the previously described phenotypic network in which all pairs of human genes are assigned a score proportional to the likelihood ratio that genes contribute to the same genetic phenotype12, 13. This ratio was calculated using a naïve Bayesian integration of various descriptors of protein function: shared Gene Ontology (GO) annotations, shared pathways in Kyoto Encyclopedia of Genes and Genomes (KEGG), protein domains from the InterPro database, tissue expression from the TiGER database, direct protein-protein interactions, shared interaction partners in a number of databases (BIND, BioGRID, DIP, HPRD, InNetDB, IntAct, BiGG, MINT and MIPS), phylogenetic profiles and chromosomal co-clustering across genomes49. The likelihood network was constructed using the carefully curated set of human genes compiled by Feldman et al., which contains 476 human genes associated with 132 different genetic phenotypes50.
The 991 genes affected by de novo mutations were mapped to the corresponding nodes of the phenotypic network. A greedy search algorithm was then used to find strongly interconnected networks among the input genes; starting from each input gene, the greedy algorithm consecutively adds the genes most strongly connected to the growing network. Networks are scored based on a weighted sum of all pairwise likelihood scores between the network genes12, 13. Each CNV event was constrained to contribute at most one gene to the network. Small networks with five or fewer genes were ignored during the network searches.
Network significance was determined by applying the same greedy search algorithm to 5000 random gene sets and comparing the score of the network obtained using real input data to the distribution of network scores obtained using random gene sets. The random gene sets used in the calculations were chosen to match the input genes in terms of network connectivity (based on the average of the top five edge scores) and protein lengths to ensure that network significance was not driven by highly connected or long genes; real data, i.e. genes affected by de novo ASD mutations, were allowed to participate in the random set. The final network P-value reflects the probability of finding a higher network score using random gene sets, while also correcting for multiple hypothesis testing due to various network sizes12, 13. The final network of 159 genes was selected as the largest network before a considerable decrease in the network significance.
FMRP and PSD enrichment among implicated gene sets
Following the approach by Iossifov et al.3 and other similar studies, we estimated the enrichments using the overlap between FMRP targets26, 30 or PSD genes31 and a set of rare synonymous mutations from a large exome sequencing study; we used for this purpose the NHLBI ESP exome sequencing study by Fu et al.29 Using the exome sequencing data of about 6500 individuals, we calculated the fractions of all rare synonymous mutations, i.e. synonymous mutations observed in the study only once, that affect FMRP target genes and genes associated with PSD. Consensus human PSD proteins from the Bayes et al. study were used. NHLBI ESP variant data was downloaded from evs.gs.washington.edu and unique entries were determined using the variant chromosomal position and allele. The calculated fractions represent the background (expected) probabilities for a random SNV mutation to occur in FMRP and PSD genes. We then compared the expected and the observed fractions of SNVs events in various ASD disease gene sets; the expected and observed fractions were compared using a two-tail binomial test (Table 2 and Supplementary Table S5). We also confirmed the FMRP enrichment results by comparing the overlap of FMRP target genes with various ASD gene sets to the overlap of FMRP target genes with random sets of human genes matched by protein length and connectivity to the genes affected by ASD mutations (Supplementary Table S6).
Hierarchical clustering of the implicated network
We used average linkage hierarchical clustering to divide the implicated network into functional clusters (Supplementary Fig. S1). The inverses of the phenotypic network likelihood scores were used as the clustering metric. In this way, gene pairs strongly connected in the phenotypic network were considered to be closer in distance.
Spatial and temporal analysis of human brain expression
Expression data across developmental stages and brain regions was obtained from the Human Brain Transcriptome database (GEO accession ID GSE25219)16; the HBT data represents quantile normalized and log2 transformed expressions values from the postmortem samples of healthy individuals.
To quantify the expression biases for each brain region, we calculated the difference between the average log2 expression of implicated genes and the average log2 expression of genes harboring SNVs in siblings. The prenatal expression bias for each brain region was quantified by calculating the difference between the average log2 of prenatal expression (embryonic to late fetal stages) and the average log2 postnatal expression (early infancy to late adulthood stages). The significance of regional and prenatal biases was evaluated using the Wilcoxon rank-sum test with the Bonferroni correction to account for multiple hypothesis testing.
Complementary to the Wilcoxon rank-sum test (PWT), we also estimated the significance of various expression differences using a method based on randomly generated expression probe sets (Supplementary Table S9). The permutation test P-values (PPT) obtained with the random trials reflect the probability to obtain a bias greater or equal to the bias in the original data. The numbers of probe sets sampled in random trials were equal to the numbers of probes sets in corresponding original datasets. Analogous to the procedure used to process the original HBT data, we computed the median probe set expression values in each of 10,000 randomly generated probe sets; expression biases were then calculated by taking the difference between the corresponding expression averages.
Cell type specific expression analysis
To evaluate expression bias of the implicated genes toward specific cell types, we considered mouse expression data obtained in the study by Doyle et al. using ribosome affinity purification (GEO accession ID GSE13379)17. The considered dataset contains expression information for 25 different cell types from the Mus. musculus central nervous system (CNS). To connect the mouse expression data and the human genetic data, we used the list of human-mouse orthologs provided by Affymetrix for the Mouse Genome 430 Array. The expression biases in probands compared to siblings were quantified for each cell type by calculating the difference between the average log2 expression of mouse orthologs for implicated human genes and the average log2 expression of mouse ortholog for human genes with de novo SNVs in unaffected siblings. We evaluated significance of the expression biases using the Wilcoxon rank-sum one-tail test and corrected for multiple hypothesis testing using the Benjamini-Hochberg procedure.
Supplementary Material
Acknowledgments
We would like to sincerely thank Michael Wigler, Matthew State, D. Geschwind, A. Packer, Gerald Fischbach, and all members of the Vitkup lab for helpful discussions. This work was supported by a grant from the Simons Foundation SFARI# 308962 to D.V., and US National Centers for Biomedical Computing (MAGNet) grant U54CA121852 to Columbia University. S.R.G. was supported in part by US NIGMS training grant T32 GM082797.
Footnotes
AUTHOR CONTRIBUTIONS
J.C., S.R.G., and A.H.C. performed computational analysis, and interpreted the results. S.J.S. contributed data, interpreted the results, and contributed to the functional analysis. D.V. designed the study, supervised the project, and interpreted the results. J.C., S.R.G., A.H.C., and D.V. wrote the manuscript.
References
- 1.Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nature reviews. Genetics. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg JM, Geschwind DH. Autism genetics: searching for specificity and convergence. Genome biology. 2012;13:247. doi: 10.1186/gb-2012-13-7-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iossifov I, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freitag CM. The genetics of autistic disorders and its clinical relevance: a review of the literature. Molecular psychiatry. 2007;12:2–22. doi: 10.1038/sj.mp.4001896. [DOI] [PubMed] [Google Scholar]
- 5.Geschwind DH. Genetics of autism spectrum disorders. Trends in cognitive sciences. 2011;15:409–416. doi: 10.1016/j.tics.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levy D, et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70:886–897. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 7.O’Roak BJ, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanders SJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veltman JA, Brunner HG. De novo mutations in human genetic disease. Nature reviews Genetics. 2012;13:565–575. doi: 10.1038/nrg3241. [DOI] [PubMed] [Google Scholar]
- 10.McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141:210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- 11.O’Roak BJ, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilman SR, et al. Diverse types of genetic variation converge on functional gene networks involved in schizophrenia. Nature neuroscience. 2012;15:1723–1728. doi: 10.1038/nn.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilman SR, et al. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron. 2011;70:898–907. doi: 10.1016/j.neuron.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelleher RJ, 3rd, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135:401–406. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 15.Zoghbi HY, Bear MF. Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harbor perspectives in biology. 2012;4 doi: 10.1101/cshperspect.a009886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang HJ, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doyle JP, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischbach GD, Lord C. The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron. 2010;68:192–195. doi: 10.1016/j.neuron.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 19.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 20.Krey JF, Dolmetsch RE. Molecular mechanisms of autism: a possible role for Ca2+ signaling. Current opinion in neurobiology. 2007;17:112–119. doi: 10.1016/j.conb.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 21.Betancur C, Sakurai T, Buxbaum JD. The emerging role of synaptic cell-adhesion pathways in the pathogenesis of autism spectrum disorders. Trends in neurosciences. 2009;32:402–412. doi: 10.1016/j.tins.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinto D, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nature reviews Genetics. 2013;14:347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santini E, et al. Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature. 2013;493:411–415. doi: 10.1038/nature11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang N, Lee I, Marcotte EM, Hurles ME. Characterising and predicting haploinsufficiency in the human genome. PLoS genetics. 2010;6:e1001154. doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Darnell JC, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huber KM, Gallagher SM, Warren ST, Bear MF. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:7746–7750. doi: 10.1073/pnas.122205699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edbauer D, et al. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu W, et al. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013;493:216–220. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ascano M, Jr, et al. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492:382–386. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bayes A, et al. Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nature neuroscience. 2011;14:19–21. doi: 10.1038/nn.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kennedy MB. The postsynaptic density at glutamatergic synapses. Trends in neurosciences. 1997;20:264–268. doi: 10.1016/s0166-2236(96)01033-8. [DOI] [PubMed] [Google Scholar]
- 33.Kennedy MB. Signal-processing machines at the postsynaptic density. Science. 2000;290:750–754. doi: 10.1126/science.290.5492.750. [DOI] [PubMed] [Google Scholar]
- 34.Ziff EB. Enlightening the postsynaptic density. Neuron. 1997;19:1163–1174. doi: 10.1016/s0896-6273(00)80409-2. [DOI] [PubMed] [Google Scholar]
- 35.Vogel-Ciernia A, et al. The neuron-specific chromatin regulatory subunit BAF53b is necessary for synaptic plasticity and memory. Nature neuroscience. 2013;16:552–561. doi: 10.1038/nn.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newschaffer CJ, et al. The epidemiology of autism spectrum disorders. Annu Rev Publ Health. 2007;28:235–258. doi: 10.1146/annurev.publhealth.28.021406.144007. [DOI] [PubMed] [Google Scholar]
- 37.Fombonne E. Epidemiology of pervasive developmental disorders. Pediatric research. 2009;65:591–598. doi: 10.1203/PDR.0b013e31819e7203. [DOI] [PubMed] [Google Scholar]
- 38.Zhao X, et al. A unified genetic theory for sporadic and inherited autism. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12831–12836. doi: 10.1073/pnas.0705803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adolphs R. What does the amygdala contribute to social cognition? Annals of the New York Academy of Sciences. 2010;1191:42–61. doi: 10.1111/j.1749-6632.2010.05445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Willsey AJ, et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013;155:997–1007. doi: 10.1016/j.cell.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samocha KE, et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lord C, Rutter M, Le Couteur A. Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. Journal of autism and developmental disorders. 1994;24:659–685. doi: 10.1007/BF02172145. [DOI] [PubMed] [Google Scholar]
- 43.Shepherd GM. Corticostriatal connectivity and its role in disease. Nature reviews Neuroscience. 2013;14:278–291. doi: 10.1038/nrn3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langen M, Durston S, Kas MJ, van Engeland H, Staal WG. The neurobiology of repetitive behavior: …and men. Neuroscience and biobehavioral reviews. 2011;35:356–365. doi: 10.1016/j.neubiorev.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 45.Burguiere E, Monteiro P, Feng G, Graybiel AM. Optogenetic stimulation of lateral orbitofronto-striatal pathway suppresses compulsive behaviors. Science. 2013;340:1243–1246. doi: 10.1126/science.1232380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rothwell PE, et al. Autism-associated neuroligin-3 mutations commonly impair striatal circuits to boost repetitive behaviors. Cell. 2014;158:198–212. doi: 10.1016/j.cell.2014.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanders SJ, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iossifov I, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen L, Vitkup D. Predicting genes for orphan metabolic activities using phylogenetic profiles. Genome biology. 2006;7:R17. doi: 10.1186/gb-2006-7-2-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feldman I, Rzhetsky A, Vitkup D. Network properties of genes harboring inherited disease mutations. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:4323–4328. doi: 10.1073/pnas.0701722105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


