Abstract
Purpose
Asthma is a complex disease caused by interplay of genes and environment on the genome of an individual. Copy number variations (CNVs) are more common compared to the other variations that disrupt genome organization. The effect of CNVs on asthma subgenome has been less studied compared to studies on the other variations. We report the assessments of CNV burden in asthma genes of normal cohorts carried out in different geographical areas of the world and discuss the relevance of the observation with respect to asthma pathogenesis.
Methods
CNV analysis was performed using Affymerix high-resolution arrays, and various bioinformatics tools were used to understand the influence of genes on asthma pathogenesis.
Results
This study identified 61 genes associated with asthma and provided various mechanisms and pathways underlying asthma pathogenesis. CCL3L1, ADAM8, and MUC5B were the most prevalent asthma genes. Among them, CCL3L1 was found across all 12 populations in varying copy number states. This study also identified the inheritance of asthma-CNVs from parents to offspring creating the latent period for manifestation of asthma.
Conclusions
This study revealed CNV burden with varying copy number states and identified susceptibility towards the disease manifestation. It can be hypothesized that primary CNVs may not be the initiating event in the pathogenesis of asthma and additional preceding mutations or CNVs may be required. The initiator or primary CNVs sensitize normal cohorts leading to an increased probability of accumulating mutations or exposure to allergic stimulating agents that can augment the development of asthma.
Keywords: Asthma, DNA copy number variation, inheritance pattern, CCL3L1, genetic markers
INTRODUCTION
Asthma is a complex disease involving a combination of genetic and environmental interactions that culminates in inflammation involving cells of the innate system enhancing the production of cytokines leading to phenotypes of bronchial hyperresponsiveness and atopy.1 The global prevalence of asthma is increasing and at present, ~235 million people are suffering from this disease.2 This widespread disease is characterized by increased IgE levels and airway hyperresponsiveness.3
Linkage studies have identified consistently replicated regions for asthma susceptibility on chromosomes 2q, 5q, 6p, 12q, and 13q, while other regions that have been reproducible in at least 2 populations include 1p, 2p, 3p, 4q, 7p, 11q, 16p-q, 17q, 19q, and 20p. These loci were identified in samples covering African-Americans, Hispanics, Dutch, Caucasians, Hutterites, Netherlands, German, and Swedish families.4,5,6 Genetic studies have identified numerous genes reproducibly associated with asthma, yet these studies have focused almost entirely on single nucleotide polymorphisms (SNPs), ignoring the contribution from the other form of large variations, such as copy number variations (CNVs).7 Differences between the genotypes of any 2 individuals occur mainly because of SNPs and CNVs.
Among the variations, CNVs are segments longer than 1 kb up to several Mbs, which contribute to a large number of nucleotide variations, compared to SNPs. CNVs are prone to cause instability and rearrangements leading to genomic disorders.8,9 Several genome-wide screenings for CNVs have demonstrated that deletions and duplications that disrupt gene functioning are more found in patients than in healthy subjects.10,11 The whole copy of gene sequence or partial gene sequence disruption leads to abnormalities in protein expression leading to disease manifestation.12 However, copy number (CN) state of a CNV plays a significant role in determining the zygosity of alleles.13 CNVs alter the genome by exon shuffling, disrupting coding sequences, modify the dosage of genes, and disturbing the regulation mechanism of gene expression. A previous study reveals the association of CNVs with asthma.14 Therefore, the present study aimed to identify CNV burden on randomly selected control cohorts and to assess the prevalence of primary hit CNVs in asthma-causing genes in normal cohorts predisposing them to asthma.
MATERIALS AND METHODS
Subjects and genotyping
The study included a total of 1,715 individuals excluding QC-failed sample sets of the populations, involving 38 Indian samples, 269 HapMap samples, 31 Tibetan samples, 155 Chinese samples, 464 of Ashkenazi Jews (AJ) replicate I (AJ-I), 480 of AJ replicate II (AJ-II), 184 individuals from Taiwan, 53 from Australia, and 41 from New World populations (Totonacs and Bolivians), were selected for CNV analysis in the genome. The HapMap 269 individuals' sample data from the 4 populations was obtained from the International HapMap Consortium, covering CEU (CEPH collection), CHB (Han Chinese in Beijing, China), JPT (Japanese in Tokyo, Japan), and YRI (Yoruba in Ibadan, Nigeria) populations.15 The raw, unprocessed data from Affymetrix Genome Wide SNP 6.0 array for all the remaining populations except India were obtained from the ArrayExpress Archive of the European Bioinformatics Institute. The Indian study group includes the members from randomly selected 12 families residing in Karnataka, India with different age group members ranging from 13 to 73 years. EDTA blood (5 mL) was collected from each member of the Indian study group, and genomic DNA was extracted using the Promega Wizard® Genomic DNA purification kit (Promega Corporation, Madison, WI, USA). The isolated DNA was quantified by Bio-photometer and gel electrophoresis. This research was approved by the University of Mysore Institutional Human Ethics review committee (IHEC). Written informed consent was obtained from all sample donors and the IHEC approved the sample consent procedure. Written informed consent was obtained from parents/guardians in the cases of participants being minors. For genotyping, CN state algorithm calling, and data analysis, we followed the methods of Veerappa et al.16
Gene ontology and pathway analysis
The selection of asthma-associated and causal genes was done through a literature survey. Genes identified under the CNVs of the study population were classified based on the biological process, molecular function, and cellular components using the EnrichR tool. EnrichR is a state-of-the-art gene set enrichment analysis, which includes application for gene-set libraries, rank-based enriched terms. EnrichR provides access to ~35 gene-set libraries, such as from the encyclopedia of DNA elements project, Epigenomics Project for histone modifications, and mouse phenotypes from the mouse genome informatics using the mammalian phenotype; structural domains; protein-protein hubs; protein complexes; kinase substrates; differentially phosphorylated proteins from the stable isotope labeling by amino acids in cell culture experiments; differentially expressed genes after approved drug perturbations; and virus-host protein interactions.17
Asthma pathway analysis was performed using WebGestalt "WEB-based GEne SeT AnaLysis Toolkit," a web-based enrichment analysis tool. This tool contains an integrated Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway. The hypergeometric statistical method with the Benjamini-Hochberg procedure was applied as a multiple test adjustment to calculate the false discovery rate (FDR) for each P value and significance level of top 10. The KEGG Pathway organizes genes based on the pathways associated with the gene set and on the number of genes in each pathway. The KEGG pathway also provides P values, indicating the significance of enrichment and each pathway map contains genes from the gene set that are highlighted in red.18
Breakpoint validations
To validate some of the recurring CNVs with the break points 195276060 bp-195446910 bp on 3q29; 34695310 bp-34857998 bp on 15q14; 14594223 bp-15101046 bp on 21q11.2 and 16377650 bp-16635603 bp on 16p13.11 that were identified from this study, PCR was performed on 400 randomly chosen individuals for 4 CNV breakpoints. Chimeric primers flanking normal and deleted/duplicated sequences were designed so as to bind only to CNV breakpoint regions. Samples that do not contain these specific CNVs would fail to amplify. PCR products were analyzed by agarose gel electrophoresis. PCR amplification was performed on 2 human genomic DNA samples using DreamTaq polymerase incubated with primers at 95℃ for 3' followed by 35 cycles of 94℃ for 30'', 62°-68℃ for 30", and 72℃ for 5' in Kyratec PCR System (Kyratec, Queensland Australia). All PCR products were analyzed on 1% agarose gels and documented with a Vilber Lourmat Imaging system (Vilber Lourmat, Marne-la-Vallée cedex France).
RESULTS
Genome-wide genotyping
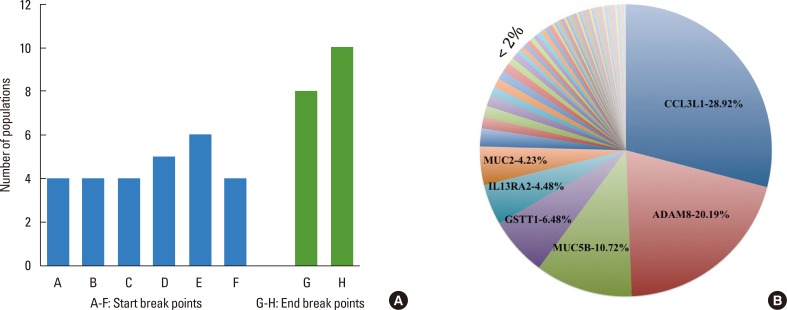
We began analysis by choosing 300 genes from the genome, which were known to be associated or causal genes for asthma. CNVs in the regions of the genome containing asthma genes were identified in 17% of subjects across the study populations. These CNVs were found in individuals across all populations ranging from 3% to 68% with the highest being identified in New World populations (68%), followed by samples from Tibet (65%), Australia (42%), YRI (30%), AJ-II (28%), JPT (22%), AJ-I (20%), Taiwan (16%), CHB (16%), China (15%), India (8%), and CEU (3%). We found 8 CNV break points that were conserved across multiple populations. Among them, 6 were start break points, and 2 were end break points on chromosomes 17 and 22 encompassing chemokine (C-C motif) ligand 3-like 1 (CCL3L1) and glutathione S-transferase theta 1 (GSTT1) genes (Fig. 1A). The 3 start break points "34435512 bp", "34439966 bp", and "34443800 bp" of CNVs encompassing CCL3L1 gene were present in 4 populations each, and 1 start break point was present in 5 populations "34437116 bp," and another was present in 6 populations "34528113 bp". Similarly, 1 start break point "24283004 bp" encompassing GSTT1 gene was present in 4 populations. An end break point "24396802 bp" with GSTT1 gene was present in 8 populations, and the other end break point "34629684 bp" embedding CCL3L1 gene was present in all the study populations. The populations AJ-I and AJ-II were considered 1 population; similarly, China and CHB were considered 1 population for this analysis because they arise from the same country.
Fig. 1.
(A) Conserved CNV break points among the study populations. Figs. A to F represent conserved start break points, and G-H represents conserved end break points. The start break points A- 34435512, B- 34439966, C- 34443800, D- 34437116, and E- 34528113 encompass CCL3L1 gene, and F- 24283004 encompasses GSTT1 gene. Similarly, the end break point G- 24396802 encompasses GSTT1 gene, and H- 34629684 encompasses CCL3L1 gene. (B) Distribution of frequency of genes obtained in the study populations. The highest percentage is observed in CCL3L1, followed by ADAM8, MUC5B, GSTT1, IL13RA2, and MUC2 in decreasing order, and the genes with frequency <2% are represented in a single slot.
Of the total 44,109 CNVs identified from 1,715 individuals, 61 singleton asthma genes were found in many individuals. These 61 genes were the singleton number of genes from 401 (includes number of same genes repeated in different individuals) genes of 294 individuals and 368 CNVs across 12 populations. Asthma genes were overembodied in duplication regions (72%) compared to deletion regions (28%) and were also distributed across populations in varying concentrations. Out of 61 asthma genes, 10 occurred in higher percentages (above 1%) among study populations; CCL3L1 was highest, followed by ADAM metallopeptidase domain 8 (ADAM8) and mucin 5B, oligomeric mucus/gel-forming (MUC5B) genes (Fig. 1B).
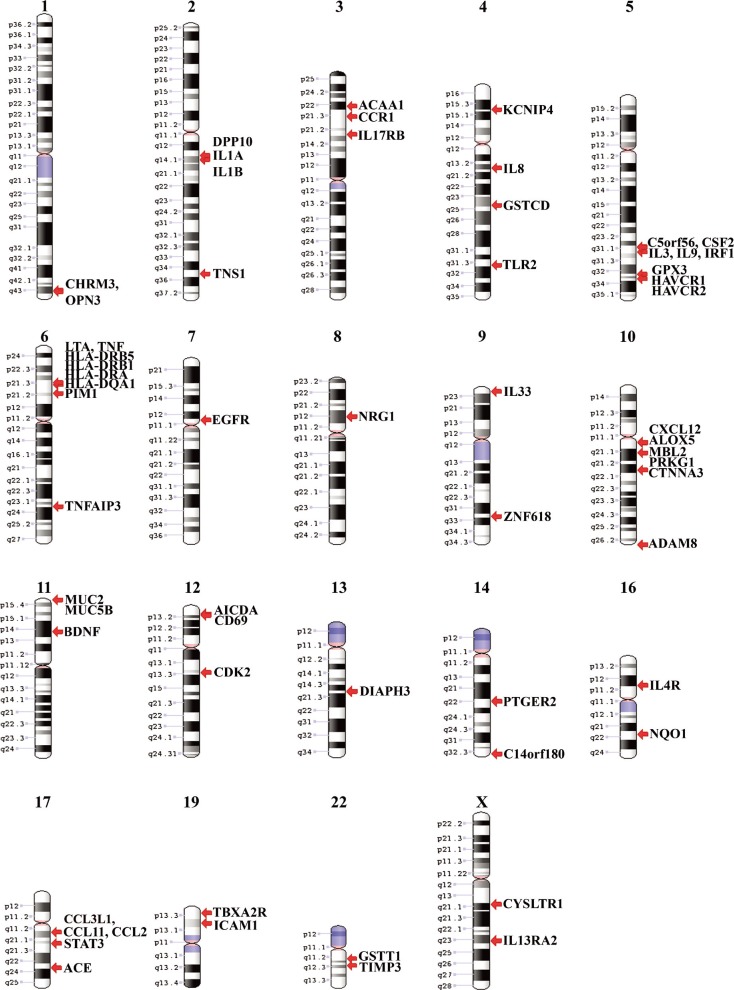
The 368 asthma-CNVs were found with an average size of 338 kb ranging from 100 to 425 kb. Variations were observed in almost all chromosomes except in 15, 18, 20, 21, and Y, and these variations were identified by ~14,000 combined SNP and CNV markers. Fig. 2 shows the singleton genes found in all 12 populations distributed in chromosomes 1-14, 16-17, 19, 22, and X. Among them, a cluster of asthma genes were embedded on chromosomes 5q, 6p, 10q, and 17q.
Fig. 2.
Karyogram indicating asthma genes in the study populations. It shows that 5q, 6p, 10q, and 17q regions are embedded with a higher percentage of asthma genes.
CNVs encompassed asthma genes both completely and partially across many genome regions; however, out of the 368 encompassing CNVs identified, ~78.66% were complete gene duplications/deletions, while ~21.33% partially truncated the coding structure. The CN state across these CNVs ranged from CN=0 to CN=4. CN state 3 was found at 60% (the highest prevalence), followed by CN=1 at 24.31%, CN=4 at 10.17%, and CN=0 at 3.22%.
Table 1 depicts the number and percentage of duplication and deletion frequency of 3 frequently occurring asthma genes under CNVs in all the populations under study. The expression levels of proteins were analyzed using the database of EBI (The European Bioinformatics Institute) expression atlas, which has baseline expression data. CCL3L1 showed a value of 0.7 in the lungs where the expression level breakdown for CCL3L1 is 1 out of 4 transcripts. CN=0 was the complete loss of protein, the CN=1 showing the dosage level of CCL3L1 protein was 0.35, CN=3 showed the expression level of 1.05, and CN=4 was present in 30 individuals with a protein level expression of 1.4 (Table 2). Similarly, ADAM8 showed the value of 8 with expression of 2 transcripts out of 11. This gene was under the burden of 2 types of CN states 0 and 3; among them CN=0 was complete loss of expression of this protein, and CN=3 was in 2 types of burden with 34 individuals carrying complete intact gene duplication with a dosage level of 12, but the remaining 46 individuals carried partial duplication of ADAM8 gene which showed a protein level of 4. Likewise, MUC5B showed a value of 15 by expressing 3 transcripts in the lungs out of 6 transcripts. This gene was under duplication burden with only 2 types of CN states 3 and 4, and all CNVs encompassed complete gene duplication. CN=3 showed a dosage level of 22.5, and CN=4 was present in 4 subjects with a dosage level of 30.
Table 1.
Duplication and deletion frequency of frequently occurring asthma genes under CNVs in 12 populations
| Population | Individuals assessed | CCL3L1 | ADAM8 | MUC5B | |||
|---|---|---|---|---|---|---|---|
| Duplication | Deletion | Duplication | Deletion | Duplication | Deletion | ||
| HapMap-YRI-Africa | 90 | 23 (25.5%) | - | - | 1 (1.1%) | - | - |
| HapMap-CEU-Europe | 90 | - | 3 (3.3%) | - | - | - | - |
| Ashkenazi Jews I | 464 | 3 (0.64%) | 9 (0.19%) | 22 (4.7%) | - | 13 (2.8%) | - |
| Ashkenazi Jews II | 480 | 2 (0.4%) | 23 (4.7%) | 38 (7.9%) | - | 29 (6.04%) | - |
| HapMap-CHB-China | 44 | 4 (9.09%) | - | - | - | - | - |
| China | 155 | 13 (8.3%) | 1 (0.64%) | - | - | - | - |
| Tibet | 31 | 7 (22.5%) | 1 (3.2%) | 5 (16.1%) | - | - | - |
| India | 38 | 1 (2.6%) | - | - | - | - | - |
| HapMap-JPT-Japan | 45 | 8 (17.7%) | - | - | - | - | - |
| Australia | 53 | 1 (1.8%) | 1 (1.8%) | 1 (1.8%) | - | 1 (1.8%) | - |
| New world | 41 | 1 (2.4%) | - | - | - | - | - |
| Taiwan | 184 | 8 (4.3%) | 7 (3.8%) | 14 (7.6%) | - | - | - |
CNV, copy number variation.
Table 2.
Impact of CNV type, copy number , and overlapping status on the protein dosage level of frequently occurring asthma genes with a cutoff value of 0.5
| Gene with baseline expression dosage in lungs | CNV type | Copy number | Number of subjects | Intact/partial | Protein dosage |
|---|---|---|---|---|---|
| CCL3L1 - 0.7 | Deletion | 0 | 1 | Intact | 0 |
| Deletion | 1 | 44 | Intact | 0.35 | |
| Duplication | 3 | 41 | Intact | 1.05 | |
| Duplication | 4 | 30 | Intact | 1.4 | |
| ADAM8 - 8 | Deletion | 0 | 1 | Intact | 0 |
| Duplication | 3 | 34 | Intact | 12 | |
| Duplication | 3 | 46 | Partial | 4 | |
| MUC5B - 15 | Duplication | 3 | 39 | Intact | 22.5 |
| Duplication | 4 | 4 | Intact | 30 |
CNV, copy number variation.
Interestingly, we also identified 67 individuals bearing multiple asthma genes under the influence of CNVs with varying CN states with either or sometimes both intact/partial CNV overlaps. CNVs that occurred with an overall frequency of less than 1% in the general population were defined as 'rare CNVs,' whereas those which were above 5% were reffered as 'common CNVs.' Asthma CNVs were both common and rare in occurrence when checked against the online database of genomic variants; further, common CNVs (91%) were highly prevalent than rare CNVs (9%). The AJ population showing the highest occurrence for common CNVs at 34%, while India and New World populations showing the highest occurrence (8%) of rare CNVs across all populations. CNV overlaps were encompassing genes either completely or partially across genomes of all populations. CNVs completely encompassing genes resulting in intact promoter and coding structure duplication/deletions were significantly higher (79%) compared to CNVs disrupting coding structure partially (21%). In addition, the CNV regions were further investigated to classify them as novel regions and breakpoints based on the CNV location and size. We identified 47 novel regions across the genome, which contained CNVs, not reported earlier in the database of genomic variants (http://dgv.tcag.ca/dgv/app/home), and also we identified 7 novel breakpoints in the recurrent CNV regions. These variations are now available in University of Mysore Genome Centre database (http://umgc.uni-mysore.ac.in). Novel regions and breakpoints were higher than novel breakpoints alone, with New World and Australia showing 15 and 12 novel regions and breakpoints, respectively. Meanwhile, HapMap, AJ-I, and Indian populations showed no CNVs in the novel regions bearing asthma genes.
Sex bias and inheritance in asthma genes under CNVs
CNVs in asthma genes were observed to be biased in male and female genomes in several populations (Supplementary Table 1). For inherited CNVs, we recognized 7 CNVs that were being inherited from parents to offspring. For this analysis, we used 3 populations, YRI, CEU, and India whose trio family data were available. YRI, with 30 children from 30 trio families were found with 7 CNVs encompassing asthma genes that were being inherited from the parents, while CEU and Indian populations did not contain inherited CNVs. All the 7 inheriting asthma CNVs were duplicative CNVs with both CN states 3 and 4. Five CNVs among them were being paternally inherited, while 2 were being maternally inherited.
Categorization of asthma genes
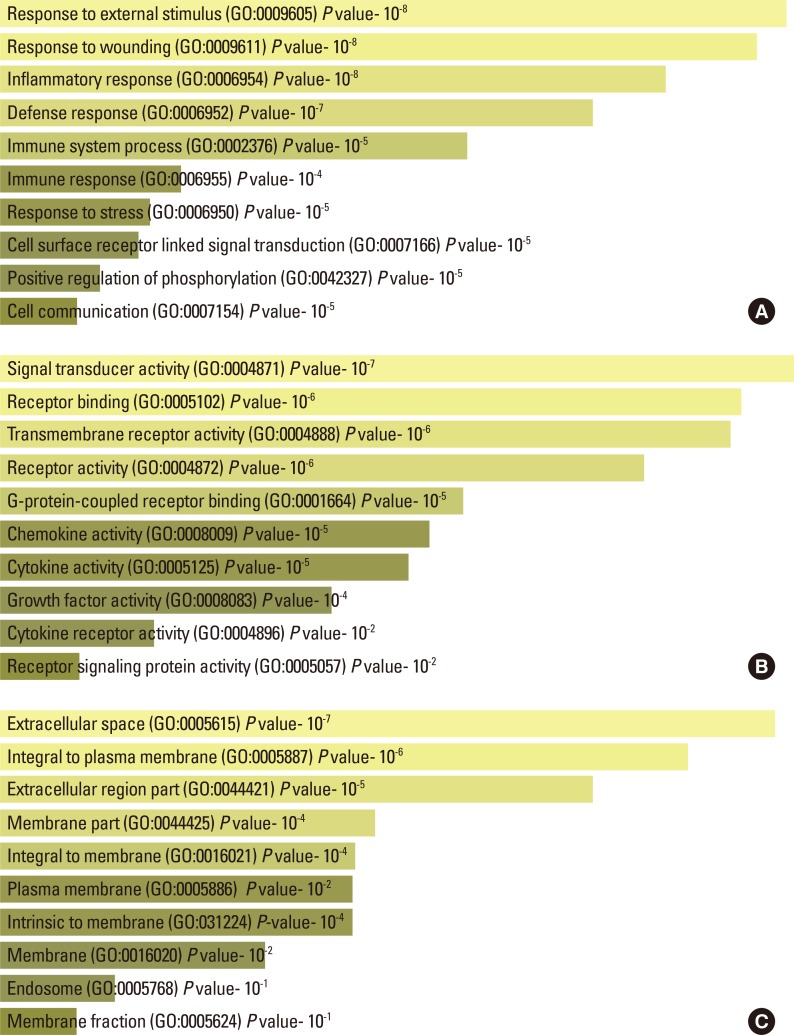
The identified 61 singleton genes were further divided into 3 major categories based on biological processes, molecular function, and cellular location using the EnrichR gene enrichment analysis tool. In biological processes, a majority of asthma genes were encoded for responses to external stimuli, responses to wound, and inflammatory/defense responses (Fig. 3A). Similarly, in the molecular function category, a majority of asthma genes were identified under signal transducer, receptor-binding, and growth factor activities (Fig. 3B), while under the cellular components the genes were coded for proteins that were located in the regions of extracellular space, integral to the plasma membrane and extracellular region (Fig. 3C).
Fig. 3.
Enrichment of asthma genes in the study populations. (A) Representation of genes under the biological process with P values. (B) Representation of genes under molecular function with P values. (C) Representation of genes under cellular components with P values.
Disease gene enrichment
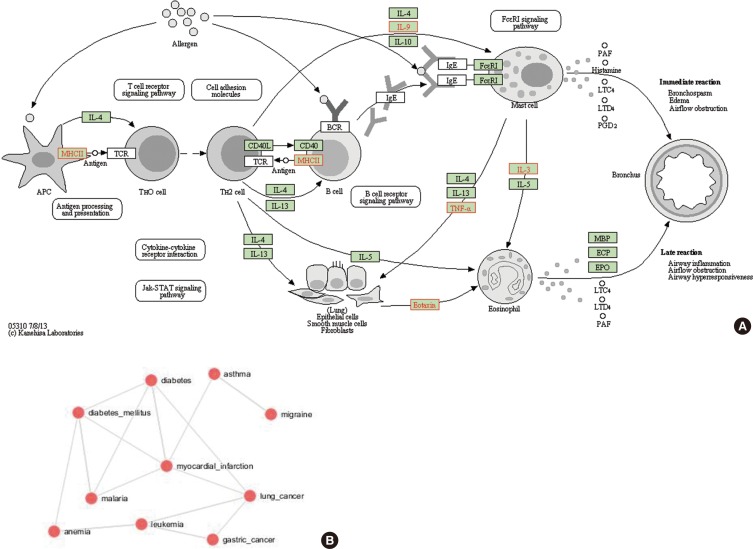
We examined identified genes to determine functional relevance and disease pathways relevant to asthma. Fig. 4A indicates the functional enrichment of pathways using the identified asthma-CNV genes. The top >20 ranked categories included functional and disease categories related to the FC-Epsilon-RI signaling pathway, and pathways for MHCII and eotaxin. These genes in these pathways, if altered, may result in asthma development. Supplementary Table 2 represents the function of genes with their common break points and CN states under study populations.
Fig. 4.
Representation of enrichment analysis. (A) Functional enrichment of the pathways in the study under the KEGG pathway for asthma. (B) CNV-asthma genes enriched for other diseases.
Genes of similar functional groups are expected to be enriched for gene ontology (Panther Classification and GORILLA annotations), share the same pathways (WikiPathways, KEGG and Pathway Commons), enabling the integrated use of these systems to establish biological, molecular, and cellular pathways based on the genes enriched in CNVs. Though the genes identified under this study were significantly enriched for asthma, they also overlapped among genes in both related and unrelated disorders. The statistical analysis to score gene enrichment on the pathway map revealed varying enrichment according to diseases. The genes identified in the study were enriched significantly for asthma, lung cancer, anemia, leukemia, diabetes, and myocardial infarction, indicating significant overlaps between genes for these diseases (Fig. 4B).
DISCUSSION
CNV burden on asthma genes
Organizational deviations that are generally occurring in the genome of individuals are CNVs. These variations occur in 2 forms, duplications and deletions conferring predominant variability than that of SNPs, inDels or chromosomal rearrangements. Large segmental duplications are presumed to be the root for CNVs, which are observed to be associated with diseases.7,19 The influence of CNV on asthma is comparatively less studied, but recent evidence strongly confirms the causality of asthma genes due to CNVs.20
In the present study, genes under duplicated CNVs were higher than those under deletion CNVs. Diseases have been shown to occur due to excess copy of dosage-sensitive genes and also by loss of gene function due to deletions.11 We assume that the genome can withstand more duplication than deletion, as deletions can be more harmful to the function and organization of the genome. A higher duplication state of the asthma genes indicates the role of high dosage of proteins playing a significant role towards the etiology of asthma pathogenesis. Similarly, CNVs completely encompassing the coding structure along with an intact promoter were found to be significantly higher compared to CNVs disrupting coding structure partially. The complete or partial CNVs in genes may affect transcription rates dynamically along with the effect on binding capacity of themselves along with other genes indirectly21 leading to disease manifestation. The presence of asthma genes under common CNVs compared to rare signifies the prevailing heavy burden of asthma CNVs in normal cohorts, indicating the presence of a latent period before being triggered by other notable variations making individuals more prone by providing a stronger effect on the phenotype leading to asthma pathogenesis.22,23
We came across 8 CNV breakpoints that are conserved in many populations in which highly frequent CNV embedding CCL3L1 has been selected to be under such conserving mechanism, YRI population being the point of origin of human evolution24 is carrying more of the conserved CNV breakpoints. Therefore, these CNVs could persist for a long period of time and probably expand to acquire further aberrations in encompassing gene or any other gene that falls under the same disease pathway.
These asthma genes are under the influence of CNVs, with varying CN states (Supplementary Table 1), which is one of the deciding factors, for the expression level of a protein. Ten genes were present at a frequency of >1% among the study populations. One of the highly abundant genes obtained in the study, CCL3L1 is under the influence of CN states-0, 1, 3, and 4. This is a chemokine and a potent regulator of inflammation, immune responses, and allergy. This gene was highest under CNVs at 28.92% with varying values of protein dosage level due to influence of different types of CN states. In AJ-I and AJ-II, out of 37 CNVs encompassing CCL3L1 gene, 31 were with CN state 1, 1 CNV with CN state 0 and the remaining 5 CNVs were with CN state 3. However, in YRI population, out of 23 CNVs, 13 were with CN state 3, and 4 CNVs were with CN state 4. The low level expression of CCL3L1 protein due to CN state 0 and 1 is associated with asthma.25 This result might help to provide one of the answers for the reason towards the susceptibility to asthma in AJ-I and AJ-II (Germany) populations compared to YRI (Nigeria). The prevalence of asthma in Nigeria is less compared to Germany.26 CCL3L1 acts by up-regulating the expression of an anti-inflammatory cytokine IL-10 in a dose dependent way, which in turn acts as a negative regulator of Th2 allergic responses during asthma pathogenesis.25 The normal level of 0.7 of CCL3L1 is required to regulate the normal level of IL-10, which is 0.5. In case of deletion in CCL3L1 with CN=1, the dosage level will be 0.35 which can regulate expression of only 0.25 level of IL-10 and hence the remaining 0.25 level of IL-10 is not expressed to keep up the normal level of IL-10. Hence, this low level of expression of IL-10 increases the incidence of asthma in individuals bearing deletion of CCL3L1 with CN=1. CNVs have been found in CCL3L1 in families with asthma,27 while deletion CNVs have been associated with asthma where the low level expression of CCL3L1 impacts the risk of asthma by modulating IL-10 expression. However, a high copy number of ≥5 is associated with decreased asthma risk. Subjects with ≤1 copy of CCL3L1 have significantly lower mRNA levels of CCL3L1 in peripheral blood cells and significantly higher serum IgE levels.25
ADAM8, which is present at a frequency of 20.19%, is a member of the disintegrin and metalloproteinase family of genes, which is high when IL-4 or IL-13 levels are increased during increased allergen-induced airway hyperresponsiveness. ADAM8 is present under CN=1 and 3 but CN=3 is in 2 forms because some individuals carry complete duplication of genes and others with partial duplication. The duplication of partial genes impacts the protein level expression in a reverse way by reducing the protein dosage rather than increasing it. However, in duplication of intact genes with CN=3, the dosage level will be 12 which is high compared to normal level of 8, thus in this duplicated level it might lead to airway hyperresponsiveness. ADAM8 gene might participate in the interplay of allergic responses with its member family genes like ADAM33.28 ADAM33 is a known candidate gene for the pathogenesis of asthma, even though the exact mechanism of interplay of this gene is not yet understood clearly but it is found to interact with multiple genes to manifest the disease.29,30 Paulissen et al.31 have shown recruitment of dendritic cells of the lung by ADAM8 and its overexpression leading to the production of CCL11 and CCL22 due to allergen exposure aiding in acute airway inflammation in asthma. Allergic asthma may also be a result of ADAM8 deregulation leading to regulatory modulation of PSGL-1 altering the expression and activation of ADAM8 during leukocyte extravasation.32 Previous studies have identified few SNPs in the ADAM8 to confer protection against allergic asthma and atopy.33 However, SNP associations in ADAM8 for asthma and atopy are found to be modest in magnitude.
Similarly, the next highest gene highest is MUC5B, which has 2 types of CN states 3 and 4, all being complete and intact gene duplications. Hence, there is a presence of high dosage level of this protein. MUC5B represents the 10.72% among the genes under the study, which is a mucin that produces mucous in small airways of the lungs and thus is involved in functional homeostatic clearance.34 With all the CNV embedding intact genes, the dosage level will be 22.5 with CN=3 and 30 with CN=4 compared to the normal level of 15. In both conditions, mucous secretion will be too high which might lead to airway thickness. These frequently occurring asthma genes can be used as biomarkers for early detection of susceptible individuals. These expression levels do not depend only on the burden of CNVs. However, CNVs may also significantly influence expression levels of these proteins. MUC5B contributes to a phenotype of distinct mucus hyper-secretion. Overexpression of MUC5B have been observed in asthma secretions, where a large volumes of MUC5B protein (form of mucus plugs) has been found in airway lumen, necks of glandular secretory ducts and in the lining of lung epithelia.35,36 The reasons for such overexpression have been mainly implicated on the presence of rare variants of MUC5B which operate by interacting with small amounts of MUC2 glycoprotein to produce mucin gel.37
We could notice 4 clusters of genes embedding on 5q, 6p, 10q, and 17q under CNV in this study. In multiple studies, 5q, 6p, and 17q are reported but 10q is less frequently associated.4 We also observed more than 1 asthma-CNV event in 67 individuals. There might be an increase in chances for the susceptibility to complex diseases upon exposure to environmental pollutants and multiple mutation incidences.38 Therefore, it can be predicted that the second hit of susceptible CNV in a person with preexisting susceptibility CNV makes the individual more prone to asthma.
Sex bias and inheritance in asthma genes under CNVs
We noticed some of the genes that are only in males of certain population and some only in females. However, these genes cannot be concluded to be gender- specific as in other populations showing balanced occurrences in both genders. As the sample size available in the present study is statistically insignificant, further studies are required to understand this phenomenon.
Studying the inheritance pattern revealed that CNVs encompassing asthma genes were inherited only in the YRI population with a frequency of 26%. We also found some asthma CNVs exhibiting the phenomenon of 1 breakpoint precisely were inherited, while the other break point was being slightly extended in the next generation, which was referred as "unknown" category of CNV inheritance.13 Previous studies indicted diverse CNV transmissions along with de novo event rates in probands for several neurodevelopmental phenotypes and monozygotic twin studies.39,40,41 Though the asthma CNV inheritance rate is comparatively lower than de novo, the role of inherited asthma CNVs towards CNV burden cannot be discounted. The "unknown" type of inheritance pattern may lead to the extension of sequences under CNVs in future generations and might cause more of gene sequences to be under CN burden. Hence, such kinds of CNV inheritance patterns are yet to be comprehended clearly to know the pathogenesis of diseases. There might also be presence of latent period in an individual who has susceptible asthma CNV that might be inherited from previous generations and due to second hit of susceptible CNV mutation or the extension of sequences of preexisting CNV might lead to manifestation of asthma.
Categorization of asthma genes
The identified asthma genes under CNV burden, which are subjected to enrichment analysis, revealed that, of the 61 genes under the biological process, majority were under the process of external stimulus followed by wounding, inflammatory, defense and immune responses. Under the cellular component, a majority of them were present in the extracellular space while some were found in various cellular locations, indicating that asthma-associated genes are not regionally specific and not biased in their location. Similarly, under the molecular function, more genes were found to participate in signal transducer activity, receptor binding, chemokine activity, and cytokine activity. Asthma depicts airway obstruction and wheezing episodes, which are the after-effects of chronic inflammation and airway narrowing. Inflammation is intensified by external stimuli and responses of smooth muscles, epithelial cells, and fibroblasts by producing different kinds of cytokines, chemokines, and proteases.42
Disease gene enrichment
There are several implicative signaling pathways for asthma disease pathogenesis. Asthma CNVs we found in the present study are involved in the FC-Epsilon-RI signaling pathway, antigen processing and presentation (MHCII), and pathway for eotaxin. The FC-Epsilon-RI signaling pathway is important to maintain the mast cell levels which are the principal component of hypersensitivity reaction.43 Antigen presenting and processing is also important in immune responses. The macrophage, which is an antigen-presenting cell, utilizes cell adhesion molecules to bind to T-cells where T-cell responses are crucial in the pathogenesis of asthma.44 Pathways, including mechanisms of eotaxin, are important for the recruitment of eosinophils to the location of allergic inflammation,45 where eosinophils are involved in inflammation of the lungs leading to bronchial hyperresponsiveness in case of asthma.46
The genes obtained in the study are significantly enriched for multiple diseases mainly for asthma followed by malaria, different types of cancers, myocardial infarction, diabetes, leukemia, anemia, migrane, alzheimer, immunodeficiency, schizophrenia, dementia, and Zellweger syndrome. This signifies that the role of CNVs burden on genes causing various types of complex disorders. These human disorders by occurring in such kind of disease network shows that they are causing due to genes of similar genetic origin and molecular function.47
Asthma is a complex disease, and its trigger is based on the susceptibility present in the genome of an individual towards environmental factors. This study helps design markers for the identification of susceptible individuals, as asthma cannot be identified unless and until the disease is manifested. CNVs alone may not be the reason for the manifestation of asthma, and the additional environmental factors or mutations may be required for the occurrence of asthma. Therefore, this study helps reveal CNV burden with varying CN states on normal cohort genome carried out in different geographical areas of the world and the relevance of the observation with respect to disease CNV burden, frequency, etiology, and pathogenesis of asthma.
Diseases have been shown to occur due to either excess or lack of copies of dosage-sensitive genes and also by loss of gene function due to deletions.11 A varying CN states of the asthma genes (ex: MUC5B in case of higher dosage and CCL3L1 in case of lower dosage) indicates the role of protein dosage playing a significant role in asthma pathogenesis. Since persons affected with deletion CNVs are more likely to have additional large CN variants than controls. The secondary CNVs are known to be associated with variable sets of outcomes, ranging from neuropsychiatric diseases to severe intellectual disability to certain autoimmune conditions and rare CN variants that are potentially pathogenic.48,49,50 Therefore, the present study was to identify the prevalence of primary hit CNVs in asthma-causing gene in normal cohorts predisposing them to asthma. It can be hypothesized that primary CNVs may not be the initiating event in the pathogenesis of asthma and additional preceding mutations or CNVs may be required. The initiator or primary CNVs sensitize the normal cohorts, leading to an increased probability of accumulating mutations or exposure to allergic stimulating agents that can augment the development of asthma.
ACKNOWLEDGMENTS
We thank the funding agencies, Indian Council of Medical Research (NCD-Ad-hoc/3/2011-12 dt. 27.4.11), New Delhi, India and Department of Science and Technology-Health Science (SR/SO/HS-103/2007), New Delhi, India; The Chairman, DOS in Zoology, University of Mysore; Prof. H.A. Ranganath and Prof. K.S. Rangappa for their support and encouragement; and JSS hospital and Medical college and University of Mysore, Mysore for providing facility to conduct this work. We also thank Dr. Kusuma Lingaiah, Ms. Megha Murthy, Mr. Raviraj VS, Mr. Somanna AN, Genetics, and Genomics lab, University of Mysore for their support.
Footnotes
There are no financial or other issues that might lead to conflict of interest.
Supplementary Materials
Supplementary Table 1. Frequency of asthma genes under CNVs in males and females of study population.
| Gene | YRI | CEU | AJ I | AJ II | CHB | China | Tibet | India | JPT | Australia | New world | Taiwan | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M | F | M | F | M | F | M | F | M | F | M | F | M | F | M | F | M | F | M | F | M | M | F | |
| ACAA1 | 1.0 | ||||||||||||||||||||||
| ACE | 0.4 | ||||||||||||||||||||||
| ADAM8 | 1.8 | 3.8 | 5.3 | 8.5 | 7.4 | 18.2 | 15 | 5.2 | 6.5 | 8.6 | |||||||||||||
| AICDA | 0.4 | 0.4 | |||||||||||||||||||||
| ALOX5 | 0.4 | ||||||||||||||||||||||
| BDNF | 9.0 | ||||||||||||||||||||||
| C14orf180 | 3 | ||||||||||||||||||||||
| C5orf56 | 2.4 | ||||||||||||||||||||||
| CCL11 | 0.4 | 0.4 | |||||||||||||||||||||
| CCL2 | 0.4 | 0.4 | |||||||||||||||||||||
| CCR1 | 4.8 | ||||||||||||||||||||||
| CCL3L1 | 23 | 30 | 5 | 2 | 3 | 2 | 5 | 5 | 14 | 5 | 11 | 7 | 55 | 10 | 5 | 13 | 23 | 6 | 2 | 4 | 50 | ||
| CD69 | 2.4 | ||||||||||||||||||||||
| CDK2 | 0.4 | ||||||||||||||||||||||
| CHRM3 | 4.5 | 6 | |||||||||||||||||||||
| CSF2 | 0.4 | ||||||||||||||||||||||
| CTNNA3 | 0.8 | 0.4 | 1.3 | ||||||||||||||||||||
| CXCL12 | 0.8 | 3 | 5.2 | ||||||||||||||||||||
| CYSLTR1 | 0.4 | ||||||||||||||||||||||
| DIAPH3 | 0.4 | ||||||||||||||||||||||
| DPP10 | 4.5 | ||||||||||||||||||||||
| EGFR | 0.4 | ||||||||||||||||||||||
| GPX3 | 0.8 | ||||||||||||||||||||||
| GSTCD | 0.4 | ||||||||||||||||||||||
| GSTT1 | 3.7 | 2.7 | 0.8 | 0.4 | 1.2 | 1.2 | 4.5 | 7.5 | 1.3 | 5 | 5.5 | 8.6 | 5.2 | 2.4 | |||||||||
| HAVCR1 | 0.4 | ||||||||||||||||||||||
| HAVCR2 | 0.4 | ||||||||||||||||||||||
| HLA-DQA1 | 0.4 | ||||||||||||||||||||||
| HLA-DRA | 5.2 | ||||||||||||||||||||||
| HLA-DRB1 | 0.4 | 0.4 | 9.0 | 5 | 5.2 | ||||||||||||||||||
| HLA-DRB5 | 0.8 | 0.4 | 9.0 | 5 | 15.7 | ||||||||||||||||||
| ICAM1 | 1.3 | ||||||||||||||||||||||
| IL13RA2 | 2.5 | 2.2 | 1.7 | 1.2 | |||||||||||||||||||
| IL17RB | 0.4 | 0.4 | |||||||||||||||||||||
| IL1A | 12.1 | ||||||||||||||||||||||
| IL1B | 12.1 | ||||||||||||||||||||||
| IL3 | 0.4 | ||||||||||||||||||||||
| IL33 | 0.4 | ||||||||||||||||||||||
| IL4R | 2.4 | ||||||||||||||||||||||
| IL8 | 7.3 | ||||||||||||||||||||||
| IL9 | 3 | ||||||||||||||||||||||
| IRF1 | 2.4 | ||||||||||||||||||||||
| KCNIP4 | 0.8 | 0.4 | 0.4 | ||||||||||||||||||||
| LTA | 4.8 | ||||||||||||||||||||||
| MBL2 | 0.4 | ||||||||||||||||||||||
| MUC2 | 0.8 | 1.3 | 2.9 | 2.0 | |||||||||||||||||||
| MUC5B | 3.3 | 2.2 | 8.1 | 3.7 | |||||||||||||||||||
| NQO1 | 0.4 | ||||||||||||||||||||||
| NRG1 | 5 | 5.5 | |||||||||||||||||||||
| OPN3 | 0.4 | 0.4 | |||||||||||||||||||||
| PIM1 | 1.3 | ||||||||||||||||||||||
| PRKG1 | 0.4 | ||||||||||||||||||||||
| PTGER2 | 0.4 | ||||||||||||||||||||||
| STAT3 | 4.8 | ||||||||||||||||||||||
| TBXA2R | 0.4 | ||||||||||||||||||||||
| TIMP3 | 3 | ||||||||||||||||||||||
| TLR2 | 0.4 | ||||||||||||||||||||||
| TNF | 4.8 | ||||||||||||||||||||||
| TNFAIP3 | 2.4 | ||||||||||||||||||||||
| TNS1 | 9 | 10.5 | |||||||||||||||||||||
| ZNF618 | 5.2 | ||||||||||||||||||||||
Supplementary Table 2. CNVs identified with asthma associated genes with their chromosomal location, break-points, CN states and gene functions.
| Gene | Chromosome location | Break-point (bp) | CN state | Gene function |
|---|---|---|---|---|
| ACAA1 | 3p22.2 | 38039344-38185174 | 3 | Acetyl-coA C-acyltransferase activity |
| ACE | 17q23.3 | 61518220-61624537 | 3 | Actin binding |
| ADAM8 | 10q26.3 | 135056464-135174110 | 0, 3 | Metalloendopeptidase activity |
| AICDA | 12p13.31 | 8529380-8763073 | 1 | Cytidinedeaminase activity |
| ALOX5 | 10q11.21 | 45827177-45940122 | 3 | Arachidonate 5-lipoxygenase activity |
| BDNF | 11p14.1 | 27636885-27789198 | 3 | Growth factor activity |
| C14orf180 | 14q32.33 | 104938308-105067722 | 3 | - |
| C5orf56 | 5q31.1 | 131721749-131825114 | 1 | - |
| CCL11 | 17q12 | 31953738-32936539 | 3 | Chemokine activity |
| CCL2 | 17q12 | 31957151-32938022 | 3 | Chemokine activity |
| CCR1 | 3p21.31 | 46129533-46281822 | 1 | Chemokine receptor activity |
| CCL3L1 | 17q12 | 34437116-34629684 | 0,1,3,4 | Chemotactic for lymphocytes and monocytes |
| CD69 | 12p13.31 | 9879084-9993452 | 1 | Transmembrane signaling receptor activity |
| CDK2 | 12q13.2 | 56297625-56413629 | 3 | Cyclin-dependent protein serine/threonine kinase activity |
| CHRM3 | 1q43 | 239917246-240298870 | 3 | G-protein coupled acetylcholine receptor activity |
| CSF2 | 5q31.1 | 131317464-131419202 | 3 | Cytokine and growth factor activity |
| CTNNA3 | 10q21.3 | 68367662-68515140 | 1, 3 | Cadherin binding |
| CXCL12 | 10q11.21 | 44860013-44975407 | 3 | Cytokine and growth factor activity |
| CYSLTR1 | Xq21.1 | 77513603-77753914 | 3 | Leukotriene receptor activity |
| DIAPH3 | 13q21.2 | 60204504-60381540 | 1 | Rho gtpase binding |
| DPP10 | 2q14.1 | 116532508-116675350 | 3 | Serine-type peptidase activity |
| EGFR | 7p11.2 | 55126900-55245507 | 3 | Epidermal growth factor-activated receptor activity |
| GPX3 | 5q33.1 | 150399114-150513847 | 3 | Glutathione peroxidase activity |
| GSTCD | 4q24 | 106447752-106760931 | 1 | - |
| GSTT1 | 22q11.23 | 24288422-24396802 | 0- 4 | Glutathione peroxidase activity |
| HAVCR1 | 5q33.3 | 156409111-156564456 | 3 | - |
| HAVCR2 | 5q33.3 | 156409111-156564456 | 3 | - |
| HLA-DQA1 | 6p21.32 | 32456671-32625083 | 1 | T cell receptor signaling pathway |
| HLA-DRA | 6p21.32 | 32408527-32530286 | 4 | T cell receptor signaling pathway |
| HLA-DRB1 | 6p21.32 | 32456671-32625083 | 1, 4 | Peptide antigen binding |
| HLA-DRB5 | 6p21.32 | 32429894-32530286 | 1, 4 | Peptide antigen binding |
| ICAM1 | 19p13.2 | 10369086-10471048 | 1 | Transmembrane signaling receptor activity |
| IL13RA2 | Xq23 | 114205618-114397728 | 2, 3 | Cytokine receptor activity |
| IL17RB | 3p21.1 | 53775447-53890848 | 3 | Cytokine receptor activity |
| IL1A | 2q13 | 113500222-113644927 | 1 | Cytokine activity |
| IL1B | 2q13 | 113512866-113644927 | 1 | Cytokine activity |
| IL3 | 5q31.1 | 131317464-131419202 | 3 | Cytokine activity |
| IL33 | 9p24.1 | 6121448-6256144 | 1 | Cytokine activity |
| IL4R | 16p12.1 | 27251081-27377242 | 1 | Interleukin-4 receptor activity |
| IL8 | 4q13.3 | 74567500-74758477 | 1 | Chemokine activity |
| IL9 | 5q31.1 | 135123627-135231195 | 3 | Cytokine activity |
| IRF1 | 5q31.1 | 131721749-131825114 | 1 | Sequence-specific DNA binding transcription factor activity |
| KCNIP4 | 4p15.2 | 21637793-21740358 | 3 | Voltage-gated ion channel activity |
| LTA | 6p21.33 | 31474688-31595882 | 1 | Cytokine activity |
| MBL2 | 10q21.1 | 53511985-57787459 | 1 | Calcium-dependent protein binding |
| MUC2 | 11p15.5 | 1073860-1332223 | 3 | Protein binding |
| MUC5B | 11p15.5 | 1150425-1291777 | 3, 4 | Extracellular matrix structural constituent |
| NQO1 | 16q22.1 | 68860698-69771914 | 1 | NAD(P)H dehydrogenase (quinone) activity |
| NRG1 | 8p12 | 32422268-32522771 | 3 | Cytokine and growth factor activity |
| OPN3 | 1q43 | 241099314-242090693 | 3 | G-protein coupled receptor activity |
| PIM1 | 6p21.2 | 37130282-37236436 | 3 | Protein serine/threonine kinase activity |
| PRKG1 | 10q21.1 | 53511985-57787459 | 1 | Calcium channel regulator activity |
| PTGER2 | 14q22.1 | 52788759-52902018 | 1 | Prostaglandin E receptor activity |
| STAT3 | 17q21.2 | 40498189-40608285 | 1 | Transcription regulatory region DNA binding |
| TBXA2R | 19p13.3 | 3554783-3700847 | 3 | Guanyl-nucleotide exchange factor activity |
| TIMP3 | 22q12.3 | 33142908-33251959 | 3 | Metalloendopeptidase inhibitor activity |
| TLR2 | 4q31.3 | 154515618-154825256 | 3 | Transmembrane signaling receptor activity |
| TNF | 6p21.33 | 31480272-31654732 | 1 | Cytokine activity |
| TNFAIP3 | 6q23.3 | 138092422-138206150 | 1 | Ubiquitin-specific protease activity |
| TNS1 | 2q35 | 218600074-218709313 | 3 | Actin and protein binding |
| ZNF618 | 9q32 | 116811184-116930194 | 3 | DNA binding |
References
- 1.Meng JF, Rosenwasser LJ. Unraveling the genetic basis of asthma and allergic diseases. Allergy Asthma Immunol Res. 2010;2:215–227. doi: 10.4168/aair.2010.2.4.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.International Union Against Tuberculosis and Lung Disease (FR) The global asthma report 2011. Paris: International Union Against Tuberculosis and Lung Disease; 2011. [Google Scholar]
- 3.Castro-Giner F, Kauffmann F, de Cid R, Kogevinas M. Gene-environment interactions in asthma. Occup Environ Med. 2006;63:776–786. 761. doi: 10.1136/oem.2004.019216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffjan S, Ober C. Present status on the genetic studies of asthma. Curr Opin Immunol. 2002;14:709–717. doi: 10.1016/s0952-7915(02)00393-x. [DOI] [PubMed] [Google Scholar]
- 5.Park HS, Kim SH, Park CS. The role of novel genes in modifying airway responses in asthma. Curr Allergy Asthma Rep. 2006;6:112–116. doi: 10.1007/s11882-006-0048-x. [DOI] [PubMed] [Google Scholar]
- 6.Zhang J, Paré PD, Sandford AJ. Recent advances in asthma genetics. Respir Res. 2008;9:4. doi: 10.1186/1465-9921-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ionita-Laza I, Rogers AJ, Lange C, Raby BA, Lee C. Genetic association analysis of copy-number variation (CNV) in human disease pathogenesis. Genomics. 2009;93:22–26. doi: 10.1016/j.ygeno.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee C, Iafrate AJ, Brothman AR. Copy number variations and clinical cytogenetic diagnosis of constitutional disorders. Nat Genet. 2007;39:S48–S54. doi: 10.1038/ng2092. [DOI] [PubMed] [Google Scholar]
- 9.Gonzaga-Jauregui C, Lupski JR, Gibbs RA. Human genome sequencing in health and disease. Annu Rev Med. 2012;63:35–61. doi: 10.1146/annurev-med-051010-162644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kitsiou-Tzeli S, Frysira H, Giannikou K, Syrmou A, Kosma K, Kakourou G, et al. Microdeletion and microduplication 17q21.31 plus an additional CNV, in patients with intellectual disability, identified by array-CGH. Gene. 2012;492:319–324. doi: 10.1016/j.gene.2011.10.023. [DOI] [PubMed] [Google Scholar]
- 13.Veerappa AM, Murthy MN, Vishweswaraiah S, Lingaiah K, Suresh RV, Nachappa SA, et al. Copy number variations burden on miRNA genes reveals layers of complexities involved in the regulation of pathways and phenotypic expression. PLoS One. 2014;9:e90391. doi: 10.1371/journal.pone.0090391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogers AJ, Chu JH, Darvishi K, Ionita-Laza I, Lehmann H, Mills R, et al. Copy number variation prevalence in known asthma genes and their impact on asthma susceptibility. Clin Exp Allergy. 2013;43:455–462. doi: 10.1111/cea.12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 16.Veerappa AM, Vishweswaraiah S, Lingaiah K, Murthy M, Manjegowda DS, Nayaka R, et al. Unravelling the complexity of human olfactory receptor repertoire by copy number analysis across population using high resolution arrays. PLoS One. 2013;8:e66843. doi: 10.1371/journal.pone.0066843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41:W77–W83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eichler EE. Copy number variation and human disease. Nat Educ. 2008;1:1. [Google Scholar]
- 20.Ferreira MA, McRae AF, Medland SE, Nyholt DR, Gordon SD, Wright MJ, et al. Association between ORMDL3, IL1RL1 and a deletion on chromosome 17q21 with asthma risk in Australia. Eur J Hum Genet. 2011;19:458–464. doi: 10.1038/ejhg.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mileyko Y, Joh RI, Weitz JS. Small-scale copy number variation and large-scale changes in gene expression. Proc Natl Acad Sci U S A. 2008;105:16659–16664. doi: 10.1073/pnas.0806239105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winchester L, Newbury DF, Monaco AP, Ragoussis J. Detection, breakpoint identification and detailed characterisation of a CNV at the FRA16D site using SNP assays. Cytogenet Genome Res. 2008;123:322–332. doi: 10.1159/000184724. [DOI] [PubMed] [Google Scholar]
- 23.Korn JM, Kuruvilla FG, McCarroll SA, Wysoker A, Nemesh J, Cawley S, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cavalli-Sforza LL. Genes, peoples, and languages. Proc Natl Acad Sci U S A. 1997;94:7719–7724. doi: 10.1073/pnas.94.15.7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee H, Bae S, Choi BW, Choi JC, Yoon Y. Copy number variation of CCL3L1 influences asthma risk by modulating IL-10 expression. Clin Chim Acta. 2011;412:2100–2104. doi: 10.1016/j.cca.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 26.Masoli M, Fabian D, Holt S, Beasley R Global Initiative for Asthma (GINA) Program. The global burden of asthma: executive summary of the GINA Dissemination Committee report. Allergy. 2004;59:469–478. doi: 10.1111/j.1398-9995.2004.00526.x. [DOI] [PubMed] [Google Scholar]
- 27.Kosta K, Sabroe I, Goke J, Nibbs RJ, Tsanakas J, Whyte MK, et al. A Bayesian approach to copy-number-polymorphism analysis in nuclear pedigrees. Am J Hum Genet. 2007;81:808–812. doi: 10.1086/520096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.King NE, Zimmermann N, Pope SM, Fulkerson PC, Nikolaidis NM, Mishra A, et al. Expression and regulation of a disintegrin and metalloproteinase (ADAM) 8 in experimental asthma. Am J Respir Cell Mol Biol. 2004;31:257–265. doi: 10.1165/rcmb.2004-0026OC. [DOI] [PubMed] [Google Scholar]
- 29.Holloway JW, Holgate ST. Identification and function of a Novel Candidate Gene for Asthma: ADAM 33. Allergol Int. 2005;54:25–30. [Google Scholar]
- 30.Mahesh PA. Unravelling the role of ADAM 33 in asthma. Indian J Med Res. 2013;137:447–450. [PMC free article] [PubMed] [Google Scholar]
- 31.Paulissen G, Rocks N, Guéders MM, Bedoret D, Crahay C, Quesada-Calvo F, et al. ADAM-8, a metalloproteinase, drives acute allergen-induced airway inflammation. Eur J Immunol. 2011;41:380–391. doi: 10.1002/eji.200940286. [DOI] [PubMed] [Google Scholar]
- 32.Domínguez-Luis M, Lamana A, Vazquez J, García-Navas R, Mollinedo F, Sánchez-Madrid F, et al. The metalloprotease ADAM8 is associated with and regulates the function of the adhesion receptor PSGL-1 through ERM proteins. Eur J Immunol. 2011;41:3436–3442. doi: 10.1002/eji.201141764. [DOI] [PubMed] [Google Scholar]
- 33.Knolle MD, Owen CA. ADAM8: a new therapeutic target for asthma. Expert Opin Ther Targets. 2009;13:523–540. doi: 10.1517/14728220902889788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evans CM, Kim K, Tuvim MJ, Dickey BF. Mucus hypersecretion in asthma: causes and effects. Curr Opin Pulm Med. 2009;15:4–11. doi: 10.1097/MCP.0b013e32831da8d3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Groneberg DA, Eynott PR, Lim S, Oates T, Wu R, Carlstedt I, et al. Expression of respiratory mucins in fatal status asthmaticus and mild asthma. Histopathology. 2002;40:367–373. doi: 10.1046/j.1365-2559.2002.01378.x. [DOI] [PubMed] [Google Scholar]
- 36.Morcillo EJ, Cortijo J. Mucus and MUC in asthma. Curr Opin Pulm Med. 2006;12:1–6. doi: 10.1097/01.mcp.0000198064.27586.37. [DOI] [PubMed] [Google Scholar]
- 37.Vinall LE, Fowler JC, Jones AL, Kirkbride HJ, de Bolós C, Laine A, et al. Polymorphism of human mucin genes in chest disease: possible significance of MUC2. Am J Respir Cell Mol Biol. 2000;23:678–686. doi: 10.1165/ajrcmb.23.5.4176. [DOI] [PubMed] [Google Scholar]
- 38.Nambiar M, Raghavan SC. Chromosomal translocations among the healthy human population: implications in oncogenesis. Cell Mol Life Sci. 2013;70:1381–1392. doi: 10.1007/s00018-012-1135-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ehli EA, Abdellaoui A, Hu Y, Hottenga JJ, Kattenberg M, van Beijsterveldt T, et al. De novo and inherited CNVs in MZ twin pairs selected for discordance and concordance on Attention Problems. Eur J Hum Genet. 2012;20:1037–1043. doi: 10.1038/ejhg.2012.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cohn L, Elias JA, Chupp GL. Asthma: mechanisms of disease persistence and progression. Annu Rev Immunol. 2004;22:789–815. doi: 10.1146/annurev.immunol.22.012703.104716. [DOI] [PubMed] [Google Scholar]
- 43.Stone KD, Prussin C, Metcalfe DD. IgE, mast cells, basophils, and eosinophils. J Allergy Clin Immunol. 2010;125:S73–S80. doi: 10.1016/j.jaci.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robinson DS. The role of the T cell in asthma. J Allergy Clin Immunol. 2010;126:1081–1091. doi: 10.1016/j.jaci.2010.06.025. [DOI] [PubMed] [Google Scholar]
- 45.Conroy DM, Williams TJ. Eotaxin and the attraction of eosinophils to the asthmatic lung. Respir Res. 2001;2:150–156. doi: 10.1186/rr52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Filipović M, Cekić S. The role of eosinophils in asthma. Facta Univ Ser Med Biol. 2001;8:6–10. [Google Scholar]
- 47.Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabási AL. The human disease network. Proc Natl Acad Sci U S A. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Itsara A, Wu H, Smith JD, Nickerson DA, Romieu I, London SJ, et al. De novo rates and selection of large copy number variation. Genome Res. 2010;20:1469–1481. doi: 10.1101/gr.107680.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walsh KM. Copy-number variation and the etiology of chronic diseases: computational detections, laboratory validations, and public health relevance [Dissertation] New Haven (CT): Yale University; 2011. [Google Scholar]
- 50.Girirajan S, Rosenfeld JA, Coe BP, Parikh S, Friedman N, Goldstein A, et al. Phenotypic heterogeneity of genomic disorders and rare copy-number variants. N Engl J Med. 2012;367:1321–1331. doi: 10.1056/NEJMoa1200395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Frequency of asthma genes under CNVs in males and females of study population.
| Gene | YRI | CEU | AJ I | AJ II | CHB | China | Tibet | India | JPT | Australia | New world | Taiwan | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M | F | M | F | M | F | M | F | M | F | M | F | M | F | M | F | M | F | M | F | M | M | F | |
| ACAA1 | 1.0 | ||||||||||||||||||||||
| ACE | 0.4 | ||||||||||||||||||||||
| ADAM8 | 1.8 | 3.8 | 5.3 | 8.5 | 7.4 | 18.2 | 15 | 5.2 | 6.5 | 8.6 | |||||||||||||
| AICDA | 0.4 | 0.4 | |||||||||||||||||||||
| ALOX5 | 0.4 | ||||||||||||||||||||||
| BDNF | 9.0 | ||||||||||||||||||||||
| C14orf180 | 3 | ||||||||||||||||||||||
| C5orf56 | 2.4 | ||||||||||||||||||||||
| CCL11 | 0.4 | 0.4 | |||||||||||||||||||||
| CCL2 | 0.4 | 0.4 | |||||||||||||||||||||
| CCR1 | 4.8 | ||||||||||||||||||||||
| CCL3L1 | 23 | 30 | 5 | 2 | 3 | 2 | 5 | 5 | 14 | 5 | 11 | 7 | 55 | 10 | 5 | 13 | 23 | 6 | 2 | 4 | 50 | ||
| CD69 | 2.4 | ||||||||||||||||||||||
| CDK2 | 0.4 | ||||||||||||||||||||||
| CHRM3 | 4.5 | 6 | |||||||||||||||||||||
| CSF2 | 0.4 | ||||||||||||||||||||||
| CTNNA3 | 0.8 | 0.4 | 1.3 | ||||||||||||||||||||
| CXCL12 | 0.8 | 3 | 5.2 | ||||||||||||||||||||
| CYSLTR1 | 0.4 | ||||||||||||||||||||||
| DIAPH3 | 0.4 | ||||||||||||||||||||||
| DPP10 | 4.5 | ||||||||||||||||||||||
| EGFR | 0.4 | ||||||||||||||||||||||
| GPX3 | 0.8 | ||||||||||||||||||||||
| GSTCD | 0.4 | ||||||||||||||||||||||
| GSTT1 | 3.7 | 2.7 | 0.8 | 0.4 | 1.2 | 1.2 | 4.5 | 7.5 | 1.3 | 5 | 5.5 | 8.6 | 5.2 | 2.4 | |||||||||
| HAVCR1 | 0.4 | ||||||||||||||||||||||
| HAVCR2 | 0.4 | ||||||||||||||||||||||
| HLA-DQA1 | 0.4 | ||||||||||||||||||||||
| HLA-DRA | 5.2 | ||||||||||||||||||||||
| HLA-DRB1 | 0.4 | 0.4 | 9.0 | 5 | 5.2 | ||||||||||||||||||
| HLA-DRB5 | 0.8 | 0.4 | 9.0 | 5 | 15.7 | ||||||||||||||||||
| ICAM1 | 1.3 | ||||||||||||||||||||||
| IL13RA2 | 2.5 | 2.2 | 1.7 | 1.2 | |||||||||||||||||||
| IL17RB | 0.4 | 0.4 | |||||||||||||||||||||
| IL1A | 12.1 | ||||||||||||||||||||||
| IL1B | 12.1 | ||||||||||||||||||||||
| IL3 | 0.4 | ||||||||||||||||||||||
| IL33 | 0.4 | ||||||||||||||||||||||
| IL4R | 2.4 | ||||||||||||||||||||||
| IL8 | 7.3 | ||||||||||||||||||||||
| IL9 | 3 | ||||||||||||||||||||||
| IRF1 | 2.4 | ||||||||||||||||||||||
| KCNIP4 | 0.8 | 0.4 | 0.4 | ||||||||||||||||||||
| LTA | 4.8 | ||||||||||||||||||||||
| MBL2 | 0.4 | ||||||||||||||||||||||
| MUC2 | 0.8 | 1.3 | 2.9 | 2.0 | |||||||||||||||||||
| MUC5B | 3.3 | 2.2 | 8.1 | 3.7 | |||||||||||||||||||
| NQO1 | 0.4 | ||||||||||||||||||||||
| NRG1 | 5 | 5.5 | |||||||||||||||||||||
| OPN3 | 0.4 | 0.4 | |||||||||||||||||||||
| PIM1 | 1.3 | ||||||||||||||||||||||
| PRKG1 | 0.4 | ||||||||||||||||||||||
| PTGER2 | 0.4 | ||||||||||||||||||||||
| STAT3 | 4.8 | ||||||||||||||||||||||
| TBXA2R | 0.4 | ||||||||||||||||||||||
| TIMP3 | 3 | ||||||||||||||||||||||
| TLR2 | 0.4 | ||||||||||||||||||||||
| TNF | 4.8 | ||||||||||||||||||||||
| TNFAIP3 | 2.4 | ||||||||||||||||||||||
| TNS1 | 9 | 10.5 | |||||||||||||||||||||
| ZNF618 | 5.2 | ||||||||||||||||||||||
Supplementary Table 2. CNVs identified with asthma associated genes with their chromosomal location, break-points, CN states and gene functions.
| Gene | Chromosome location | Break-point (bp) | CN state | Gene function |
|---|---|---|---|---|
| ACAA1 | 3p22.2 | 38039344-38185174 | 3 | Acetyl-coA C-acyltransferase activity |
| ACE | 17q23.3 | 61518220-61624537 | 3 | Actin binding |
| ADAM8 | 10q26.3 | 135056464-135174110 | 0, 3 | Metalloendopeptidase activity |
| AICDA | 12p13.31 | 8529380-8763073 | 1 | Cytidinedeaminase activity |
| ALOX5 | 10q11.21 | 45827177-45940122 | 3 | Arachidonate 5-lipoxygenase activity |
| BDNF | 11p14.1 | 27636885-27789198 | 3 | Growth factor activity |
| C14orf180 | 14q32.33 | 104938308-105067722 | 3 | - |
| C5orf56 | 5q31.1 | 131721749-131825114 | 1 | - |
| CCL11 | 17q12 | 31953738-32936539 | 3 | Chemokine activity |
| CCL2 | 17q12 | 31957151-32938022 | 3 | Chemokine activity |
| CCR1 | 3p21.31 | 46129533-46281822 | 1 | Chemokine receptor activity |
| CCL3L1 | 17q12 | 34437116-34629684 | 0,1,3,4 | Chemotactic for lymphocytes and monocytes |
| CD69 | 12p13.31 | 9879084-9993452 | 1 | Transmembrane signaling receptor activity |
| CDK2 | 12q13.2 | 56297625-56413629 | 3 | Cyclin-dependent protein serine/threonine kinase activity |
| CHRM3 | 1q43 | 239917246-240298870 | 3 | G-protein coupled acetylcholine receptor activity |
| CSF2 | 5q31.1 | 131317464-131419202 | 3 | Cytokine and growth factor activity |
| CTNNA3 | 10q21.3 | 68367662-68515140 | 1, 3 | Cadherin binding |
| CXCL12 | 10q11.21 | 44860013-44975407 | 3 | Cytokine and growth factor activity |
| CYSLTR1 | Xq21.1 | 77513603-77753914 | 3 | Leukotriene receptor activity |
| DIAPH3 | 13q21.2 | 60204504-60381540 | 1 | Rho gtpase binding |
| DPP10 | 2q14.1 | 116532508-116675350 | 3 | Serine-type peptidase activity |
| EGFR | 7p11.2 | 55126900-55245507 | 3 | Epidermal growth factor-activated receptor activity |
| GPX3 | 5q33.1 | 150399114-150513847 | 3 | Glutathione peroxidase activity |
| GSTCD | 4q24 | 106447752-106760931 | 1 | - |
| GSTT1 | 22q11.23 | 24288422-24396802 | 0- 4 | Glutathione peroxidase activity |
| HAVCR1 | 5q33.3 | 156409111-156564456 | 3 | - |
| HAVCR2 | 5q33.3 | 156409111-156564456 | 3 | - |
| HLA-DQA1 | 6p21.32 | 32456671-32625083 | 1 | T cell receptor signaling pathway |
| HLA-DRA | 6p21.32 | 32408527-32530286 | 4 | T cell receptor signaling pathway |
| HLA-DRB1 | 6p21.32 | 32456671-32625083 | 1, 4 | Peptide antigen binding |
| HLA-DRB5 | 6p21.32 | 32429894-32530286 | 1, 4 | Peptide antigen binding |
| ICAM1 | 19p13.2 | 10369086-10471048 | 1 | Transmembrane signaling receptor activity |
| IL13RA2 | Xq23 | 114205618-114397728 | 2, 3 | Cytokine receptor activity |
| IL17RB | 3p21.1 | 53775447-53890848 | 3 | Cytokine receptor activity |
| IL1A | 2q13 | 113500222-113644927 | 1 | Cytokine activity |
| IL1B | 2q13 | 113512866-113644927 | 1 | Cytokine activity |
| IL3 | 5q31.1 | 131317464-131419202 | 3 | Cytokine activity |
| IL33 | 9p24.1 | 6121448-6256144 | 1 | Cytokine activity |
| IL4R | 16p12.1 | 27251081-27377242 | 1 | Interleukin-4 receptor activity |
| IL8 | 4q13.3 | 74567500-74758477 | 1 | Chemokine activity |
| IL9 | 5q31.1 | 135123627-135231195 | 3 | Cytokine activity |
| IRF1 | 5q31.1 | 131721749-131825114 | 1 | Sequence-specific DNA binding transcription factor activity |
| KCNIP4 | 4p15.2 | 21637793-21740358 | 3 | Voltage-gated ion channel activity |
| LTA | 6p21.33 | 31474688-31595882 | 1 | Cytokine activity |
| MBL2 | 10q21.1 | 53511985-57787459 | 1 | Calcium-dependent protein binding |
| MUC2 | 11p15.5 | 1073860-1332223 | 3 | Protein binding |
| MUC5B | 11p15.5 | 1150425-1291777 | 3, 4 | Extracellular matrix structural constituent |
| NQO1 | 16q22.1 | 68860698-69771914 | 1 | NAD(P)H dehydrogenase (quinone) activity |
| NRG1 | 8p12 | 32422268-32522771 | 3 | Cytokine and growth factor activity |
| OPN3 | 1q43 | 241099314-242090693 | 3 | G-protein coupled receptor activity |
| PIM1 | 6p21.2 | 37130282-37236436 | 3 | Protein serine/threonine kinase activity |
| PRKG1 | 10q21.1 | 53511985-57787459 | 1 | Calcium channel regulator activity |
| PTGER2 | 14q22.1 | 52788759-52902018 | 1 | Prostaglandin E receptor activity |
| STAT3 | 17q21.2 | 40498189-40608285 | 1 | Transcription regulatory region DNA binding |
| TBXA2R | 19p13.3 | 3554783-3700847 | 3 | Guanyl-nucleotide exchange factor activity |
| TIMP3 | 22q12.3 | 33142908-33251959 | 3 | Metalloendopeptidase inhibitor activity |
| TLR2 | 4q31.3 | 154515618-154825256 | 3 | Transmembrane signaling receptor activity |
| TNF | 6p21.33 | 31480272-31654732 | 1 | Cytokine activity |
| TNFAIP3 | 6q23.3 | 138092422-138206150 | 1 | Ubiquitin-specific protease activity |
| TNS1 | 2q35 | 218600074-218709313 | 3 | Actin and protein binding |
| ZNF618 | 9q32 | 116811184-116930194 | 3 | DNA binding |