Abstract
Purpose
Current diagnostic tests for diffuse large B-cell lymphoma use the updated WHO criteria based on biologic, morphologic, and clinical heterogeneity. We propose a refined classification system based on subset-specific B-cell–associated gene signatures (BAGS) in the normal B-cell hierarchy, hypothesizing that it can provide new biologic insight and diagnostic and prognostic value.
Patients and Methods
We combined fluorescence-activated cell sorting, gene expression profiling, and statistical modeling to generate BAGS for naive, centrocyte, centroblast, memory, and plasmablast B cells from normal human tonsils. The impact of BAGS-assigned subtyping was analyzed using five clinical cohorts (treated with cyclophosphamide, doxorubicin, vincristine, and prednisone [CHOP], n = 270; treated with rituximab plus CHOP [R-CHOP], n = 869) gathered across geographic regions, time eras, and sampling methods. The analysis estimated subtype frequencies and drug-specific resistance and included a prognostic meta-analysis of patients treated with first-line R-CHOP therapy.
Results
Similar BAGS subtype frequencies were assigned across 1,139 samples from five different cohorts. Among R-CHOP–treated patients, BAGS assignment was significantly associated with overall survival and progression-free survival within the germinal center B-cell–like subclass; the centrocyte subtype had a superior prognosis compared with the centroblast subtype. In agreement with the observed therapeutic outcome, centrocyte subtypes were estimated as being less resistant than the centroblast subtype to doxorubicin and vincristine. The centroblast subtype had a complex genotype, whereas the centrocyte subtype had high TP53 mutation and insertion/deletion frequencies and expressed LMO2, CD58, and stromal-1–signature and major histocompatibility complex class II–signature genes, which are known to have a positive impact on prognosis.
Conclusion
Further development of a diagnostic platform using BAGS-assigned subtypes may allow pathogenetic studies to improve disease management.
INTRODUCTION
Diffuse large B-cell lymphoma (DLBCL) is considered to be derived from germinal center cells based on their histologic features, immunohistochemical characteristics, and immunoglobulin gene rearrangements. Global gene expression profiling allows alternative determination of cell of origin that reflects normal B-cell function and differentiation. Thus, cell-of-origin assignments enable the categorization of DLBCL into activated B-cell–like (ABC) and germinal center B-cell–like (GCB) subclasses, each differing in pathogenesis, signaling pathway regulation, genetic abnormalities, and survival outcomes.1 However, this classification is based on only a fraction of naturally occurring B-cell subsets, namely germinal center cells containing centroblasts and centrocytes or in vitro–activated B cells from peripheral blood.2 In addition, the ABC and GCB subclasses do not allow direct identification and isolation of malignant B cells for the study of heterogeneity and pathogenesis. However, sorting malignant DLBCL cells based on B-cell differentiation markers indirectly supports a hierarchical classification3 that recapitulates the spectrum of physiologic processes, including lineage differentiation and the transcriptional programs of normal lymphopoiesis.
We recently described a procedure for investigating the gene expression of immunophenotype-based flow-sorted naive, centroblast, centrocyte, memory, and plasmablast B cells from human tonsils.4,5 With statistical modeling, the gene expression profiles can define subset-specific B-cell–associated gene signatures (BAGS) that, when applied to clinical tumor samples, allow examination of the association between DLBCL subtypes and prognosis. This strategy extends the current cell-of-origin classification and provides a new tool for generating insight into the stages of clonal differentiation and oncogenesis6,7 and may offer several diagnostic and prognostic advantages, representing important steps toward individualized therapy.
PATIENTS AND METHODS
Normal and Malignant Tissue
Tonsils from eight healthy donors (Health Research Ethics Committee for North Denmark Region Approval N-20080062MCH; Data Supplement) were sorted by fluorescence-activated cell sorting into five distinct B-cell subsets (Data Supplement). RNA in the B-cell subset was labeled and hybridized to Affymetrix GeneChip Human Genome U133 Plus 2.0 Arrays (Affymetrix, Santa Clara, CA; Data Supplement) and is referred to as the tonsil data (Data Supplement).
Pretreatment lymphoma tissues (n = 89) were collected during the diagnostic procedure in accordance with the research protocol accepted by the Health Research Ethics Committee for North Denmark Region (Approval N-20100059). The clinical, staging, therapy, and outcome data were registered in the National Clinical Quality Database for Malignant Lymphoma8,9 (Data Supplement). Total RNA from tumors was labeled and hybridized to Affymetrix GeneChip Human Genome U133 Plus 2.0 Arrays (Affymetrix; Data Supplement) and is referred to as CHEPRETRO (Chemotherapy Prediction in Retrospective Samples).
The CEL files from both the tonsil data and CHEPRETRO were deposited in the National Center for Biotechnology Information Gene Expression Omnibus repository (GSE56315) and comply with Minimum Information About a Microarray Experiment requirements.10 In addition to CHEPRETRO, we used the following four online data sets: Lymphoma/Leukemia Molecular Profiling Project (LLMPP) cyclophosphamide, doxorubicin, vincristine, and prednisone (CHOP) and LLMPP rituximab plus CHOP (R-CHOP)11; International DLBCL Rituximab-CHOP Consortium MD Anderson Project (IDRC)12; and Mayo Clinic, Brigham and Women's Hospital, and Dana-Farber Cancer Institute Project (MDFCI)13 (Data Supplement).
Statistical Analysis
All statistical analyses were performed with R version 3.1.114 using Bioconductor packages.15 The statistical analysis is summarized here; for full documentation, see the Data Supplement.16 Before the statistical analysis, the arrays were background corrected, normalized, and summarized cohort-wise by robust multichip average.17
The BAGS classification was based on median-centered probe sets from the tonsil data using regularized multinomial regression with five discrete outcomes representing B-cell subtypes and the elastic net penalty.18 The regularization parameters were chosen by cross validation. To compensate for cohort-wise technical batch effects, each clinical cohort was median centered and adjusted probe set–wise to have the same variance as in the tonsil data. Each patient underwent BAGS classification according to the highest predicted probability score of more than 0.45 or was otherwise unclassified. Both the robustness of the probe set–wise scaling and the probability cutoff were tested (Data Supplement).
Survival analyses were performed using the Kaplan-Meier method, log-rank tests, and simple and multiple covariate Cox proportional hazards regression. The cohorts were combined into a meta-data set to increase the power of the study. BAGS was investigated as an independent explanatory variable in the meta-data set using a Cox proportional hazards model with BAGS, ABC/GCB classes, the International Prognostic Index (IPI; dichotomized into low [IPI, 0 to 1] and high [IPI, 2 to 5]), and cohort as potential confounders. Cohort was not significant and was omitted from the analysis.
Each sample was assigned a drug resistance probability for cyclophosphamide (mafosfamide), doxorubicin, and vincristine using resistance gene signature (REGS) classifiers19–21 (Falgreen et al, submitted for publication). Briefly, 26 B-cell cancer cell lines were categorized as resistant, intermediate, and sensitive by systematic in vitro dose-response drug screens.22 REGS classification was based on cell line gene expression profiles using logistic regression, with outcomes representing resistant and sensitive cell lines regularized by the elastic net penalty.18 The regularization parameters were chosen by leave-one-out cross validation. The chance of complete remission (CR) between subgroups was compared using relative risk.
The significance level was set to P = .05, and effect estimates were provided with 95% CIs. P values were adjusted by Holm's method.23
Biologic Phenotyping
Mutational analysis of MYD88 (L265), CD79B (Y196), and EZH2 (Y641) included polymerase chain reaction amplification of purified DNA and sequencing using previously described primers.24,25 The genotypes of BAGS subtypes were studied using the Affymetrix Genome-Wide Human SNP Array 6.0 data (Affymetrix)13 of MDFCI CEL files imported into the Partek Genomics Suite's Copy Number work flow (Partek, St Louis, MO), and the results were exported as a text file and subsequently loaded into R.14
RESULTS
BAGS Classifier Generation and Clinical Sample Assignment
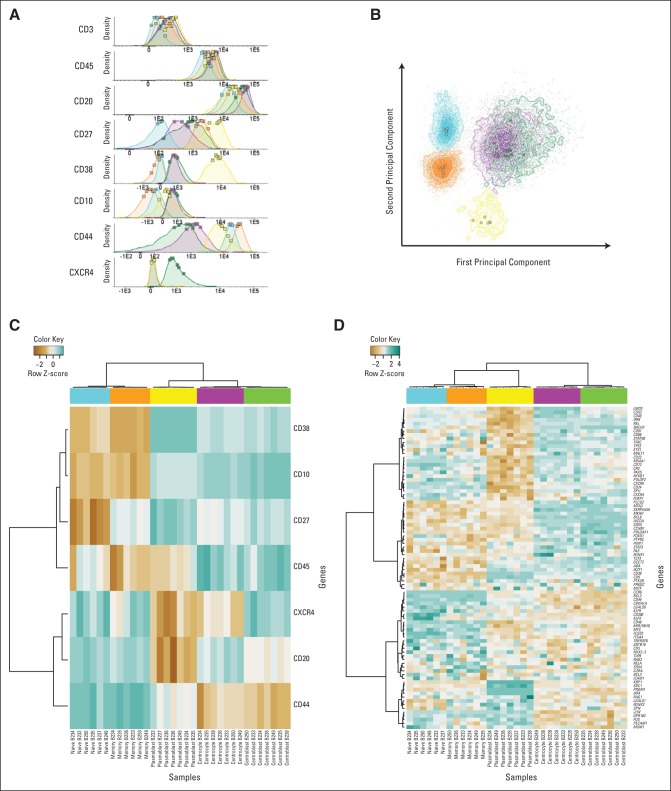
The B-cell subset identity was validated by density plots (Fig 1A) and principal component analysis (Fig 1B) of the intensities of the CD markers used for fluorescence-activated cell sorting and unsupervised cluster analysis of gene expression data for classical differentiation and transcription factor genes (Figs 1C and 1D).26–29 Subset-specific segregation was documented by principal component analysis of the tonsil data set (Appendix Fig A1, online only).
Fig 1.
Expression of CD markers, transcription factors, and B-cell subset–specific genes. Tonsil B cells were defined as CD20+, CD45+, and CD3– and were further subdivided according to differential expression of CD10, CD27, CD38, CD44, and CXCR4 into naive cells (blue), centrocytes (purple), centroblasts (green), memory cells (orange), and plasmablasts (yellow). (A) Normalized histograms of the fluorescence intensities of CD markers based on merged multiparametric flow cytometry reanalysis of pure sorted populations resulting from five independent sorting procedures. The squares are mean values of the fluorescence intensities for each single sorted B-cell subset. (B) Principal component analysis of the fluorescence intensities for each sorted cell in all samples. Cells are color coded according to their original subset. The squares are mean values for each sorted B-cell subset included in the principal component analysis. Surface markers, transcription factors, and B-cell differentiation–specific genes were identified through a literature review, and their expression across subsets was evaluated. The most varying probe sets were included in an unsupervised hierarchical clustering analysis (C) of the sorting markers and (D) of transcription factors, surface markers, and B-cell differentiation–specific genes. The color scale (top) indicates the relative gene expression in each sample, with blue illustrating high gene expression and brown low gene expression.
The B-cell subset classifier with the smallest deviance determined by cross validation consisted of 327 probe sets representing 223 different genes (Data Supplement). Each B-cell subset signature contained 54 to 93 probe sets, of which 37 to 76 were unique, ensuring comparable gene representation for all subsets in the BAGS classifier.
Lymphoma samples from five independent cohorts (N = 1,139) of patients with de novo DLBCL8,11–13 were classified into BAGS subtypes (Data Supplement). The five BAGS subtype frequencies did not vary significantly between the cohorts (Table 1). We chose 15% of samples within each cohort to be unclassified, resulting in a probability cutoff of approximately 0.45.
Table 1.
BAGS Classification of Clinical Samples
| Clinical Cohort and ABC/GCB Class | Naive |
Centroblast |
Centrocyte |
Memory |
Plasmablast |
Unclassified |
Total |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of Samples | % | No. of Samples | % | No. of Samples | % | No. of Samples | % | No. of Samples | % | No. of Samples | % | No. of Samples | % | |
| Clinical cohort | ||||||||||||||
| CHEPRETRO, CHOP | 2 | 2 | 18 | 20 | 35 | 39 | 4 | 4 | 16 | 18 | 14 | 16 | 89 | 100 |
| LLMPP, CHOP | 10 | 6 | 39 | 22 | 59 | 33 | 25 | 14 | 21 | 12 | 27 | 15 | 181 | 100 |
| LLMPP, R-CHOP | 14 | 6 | 41 | 18 | 89 | 38 | 22 | 9 | 32 | 14 | 35 | 15 | 233 | 100 |
| IDRC, R-CHOP | 32 | 7 | 111 | 24 | 161 | 34 | 44 | 9 | 49 | 10 | 71 | 15 | 468 | 100 |
| MDFCI, R-CHOP | 13 | 8 | 38 | 23 | 61 | 36 | 9 | 5 | 21 | 12 | 26 | 15 | 168 | 100 |
| Total | 71 | 6 | 247 | 22 | 405 | 36 | 104 | 9 | 139 | 12 | 173 | 15 | 1139 | 100 |
| ABC/GCB class | ||||||||||||||
| ABC | 41 | 9 | 62 | 14 | 96 | 21 | 75 | 16 | 82 | 18 | 103 | 22 | 459 | 100 |
| GCB | 20 | 4 | 160 | 30 | 264 | 50 | 10 | 2 | 28 | 5 | 45 | 9 | 527 | 100 |
| Unclassified | 9 | 6 | 23 | 15 | 45 | 30 | 19 | 13 | 29 | 19 | 25 | 17 | 150 | 100 |
NOTE. First, the distributions and frequencies of assigned BAGS subtypes across five different clinical cohorts are shown. Tests for significantly different distributions across data sets were done using Pearson's χ2 (χ2 = 21, df = 20, and P = .4). Second, BAGS assignment in the five clinical data sets is stratified according to ABC or GCB classification.
Abbreviations: ABC, activated B-cell–like; BAGS, B-cell–associated gene signature; CHEPRETRO, Chemotherapy Prediction in Retrospective Samples; CHOP, cyclophosphamide, doxorubicin, vincristine, and prednisone; GCB, germinal center B-cell–like; IDRC, International Diffuse Large B-Cell Lymphoma Rituximab-CHOP Consortium MD Anderson Project; LLMPP, Lymphoma/Leukemia Molecular Profiling Project; MDFCI, Mayo Clinic, Brigham and Women's Hospital, and Dana-Farber Cancer Institute Project; R-CHOP, rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone.
The overlap or correspondence between the ABC/GCB classes and the BAGS subtypes is shown in Table 1. High percentages of GCB subclasses were assigned as centroblast subtype (30%) or centrocyte subtype (50%), whereas ABC did not cluster into specific subtypes and had a relatively high frequency of unclassified samples (22%).
Prognostic Impact of Assigned BAGS Subtypes
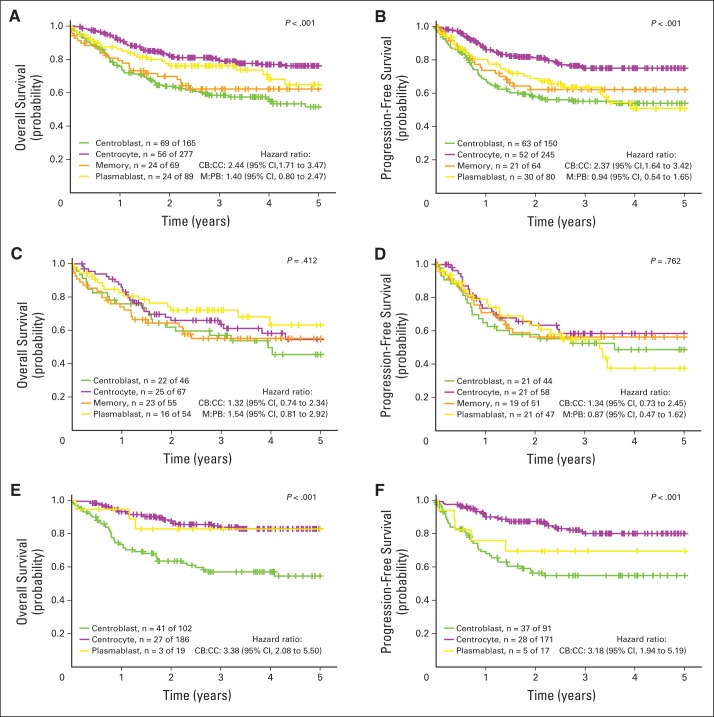
The BAGS-assigned subtypes in LLMPP, IDRC, and MDFCI were analyzed collectively in a meta-analysis of the association with outcome after R-CHOP treatment.30 BAGS assignment had significant prognostic associations with overall survival (OS) and progression-free survival (PFS; Fig 2 and Table 2).
Fig 2.
Meta-analysis of the prognostic impact of assigned B-cell–associated gene signature (BAGS) subtypes. (A, C, E) Overall survival and (B, D, F) progression-free survival were compared between BAGS subtypes for patients treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP). For overall survival, the following three cohorts were used: Lymphoma/Leukemia Molecular Profiling Project (LLMPP) R-CHOP, International Diffuse Large B-Cell Lymphoma Rituximab-CHOP Consortium MD Anderson Project (IDRC), and Mayo Clinic, Brigham and Women's Hospital, and Dana-Farber Cancer Institute Project (MDFCI). For progression-free survival, only LLMPP R-CHOP and IDRC were used. The comparisons were performed (A, B) overall and according to the (C, D) activated B-cell–like and (E, F) germinal center B-cell–like subclasses based on Kaplan-Meier survival curves, log-rank test P values, and hazard ratios. For clarity, the naive and unclassified cells were excluded from the Kaplan-Meier analysis. CB, centroblast; CC, centrocyte; M, memory; PB, plasmablast.
Table 2.
BAGS Assignment and Outcome
| Outcome | Simple Cox Regression |
Multiple Cox Regression |
||||
|---|---|---|---|---|---|---|
| Hazard Ratio | 95% CI | P | Hazard Ratio | 95% CI | P | |
| OS: IDRC, MDFCI, and LLMPP R-CHOP (n = 506, No. of events = 147) | ||||||
| BAGS subtype | ||||||
| Centroblast | 1 | 1 | ||||
| Centrocyte | 0.41 | 0.28 to 0.60 | < .001 | 0.48 | 0.33 to 0.71 | < .001 |
| Memory | 0.74 | 0.44 to 1.24 | .26 | 0.58 | 0.34 to 0.99 | .047 |
| Plasmablast | 0.56 | 0.34 to 0.94 | .028 | 0.52 | 0.31 to 0.88 | .016 |
| ABC/GCB class | ||||||
| ABC | 1 | 1 | ||||
| GCB | 0.53 | 0.37 to 0.75 | < .001 | 0.62 | 0.43 to 0.90 | .013 |
| Unclassified | 0.63 | 0.36 to 1.10 | .1 | 0.66 | 0.38 to 1.15 | .14 |
| IPI | ||||||
| 0-1 | 1 | 1 | ||||
| 2-5 | 4.23 | 2.73 to 6.56 | < .001 | 3.76 | 2.42 to 5.86 | < .001 |
| PFS: IDRC and LLMPP R-CHOP (n = 456, No. of events = 145) | ||||||
| BAGS subtype | ||||||
| Centroblast | 1 | 1 | ||||
| Centrocyte | 0.47 | 0.32 to 0.69 | < .001 | 0.55 | 0.37 to 0.82 | .0029 |
| Memory | 0.79 | 0.47 to 1.33 | .37 | 0.63 | 0.37 to 1.08 | .095 |
| Plasmablast | 0.77 | 0.47 to 1.25 | .29 | 0.73 | 0.44 to 1.20 | .21 |
| ABC/GCB class | ||||||
| ABC | 1 | 1 | ||||
| GCB | 0.50 | 0.36 to 0.71 | < .001 | 0.65 | 0.44 to 0.95 | .026 |
| Unclassified | 0.59 | 0.33 to 1.07 | .083 | 0.63 | 0.35 to 1.13 | .12 |
| IPI | ||||||
| 0-1 | 1 | 1 | ||||
| 2-5 | 3.54 | 2.36 to 5.33 | < .001 | 3.13 | 2.07 to 4.74 | < .001 |
NOTE. BAGS assignment and outcome were analyzed by Cox proportional hazards regression analysis for OS and PFS. For OS, the three cohorts of LLMPP R-CHOP, IDRC, and MDFCI were used, whereas only LLMPP R-CHOP and IDRC were used for PFS. The analysis excluded the naive and unclassified subsets but included the unclassified cases from the ABC/GCB classification.
Abbreviations: ABC, activated B-cell–like; BAGS, B-cell–associated gene signature; GCB, germinal center B-cell–like; IDRC, International Diffuse Large B-Cell Lymphoma Rituximab-CHOP Consortium MD Anderson Project; IPI, International Prognostic Index; LLMPP, Lymphoma/Leukemia Molecular Profiling Project; MDFCI, Mayo Clinic, Brigham and Women's Hospital, and Dana-Farber Cancer Institute Project; OS, overall survival; PFS, progression-free survival; R-CHOP, rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone.
The prognostic impact of BAGS subtype was evaluated separately in the GCB and ABC subclasses. BAGS subtyping only had an impact within the GCB subclass (P < .001 for OS and PFS), and the hazard ratios (HRs) for centrocytes and centroblasts were distinct (OS: HR, 3.38; 95% CI, 2.08 to 5.50; PFS: HR, 3.18; 95% CI, 1.94 to 5.19). BAGS assignment conferred new superior prognostic information to 23% of patients (264 of 1,139 samples) classified as GCB-centrocyte (GCB-CC) and had an adverse prognostic impact on 14% of patients (160 of 1,139 samples) classified as GCB-centroblast (GCB-CB) as evaluated in the 869 R-CHOP–treated patients in LLMPP, IDRC, and MDFCI.
Cox proportional hazards meta-analysis showed that the BAGS subtypes added significant prognostic information to the ABC/GCB subclasses and high and low IPI (Table 2). This finding indicated that distinct pathogenetic and prognostic knowledge not already explained by the ABC/GCB classification or IPI could be captured by the BAGS subtypes. The robustness of the BAGS association with outcome was evaluated successfully for a wide range of probability cutoffs for the percentage of unclassified samples (Data Supplement).
Predicting Drug Resistance in BAGS-Assigned Subtypes
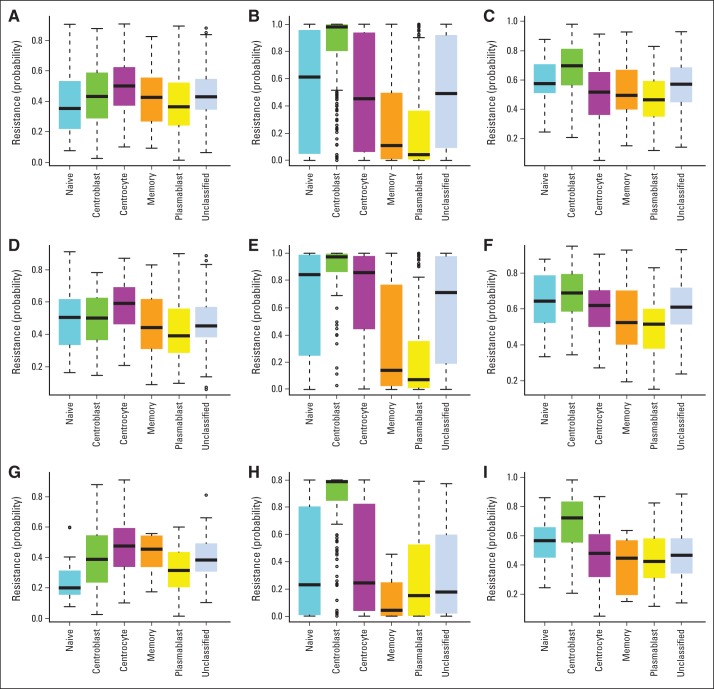
REGS for cyclophosphamide, doxorubicin, and vincristine were used to assign a probability of drug-specific resistance to individual samples. The probability of cyclophosphamide-, doxorubicin-, and vincristine-specific drug resistance was estimated for normal B-cell subsets (Table 3 and Appendix Fig A2, online only); centroblasts were predicted to be more resistant to doxorubicin and vincristine than centrocytes. When tested in all five clinical cohorts in a meta-analysis (Table 3 and Appendix Fig A3, online only), the centrocyte and centroblast subtypes had resistance patterns as seen in normal B-cell subsets. In concordance with the survival analysis (Fig 2), the major differences in estimated doxorubicin and vincristine resistance were observed between GCB-CB and GCB-CC (Table 3 and Appendix Figs A3H and A3I). The predicted drug response was supported by a 37% greater chance of CR30 in GCB-CC than in GCB-CB (relative risk, 1.37; 95% CI, 1.14 to 1.65) as evaluated in LLMPP and IDRC.
Table 3.
Difference in Drug Resistance Between Centrocyte and Centroblast Subtypes
| Drug and Sample | Probability of Resistance |
P | |||
|---|---|---|---|---|---|
| Centroblast |
Centrocyte |
||||
| Mean | 95% CI | Mean | 95% CI | ||
| Cyclophosphamide | |||||
| Normal tonsil | 0.48 | 0.39 to 0.56 | 0.65 | 0.56 to 0.74 | .0016 |
| Clinical | 0.42 | 0.39 to 0.45 | 0.50 | 0.48 to 0.52 | < .001 |
| Clinical, ABC | 0.49 | 0.44 to 0.53 | 0.59 | 0.55 to 0.62 | < .001 |
| Clinical, GCB | 0.37 | 0.34 to 0.41 | 0.47 | 0.45 to 0.50 | < .001 |
| Doxorubicin | |||||
| Normal tonsil | 0.94 | 0.82 to 0.98 | 0.76 | 0.63 to 0.85 | .05 |
| Clinical | 0.99 | 0.98 to 0.99 | 0.53 | 0.44 to 0.63 | < .001 |
| Clinical, ABC | 0.98 | 0.95 to 0.99 | 0.88 | 0.78 to 0.93 | .0013 |
| Clinical, GCB | 0.99 | 0.98 to 1.00 | 0.34 | 0.25 to 0.46 | < .001 |
| Vincristine | |||||
| Normal tonsil | 0.74 | 0.67 to 0.8 | 0.65 | 0.57 to 0.73 | .015 |
| Clinical | 0.70 | 0.68 to 0.72 | 0.51 | 0.49 to 0.53 | < .001 |
| Clinical, ABC | 0.70 | 0.66 to 0.73 | 0.63 | 0.59 to 0.66 | .0088 |
| Clinical, GCB | 0.71 | 0.67 to 0.74 | 0.46 | 0.44 to 0.49 | < .001 |
NOTE. The data represent mean (with 95% CI) probability of tissue being resistant to the drugs cyclophosphamide, doxorubicin, and vincristine for the centrocyte and centroblast subtypes in the tonsil data set and for B-cell–associated gene signature–defined centrocyte and centroblast subtypes based on all five cohorts. The P values are based on two-sample t tests for the logit-transformed probabilities. Effect estimates and CIs are obtained by back transformation from the logit scale.
Abbreviations: ABC, activated B-cell–like; GCB, germinal center B-cell–like.
Characterization of the Genotypes of BAGS Subtypes
The overall ratios of copy number alterations were examined across all BAGS subtypes and ABC/GCB/unclassified subclasses in the MDFCI data set by counting the base pairs affected by deletions or amplifications in each sample (Appendix Table A1, online only). Focusing on the germinal center subtypes, GCB-CB had more alterations than GCB-CC (11.9% v 6.9%, respectively).
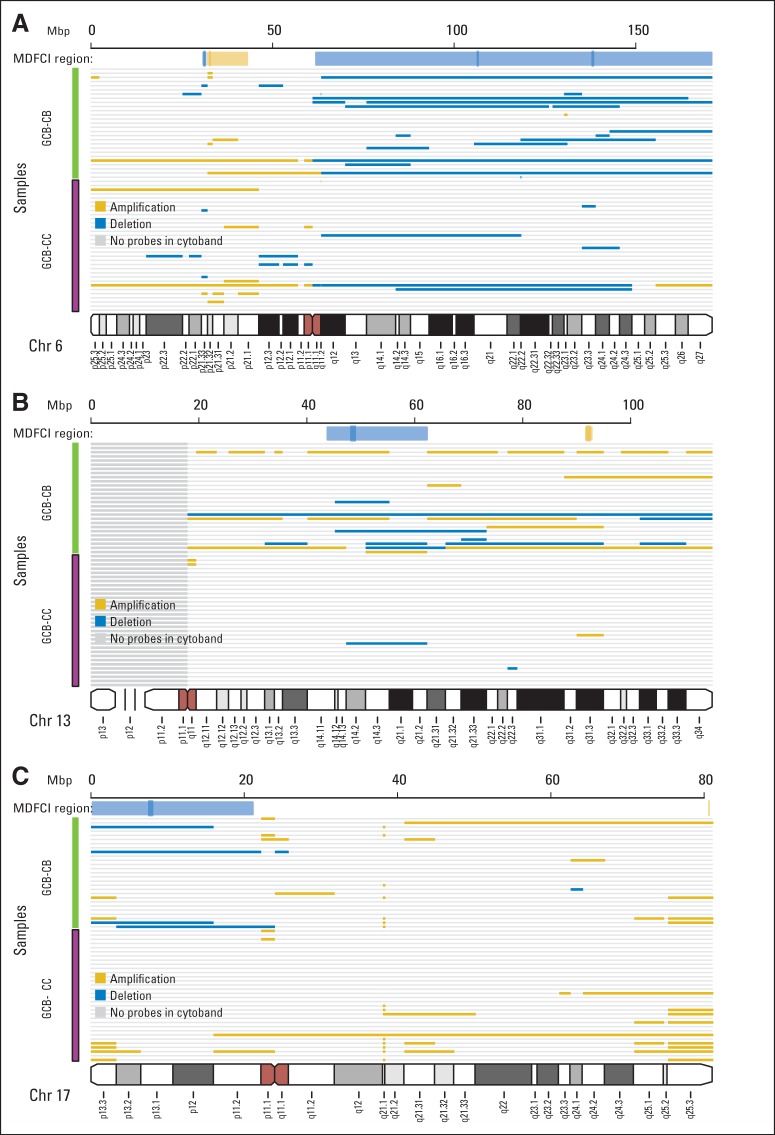
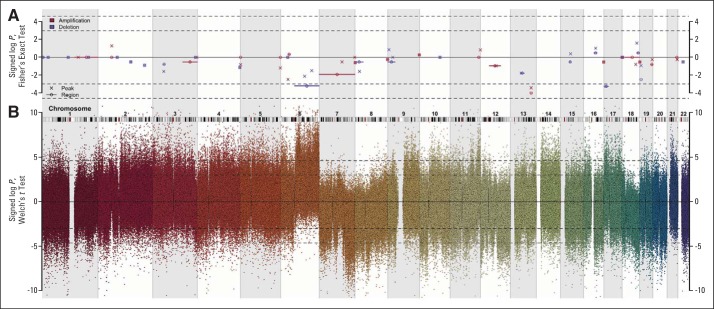
Monti et al13 used the Genomic Identification of Significant Targets in Cancer algorithm to identify recurrent chromosomal peaks and regions with copy number alterations. The distribution of alterations between these chromosomal positions and the assigned GCB-CC and GCB-CB subtypes in the MDFCI data set (Fig 3 and Appendix Fig A4, online only) resulted in an overrepresentation of aberrations in GCB-CB affecting Chr6q, Chr13q14.2, and Chr17p13.1 (Fig 3). Interestingly, Chr17p13.1 containing TP53, KDM6B, and RPL26 was deleted more frequently in GCB-CB than in GCB-CC (P = .039, Fig 3C).
Fig 3.
Characterization of the genotypes of B-cell–associated gene signature (BAGS) subtypes. Copy number aberrations (CNAs) in the assigned subtypes were found at various chromosome positions when studied in 165 samples from Mayo Clinic, Brigham and Women's Hospital, and Dana-Farber Cancer Institute Project (MDFCI) with Affymetrix Genome-Wide Human SNP Array 6.0 and Affymetrix Gene Chip Human Genome U133 Plus 2.0 Arrays or U133A+B Arrays. (A) Graphical representation of CNAs on chromosome 6, (B) chromosome 13, and (C) chromosome 17 with copy numbers (CNs) averaged over each cytoband. A deletion was defined as an average CN of less than 1.62, and an amplification was defined as an average CN of more than 2.46. Gold indicates amplification, and blue indicates deletion. The subtype identity is color coded as follows: germinal center B-cell–like (GCB) centrocyte (CC) is purple, and GCB centroblast (CB) is green. Above each chromosome, the extents of the MDFCI peaks (dark gold for amplification and dark blue for deletion) and region (light gold for amplification and light blue for deletion) are shown.
Additional genetic support for BAGS classification was observed after the application of clean/complex genotypes, which were defined by the absence or presence of copy number aberrations that alter the p53 pathway and cell cycle components,13 because the centroblast subtype never (zero of 13 samples) had a clean genotype (Appendix Table A1). TP53 mutation and insertion/deletion status also correlated with BAGS subtype (Appendix Table A1); centrocytes were targeted most frequently (P = .043). The CHEPRETRO data set was analyzed for mutations in CD79B, MYD88, and EZH2, confirming the distribution within the ABC/GCB subclasses24,25,31–36 but with no association with specific BAGS subtypes (Appendix Table A1).
Characterization of Gene Expression in BAGS-Assigned Subtypes
Determining the differential expression of individual probe sets by comparing GCB-CC versus GCB-CB, and GCB-CC versus GCB-CB, ABC-CC, and ABC-CB within IDRC, revealed known strong predictors of superior outcome (Data Supplement), particularly LMO2,37 which was most significantly upregulated in GCB-CC (P < .001). Loss of gene expression within the major histocompatibility complex class II signature was associated with unfavorable prognosis,38 and several members of the HLA family (Data Supplement) were downregulated in GCB-CB compared with GCB-CC. Also CD58, an important marker of immune surveillance through T cells and natural killer cells, was highly expressed in GCB-CC. Less directly, GCB-CC had high expression levels of IL13RA1, a gene known to be highly expressed in CD30+ DLBCLs with favorable prognosis.39,40 The unfavorable prognostic marker FOXP1 was significantly downregulated in GCB-CCs, and several members of the stromal-1 favorable-prognosis gene signature were upregulated in GCB-CC11 (Data Supplement). Notably, GCB-CC had significantly higher expression of MS4A1 (CD20), the target of rituximab, than GCB-CB.
DISCUSSION
Here, we combined methodologies to visualize the distinct cellular subsets of the tonsil and generated a classifier that assigns individual DLBCLs into BAGS-defined naive, centroblast, centrocyte, memory, or plasmablast subtypes. We hypothesized that this assignment of new subtypes would allow hierarchical DLBCL classification that associates normal B-cell phenotype subsets with prognosis.
The BAGS classifier assigned identical frequencies to individual DLBCL samples from five cohorts across geographic regions, time eras, and sampling methods, including both formalin-fixed12 paraffin-embedded and optimal cutting temperature compound.8,11,13 In a meta-analysis of 871 patients with DLBCL treated with R-CHOP, the BAGS classification demonstrated a significant prognostic association with OS and PFS adjusted for ABC/GCB classification and IPI. When the prognostic impact of BAGS subtype was evaluated separately in the ABC and GCB subclasses, only the GCB subclass achieved prognostic stratification, conferring superior prognosis to GCB-CC compared with GCB-CB. In agreement with the therapeutic outcome, the REGS classification for doxorubicin and vincristine indicated that centroblasts are more resistant than centrocytes. Again, the discrepancies in drug efficiency were most pronounced within the GCB subclass. Consistently, the CR rate after R-CHOP was higher for centrocytes than centroblasts (81% v 59%, respectively).
The unfavorable GCB-CB subtype had high total copy number aberrations and a complex genotype associated with deregulated p53 and cell cycle activity.13 GCB-CC had low total copy number aberrations, high TP53 mutation and insertion/deletion frequencies, and high expression of LMO2, CD58, and stromal-1–signature and major histocompatibility complex class II–signature genes with known favorable prognostic impact.11,37,38,41
Collectively, the results indicate that the BAGS classes have different clinical courses, drug resistance mechanisms, and molecular pathogenesis. However, further studies are needed to confirm the observations related to diagnostic phenotyping and individual therapy in DLBCL, because they involve a range of statistical, biologic, and clinical considerations.
In this study, restricted multinomial regression was used to estimate the probability of each sample being one of five BAGS subtypes. Samples with low classification probabilities were considered unclassified. The frequency of unclassified samples in other gene expression–based cell-of-origin classifications is approximately 15%.11 To ensure that 85% of the samples were classified by BAGS, a pragmatic probability cutoff of 0.45 was used, which is well above the random assignment probability of one out of five. The robustness of the BAGS association with outcome was evaluated successfully for a wide range of probability cutoffs.
This study followed various guidelines of omics-directed medicine10,42,43 whenever possible. However, the BAGS classifier used cohort-based normalization, median centering, and scaling, implying that it cannot easily be applied in a clinical setting with one patient at a time. Solutions to this problem have been suggested elsewhere44 and were not pursued in this study. Moreover, the median centering and scaling transformation could raise concerns that the BAGS-specific probe sets, which are not expressed in the clinical samples, are generally magnified and blurring the picture. We investigated this concern by comparing the empirical distribution of the probe sets in the BAGS classifier in the tonsil data set with the clinical data sets by two-cohort quantile-quantile plots (Data Supplement). The skewness of the IDRC data set did not raise concerns because it had the same underlying BAGS subtype frequency distribution. The entire workflow was analyzed without the probe set–wise scaling, resulting in a classification accuracy of 89%, further confirming the robustness of the proposed BAGS classification workflow.
The BAGS definition and assignment of malignant subtypes to DLBCL may explain interindividual disease heterogeneity, which likely reflects the often overlooked association between cellular differentiation and oncogenesis.6,7 Recently, a standardized n-dimensional flow cytometric immunophenotyping of hematologic malignancies, including germinal and postgerminal malignancies, illustrated the potential clinical use of surface-expressed markers to identify diagnostic tumor clones.45 Accordingly, an integrative analysis of sorted malignant DLBCL cells based on B-cell differentiation markers and gene expression profiling documented normal B-lineage differentiation and disease-specific changes.3
From gene expression–based ABC/GCB classification of DLBCL, multiple genetic lesions of pathogenetic significance have been described, illustrating subclasses that rely on distinct oncogenic mechanisms.33,46,47 The stratification of GCB subclasses into naturally occurring centrocyte and centroblast subtypes may similarly contribute to the pathogenic understanding of DLBCL.
In addition to the possible genetic variations between GCB-CC and GCB-CB, the GCB-CC expression of LMO2 was high and that of LEF1 low compared with GCB-CB. High LMO2 expression in GCB-CC could suppress recycling of centrocytes to centroblasts via LEF1,48,49 with high proliferation rates and decreased sensing of and response to DNA damage,50,51 or final differentiation of centrocytes into memory B cells or plasma cells.27 Nuclear LMO2 is part of Tally's immunohistochemical classification of GCB/non-GCB,52 and it could be relevant to incorporate LMO2 in established immunohistochemical classifiers of DLBCL12,53–55 to separate GCB-CC from GCB-CB.
R-CHOP treatment still forms the backbone of DLBCL treatments. Patients with relapsed or refractory disease have a cure rate of less than 50%, indicating a large unmet need for novel therapies in this setting.56–58 Therefore, increasing the efficacy of first-line treatment and decreasing the risk of early treatment failure are important from a clinical perspective. In a prospective phase I/II study with bortezomib plus standard R-CHOP in previously untreated patients with DLBCL, the lack of a difference in outcome between the GCB and non-GCB subclasses suggested that the bortezomib combination preferentially benefits patients with non-GCB DLBCL.59 In patients with relapsed or refractory CD20+ DLBCL, the response to salvage therapy with rituximab, dexamethasone, cytarabine, and cisplatin was better in GCB-like DLBCL compared with the response to rituximab, ifosfamide, carboplatin, and etoposide (the Collaborative Trial in Relapsed Aggressive Lymphoma [bio-CORAL] study).57 Because the prognostic impact of BAGS cannot be explained by association with ABC/GCB or IPI alone, utilization of the prognostic information residing in the individual BAGS subtypes at the time of diagnosis may be exploited to guide future patient treatment in the GCB subclass.
We think that our results support the future inclusion of immunophenotyping and gene expression profiling in randomized prospective clinical trials aimed at improving DLBCL treatment. Our hypothesis is that classification of the phenotypic cell of origin after gene signature assignment in malignant B-cell disorders should be assessed for clinical impact by end points, including diagnosis, prognosis, and prediction of therapeutic outcome. Future studies will attempt to prove this concept by implementing BAGS in studies of prognostic and predictive impact using clinical data sets from national trials.
Supplementary Material
Acknowledgment
Technicians Ann-Maria Jensen, Louise Hvilshøj Madsen, and Helle Høholt and bioengineer Anette Mai Tramm participated in all aspects of the laboratory work.
Appendix
Table A1.
Characterization of the Genotypes of BAGS Subtypes
| Genetic Analysis | Naive | Centroblast | Centrocyte | Memory | Plasmablast | Unclassified | Total |
|---|---|---|---|---|---|---|---|
| Base pairs affected by CNA in MDFCI (n = 165), %* | |||||||
| GCB | 1.1 | 11.9 | 6.9 | 1 | 4 | 8.3 | |
| ABC | 9.4 | 10.1 | 14.2 | 12.5 | 4.2 | 8.6 | |
| Unclassified | 3.6 | 8.3 | 9.7 | 2.8 | 2.4 | 5 | |
| Total | 6.4 | 11.2 | 9.1 | 8 | 3.8 | 7.7 | |
| All samples in MDFCI (n = 165), No. | |||||||
| Clean | 5 | 0 | 7 | 0 | 7 | 4 | 23 |
| Complex | 3 | 13 | 20 | 3 | 1 | 4 | 44 |
| NA | 4 | 23 | 34 | 6 | 13 | 18 | 98 |
| GCB samples in MDFCI (n = 75), No. | |||||||
| Clean | 1 | 0 | 4 | 0 | 1 | 0 | 6 |
| Complex | 0 | 9 | 8 | 0 | 0 | 0 | 17 |
| NA | 2 | 18 | 20 | 1 | 3 | 8 | 52 |
| ABC samples in MDFCI (n = 53), No. | |||||||
| Clean | 3 | 0 | 1 | 0 | 6 | 3 | 13 |
| Complex | 3 | 2 | 7 | 3 | 1 | 3 | 19 |
| NA | 1 | 2 | 4 | 2 | 6 | 6 | 21 |
| TP53 status in MDFCI samples (n = 165), No. | |||||||
| Absent | 9 | 32 | 45 | 6 | 19 | 23 | 134 |
| Indel | 1 | 0 | 2 | 0 | 0 | 0 | 3 |
| Mutant | 0 | 2 | 12 | 2 | 2 | 3 | 21 |
| NA | 2 | 2 | 2 | 1 | 0 | 0 | 7 |
| Mutation status in CHEPRETRO samples (n = 61-89), No. | |||||||
| MYD88 mutation | 1 | 2 | 3 | 0 | 2 | 2 | 10 |
| MYD88 wt | 1 | 11 | 22 | 3 | 7 | 7 | 51 |
| EZH2 mutation | 0 | 4 | 3 | 0 | 0 | 0 | 7 |
| EZH2 wt | 2 | 14 | 32 | 4 | 16 | 14 | 82 |
| CD79B mutation | 1 | 0 | 2 | 1 | 3 | 1 | 8 |
| CD79B wt | 1 | 17 | 29 | 3 | 12 | 11 | 73 |
NOTE. The U133 and SNP6 paired samples (n = 165) from MDFCI data set19 were reanalyzed and assigned to BAGS.
Abbreviations: ABC, activated B-cell–like; BAGS, B-cell–associated gene signature; CHEPRETRO, Chemotherapy Prediction in Retrospective Samples; CNA, copy number alteration; GCB, germinal center B-cell–like; MDFCI, Mayo Clinic, Brigham and Women's Hospital, and Dana-Farber Cancer Institute Project; NA, not analyzed; wt, wild type.
The average percentage of base pairs affected by copy number changes stratified against ABC/GCB classification and BAGS subtype. The human genome was set to 3 × 109 base pairs.
Fig A1.
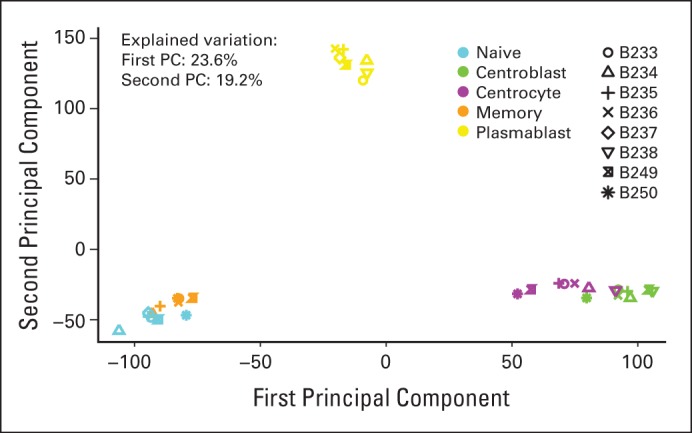
Principal component (PC) analysis identified distinct B-cell subset clusters. PC analysis was performed based on all probe sets for all samples (n = 8). The second PC is plotted versus the first PC. All subsets segregated into distinct clusters, albeit with relatively minor separation between the centroblast and centrocyte clusters and between the naive and memory clusters. The first and second PCs explained 23.6% and 19.2% of the variation, respectively. Notably, the analysis was conducted on all probe sets, resulting in a large number of noise probes, which explains the large proportion of the variation explained by the first PC.
Fig A2.
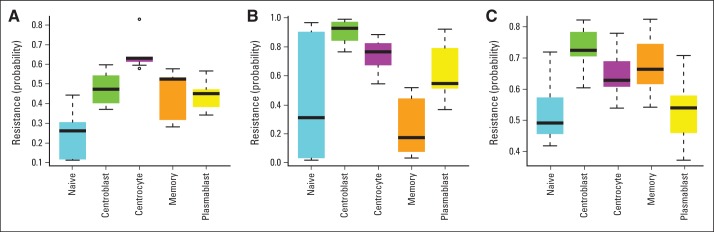
Drug-specific resistance of B-cell–associated gene signature subtypes. The probability of resistance in normal tonsil subsets was sorted and analyzed by microarray. Box plots indicate the estimated probabilities of resistance to (A) cyclophosphamide/mafosfamide, (B) doxorubicin, and (C) vincristine.
Fig A3.
Drug-specific resistance of B-cell–associated gene signature (BAGS) subtypes in all clinical cohorts. Box plots show the probability of resistance versus BAGS subtype for all clinical cohorts: (A, D, G) cyclophosphamide, (B, E, H) doxorubicin, and (C, F, I) vincristine in (A, B, C) all clinical cohorts, (D, E, F) activated B-cell–like subclass, and (G, H, I) germinal center B-cell–like subclass.
Fig A4.
Characterization of the genotypes of B-cell–associated gene signature subtypes. Copy number aberrations (CNAs) in the assigned subtypes were found at various chromosome positions when studied in 165 samples from Mayo Clinic, Brigham and Women's Hospital, and Dana-Farber Cancer Institute Project (MDFCI) with Affymetrix Genome-Wide Human SNP Array 6.0 and Affymetrix Gene Chip Human Genome U133 Plus 2.0 Arrays or U133A+B Arrays. (A) Signed log P values calculated using Fisher's exact test for the hypothesis of equal proportions of CNA in germinal center B-cell–like (GCB) centrocyte (CC; n = 32) versus GCB-centroblast (CB; n = 23) in peaks and regions (as defined by Monti et al13) are plotted against the chromosomal position of the region. Crosses and circles denote genomic peaks and regions, respectively. Color indicates the type of CNA investigated, as follows: amplification (red) or deletion (blue). Values greater than zero indicate more frequent CNAs in GCB-CC compared with GCB-CB, and vice versa for negative values. The dashed horizontal lines indicate the 95% and 99% acceptance intervals. (B) Signed log P values calculated based on t test for the hypothesis of no difference in copy number for each probe set in GCB-CC and GCB-CB samples are plotted against the chromosomal position of that probe set. Values greater than zero indicate amplification in GCB-CC or deletion in GCB-CB, and vice versa for values less than zero.
Footnotes
Supported by research funds from the European Union Sixth Framework Programme to MSCNET (Grant No. LSHC-CT-2006-037602), the Danish Cancer Society, the Danish Research Agency to CHEPRE (Grant No. 2101-07-0007), the KE Jensen Foundation (2006 to 2013) to H.E.J. and K.D., and the Obelske Family Foundation (2006 to 2013) to H.E.J. and M.B. K.H.Y. is supported by the MD Anderson Institution Fund, a Specialized Programs of Research Excellence Research Development Program Award, and, in part, by National Cancer Institute and National Institutes of Health Grants No. R01CA138688 and R01CA187415. R.J.S. acknowledges the support of an Engineering and Physical Sciences Research Council fellowship.
Authors' disclosures of potential conflicts of interest are found in the article online at www.jco.org. Author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Disclosures provided by the authors are available with this article at www.jco.org.
AUTHOR CONTRIBUTIONS
Conception and design: Karen Dybkær, Martin Bøgsted, Hans E. Johnsen
Collection and assembly of data: Karen Dybkær, Martin Bøgsted, Steffen Falgreen, Julie S. Bødker, Malene K. Kjeldsen, Alexander Schmitz, Zijun Y. Xu-Monette, Ling Li, Kim S. Bergkvist, Maria B. Laursen, Maria Rodrigo-Domingo, Sara C. Marques, Sophie B. Rasmussen, Mette Nyegaard, Michael Gaihede, Michael B. Møller, Preben Johansen, Tarec C. El-Galaly, Ken H. Young, Hans E. Johnsen
Data analysis and interpretation: Karen Dybkær, Martin Bøgsted, Steffen Falgreen, Julie S. Bødker, Malene K. Kjeldsen, Alexander Schmitz, Anders E. Bilgrau, Richard J. Samworth, Rajen D. Shah, Tarec C. El-Galaly, Ken H. Young, Hans E. Johnsen
Manuscript writing: All authors
Final approval of manuscript: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Diffuse Large B-Cell Lymphoma Classification System That Associates Normal B-Cell Subset Phenotypes With Prognosis
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or jco.ascopubs.org/site/ifc.
Karen Dybkær
No relationship to disclose
Martin Bøgsted
No relationship to disclose
Steffen Falgreen
No relationship to disclose
Julie S. Bødker
No relationship to disclose
Malene K. Kjeldsen
No relationship to disclose
Alexander Schmitz
No relationship to disclose
Anders E. Bilgrau
No relationship to disclose
Zijun Y. Xu-Monette
No relationship to disclose
Ling Li
No relationship to disclose
Kim S. Bergkvist
No relationship to disclose
Maria B. Laursen
No relationship to disclose
Maria Rodrigo-Domingo
No relationship to disclose
Sara C. Marques
No relationship to disclose
Sophie B. Rasmussen
Employment: Novo Nordisk
Mette Nyegaard
Patents, Royalties, Other Intellectual Property: MN is an inventor of a patent relating to genetic diagnostics in the cardiac arrhythmia field
Michael Gaihede
No relationship to disclose
Michael B. Møller
No relationship to disclose
Richard J. Samworth
No relationship to disclose
Rajen D. Shah
No relationship to disclose
Preben Johansen
No relationship to disclose
Tarec C. El-Galaly
No relationship to disclose
Ken H. Young
No relationship to disclose
Hans E. Johnsen
No relationship to disclose
REFERENCES
- 1.Campo E, Swerdlow SH, Harris NL, et al. The 2008 WHO classification of lymphoid neoplasms and beyond: Evolving concepts and practical applications. Blood. 2011;117:5019–5032. doi: 10.1182/blood-2011-01-293050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 3.Andréasson U, Dictor M, Jerkeman M, et al. Identification of molecular targets associated with transformed diffuse large B cell lymphoma using highly purified tumor cells. Am J Hematol. 2009;84:803–808. doi: 10.1002/ajh.21549. [DOI] [PubMed] [Google Scholar]
- 4.Kjeldsen MK, Perez-Andres M, Schmitz A, et al. Multiparametric flow cytometry for identification and fluorescence activated cell sorting of five distinct B-cell subpopulations in normal tonsil tissue. Am J Clin Pathol. 2011;136:960–969. doi: 10.1309/AJCPDQNP2U5DZHVV. [DOI] [PubMed] [Google Scholar]
- 5.Perez-Andres M, Paiva B, Nieto WG, et al. Human peripheral blood B-cell compartments: A crossroad in B-cell traffic. Cytometry B Clin Cytom. 2010;78(suppl 1):S47–S60. doi: 10.1002/cyto.b.20547. [DOI] [PubMed] [Google Scholar]
- 6.Visvader JE. Cells of origin in cancer. Nature. 2011;469:314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 7.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gang AO, Strøm C, Pedersen M, et al. R-CHOEP-14 improves overall survival in young high-risk patients with diffuse large B-cell lymphoma compared with R-CHOP-14: A population-based investigation from the Danish Lymphoma Group. Ann Oncol. 2012;23:147–153. doi: 10.1093/annonc/mdr058. [DOI] [PubMed] [Google Scholar]
- 9.Mårtensson S, Frederiksen BL, De Nully Brown P, et al. Differential case reporting in a national clinical quality database: An analysis of the impact on comparison of outcome between departments. Qual Manag Health Care. 2012;21:278–285. doi: 10.1097/QMH.0b013e31826d1f6d. [DOI] [PubMed] [Google Scholar]
- 10.Brazma A, Hingamp P, Quackenbush J, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 11.Lenz G, Wright G, Dave SS, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008;359:2313–2323. doi: 10.1056/NEJMoa0802885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Visco C, Li Y, Xu-Monette ZY, et al. Comprehensive gene expression profiling and immunohistochemical studies support application of immunophenotypic algorithm for molecular subtype classification in diffuse large B-cell lymphoma: A report from the International DLBCL Rituximab-CHOP Consortium. Leukemia. 2012;26:2103–2113. doi: 10.1038/leu.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monti S, Chapuy B, Takeyama K, et al. Integrative analysis reveals an outcome-associated and targetable pattern of p53 and cell cycle deregulation in diffuse large B cell lymphoma. Cancer Cell. 2012;22:359–372. doi: 10.1016/j.ccr.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2010. [Google Scholar]
- 15.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rossini AJ, Lumley T, Leisch F. On the edge: Statistics & computing—Reproducible statistical research. Chance. 2012;16:41–45. [Google Scholar]
- 17.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 18.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 19.Laursen MB, Falgreen S, Bødker JS, et al. Human B-cell cancer cell lines as a preclinical model for studies of drug effect in diffuse large B-cell lymphoma and multiple myeloma. Exp Hematol. 2014;42:927–938. doi: 10.1016/j.exphem.2014.07.263. [DOI] [PubMed] [Google Scholar]
- 20.Boegsted M, Holst JM, Fogd K, et al. Generation of a predictive melphalan resistance index by drug screen of B-cell cancer cell lines. PLoS One. 2011;6:e19322. doi: 10.1371/journal.pone.0019322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bøgsted M, Bilgrau AE, Wardell CP, et al. Proof of the concept to use a malignant B cell line drug screen strategy for identification and weight of melphalan resistance genes in multiple myeloma. PLoS One. 2013;8:e83252. doi: 10.1371/journal.pone.0083252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falgreen S, Laursen MB, Bødker JS, et al. Exposure time independent summary statistics for assessment of drug dependent cell line growth inhibition. BMC Bioinformatics. 2014;15:168. doi: 10.1186/1471-2105-15-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat. 1979;6:65–70. [Google Scholar]
- 24.Davis RE, Ngo VN, Lenz G, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463:88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Capello D, Gloghini A, Martini M, et al. Mutations of CD79A, CD79B and EZH2 genes in immunodeficiency-related non-Hodgkin lymphomas. Br J Haematol. 2011;152:777–780. doi: 10.1111/j.1365-2141.2010.08445.x. [DOI] [PubMed] [Google Scholar]
- 26.Andréasson U, Edén P, Peterson C, et al. Identification of uniquely expressed transcription factors in highly purified B-cell lymphoma samples. Am J Hematol. 2010;85:418–425. doi: 10.1002/ajh.21701. [DOI] [PubMed] [Google Scholar]
- 27.Klein U, Dalla-Favera R. Germinal centres: Role in B-cell physiology and malignancy. Nat Rev Immunol. 2008;8:22–33. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- 28.Matthias P, Rolink AG. Transcriptional networks in developing and mature B cells. Nat Rev Immunol. 2005;5:497–508. doi: 10.1038/nri1633. [DOI] [PubMed] [Google Scholar]
- 29.McHeyzer-Williams M, Okitsu S, Wang N, et al. Molecular programming of B cell memory. Nat Rev Immunol. 2012;12:24–34. doi: 10.1038/nri3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheson BD, Pfistner B, Juweid ME, et al. Revised response criteria for malignant lymphoma. J Clin Oncol. 2007;25:579–586. doi: 10.1200/JCO.2006.09.2403. [DOI] [PubMed] [Google Scholar]
- 31.Lohr JG, Stojanov P, Lawrence MS, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109:3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J, Grubor V, Love CL, et al. Genetic heterogeneity of diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A. 2013;110:1398–1403. doi: 10.1073/pnas.1205299110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pasqualucci L, Trifonov V, Fabbri G, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43:830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morin RD, Mungall K, Pleasance E, et al. Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood. 2013;122:1256–1265. doi: 10.1182/blood-2013-02-483727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morin RD, Johnson NA, Severson TM, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rasmussen SB. Cytogenetic profiling of B-cell lymphomas. http://projekter.aau.dk/projekter/files/58649577/Cytogenetic_Profiling_of_B_Cell_Lymphomas.pdf.
- 37.Alizadeh AA, Gentles AJ, Alencar AJ, et al. Prediction of survival in diffuse large B-cell lymphoma based on the expression of 2 genes reflecting tumor and microenvironment. Blood. 2011;118:1350–1358. doi: 10.1182/blood-2011-03-345272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rimsza LM, Roberts RA, Miller TP, et al. Loss of MHC class II gene and protein expression in diffuse large B-cell lymphoma is related to decreased tumor immunosurveillance and poor patient survival regardless of other prognostic factors: A follow-up study from the Leukemia and Lymphoma Molecular. Blood. 2004;103:4251–4258. doi: 10.1182/blood-2003-07-2365. [DOI] [PubMed] [Google Scholar]
- 39.Hu S, Xu-Monette ZY, Balasubramanyam A, et al. CD30 expression defines a novel subgroup of diffuse large B-cell lymphoma with favorable prognosis and distinct gene expression signature: A report from the International DLBCL Rituximab-CHOP Consortium Program Study. Blood. 2013;121:2715–2724. doi: 10.1182/blood-2012-10-461848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu S, Xu-Monette ZY, Tzankov A, et al. MYC/BCL2 protein coexpression contributes to the inferior survival of activated B-cell subtype of diffuse large B-cell lymphoma and demonstrates high-risk gene expression signatures: A report from The International DLBCL Rituximab-CHOP Consortium Program. Blood. 2013;121:4021–4031. doi: 10.1182/blood-2012-10-460063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Challa-Malladi M, Lieu YK, Califano O, et al. Combined genetic inactivation of β2-microglobulin and CD58 reveals frequent escape from immune recognition in diffuse large B cell lymphoma. Cancer Cell. 2011;20:728–740. doi: 10.1016/j.ccr.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McShane LM, Cavenagh MM, Lively TG, et al. Criteria for the use of omics-based predictors in clinical trials: Explanation and elaboration. BMC Med. 2013;11:220. doi: 10.1186/1741-7015-11-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McShane LM, Altman DG, Sauerbrei W, et al. Reporting recommendations for tumor marker prognostic studies. J Clin Oncol. 2005;23:9067–9072. doi: 10.1200/JCO.2004.01.0454. [DOI] [PubMed] [Google Scholar]
- 44.Care MA, Barrans S, Worrillow L, et al. A microarray platform-independent classification tool for cell of origin class allows comparative analysis of gene expression in diffuse large B-cell lymphoma. PLoS One. 2013;8:e55895. doi: 10.1371/journal.pone.0055895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Dongen JJ, Lhermitte L, Böttcher S, et al. EuroFlow antibody panels for standardized n-dimensional flow cytometric immunophenotyping of normal, reactive and malignant leukocytes. Leukemia. 2012;26:1908–1975. doi: 10.1038/leu.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shaffer AL, 3rd, Young RM, Staudt LM. Pathogenesis of human B cell lymphomas. Annu Rev Immunol. 2012;30:565–610. doi: 10.1146/annurev-immunol-020711-075027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lenz G, Wright GW, Emre NC, et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc Natl Acad Sci U S A. 2008;105:13520–13525. doi: 10.1073/pnas.0804295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Natkunam Y, Zhao S, Mason DY, et al. The oncoprotein LMO2 is expressed in normal germinal-center B cells and in human B-cell lymphomas. Blood. 2007;109:1636–1642. doi: 10.1182/blood-2006-08-039024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cubedo E, Gentles AJ, Huang C, et al. Identification of LMO2 transcriptome and interactome in diffuse large B-cell lymphoma. Blood. 2012;119:5478–5491. doi: 10.1182/blood-2012-01-403154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432:635–639. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- 51.Caron G, Le Gallou S, Lamy T, et al. CXCR4 expression functionally discriminates centroblasts versus centrocytes within human germinal center B cells. J Immunol. 2009;182:7595–7602. doi: 10.4049/jimmunol.0804272. [DOI] [PubMed] [Google Scholar]
- 52.Meyer PN, Fu K, Greiner TC, et al. Immunohistochemical methods for predicting cell of origin and survival in patients with diffuse large B-cell lymphoma treated with rituximab. J Clin Oncol. 2011;29:200–207. doi: 10.1200/JCO.2010.30.0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 54.Choi WW, Weisenburger DD, Greiner TC, et al. A new immunostain algorithm classifies diffuse large B-cell lymphoma into molecular subtypes with high accuracy. Clin Cancer Res. 2009;15:5494–5502. doi: 10.1158/1078-0432.CCR-09-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coutinho R, Clear AJ, Owen A, et al. Poor concordance among nine immunohistochemistry classifiers of cell-of-origin for diffuse large B-cell lymphoma: Implications for therapeutic strategies. Clin Cancer Res. 2013;19:6686–6695. doi: 10.1158/1078-0432.CCR-13-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Friedberg JW. Relapsed/refractory diffuse large B-cell lymphoma. Hematology Am Soc Hematol Educ Program. 2011;2011:498–505. doi: 10.1182/asheducation-2011.1.498. [DOI] [PubMed] [Google Scholar]
- 57.Thieblemont C, Briere J, Mounier N, et al. The germinal center/activated B-cell subclassification has a prognostic impact for response to salvage therapy in relapsed/refractory diffuse large B-cell lymphoma: A bio-CORAL study. J Clin Oncol. 2011;29:4079–4087. doi: 10.1200/JCO.2011.35.4423. [DOI] [PubMed] [Google Scholar]
- 58.Vitolo U, Chiappella A, Franceschetti S, et al. Lenalidomide plus R-CHOP21 in elderly patients with untreated diffuse large B-cell lymphoma: Results of the REAL07 open-label, multicentre, phase 2 trial. Lancet Oncol. 2014;15:730–737. doi: 10.1016/S1470-2045(14)70191-3. [DOI] [PubMed] [Google Scholar]
- 59.Ruan J, Martin P, Furman RR, et al. Bortezomib plus CHOP-rituximab for previously untreated diffuse large B-cell lymphoma and mantle cell lymphoma. J Clin Oncol. 2011;29:690–697. doi: 10.1200/JCO.2010.31.1142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.