Abstract
A study was conducted to evaluate the risk of Salmonella contamination and its survival at different stages in chill ready-to-eat poultry meat products chain. Samples (n = 181) were collected and examined for the presence of Salmonella species. The bacteria were initially identified against polyvalent antisera “O” and “H”, followed by confirmation with 16 s rDNA. The single Salmonella, isolate from the tested food samples showed 99.8 % phylogenetic similarity with Salmonella enterica. It was further evaluated for antibiotic sensitivity pattern and found resistant to four antibiotics including ampicillin, chloramphenicol, tetracycline and nalidixic acid. Salmonella associated with ready-to-eat poultry meat products were found active at storage temperature ≥4 °C in a challenge study. It was revealed that shape and weight of the meat pieces have direct influence on the reduction of pathogens during microwave heating. The 30 and 60 s microwaving (with radiation power fixed at 900 W) was found ineffective for the elimination of target bacteria (106-107 CFU/g) in meat pieces with weight ≥90 g. Salmonella enterica was able to survive in simulated gastric fluid. The storage temperature and microwaving were found critical point for the transfer of pathogens through ready-to-eat poultry meat products to consumer in chill ready-to-eat poultry meat chain.
Keywords: Food safety, Risk assessment, Foodborne pathogens, Enteric fever, Microwaving, Salmonellosis, Poultry meat
Introduction
Salmonella is an important foodborne pathogen, currently containing 2,587 serotypes (Fashae et al. 2010). Poultry meat is one of the frequent vehicles of Salmonellosis as zoonotic infection and is a major concern of public health (Hur et al. 2011; Fashae et al. 2010). Salmonella enterica is one of the prominent reasons of enteric diseases worldwide. It causes great number of illness and substantial economic losses in both developing and developed countries. It is more often associated with foodborne Salmonellosis (Akbar and Anal 2013; Fernandez et al. 2012; Hur et al. 2011), and human gastroenteritis (Skov et al. 2007). Consumption of animal food products are thought to be the major cause of the Salmonella outbreaks (Thai et al. 2012). Salmonella were found responsible for 1,722 outbreaks of foodborne infections in the European countries in 2009 (Fernandez et al. 2012). An estimated number of 1.8 million people died due to foodborne diarrhoeal infections each year in developing countries (Akbar and Anal 2011). Every year approximately one million diarrheal and 120,000 cases of food poisoning reported from Thailand (Chomvarin et al. 2006), causing a serious public health concern. Infections with drug resistant Salmonella are more fatal and take longer time for recovery as compare to drug susceptible Salmonella species (Skov et al. 2007). Excessive and misuse of antibiotics in animals and human is the leading cause of drug resistance development in pathogens (Thai et al. 2012). Food contaminated with drug resistant bacteria is a major threat to public health as the antibiotic resistance can transfer to other bacteria (Akbar and Anal 2011). Monitoring the presence and antimicrobial resistance of bacteria are necessary to understand the trends and magnitude of food related pathogens, and to plan an effective intervention (Fernandez et al. 2012). Epidemiological data related to Salmonella prevalence and its antimicrobial drug resistance pattern is desirable in order to develop an efficient mechanism towards its control at every level of food processing and production, to ensure food safety and public health (Angkititrakul et al. 2005). Antibiogram profile and genetic typing is a useful tool for infections sources determination (Aarestrup et al. 2007).
Assessment of risk associated to foodborne infection is an important approach for designing food safety plans and programs to address emerging foodborne diseases. Risk assessment is describing a system through which the flow of microbial hazards reaches its host and causes harm (Lammerding and Paoli 1997). Presence of Salmonella in ready-to-eat (RTE) food is a great risk to consumer health, there is zero tolerance towards its presence in RTE products. Consumer safety remains the prime concern for food production industries, it is always a challenge to eliminate the food safety risk and reduce the chances of foodborne pathogens contamination and flow in food chain (Akbar and Anal 2014a). It has been estimated that 103-109 number of Salmonella enterica can be an infectious dose for human being depending on their immunity (Parry 2006). Handling, processing and storage are some of the factors affecting the microbial status of RTE foods (Akbar and Anal 2014b; Roy et al. 2011). The existing data related to Salmonella presence in RTE chilled poultry meat products is limited.
The objective of this study was to investigate the prevalence of Salmonella species in chilled RTE poultry meat products, and to understand the effects of different hurdles (low temperature, microwaving and simulated gastric juices) on its survival. Challenge studies for the Salmonella survival in RTE poultry meat at low temperature storage, microwaving and in simulated gastric fluid (SGF) was conducted for better understanding of possible exposure of consumer to the pathogen present in RTE poultry meat products.
Materials and methods
Sampling and isolation of Salmonella
The RTE poultry meat samples (181) were collected from departmental stores and transferred to the laboratory for further processing. All the samples were examined within 4–6 h of its collection time. Sample (25 g) were added to 225 mL of buffered peptone water (BPW) and mixed well with the help of homogenizer (IKA Labortechnik, Germany), incubated at 37 °C for 18–24 h. Pre-inoculated BPW were transferred to 10 mL of selective enrichment media rappaport vassiliadis soyabean meal broth (RVSM) (BioMark, India) and tetrathionate (TT) broth (BioMark) and incubated further at 37 °C for 24 h. On the following day, two loopful of RVSM and TT broth were transferred to bismuth sulfite agar (BSA) (Himedia, India) and xylose lysine desoxycholate Agar (XLD) (Difco, USA), and further incubated at 37 °C for 24–48 h to isolate visible colonies of Salmonella species. Pink colonies with black centre on XLD agar and brown, gray or black colonies with metallic sheen on BSA were presumptively counted as Salmonella. Positive colonies were then inoculated on triple sugar iron agar (Himedia) and lysine iron agar (Himedia) and further confirmed by submitting to API 20E kit (Biomerieux, France) along API web and with the help of polyvalent antisera “O” and “H” (Serosystem, Clinag, Thailand). The isolated bacteria were then subjected to molecular confirmation.
Molecular identification of Salmonella
The “Genomic DNA mini kit (Blood/culture cell)” (Geneaid Biotech Ltd., Taiwan) was used to prepare DNA templates for PCR amplification. Coding of DNA for 16S rRNA regions was amplified by PCR with Taq polymerase, following the detail methodology described by Buranarom (2010).
Sequence and phylogenetic analysis
The nucleotide sequences obtained were assembled using Cap contig assembly program, BioEdit (Biological sequence alignment editor) Program. The phylogenetic neighbors were identified by BLAST (Altschul et al. 1997) and mega BLAST (Zhang et al. 2000). Fifty neighboring sequences were selected for the calculation of pair wise sequence similarity using global alignment algorithm, which was implemented at the EzTaxon server (Chun et al. 2007). The DNA sequences obtained from databases were aligned with CLUSTAL X (version 1.8) (Thompson et al. 1997) in BioEdit Program (Hall 1999). The evolutionary distances were computed using the maximum composite likelihood method (Tamura et al. 2004). Phylogenetic trees were constructed by neighbor-joining method of Saitou and Nei (1987). The robustness for individual branches was estimated by 1,000 replications bootstrapping with MEGA Version 4.0 (Tamura et al. 2011).
Antibiogram of Salmonella
Antibiogram of Salmonella were determined by using disk diffusion assay following the guidelines of clinical and laboratory standard institute. Pre-incubated 24 h cultures of Salmonella were diluted to 108 CFU/mL in sterile buffer peptone water and spread over mueller-hinton agar (Merck, Germany). The antibiotic discs were placed over the lawn and incubated at 37 °C for 18–24 h. The clear zone around each antibiotic disc was measured on the following day. The antibiotic used were Ampicillin (10 μg), Cefotaxime (30 μg), Norfloxacin (10 μg), Chloramphenicol (30 μg), Tetracycline (30 μg), Erythromycin (5 μg), Trimethoprim / Sulfamethoxazole (25 μg) Nalidixic acid (30 μg) (Oxoid Ltd. UK).
Low temperature storage effects on Salmonella in RTE poultry meat (challenge study)
Salmonella free (laboratory confirmed) RTE poultry meat sausages were used to study the survival of Salmonella under low temperature condition. The S. enterica, isolated from RTE poultry meat products were used as test bacteria in this challenge study. The RTE poultry meat sausages were cut in 4 cm2 pieces, approximately equal to 8–10 g weight after 15 days of its storage under freezing temperature. The meat pieces were exposed to ultraviolet radiation for 2 h equally on each side to eliminate the number of surface bacteria. Fresh culture (50 mL) of S. enterica (104–105 CFU/mL) washed twice with sterile normal saline were introduced to the surface of the meat pieces and spread evenly with the help of hand operated bottle. Initial count of the inoculated bacteria in the meat pieces were obtained with the help of standard plate count (SPC) method using XLD agar. The inoculated meat pieces were then kept at two different temperatures conditions (2 ± 2 °C and 6 ± 2 °C) and bacterial count were made at specific time interval using standard plate count (SPC) method.
Microwaving effects on Salmonella in RTE poultry meat (challenge study)
A challenges study for Salmonella contaminated RTE poultry meat products with microwave were conducted for specific durations of time in order to know the possible effect of microwaving on the pathogens present in the meat products.
Pre-treatment of the samples were done in a similar way as previously described in for low temperature storage effects on Salmonella. Salmonella enterica, isolated from RTE poultry meat were introduced to the surfaces of three set of experiments with meat pieces of different weight ranges, roughly equal to 30–50 g, 60–90 g and 100–150 g by spraying and injecting with sterile needles to obtain 106–107 CFU/g in meat for challenge study. The inoculated meat pieces were then exposed to microwave using microwave oven (Electrolux, China) with radiation power fixed at 900 W for 30, 60 and 90 s. Enumeration of the target bacteria were conducted before treatment and after treatment with SPC method using XLD agar.
Survival of Salmonella enterica in simulated gastric fluids
Consumption of S. enterica with food at the rate of 103 or above can be an infectious dose for human being (Parry 2006). Contaminated foods are the main source of such incidence. The effect of gastric fluid on the survival of Salmonella enterica was studied using simulated gastric fluids (SGF) following (Zarate et al. 2004). The effect of SGF with pH 2, 3, 4 and neutral pH 7 (control) was observed. Salmonella enterica (1 mL) at the rate of (106–107 CFU/mL) were introduced to 9 mL to SGF of different pH, after washing twice with sterile normal saline. The tubes were incubated in water bath at 37 °C for 3 h. The bacteria in SGF were enumerated at 0 h, 1 h and 3 h with the help of SPC technique using XLD agar.
Results
In this study a series of experiments were conducted for isolation of Salmonella species and its survivability at different low temperature, microwaving and in SGF. Out of all tested (181) samples, only one (0.55 %) was found contaminated with Salmonella, as initially identified with the help of biochemical, serological tests and confirmed by molecular detection method. The isolate showed 99.80 % phylogenetic similarity to Salmonella enterica species. The phylogenetic relationship of the Salmonella reveled that it has a common boot strip value of 64 % with the species S. enterica subsp. enterica LT2 (T) AE006468 and S. enterica subsp. salamae. The S. enterica subsp. enterica LT2(T) AE006468 and S. enterica subsp. diarizonae DSM 14847(T) EU014688 showed 98.88 % similarity with the isolate, whereas the subsp. enterica sharing the same node. Salmonella enterica subsp. indica DSM 14848(T) EU014680 showed 98.94 % similarity (Fig. 1). The isolate showed resistance to four antibiotics including ampicillin, chloramphenicol, tetracycline and nalidixic acid, whereas it was susceptible to cefotaxime, norfloxacin, erythromycin and sulfamethoxazole-trimethoprim.
Fig. 1.
Phylogenetic tree of Salmonella (RBC19) isolated from ready-to-eat poultry meat based on 5’ end of 16S rDNA
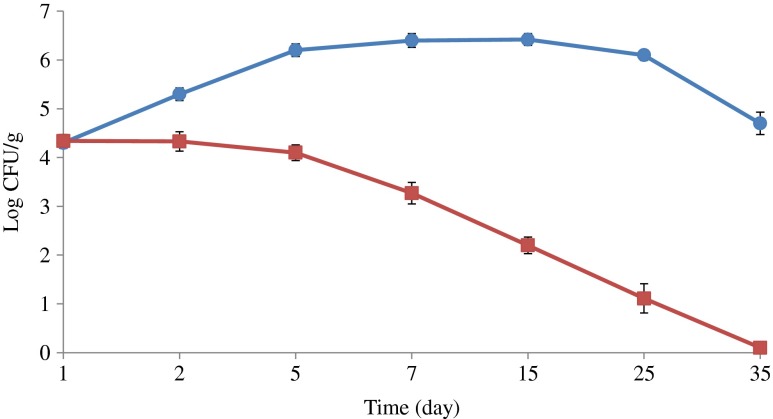
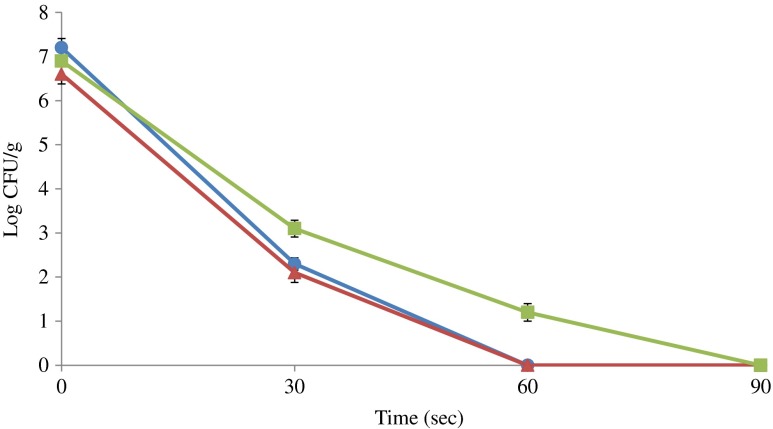
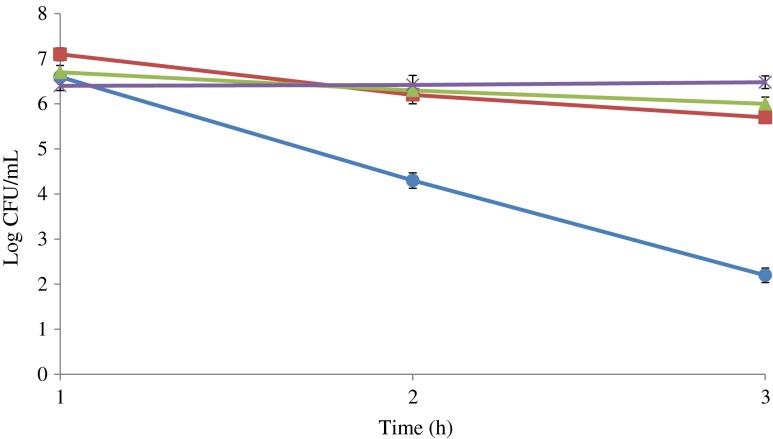
Storage temperature greatly affected the growth of S. enterica during the experiments. The number of inoculated S. enterica increased by 2 log cycle at storage temperature 6 ± 2 °C during the first 7 days, and decreased slowly with increasing storage time to a number 104 CFU/mL equals to the initial inocula after 35 days of storage (Fig. 2). Salmonella enterica was able to retain its initial number intact for first 2 days in the meat samples kept at temperature 2 ± 2 °C, but decreased gradually (log 4 - log 1 CFU/mL) with the passage of time (1–25 days) respectively and was undetectable at 35th day of storage. During the course of microwaving, it was found that 30 s treatment is not able to destroy the total number of S. enterica in inoculated RTE meat products. The number of inoculated bacteria in RTE poultry meat with weight ≤90 g was reduced to undetectable number during 60 s microwaving. The same treatment for higher weight 100–150 g was found ineffective and needed 90 s time in order to eliminate the risk of Salmonella in contaminated samples (Fig. 3). It was found that S. enterica in SGF have significant growth stability in the SGF of pH 7, 4 and 3 respectively during the challenge study. The highest number of inoculated bacterial number reduction was observed in SGF of pH 2 from log 6 to log 2 CFU/mL (Fig. 4). There was no significant reduction in number of inoculated S. enterica observed in SGF with pH 3, 4 and 7. It was noted that S. enterica has the potential to reach the intestinal mucosa of human along with contaminated food during the consumption of contaminated food materials.
Fig. 2.
Low temperature challenge study for the growth of Salmonella enterica, (black circle) temperature 6 ± 2 °C and (black square) temperature 2 ± 2 °C
Fig. 3.
Microwave treatment of Salmonella enterica contaminated ready-to-eat poultry meat, (black circle) 30–50 g, (black up-pointing triangle) 60–90 g and (black square) 100–150 g
Fig. 4.
Survival rate of Salmonella enterica in simulated gastric fluids, (black circle) pH 2, (black square) pH3, (black up-pointing triangle) pH4 and (x) pH 7
Discussion
Salmonella enterica is an important pathogen with a huge contribution in Salmonellosis globally and its prevalence is mostly associated to the food of animal origin (Aslama et al. 2012). Classical culturing methods assisted with DNA based confirmation is a gold tool for pathogen’s identification. Detection based on 16 s rDNA is a reference method for bacterial identification and taxonomic studies (Mignard and Flandrois 2006). Chomvarin et al. (2006) reported that 78 % of RTE food tested in Thailand did not qualify the microbiological standard. They also reported that 4.3 % of the samples were positive for Salmonella. Boonmar et al. (1998) reported that the most common serovar in RTE foods of poultry origin was S. enteritidis, prevalent 17.1 % in year 1993, 33.8 % in 1994, 29.6 % in 1995 and 15 % in 1996 of the total isolates from frozen poultry meats. The serovars isolated from RTE Thai food including S. anatum and S. derby, were also detected in frozen chicken meat. The results from this study are in agreement. Cabedo et al. (2008) reported 1.5 % Salmonella prevalence in RTE frozen chicken croquettes from Spain. Angkititrakul et al. (2005) reported 75 % Salmonella prevalence in retail chicken meat samples in Khon Kaen Provence of Thailand. Khaitsa et al., (2007) reported 1.1 % Salmonella contamination in RTE turkey meat products from USA. Monitoring and control of Salmonella in RTE meat products are an important task, as the results of the present and all reported studies confirming the presence of different Salmonella species from RTE food products in Thailand and other region of the world.
Sulfamethoxazole-trimethoprim previously considered as a drug of choice for diarrheal infection are encountered ineffective against some Salmonella isolated from Salmonellosis patients. Such resistance is the cause of therapeutic failure against the pathogen, and increase infection severity and recovery time (Angkititrakul et al. 2005). Aarestrup et al. (2007) reported 89 % of Salmonella isolates from chicken meat in Thailand, were found resistance to nalidixic acid. The S. enterica isolate in the current study also showed resistance against nalidixic acid. Angkititrakul et al. (2005) reported in a study from Thailand, that none of the Salmonella isolates were found resistant to norfloxacin and ciprofloxacin. All isolates from chicken sources were found resistant to streptomycin, sulfamethoxazole and tetracycline. In a similar study, Addis et al. (2011) found 100 % resistance of Salmonella towards ampicillin. They found 66.7 %, and 58.3 % resistant to streptomycin and nitrofurantoine respectively. Emergence of multidrug resistance Salmonella is thought to be the result of excessive antimicrobials use in animal farming (Aslama et al. 2012). Extensive use, over the counter availability and unrestricted purchase of common antimicrobials in Thailand for human and animals uses are contributing in creating resistant strains (Angkititrakul et al. 2005). The single pathogenic strain isolated from poultry meat in this study showed 50 % resistance against the antibiotics used. Prevalence of drug resistant foodborne pathogens are of great concern and need more attention towards its control.
Usual practices of showcase temperature (4–6 °C) for RTE products in super markets were observed during the collection of samples for the prevalence study. Storage temperature is a critical point for contaminated meat products, the pathogens can multiply at temperature above 4 °C. As a 2 log cycle increase of S. enterica was observed in this study at temperature 6 ± 2 °C. The storage temperature below 4 °C is not only stunting the growth of pathogens, but also decreases the number with passage of time. Matches and Liston (1968) reported the minimum growth temperature 5.3 °C for S. Heidelberg in nutrient broth, whereas 6.2 and 6.9 °C for S. typhimurium and S. derby respectively after 19 days of incubation. Morey and Singh (2012) reported that the survival of Salmonella at 4 °C and 10 °C was different and the near refrigeration temperature can be helpful for Salmonella reduction in chicken carcases during storage. Bhat et al. (2013) reported that, the total aerobic microbial load increased (p < 0.05) in RTE chicken seekh kebabs stored for 21 days at refrigerating temperature (4 ± 1 °C). These finding are in agreement with our study. Rajan et al. (2014) reported that, no E. coli, Salmonella spp, Clostridium spp, Staphylococci spp, Streptococci spp, yeast and mould were detected in Chettinad chicken processed at high temperature (121.1 °C) during its storage at ambient temperature (35 ± 1 °C).
Re-heating of chilled RTE meat products with microwave oven becomes usual practices at home and departmental stores. It is desirable in case of chilled RTE meat products. It not only affects the physical nature of the meat products but can also reduce the number of unwanted bacteria. Jamshidi et al. (2009) reported that 35 s microwave (850 W) treatment is effective against S. typhimurium inoculated to chicken meat. Woo et al. (2000) reported 5 logs cycle reduction of E. coli suspension exposed to microwave radiation (600 W) for 80 s. The microwave radiation which produces an internal heat of 85 °C could eliminate S. typhimurium (Schnepf and Barbeau 2007). The reduction of pathogen in microwave treatment is greatly associated with the shape, size of materials and wattage of microwave oven (Apostolou et al. 2005). Our findings are in agreement with their study. Similar relation among shape, size, microwaving time and microbial dose were observed in our study. Salmonella enterica showed significant growth stability in the SGF of low pH in this study. The growth reduction of S. enterica to some extent was observed only in SGF with pH 2.
Salmonella is known to be an enteric pathogen with a great ability of causing enteric fever worldwide (Akbar and Anal 2013). Oliveiraa et al. (2011) reported 8 % reduction of S. typhimurium DT104 population after 30 min of incubation in SGF of pH 3.5. Perez et al. (2010) reported fast reduction (5.5 logs in 5 min) of S. typhimurium DT104 in SGF with pH 1.5. Intestinal pH and environment can vitally affect the number of pathogens ingested. The present study revealed that the Salmonella enterica isolated from RTE poultry meat can survive during the gastrointestinal passage along with contaminated food materials.
Conclusion
Investigating the presence of pathogenic bacteria in food products particularly on the consumer counter becomes essential to control its presence and to establish effective prevention mechanisms. The presence of Salmonella in RTE chilled poultry meat was confirmed in the study. It is a potential threat to consumer health in case of its presence in RTE chilled foods particularly and other generally. As such foods are of great demand because of its easy use and time saving for consumption. Such foods are nominally heated before consumption, which may not be appropriate for elimination of the pathogenic bacteria load in contaminated products. The reported prevalence rate of Salmonella in chilled RTE poultry meat is still low but need immediate attention due to its vulnerability and emerging resistance patterns to antibiotics. Salmonella enterica isolated from RTE poultry meat exhibits drug resistant towards commonly used antibiotics, the impact of irregular use of antimicrobials. It is concluded that showcasing and storage temperature is critical for pathogens growth. The 90 s or above microwaving time for re-heating RTE poultry meat products is necessary for Salmonella risk reduction.
Acknowledgments
The authors acknowledge University of Balochistan, Pakistan and Asian Institute of Technology, Thailand for providing fund to one of the author Ali Akbar for conducting this work.
References
- Aarestrup MF, Hendriksen SR, Lockett J, Gay K, Teates K, McDermott FP, White GD, Hasman H, Sørensen G, Bangtrakulnonth A, Pornreongwong S, Pulsrikarn C, Angulo JF, Smidt GP. International spread of multidrug-resistant Salmonella Schwarzengrund in food products. Emerg Infect Dis. 2007;13(5):726–731. doi: 10.3201/eid1305.061489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addis Z, Kebede N, Sisay Z, Alemayehu H, Yirsaw A, Kassa T. Prevalence and antimicrobial resistance of Salmonella isolated from lactating cows and in contact humans in dairy farms of Addis Ababa: a cross sectional study. BMC Infect Dis. 2011;11:222. doi: 10.1186/1471-2334-11-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbar A, Anal KA. Food safety concerns and food-borne pathogens, Salmonella, Escherichia coli and Campylobacter. FUUAST J Biol. 2011;1(1):5–17. [Google Scholar]
- Akbar A, Anal KA. Prevalence and antibiogram study of Salmonella and Staphylococcus aureus in poultry meat. Asian Pac J Trop Biomed. 2013;3(2):163–168. doi: 10.1016/S2221-1691(13)60043-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbar A, Anal KA. Occurrence of Staphylococcus aureus and evaluation of anti-staphylococcal activity of Lactococcus lactis subsp. lactis in ready-to-eat poultry meat. Ann Microbiol. 2014;64(1):131–138. doi: 10.1007/s13213-013-0641-x. [DOI] [Google Scholar]
- Akbar A, Anal KA. Zinc oxide nanoparticles loaded active packaging a challenge study against Salmonella typhimurium and Staphylococcus aureus in ready- to- eat poultry meat. Food Cont. 2014;38:88–95. doi: 10.1016/j.foodcont.2013.09.065. [DOI] [Google Scholar]
- Altschul SF, Madden TL, Schaeffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angkititrakul S, Chomvarin C, Chaita T, Kanistanon K, Waethewutajarn S. Epidemiology of antimicrobial resistance in Salmonella isolated from pork, chicken meat and humans in Thailand. Southeast Asian J Trop Med Public Health. 2005;36(6):510–1515. [PubMed] [Google Scholar]
- Apostolou I, Papadopoulou C, Levidiotou S, Ioannides K. The effect of short-time microwave exposures on Escherichia coli O157:H7 inoculated onto chicken meat portions and whole chickens. Int J Food Microbiol. 2005;101:105–110. doi: 10.1016/j.ijfoodmicro.2004.10.043. [DOI] [PubMed] [Google Scholar]
- Aslama M, Checkley S, Avery B, Chalmers G, Bohaychuk V, Gensler G, Smith R, Richard BP. Phenotypic and genetic characterization of antimicrobial resistance in Salmonella serovars isolated from retail meats in Alberta Canada. Food Microbiol. 2012;32:110–117. doi: 10.1016/j.fm.2012.04.017. [DOI] [PubMed] [Google Scholar]
- Bhat ZF, Pathak V, Fayaz H. Effect of refrigerated storage on the quality characteristics of microwave cooked chicken seekh kababs extended with different non-meat proteins. J Food Sci Tech. 2013;50(5):926–933. doi: 10.1007/s13197-011-0410-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonmar S, Bangtrakulnonth A, Pornrunangwong S, Marnrim N, Kaneko K, Ogawa M. Predominant serovars of Salmonella in humans and foods from Thailand. J Vet Med Sci. 1998;60(7):877–80. doi: 10.1292/jvms.60.877. [DOI] [PubMed] [Google Scholar]
- Buranarom A (2010) Proteomic analysis of lipase producing soil-isolated bacteria. Thesis report Chiang Mai University Thailand
- Cabedo L, Picart i Barrot L, Teixidó i Canelles A. Prevalence of Listeria monocytogenes and Salmonella in ready-to-eat food in Catalonia, Spain. J Food Prot. 2008;71(4):855–9. doi: 10.4315/0362-028x-71.4.855. [DOI] [PubMed] [Google Scholar]
- Chomvarin C, Chantarasuk Y, Srigulbutr S, Chareonsudjai S, Chaicumpar K. Enteropathogenic bacteria and enterotoxin-producing Staphylococcus aureus isolated from ready-to-eat foods in Khon Kaen, Thailand. Southeast Asian J Trop Med Public Health. 2006;37(5):983–990. [PubMed] [Google Scholar]
- Chun J, Lee J-H, Jung Y, Kim M, Kim S, Kim BK, Lim YW. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol. 2007;57:2259–2261. doi: 10.1099/ijs.0.64915-0. [DOI] [PubMed] [Google Scholar]
- Fashae K, Ogunsola F, Aarestrup MF, Hendriksen SR. Antimicrobial susceptibility and serovars of Salmonella from chickens and humans in Ibadan, Nigeria. J Infect Dev Countr. 2010;4(8):484–494. doi: 10.3855/jidc.909. [DOI] [PubMed] [Google Scholar]
- Fernandez ÁE, Calleja AC, Fernández GC, Capita R. Prevalence and antimicrobial resistance of Salmonella serotypes isolated from poultry in Spain: Comparison between 1993 and 2006. Int J Food Microbiol. 2012;153:281–287. doi: 10.1016/j.ijfoodmicro.2011.11.011. [DOI] [PubMed] [Google Scholar]
- Hall TA. Bio Edit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hur J, Kim HJ, Park HJ, Lee JY, Lee HJ. Molecular and virulence characteristics of multi-drug resistant Salmonella enteritidis strains isolated from poultry. Vet J. 2011;189:306–311. doi: 10.1016/j.tvjl.2010.07.017. [DOI] [PubMed] [Google Scholar]
- Jamshidi A, Ghasemi A, Mohammadi A. The effect of short-time microwave exposures on Salmonella typhimurium inoculated onto chicken drumettes. Iran J Vet Res. 2009;10(4):378–382. [Google Scholar]
- Khaitsa ML, Kegode RB, Doetkott DK. Occurrence of antimicrobial-resistant Salmonella species in raw and ready to eat turkey meat products from retail outlets in the midwestern United States. Foodborne Pathog Dis. 2007;4(4):517–25. doi: 10.1089/fpd.2007.0010. [DOI] [PubMed] [Google Scholar]
- Lammerding MA, Paoli MG. Quantitative risk assessment: An emerging tool for emerging foodborne pathogens. Emerg Infect Dis. 1997;3(4):483–487. doi: 10.3201/eid0304.970411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matches JR, Liston J. Low temperature growth of Salmonella. J Food Sci. 1968;33:641–645. doi: 10.1111/j.1365-2621.1968.tb09092.x. [DOI] [Google Scholar]
- Mignard S, Flandrois JP. 16S rRNA sequencing in routine bacterial identification: a 30-month experiment. J Microbiol Methods. 2006;67:574–581. doi: 10.1016/j.mimet.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Morey A, Singh M. Low-temperature survival of Salmonella spp. in a model food system with natural microflora. Foodborne Pathog Dis. 2012;9(3):218–23. doi: 10.1089/fpd.2011.1016. [DOI] [PubMed] [Google Scholar]
- Oliveiraa M, Wijnands L, Abadias M, Aarts H, Franz E. Pathogenic potential of Salmonella typhimurium DT104 following sequential passage through soil, packaged fresh-cut lettuce and a model gastrointestinal tract. Int J Food Microbiol. 2011;148:149–155. doi: 10.1016/j.ijfoodmicro.2011.05.013. [DOI] [PubMed] [Google Scholar]
- Parry CM. Epidemiological and clinical aspects of human typhoid fever: in Salmonella infections clinical, immunological and molecular aspects. In: Mastroeni P, Maskell D, editors. Advances in molecular and cellular microbiology 9. New York: Cambridge University Press; 2006. pp. 1–17. [Google Scholar]
- Perez KJ, Ceccon RV, Da Silva MP, Jong EV, Cesar TE. Influence of acid adaptation on the survival of Salmonella enteritidis and Salmonella typhimurium in simulated gastric fluid and in Rattus Norvegicus intestine infection. J Food Safety. 2010;30:398–414. doi: 10.1111/j.1745-4565.2010.00215.x. [DOI] [Google Scholar]
- Rajan S, Kulkarni VV, Chandirasekaran V. Preparation and storage stability of retort processed Chettinad chicken. J Food Sci Tech. 2014;51(1):173–177. doi: 10.1007/s13197-011-0477-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Moktan B, Sarkar PK. Survival and growth of foodborne bacterial pathogens in fermenting dough of wadi, a legume-based indigenous food. J Food Sci Tech. 2011;48(4):506–509. doi: 10.1007/s13197-010-0185-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schnepf M, Barbeau W. Survival of Salmonella typhimurium in roasting chickens cooked in a microwave, convection microwave, and a conventional electric oven. J Food Safety. 2007;9:245–252. doi: 10.1111/j.1745-4565.1989.tb00524.x. [DOI] [Google Scholar]
- Skov NM, Andersen SJ, Aabo S, Ethelberg S, Aarestrup MF, Sørensen HA, Sørensen G, Pedersen K, Nordentoft S, Olsen EPK, Smidt GP, Baggesen LD. Antimicrobial drug resistance of Salmonella isolates from meat and humans, Denmark. Emerg Infect Dis. 2007;13(4):638–641. doi: 10.3201/eid1304.060748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 2004;101:1030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Neim M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai HT, Hirai T, Lan TN, Yamaguchi R. Antibiotic resistance profiles of Salmonella serovars isolated from retail pork and chicken meat in North Vietnam. Int J Food Microbiol. 2012;156:147–151. doi: 10.1016/j.ijfoodmicro.2012.03.016. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo IS, Rhee IK, Park HD. Differential damage in bacterial cells by microwave radiation on the basis of cell wall structure. Appl Environ Microbiol. 2000;66:2243–2247. doi: 10.1128/AEM.66.5.2243-2247.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarate G, Gonzalez S, Chaia PA. Assessing survival of dairy propioibacteria in gastrointestinal conditions and adherence to intestinal epithelia. In: Spencer JFT, Ragout de Spencer AL, editors. Public health microbiology: methods and protocols. New Jersey: Humana Press Inc; 2004. pp. 423–432. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]