Abstract
Purpose
Considerable molecular heterogeneity exists among human epidermal growth factor receptor 2 (HER2) –positive breast cancer regarding gene expression and mutation profiling. Evidence from preclinical, clinical neoadjuvant, and metastatic clinical trials suggested that PIK3CA mutational status and PAM50 intrinsic subtype of a tumor were markers of response to anti-HER2 therapies. We evaluated the predictive value of these two biomarkers in the adjuvant setting using archived tumor blocks from National Surgical Adjuvant Breast and Bowel Project (NSABP) trial B-31.
Patients and Methods
Expression data for 49 genes using the nCounter platform were used to generate PAM50 intrinsic subtypes for 1,578 archived tumor blocks from patients in the B-31 trial. Six PIK3CA hotspot mutations were examined by mass spectrometry of the primer extension products in a randomly selected subset (n = 671). We examined the heterogeneity of trastuzumab treatment effect across different subsets defined by each marker using Cox regression and disease-free survival as the end point.
Results
Seven hundred forty-one (47.0%) of 1,578 tumors were classified as HER2-enriched (HER2E) subtype, and 166 (24.7%) of 671 tumors had PIK3CA mutations. Hazard ratios (HRs) for trastuzumab in HER2E and other subtypes were 0.44 (95% CI, 0.34 to 0.58; P < .001) and 0.47 (95% CI, 0.35 to 0.62; P < .001), respectively (interaction P = .67). HRs for trastuzumab in PIK3CA wild-type and mutated tumors were 0.51 (95% CI, 0.37 to 0.71; P < .001) and 0.44 (95% CI, 0.24 to 0.82; P = .009), respectively (interaction P = .64).
Conclusion
Unlike results seen in the metastatic and neoadjuvant clinical trials, PIK3CA and PAM50 intrinsic subtypes were not predictive biomarkers for adjuvant trastuzumab in NSABP B-31. These data suggest that results from the metastatic and neoadjuvant setting may not be always applicable to the adjuvant setting.
INTRODUCTION
Randomized clinical trials have demonstrated the clinical efficacy of trastuzumab added to chemotherapy in treatment of human epidermal growth factor receptor 2 (HER2) –positive stage II and III breast cancer with approximately 50% reduction in recurrence rate.1
Considerable molecular heterogeneity exists among HER2-positive breast cancer in regard to gene expression and mutation spectrum, and tumor response to trastuzumab may also be heterogeneous accordingly. Using an unsupervised approach not based on trastuzumab response data, the Cancer Genome Atlas data demonstrated that only approximately 50% of clinically HER2-positive breast cancers were classified as the HER2-enriched (HER2E) intrinsic subtype by the PAM50 algorithm, with the rest classified largely as luminal subtypes.2,3 In the neoadjuvant therapy setting (Cancer and Leukemia Group B 40601 trial), 80% of HER2E tumors had pathologic complete response (pCR) on neoadjuvant chemotherapy plus HER2-targeted therapies, whereas the pCR rate was 25% to 32% in the other subtypes, which was similar to the pCR rate in the chemotherapy arm.4 Thus, the PAM50 intrinsic subtype may identify a much larger subset (approximately 50%) of patients with HER2-positive breast cancer who may not benefit from adjuvant trastuzumab.4 This is in contrast to our recently reported 8-gene (ESR1, CA12, GATA3, IGFR1, NAT1, ERBB2, GRB7, C17orf37) predictive signature developed from a supervised approach using clinical data from the National Surgical Adjuvant Breast and Bowel Project (NSABP) B-31 trial, which classified only 10% of HER2-positive patients as a no-benefit group.5
Another proposed marker of trastuzumab response is PIK3CA mutational status. In vitro data suggested that PIK3CA mutations present in approximately 25% of HER2-positive tumors were a marker of resistance to trastuzumab.6 Activating mutations in the PIK3CA gene and loss of PTEN expression have been proposed as the most important markers of PI3K pathway activation that lead to intrinsic resistance to trastuzumab.7 However, whereas several clinical studies in the metastatic or neoadjuvant setting demonstrated such an association,8,9 other studies failed to replicate the association.10 A recent meta-analysis of published studies failed to confirm the interaction between PIK3CA mutation or PTEN loss and resistance to trastuzumab.9 Furthermore, retrospective analyses of the Finland Herceptin (FINHER)10 and North Central Cancer Treatment Group (NCCTG) N9831 trials,11 two randomized clinical trials of adjuvant trastuzumab, found no association of PIK3CA mutation (FINHER) or PTEN loss (N9831) with trastuzumab resistance. To test their worth as predictive markers for adjuvant trastuzumab, we have assayed PAM50 intrinsic subtypes and PIK3CA mutations using archived tumor blocks from the NSABP B-31 trial, which tested the worth of adding 1 year of trastuzumab to adjuvant chemotherapy in the treatment of HER2-positive stage III breast cancer.1
PATIENTS AND METHODS
Study Design and Patients
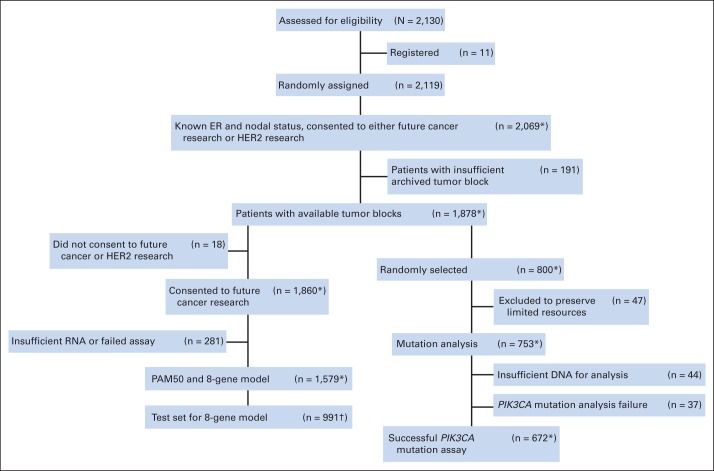
The general design of this study and description of available materials are presented in Figure 1. Among patients who participated in the B-31 trial (N = 2,130), 1,878 patients signed informed consent to permit use of banked tissue and had clinical follow-up data, available tumor blocks, and data on estrogen receptor (ER) status and number of positive nodes. The discovery cohort (n = 800) was selected by random sampling stratified by treatment and tested to ensure that it had a similar benefit from trastuzumab as did the entire cohort.
Fig 1.
CONSORT diagram for National Surgical Adjuvant Breast and Bowel Project B-31 trial. (*) One patient retracted consent in 2012 after the work of Pogue-Geile et al5 had been completed. (†) Used to test 8-gene model. ER, estrogen receptor; HER2, human epidermal growth factor receptor 2.
As previously reported, nCounter (nanoString Technologies, Seattle, WA) gene expression profiling for 462 genes was performed on 1,579 cases, and the data will be available through the Database of Genotypes and Phenotypes of the National Institutes of Health.5 (However, one patient recently has retracted consent after the work on Pogue-Geile et al5 had been completed). Among 1,878 cases, 800 were randomly selected and used for evaluation of PIK3CA. (One was excluded because of removal of consent.) Of these, 47 with thin blocks were excluded to preserve precious resources, leaving 753 for mutation analyses. The clinical and pathologic characteristics of the study cohort were similar to the original B-31 trial cohort (Data Supplement).
PAM50 Intrinsic Subtype Classification.
The nCounter platform-based gene expression profiling of archived tumor blocks from NSABP B-31 has been previously published.5 Among 462 genes examined using the nCounter platform, 49 from the PAM50 gene list were represented. One gene, CCNE1, was unintentionally excluded from the code set, but this exclusion had little impact on the results. The misclassification rate within the Parker et al3 data set of 200 cases was only 0.5% when the 49 genes were used (Data Supplement).
To identify intrinsic subtype, we first normalized B-31 nCounter gene expression data to remove technical platform differences and patient population differences between B-31 (who were mainly HER2 positive) and the original PAM50 development cohort published by Parker et al3 (which included all breast cancer). Details and R code used for normalization are included in the Data Supplement. Based on the published PAM50 algorithm, we then identified the following intrinsic subtypes: luminal A, luminal B, HER2E, basal like, and normal like.
PIK3CA Mutation
Two 5-μm sections were cut from formalin-fixed, paraffin-embedded blocks, tumor areas with 80% or more tumor cellularity were macrodissected by comparison to matching hematoxylin and eosin–stained slides, and DNA was isolated using the Mag-Bind FFPE DNA kit (Omega Bio-Tek, Norcross, GA). DNA was then subjected to mutation detection using TypePlex chemistry and the Mass-Array platform (Sequenom, San Diego, CA), as described previously.12 In brief, the method involves polymerase chain reaction amplification of the target region followed by a single base extension reaction and resolution of alleles by matrix-assisted laser desorption ionization–time of flight mass spectrometry. In this study, we evaluated the following amino acid changes at six hotspots in PIK3CA: N345K, C420R, E542K, E545K, H1047L, and H1047R. These hotspots cover 92% of PIK3CA mutations (1,918 of 2,075 mutations) reported in the Catalogue of Somatic Mutations in Cancer database13 and have been functionally validated as strongly activating mutations.14,15 Additional methodologic details are included in the Data Supplement.
Statistical Analysis
Follow-up information was included up to October 2010. Patients from the control arm who crossed over to receive trastuzumab were censored at the time of crossover. Among 1,578 patients who were profiled with nCounter assays, 265 of the 796 patients who were on the doxorubicin and cyclophosphamide followed by paclitaxel arm crossed over to receive trastuzumab. For 671 patients who were successfully profiled for PIK3CA mutational status, 112 of the 341 patients who were on the doxorubicin and cyclophosphamide followed by paclitaxel arm crossed over to receive trastuzumab. The definition of the primary end point for this analysis, disease-free survival (DFS), has been previously described.1 All experiments were performed blinded to clinical data before data anonymization by the NSABP Biostatistical Center.
For analysis of each marker's association with time to event, we first compared DFS rates among subgroups using the Kaplan-Meier method. Cox proportional hazard models were then used to assess the associations of each marker with the end point and the marker-treatment interaction simultaneously controlling for treatment, nodal status (one to three, four to nine, or ≥ 10 positive nodes), ER status (negative or positive), age (< or ≥ 60 years), and tumor size (≤ 2, 2 to 5, or > 5 cm). All reported P values are two-sided. All statistical analyses were performed in R (http://www.r-project.org).
We calculated the power of observing a treatment-group interaction for both PAM50 (HER2E v other subtypes) and PIK3CA (mutants v wild type [WT]). For PAM50, with the current sample size (n = 1,578), the proportion of benefit versus nonbenefit, with an α level of .05, the hazard ratio (HR) between HER2E subtype versus other subtypes must be less than 0.5 to have 80% power. Similarly, for analysis of PIK3CA mutational status, to have 80% power, the HR between mutation versus WT must be greater than 2.9 (details shown in the Data Supplement).
RESULTS
All PAM50 Intrinsic Subtypes Receive Benefit From Trastuzumab in B-31
Before examining potential treatment interaction between intrinsic subtypes and trastuzumab, it was important to ensure that our research PAM50 assay resulted in accurate intrinsic subtype classification. The association between PAM50 intrinsic subtypes and clinical ER and HER2 status is shown in Table 1. The distribution of clinical ER and HER2 status among subtypes was similar to that seen in other studies; 741 tumors (47.0%) were classified as HER2E subtype, 103 (6.5%) as basal-like, 435 (27.6%) as luminal A, 223 (14.1%) as luminal B, and 76 (4.8%) as normal-like subtypes.16,17 As expected, 516 (74.9%) of 689 of the ER-negative/HER2-positive tumors were classified as the HER2E subtype, whereas 97 (91.5%) of 106 of the ER-positive/HER2-negative tumors were classified as luminal subtypes. To further verify that our research PAM50 assay was able to correctly subtype B-31 tumors, we checked expression levels of ESR1 and ERBB2 mRNAs according to assigned subtypes. As expected, HER2 mRNA expression was highest in the HER2E subtype, whereas ESR1 mRNA expression was highest in luminal subtypes (Data Supplement). Also as expected, luminal B had a higher proliferation score than did luminal A (Data Supplement).
Table 1.
Distribution of PAM50 Intrinsic Subtypes With Clinical ER and HER2 Status in NSABP B-31
| ER and HER2 Status | No. of Patients |
||||
|---|---|---|---|---|---|
| Basal Like | HER2 Enriched | Luminal A | Luminal B | Normal Like | |
| ER negative/HER2 negative | 29 | 5 | 3 | 2 | 1 |
| ER negative/HER2 positive | 64 | 516 | 44 | 10 | 55 |
| ER negative/HER2 NA | 2 | 4 | 1 | 0 | 1 |
| ER positive/HER2 negative | 3 | 3 | 66 | 31 | 3 |
| ER positive/HER2 positive | 4 | 213 | 318 | 179 | 14 |
| ER positive/HER2 NA | 1 | 0 | 3 | 1 | 2 |
| Total (N = 1,578) | 103 | 741 | 435 | 223 | 76 |
| % | 6.5 | 47.0 | 27.6 | 14.1 | 4.8 |
Abbreviations: ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; NA, data not available; NSABP, National Surgical Adjuvant Breast and Bowel Project.
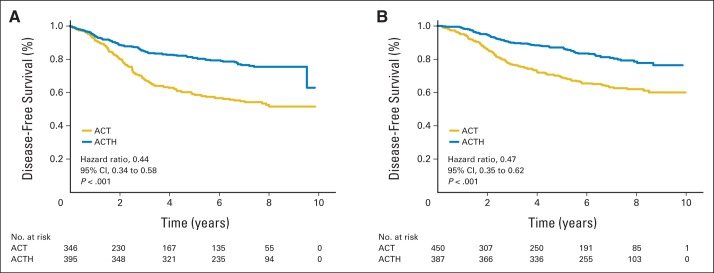
Having confirmed that research PAM50 subtyping performed as expected, we then checked whether there was heterogeneity of trastuzumab benefit among intrinsic subtypes. For DFS, which was the primary end point of the B-31 trial, there was no significant interaction between HER2E versus other subtypes and degree of benefit from trastuzumab (interaction P = .67). The HR for trastuzumab treatment in the HER2E subtype was 0.44 (95% CI, 0.34 to 0.58; P < .001), and the HR for other subtypes was 0.47 (95% CI, 0.35 to 0.62; P < .001). The Kaplan-Meier plots clearly show that both the HER2E subtype and the non-HER2E subtype receive benefit from trastuzumab (Fig 2).
Fig 2.
Kaplan-Meier plots for disease-free survival of (A) human epidermal growth factor receptor 2–enriched subtype and (B) other PAM50 intrinsic subtypes (including basal-like, luminal A, luminal B, and normal-like subtypes) by treatment received. The numbers of patients at risk are provided below the figure. ACT, doxorubicin and cyclophosphamide followed by paclitaxel; ACTH, doxorubicin and cyclophosphamide followed by paclitaxel plus trastuzumab.
Within each non-HER2E subtype, there was a trend for benefit from trastuzumab, although P values for basal-like and normal-like subtypes did not reach statistical significance because of small sample size. The HRs for luminal A, luminal B, basal-like, and normal-like subtypes were 0.42 (95% CI, 0.28 to 0.63; P < .001), 0.62 (95% CI, 0.37 to 1.04; P = .072), 0.43 (95% CI, 0.20 to 0.93; P = .033), and 0.45 (95% CI, 0.17 to 1.21; P = .114), respectively (Data Supplement). On the basis of these findings, we concluded that PAM50 intrinsic subtyping cannot identify a subset with no benefit from trastuzumab added to adjuvant chemotherapy. We also examined the PAM50 supervised predictors risk of recurrence and risk of recurrence subtype (RORS) and risk of recurrence proliferation (RORP) described by Parker et al3 (Data Supplement). All subsets defined by RORS and RORP received statistically significant benefit from trastuzumab. Thus, these predictors also failed to identify a subset that did not benefit from trastuzumab.
Relationship Between PAM50 Intrinsic Subtypes and the 8-Gene Trastuzumab Predictive Signature
We have previously published an 8-gene predictive signature for the degree of benefit from trastuzumab.5 Because these eight genes included ER- and HER2-related genes that play a major role in PAM50 intrinsic subtyping algorithm, we tested the relationship between the two signatures. As shown in the heat map of clustering results (Fig 3), the subset with no benefit defined by the 8-gene predictive signature consisted mostly of luminal subtypes defined by PAM50. However, all five PAM50 subtypes, including both HER2E and non-HER2E subtypes, are represented in the high- and moderate-benefit groups based on the 8-gene model.
Fig 3.
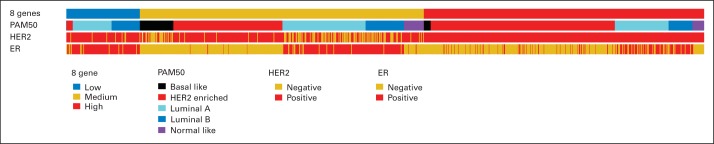
The distribution of subtypes based on the 8-gene predictive signature, PAM50, and estrogen receptor (ER) and clinical human epidermal growth factor receptor 2 (HER2) status is shown.5
PIK3CA Mutations Are Not Associated With Resistance to Adjuvant Trastuzumab
Of the randomly selected subset of 800 cases for PIK3CA mutation screening, 47 with thin blocks were excluded to preserve precious resources, leaving 753 for mutation analyses. From these, sufficient tumor DNA was obtained for 709 cases, and complete mutation results were obtained for 672 patients. Thirty-seven cases were excluded because of assay failures based on preset criteria of the presence of unextended primers in more than 25% of the area under the peaks.
The frequency and location of the PIK3CA mutations detected in B-31 tumors are similar to other studies of breast cancer PIK3CA mutations and in close agreement with other studies18,19 (Data Supplement). Mutations in exons 9 and 20 were not statistically significantly associated with any of these clinical variables (Table 2). PIK3CA mutations were also not statistically significantly associated with clinical variables and HER2 status but were associated with age, although the absolute difference was small. In addition, PIK3CA mutations were significantly over-represented in the luminal A subtype compared with WT tumors (Table 2).
Table 2.
Association of Clinical Variables With PIK3CA Mutations in NSABP B-31
| Variable |
PIK3CA Gene |
Exon 9 |
Exon 20 |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT |
MT |
P | WT |
MT |
P | WT |
MT |
P* | |||||||
| No. of Patients | % | No. of Patients | % | No. of Patients | % | No. of Patients | % | No. of Patients | % | No. of Patients | % | ||||
| Total | 505 | — | 166 | — | 624 | — | 49 | — | 573 | — | 104 | — | |||
| Age, years | .81 | .35 | .76 | ||||||||||||
| < 60 | 423 | 83.8 | 141 | 84.9 | 522 | 83.7 | 44 | 89.8 | 484 | 84.5 | 86 | 82.7 | |||
| ≥ 60 | 82 | 16.2 | 25 | 15.1 | 102 | 16.3 | 5 | 10.2 | 89 | 15.5 | 18 | 17.3 | |||
| Tumor size, cm | .03 | 0 | .39 | ||||||||||||
| ≤ 2 | 194 | 38.4 | 52 | 31.3 | 233 | 37.3 | 14 | 28.6 | 216 | 37.7 | 31 | 29.8 | |||
| 2.1 to 5 | 260 | 51.5 | 102 | 61.4 | 331 | 53 | 32 | 65.3 | 302 | 52.7 | 64 | 61.5 | |||
| > 5 | 51 | 10.1 | 11 | 6.6 | 60 | 9.6 | 2 | 4.1 | 54 | 9.4 | 9 | 8.7 | |||
| NA | 0 | 0 | 1 | 0.6 | 0 | 0 | 1 | 2 | 1 | 0.2 | 0 | 0 | |||
| No. of nodes | .37 | .7 | .53 | ||||||||||||
| 1 to 3 | 282 | 55.8 | 103 | 62 | 356 | 57.1 | 31 | 63.3 | 325 | 56.7 | 65 | 62.5 | |||
| 4 to 9 | 147 | 29.1 | 41 | 24.7 | 176 | 28.2 | 12 | 24.5 | 164 | 28.6 | 25 | 24.0 | |||
| ≥ 10 | 76 | 15 | 22 | 13.3 | 92 | 14.7 | 6 | 12.2 | 84 | 14.7 | 14 | 13.5 | |||
| ER | .45 | .26 | .8 | ||||||||||||
| Negative | 244 | 48.3 | 74 | 44.6 | 301 | 48.2 | 19 | 38.8 | 270 | 47.1 | 51 | 49.0 | |||
| Positive | 261 | 51.7 | 92 | 55.4 | 323 | 51.8 | 30 | 61.2 | 303 | 52.9 | 53 | 51.0 | |||
| HER2 | .45 | .65 | .83 | ||||||||||||
| Negative | 42 | 8.3 | 18 | 10.8 | 54 | 8.7 | 6 | 12.2 | 51 | 8.9 | 9 | 8.7 | |||
| Positive | 461 | 91.3 | 148 | 89.2 | 568 | 91 | 43 | 87.8 | 520 | 90.8 | 95 | 91.3 | |||
| NA | 2 | 0.4 | 0 | 0 | 2 | 0.3 | 0 | 0 | 2 | 0.3 | 0 | 0 | |||
| PAM50† | .05 | .17 | .52 | ||||||||||||
| Basal like | 37 | 9.1 | 3 | 2.3 | 40 | 8 | 0 | 0 | 37 | 8 | 3 | 3.8 | |||
| HER2 enriched | 193 | 47.3 | 61 | 47.7 | 238 | 47.8 | 16 | 42.1 | 216 | 46.9 | 40 | 50.6 | |||
| Luminal A | 101 | 24.8 | 41 | 32 | 130 | 26.1 | 12 | 31.6 | 119 | 25.8 | 24 | 30.4 | |||
| Luminal B | 57 | 14 | 20 | 15.6 | 68 | 13.7 | 9 | 23.7 | 68 | 14.8 | 10 | 12.7 | |||
| Normal like | 20 | 4.9 | 3 | 2.3 | 22 | 4.4 | 1 | 2.6 | 21 | 4.6 | 2 | 2.5 | |||
| DFS | .77 | .44 | .76 | ||||||||||||
| Censored | 360 | 71.3 | 121 | 72.9 | 445 | 71.3 | 38 | 77.6 | 414 | 72.3 | 73 | 70.2 | |||
| Event | 145 | 28.7 | 45 | 27.1 | 179 | 28.7 | 11 | 22.4 | 159 | 27.7 | 31 | 29.8 | |||
Abbreviations: DFS, disease-free survival; ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; MT, mutant; NA, data not available; NSABP, National Surgical Adjuvant Breast and Bowel Project; WT, wild type.
P value was calculated using χ2 test.
Not all cases with PIK3CA mutation results have been profiled for PAM50.
In univariate analyses, the DFS benefit from trastuzumab in PIK3CA mutant tumors (HR, 0.44; 95% CI, 0.24 to 0.82; P = .009) was similar to the benefit in PIK3CA WT tumors (HR, 0.51; 95% CI, 0.37 to 0.71; P < .001), as illustrated in Figure 4. These results did not change when adjusted for nodal status, ER status, age, and tumor size. In a Cox proportional hazards model controlling for nodal status, ER status, and tumor size, PIK3CA did not show a statistically significant interaction with trastuzumab benefit for DFS (interaction P = .77). We repeated the analyses with exclusion of central assay HER2-negative cases, and the results were nearly identical (data not shown).
Fig 4.
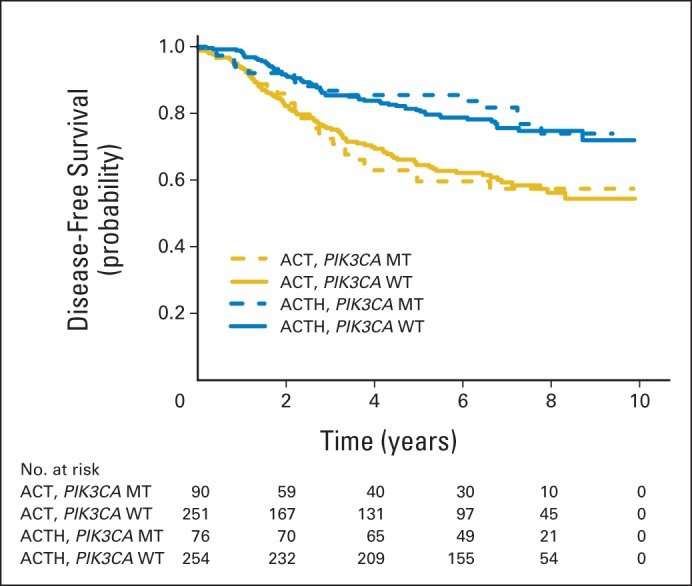
Disease-free survival for PIK3CA wild-type (WT) and mutant (MT) tumors for patients treated with doxorubicin and cyclophosphamide followed by paclitaxel (ACT) versus those treated with doxorubicin and cyclophosphamide followed by paclitaxel plus trastuzumab (ACTH). The numbers of patients at risk are provided below the figure.
Comparison of the 8-Gene Trastuzumab Predictive Model With Other Signatures
Forest plots of all markers that were tested (PAM50, RORS, RORP, PIK3CA mutation status, and the 8-gene predictive model, using only the validation cohort) with univariate and multivariate analyses are shown in Figure 5 and the Data Supplement, respectively. Perhaps it is not surprising that only the 8-gene model was able to identify a subset of patients with no benefit from trastuzumab in univariate and multivariate analyses, given the fact that the signature was developed to do just that within a discovery cohort of B-31.
Fig 5.
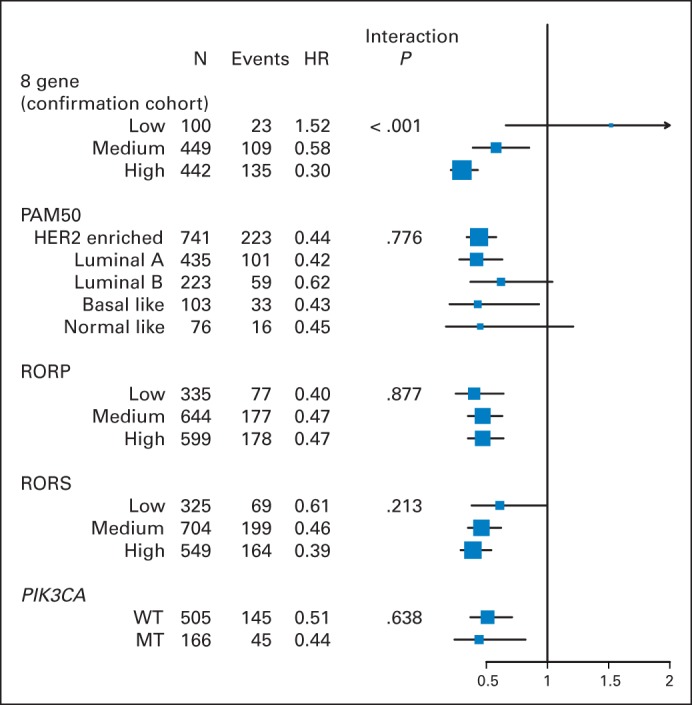
Forest plot of hazard ratios (HRs) for subtypes identified by the 8-gene predictive model,5 PAM50, risk of recurrence proliferation (RORP), risk of recurrence subtype (RORS), and PIK3CA mutation status. HR and interaction P value were determined by univariate analysis. With multiple hypothesis adjustment, only the 8-gene model, which was analyzed in the confirmation cohort only, has significant treatment-group interaction. HER2, human epidermal growth factor receptor 2; MT, mutant; WT, wild type.
Molecular Profiling Does Not Identify a Subset With Poor Prognosis After Adjuvant Trastuzumab
Recently, pertuzumab received accelerated approval based on the doubling of pCR compared with trastuzumab alone in the NeoSphere trial (NCT00545688),20 but final approval is contingent on demonstration of DFS benefit in the APHINITY (Study of Pertuzumab in Addition to Chemotherapy and Herceptin [trastuzumab] As Adjuvant Therapy in Patients With HER2-Positive Primary Breast Cancer; NCT01358877) trial. Even if the trial shows positive results, absolute reduction in event rate is expected to be small. Therefore, it would be prudent to develop a prognostic model to identify patients with high residual risk after adjuvant trastuzumab who may need a combination of trastuzumab and pertuzumab. We examined prognosis based on PAM50 intrinsic subtypes, RORS and RORP algorithms, and PIK3CA mutations in trastuzumab-treated patients. We could not identify a subtype with a particularly worse prognosis after trastuzumab, although luminal A and normal-like subtypes trended to have a more favorable residual risk (see Data Supplement for Kaplan-Meir plots). Therefore, other molecular assays or clinical parameters such as anatomic burden may be required to identify patients who require additional treatment such as pertuzumab.
DISCUSSION
Although gene expression profiling confirmed molecular heterogeneity of HER2-positive tumors in which the PAM50 intrinsic subtypes were readily identified, there was no difference in the degree of benefit from adjuvant trastuzumab based on intrinsic subtypes. This contrasts with the data from the Cancer and Leukemia Group B 40601 trial, in which 80% of HER2E tumors had pCR on neoadjuvant chemotherapy with trastuzumab compared with a pCR rate of 25% to 32% in the other subtypes, which was similar to the pCR rate in the chemotherapy arm.4
In the NeoSphere, Neoadjuvant Lapatinib and/or Trastuzumab Treatment Optimisation (Neo-ALTTO; NCT00553358), and Gepar Quinto (NCT00567554) and Sixto (NCT01426880) neoadjuvant trials, which compared the pCR rate of PIK3CA mutant versus WT tumors in patients treated with anti-HER2 therapies, the pCR rate was lower in those with PIK3CA-mutated tumors than in those with WT PIK3CA.21,22 In the metastatic setting, the CLEOPATRA (Clinical Evaluation of Pertuzumab and Trastuzumab) trial (NCT00567190) demonstrated that PIK3CA mutations were associated with a poorer prognosis for patients treated with anti-HER2 therapy, based on progression-free survival (PFS); however, both PIK3CA mutant and WT tumors showed a trend for benefit from pertuzumab.
In contrast, we have demonstrated that there was no interaction between PIK3CA hotspot activating mutations and the degree of benefit from trastuzumab in the adjuvant setting, confirming the report from the retrospective analysis of the FINHER trial by Loi et al.10 Our results, together with the report by Perez et al,11 which showed the lack of interaction between PTEN loss and the degree of benefit from trastuzumab in the NCCTG N9831 trial,11 do not support the prevailing hypothesis that PI3K pathway activation either by PIK3CA mutation or PTEN loss results in a lack of benefit from trastuzumab in the adjuvant setting.
These data support our hypothesis that there is a fundamental difference between macro diseases (advanced and neoadjuvant) and the minimal residual disease setting for trastuzumab response.5 End points for the evaluation of neoadjuvant and metastatic settings (pCR, PFS) require that large numbers of cancer cells be eliminated in the tumor. In contrast, in the adjuvant setting where the tumor has been surgically removed, improvement in DFS may largely depend on the elimination of a small number of cancer stem cells (CSCs). CSCs have been shown to depend on HER2 signaling in the absence of HER2 gene amplification.23 HER2-amplified disseminated tumor cells have been detected in clinically HER2-negative patients.24 CSCs isolated from luminal subtype breast cancer cells have been found to express high levels of HER2 protein and were responsive to trastuzumab therapy.23 Furthermore, bone metastases of breast cancer frequently overexpressed HER2 protein in the absence of ERBB2 gene amplification.23 Therefore, disseminated CSCs or bone micrometastatic cells may be responsive to trastuzumab treatment regardless of HER2 status of the primary tumor. Thus, patients with HER2-negative tumors could benefit from trastuzumab in the adjuvant setting but not in the neoadjuvant or metastatic setting when pCR or PFS is the clinical end point. Benefit from trastuzumab in patients with HER2-nonamplified early breast cancer will be assessed in the ongoing NSABP B-47 trial.
However, the fact that PIK3CA mutations were not shown to be resistance markers to trastuzumab in NSBAP B-31 should not be interpreted as a contraindication to the clinical development of PI3K pathway inhibitors in HER2-positive breast cancer. A study using a global knockout screen of a single HER2-amplified breast cancer cell line demonstrated PIK3CA mutation or PTEN loss as a potential mechanism of resistance to trastuzumab.6 However, many ERBB2-amplified cell lines were highly sensitive to PIK3CA small interfering RNA, regardless of the presence or absence of mutations in the PIK3CA gene.25 The additive effect of combining trastuzumab with a PIK3CA inhibitor was observed in both trastuzumab-sensitive and -resistant cell lines,26 suggesting that signaling through multiple pathways converge on PI3K in HER2-positive cancer cells and supporting the clinical development of PI3K pathway (PIK3CA, AKT, mTOR) inhibitors in HER2-positive breast cancer regardless of PIK3CA mutation status.
In conclusion, failure of PAM50 and PIK3CA mutations to define subsets of patients with differential benefit from trastuzumab in the adjuvant setting suggests that the predictive markers from metastatic or neoadjuvant studies cannot be applied directly to adjuvant studies, at least for HER2-positive breast cancer.
Supplementary Material
Acknowledgment
We thank Hanna Bandos, PhD, for help with patient tracking for the CONSORT diagram; Melanie Prior for data and tissue block management; William J. Hiller and Theresa A. Oeler for histology; Teresa L. Bradley, PhD, Ethan Barry, and Joyce Mull for regulatory affairs; Barbara Harkins and Frances Fonzi for protocol development; Barbara C. Good, PhD, and Wendy L. Rea for manuscript editing and submission; and Chris I. Ruddock for graphics. We also thank National Surgical Adjuvant Breast and Bowel Project members who contributed tissue blocks and the patients enrolled onto the study.
Glossary Terms
- estrogen receptor (ER):
ligand-activated nuclear proteins, belonging to the class of nuclear receptors, present in many breast cancer cells that are important in the progression of hormone-dependent cancers. After binding, the receptor-ligand complex activates gene transcription. There are two types of estrogen receptors (ERα and ERβ). ERα is one of the most important proteins controlling breast cancer function. ERβ is present in much lower levels in breast cancer, and its function is uncertain. Estrogen receptor status guides therapeutic decisions in breast cancer.
- neoadjuvant therapy:
the administration of chemotherapy prior to surgery. Induction chemotherapy is generally designed to decrease the size of the tumor prior to resection and to increase the rate of complete (R0) resections.
- PIK3CA:
the catalytic subunit of phosphatidylinositol 3-kinase involved in the generation of PIP3 which, in turn, leads to the activation of AKT and other oncogenic kinases. Mutations in the PIK3CA gene have been found in several cancers, including ovarian, breast, colon, and lung carcinomas. See PI3K and AKT/PKB.
Footnotes
Supported by Grants No. U10-CA-12027, U10-CA-69651, U10-CA-37377, and U10-CA-69974 from the National Cancer Institute, Department of Health and Human Services, Public Health Service; by a grant from the Pennsylvania State Department of Health; and by Genentech.
Terms in blue are defined in the glossary, found at the end of this article and online at www.jco.org.
The study sponsor played no role in the design of the study, collection of data, analysis or interpretation of the study, writing of the article, or decision to submit the article for publication. The Pennsylvania State Department of Health specifically disclaims responsibility for any analysis, interpretations, or conclusions.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
Clinical trial information: NCT00004067.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) and/or an author's immediate family member(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: Sandra M. Swain, Genentech/Roche (C); Charles E. Geyer Jr, Genentech (C) Stock Ownership: None Honoraria: Eleftherios P. Mamounas, Genentech/Roche; Sandra M. Swain, Genentech/Roche Research Funding: Sandra M. Swain, Genentech/Roche, Pfizer, sanofi-aventis, Bristol-Myers Squibb Expert Testimony: None Patents, Royalties, and Licenses: Norman Wolmark, Adjuvant Cancer Therapy - PCT/US2010/031740 Other Remuneration: Sandra M. Swain, Genentech/Roche
AUTHOR CONTRIBUTIONS
Conception and design: Katherine L. Pogue-Geile, Nan Song, Eleftherios P. Mamounas, Norman Wolmark, Soonmyung Paik
Financial support: Soonmyung Paik
Administrative support: Joseph P. Costantino, Norman Wolmark, Soon Paik
Provision of study materials or patients: Louis Fehrenbacher, Eleftherios P. Mamounas, Soonmyung Paik
Collection and assembly of data: Katherine L. Pogue-Geile, Patrick G. Gavin, Seong-Rim Kim, Nicole L. Blackmon, Melanie Finnigan, Priya Rastogi, Louis Fehrenbacher, Charles E. Geyer Jr, Joseph P. Costantino, Soonmyung Paik
Data analysis and interpretation: Katherine L. Pogue-Geile, Nan Song, Jong-Hyeon Jeong, Patrick G. Gavin, Priya Rastogi, Louis Fehrenbacher, Eleftherios P. Mamounas, Sandra M. Swain, D. Lawrence Wickerham, Charles E. Geyer Jr, Joseph P. Costantino, Norman Wolmark, Soonmyung Paik
Manuscript writing: All authors
Final approval of manuscript: All authors
REFERENCES
- 1.Romond EH, Perez EA, Bryant J, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353:1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carey LA, Berry DA, Ollila D, et al. Clinical and translational results of CALGB 40601: A neoadjuvant phase III trial of weekly paclitaxel and trastuzumab with or without lapatinib for HER2-positive breast cancer. J Clin Oncol. 2013;(suppl):31. abstr 500. [Google Scholar]
- 5.Pogue-Geile KL, Kim C, Jeong JH, et al. Predicting degree of benefit from adjuvant trastuzumab in NSABP trial B-31. J Natl Cancer Inst. 2013;105:1782–1788. doi: 10.1093/jnci/djt321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berns K, Horlings HM, Hennessy BT, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 7.Rexer BN, Arteaga CL. Optimal targeting of HER2-PI3K signaling in breast cancer: Mechanistic insights and clinical implications. Cancer Res. 2013;73:3817–3820. doi: 10.1158/0008-5472.CAN-13-0687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dave B, Migliaccio I, Gutierrez MC, et al. Loss of phosphatase and tensin homolog or phosphoinositol-3 kinase activation and response to trastuzumab or lapatinib in human epidermal growth factor receptor 2-overexpressing locally advanced breast cancers. J Clin Oncol. 2011;29:166–173. doi: 10.1200/JCO.2009.27.7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Liu Y, Du Y, et al. The predictive role of phosphatase and tensin homolog (PTEN) loss, phosphoinositol-3 (PI3) kinase (PIK3CA) mutation, and PI3K pathway activation in sensitivity to trastuzumab in HER2-positive breast cancer: A meta-analysis. Curr Med Res Opin. 2013;29:633–642. doi: 10.1185/03007995.2013.794775. [DOI] [PubMed] [Google Scholar]
- 10.Loi S, Michiels S, Lambrechts D, et al. Somatic mutation profiling and associations with prognosis and trastuzumab benefit in early breast cancer. J Natl Cancer Inst. 2013;105:960–967. doi: 10.1093/jnci/djt121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perez EA, Dueck AC, McCullough AE, et al. Impact of PTEN protein expression on benefit from adjuvant trastuzumab in early-stage human epidermal growth factor receptor 2-positive breast cancer in the North Central Cancer Treatment Group N9831 Trial. J Clin Oncol. 2013;31:2115–2122. doi: 10.1200/JCO.2012.42.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fumagalli D, Gavin PG, Taniyama Y, et al. A rapid, sensitive, reproducible and cost-effective method for mutation profiling of colon cancer and metastatic lymph nodes. BMC Cancer. 2010;10:101. doi: 10.1186/1471-2407-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forbes SA, Bindal N, Bamford S, et al. COSMIC: Mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gymnopoulos M, Elsliger MA, Vogt PK. Rare cancer-specific mutations in PIK3CA show gain of function. Proc Natl Acad Sci U S A. 2007;104:5569–5574. doi: 10.1073/pnas.0701005104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikenoue T, Kanai F, Hikiba Y, et al. Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer Res. 2005;65:4562–4567. doi: 10.1158/0008-5472.CAN-04-4114. [DOI] [PubMed] [Google Scholar]
- 16.Gomez Pardo P, Prat A, Bianchini G, et al. PAM50 intrinsic subtyping and pathologic responses to neoadjuvant trastuzumab-based chemotherapy in HER2-positive breast cancer. J Clin Oncol. 2011;(suppl):29. abstr 554. [Google Scholar]
- 17.Cheang MC, Voduc KD, Tu D, et al. Responsiveness of intrinsic subtypes to adjuvant anthracycline substitution in the NCIC.CTG MA.5 randomized trial. Clin Cancer Res. 2012;18:2402–2412. doi: 10.1158/1078-0432.CCR-11-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saal LH, Holm K, Maurer M, et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65:2554–2559. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- 19.Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008;68:6084–6091. doi: 10.1158/0008-5472.CAN-07-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gianni L, Pienkowski T, Im YH, et al. Efficacy and safety of neoadjuvant pertuzumab and trastuzumab in women with locally advanced, inflammatory, or early HER2-positive breast cancer (NeoSphere): A randomised multicentre, open-label, phase 2 trial. Lancet Oncol. 2012;13:25–32. doi: 10.1016/S1470-2045(11)70336-9. [DOI] [PubMed] [Google Scholar]
- 21.Loibl S. PIK3CA mutation predicted pCR in HER-2 positive breast cancer. San Antonio Breast Cancer Symposium; December 10-14, 2013; San Antonio, TX. (abstr S4-06) [Google Scholar]
- 22.Baselga J, Majewski I, Nuciforo PG, et al. PI3KCA mutations and correlation with pCR in the NeoALTTO trial (BIG 01-06). European Cancer Congress 2013; September 27-October 1, 2013; Amsterdam, the Netherlands. (abstr 1859) [Google Scholar]
- 23.Ithimakin S, Day KC, Malik F, et al. HER2 drives luminal breast cancer stem cells in the absence of HER2 amplification: Implications for efficacy of adjuvant trastuzumab. Cancer Res. 2013;73:1635–1646. doi: 10.1158/0008-5472.CAN-12-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartkopf AD, Banys M, Meier-Stiegen F, et al. The HER2 status of disseminated tumor cells in the bone marrow of early breast cancer patients is independent from primary tumor and predicts higher risk of relapse. Breast Cancer Res Treat. 2013;138:509–517. doi: 10.1007/s10549-013-2470-9. [DOI] [PubMed] [Google Scholar]
- 25.Brough R, Frankum JR, Sims D, et al. Functional viability profiles of breast cancer. Cancer Discov. 2011;1:260–273. doi: 10.1158/2159-8290.CD-11-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chakrabarty A, Bhola NE, Sutton C, et al. Trastuzumab-resistant cells rely on a HER2-PI3K-FoxO-survivin axis and are sensitive to PI3K inhibitors. Cancer Res. 2013;73:1190–1200. doi: 10.1158/0008-5472.CAN-12-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.