Abstract
The group of avian sarcoma and leukosis virus (ASLV) in chickens contains six highly related subgroups, A to E and J. Four genetic loci, tva, tvb, tvc and tvj, encode for corresponding receptors that determine the susceptibility to the ASLV subgroups. The prevalence of ASLV in hosts may have imposed strong selection pressure toward resistance to ASLV infection, and the resistant alleles in all four receptor genes have been identified. In this study, two new alleles of the tva receptor gene, tvar5 and tvar6, with similar intronic deletions were identified in Chinese commercial broilers. These natural mutations delete the deduced branch point signal within the first intron, disrupting mRNA splicing of the tva receptor gene and leading to the retention of intron 1 and introduction of premature TGA stop codons in both the longer and shorter tva isoforms. As a result, decreased susceptibility to subgroup A ASLV in vitro and in vivo was observed in the subsequent analysis. In addition, we identified two groups of heterozygous allele pairs which exhibited quantitative differences in host susceptibility to ASLV-A. This study demonstrated that defective splicing of the tva receptor gene can confer genetic resistance to ASLV subgroup A in the host.
Entry of retroviruses into the host cell represents one of the most important steps in the viral life cycle1. The process is mediated by the interaction of retroviral envelope glycoproteins with specific cell surface receptors2. This interaction, as well as subsequent phases of virus entry, requires a perfect match of envelope glycoproteins and receptors3,4. Despite the strict structural requirements for these interactions, hypervariability of retroviral glycoproteins can change the receptor usage and broaden the host range. Avian sarcoma and leukosis viruses (ASLVs) in chickens are a closely related group of retroviruses thought to have evolved from a common viral ancestor into six subgroups, A to E and J, which utilize four different cell surface receptors encoded by four genetic loci, tva, tvb, tvc and tvj1,4,5,6. The susceptibility of chicken cells to subgroup A ASLV is determined by the tva locus, which encodes a protein belonging to the family of low-density lipoprotein receptors7. The susceptibility to the subgroup B, D and E ASLVs are determined by the tvb locus, which encodes the tumor necrosis factor receptor-related protein8,9,10. The Tvc protein encoded by the tvc locus, closely related to the mammalian butyrophilins, serves as the receptor for subgroup C ASLV11. The receptor for subgroup J ASLV was identified as chicken Na+/H+ exchanger type 1 (chNHE1), encoded by the tvj locus6.
The complete resistance or decreased susceptibility of host cells to a particular ASLV subgroup can be caused by genetic variations of the tva, tvb, tvc or tvj locus, resulting in the complete lack of receptor protein expression or the expression of an aberrant protein not suitable as a viral receptor. Genetic variations that confer host resistance to infection by specific ASLVs, tvar, tvbr and tvcr alleles, have been identified in some inbred lines of White Leghorn chickens. The mutations found in the resistant alleles result in premature termination or a frameshift in the receptor-encoding loci11,12,13, decreased receptor expression and display14, and even single amino acid substitutions in the receptor gene sequence13,15. In addition, the absence of tryptophan 38 of the cell surface receptor abrogates binding of the subgroup J envelope glycoprotein to ASLV-J-resistant cells, which discriminates sensitive from resistant avian species16.
Although research on host genetic resistance to specific ASLVs has progressed in recent years, the current status of host resistance to infection by specific ASLVs in Chinese chickens is unknown. In order to identify additional resistance alleles of the tva receptor gene, we screened a panel of Chinese commercial broiler lines, which have undergone strict virus eradication management. Here, we characterized two alleles of the tva receptor gene, named tvar5 and tvar6, respectively, with similar intronic deletions encompassing the deduced branch point signal within the first intron and leading to defective splicing of tva mRNA. We also identified two groups of heterozygous allele pairs which exhibited discrepant susceptibility to subgroup A ASLV. To our knowledge, this study is the first to report genetic variations within the tva receptor gene that result in a quantitative effect on ASLV-A susceptibility and pathogenesis in Chinese chickens.
Results
Polymorphisms in the first intron of tva receptor gene and genotyping
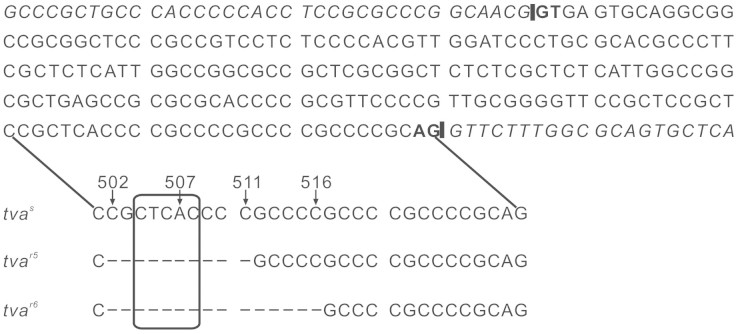
To dissect genetic variations within the tva receptor gene in a panel of Chinese commercial broiler lines, the genomic region of the tva gene in each bird was amplified and sequenced. Four novel variants in the same region within the first intron of the tva gene were identified in the Chinese chickens surveyed. In addition to the tvar3 and tvar4 alleles14, sequencing revealed one new variant with a deletion of the sequence CGCTCACCCC (nucleotides 502 to 511) and another new variant with a deletion of the sequence CGCTCACCCCGCCCC (nucleotides 502 to 516) (Fig. 1). We designated these two novel variants as tvar5 and tvar6 alleles. These four variants in the tva receptor gene (tvar3, tvar4, tvar5 and tvar6) together may be considered a multiple allele, denoted here as tvamut502–516. The genotypic frequencies of the tvamut502–516 multiple allele within the tva receptor gene in the Chinese commercial broiler lines surveyed are presented in Table 1. All genotypes of the tvamut502–516 multiple allele were identified, including the tvas/s, tvar3/r3, tvar4/r4, tvar5/r5 and tvar6/r6 homozygotes, as well as two groups of heterozygous genotypes. One group of heterozygous genotypes is sensitive to ALSV-A, included tvas/r3, tvas/r4, tvas/r5 and tvas/r6, while the other group is resistant to ALSV-A, comprised of tvar3/r4, tvar3/r5, tvar3/r6, tvar4/r5, tvar4/r6 and tvar5/r6. This difference between the two groups of heterozygous genotypes was identified in the subsequent analysis. The genotypes and their frequencies in Chinese commercial broiler lines were obviously different (Table 1).
Figure 1. Alleles tvar5 and tvar6 in Chinese commercial broiler lines containing deletions in intron 1 of tva receptor gene.
Partial genomic sequence showing intron 1 and junctions with exons 1 and 2 in the tva gene. The regions of the branch point with corresponding deletions in the tvar5 and tvar6 alleles are indicated below, and the deleted bases are represented by dashes. The deduced branch point signal is boxed, and the GT-AG intron termini are shown in bold. The underlined sequences indicate the putative alternative branch point signals. The intron-exon junctions are indicated by vertical bars, while exon sequences are indicated in italics. The nucleotide numbering based on the published tvas allele genomic sequence (GenBank accession number: AY531262.1) is provided for reference.
Table 1. Genotypic frequency of tvamut502–516 multiple allele1 in Chinese commercial broiler lines.
| Line | No. | tvamut502–516 genotype 2 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| tvas/s | tvas/r3 | tvas/r4 | tvas/r5 | tvas/r6 | tvar3/r3 | tvar4/r4 | tvar5/r5 | tvar6/r6 | tvar3/r4 | tvar3/r5 | tvar3/r6 | tvar4/r5 | tvar4/r6 | tvar5/r6 | ||
| 202 | 44 | 0 | 0 | 0.12 | 0 | 0 | 0.07 | 0.52 | 0 | 0 | 0.02 | 0.11 | 0 | 0.11 | 0 | 0.05 |
| 203 | 29 | 0.31 | 0 | 0 | 0 | 0 | 0.10 | 0.41 | 0 | 0 | 0 | 0.14 | 0 | 0.03 | 0 | 0 |
| 204 | 30 | 0.23 | 0 | 0 | 0 | 0 | 0.17 | 0.27 | 0.03 | 0 | 0.13 | 0.10 | 0 | 0.07 | 0 | 0 |
| 205 | 30 | 0.57 | 0 | 0 | 0 | 0 | 0.17 | 0 | 0.13 | 0 | 0 | 0 | 0 | 0.10 | 0 | 0.03 |
| 207 | 29 | 0 | 0 | 0 | 0 | 0.08 | 0.03 | 0 | 0.26 | 0.10 | 0 | 0.03 | 0 | 0 | 0 | 0.41 |
| 208 | 27 | 0.22 | 0 | 0 | 0 | 0 | 0.19 | 0.11 | 0.37 | 0 | 0.04 | 0.04 | 0 | 0.04 | 0 | 0 |
| 209 | 28 | 0.18 | 0 | 0 | 0 | 0 | 0.21 | 0.00 | 0.43 | 0 | 0.07 | 0.04 | 0 | 0.04 | 0 | 0.04 |
| 301 | 41 | 0.20 | 0 | 0 | 0 | 0 | 0.20 | 0.02 | 0.34 | 0 | 0.02 | 0.17 | 0 | 0.02 | 0 | 0.02 |
| 306 | 31 | 0 | 0 | 0 | 0.13 | 0 | 0.16 | 0 | 0.55 | 0 | 0 | 0.10 | 0.03 | 0 | 0 | 0.03 |
| 406 | 26 | 0.27 | 0.04 | 0 | 0 | 0 | 0.31 | 0.08 | 0 | 0 | 0.12 | 0.08 | 0 | 0.08 | 0.04 | 0 |
| 407 | 40 | 0.10 | 0.05 | 0.05 | 0 | 0 | 0 | 0.05 | 0.35 | 0 | 0.05 | 0.05 | 0 | 0.30 | 0 | 0 |
| 408 | 30 | 0.07 | 0 | 0 | 0.07 | 0 | 0.10 | 0.27 | 0 | 0 | 0.27 | 0.17 | 0 | 0.07 | 0 | 0 |
| 411 | 27 | 0.04 | 0 | 0 | 0 | 0 | 0.15 | 0.30 | 0.11 | 0.04 | 0.22 | 0.07 | 0.04 | 0 | 0.04 | 0 |
| 413 | 29 | 0.38 | 0.03 | 0.03 | 0 | 0 | 0.28 | 0 | 0 | 0 | 0.21 | 0.03 | 0 | 0.03 | 0 | 0 |
| 417 | 42 | 0.14 | 0 | 0 | 0 | 0 | 0.14 | 0.29 | 0.05 | 0 | 0.19 | 0.05 | 0 | 0.14 | 0 | 0 |
| 418 | 28 | 0.14 | 0.04 | 0 | 0 | 0 | 0.25 | 0.07 | 0.04 | 0 | 0.18 | 0.25 | 0 | 0.04 | 0 | 0 |
| 419 | 46 | 0.13 | 0 | 0 | 0 | 0 | 0.04 | 0.33 | 0.04 | 0.02 | 0.04 | 0.17 | 0 | 0.04 | 0.04 | 0.13 |
| 501 | 29 | 0.17 | 0 | 0 | 0 | 0 | 0.17 | 0.07 | 0 | 0 | 0.34 | 0.17 | 0 | 0.07 | 0 | 0 |
| 502 | 28 | 0.21 | 0 | 0 | 0.04 | 0.04 | 0.21 | 0.11 | 0 | 0 | 0.04 | 0.21 | 0 | 0.04 | 0.11 | 0 |
| 505 | 30 | 0.13 | 0 | 0 | 0 | 0 | 0.07 | 0.07 | 0.47 | 0 | 0 | 0.07 | 0 | 0.20 | 0 | 0 |
| 511 | 38 | 0.32 | 0 | 0 | 0 | 0 | 0.11 | 0.11 | 0.16 | 0 | 0 | 0.05 | 0 | 0.16 | 0 | 0.11 |
| 603 | 30 | 0.20 | 0.03 | 0.03 | 0.07 | 0 | 0.13 | 0.20 | 0.03 | 0 | 0.07 | 0.10 | 0.07 | 0.03 | 0 | 0.03 |
1tvamut502–516 = multiple allele of tvar3, tvar4, tvar5 and tvar6 at the tva locus.
2tvas/s = susceptible homozygote of tva receptor gene; tvas/r3 = heterozygote of deleted ACCCCGCCCC; tvas/r4 = heterozygote of deleted ACCCC; tvas/r5 = heterozygote of deleted CGCTCACCCC; tvas/r6 = heterozygote of deleted CGCTCACCCCGCCCC; tvar3/r3 = homozygote of deleted ACCCCGCCCC; tvar4/r4 = homozygote of deleted ACCCC; tvar5/r5 = homozygote of deleted CGCTCACCCC; tvar6/r6 = homozygote of deleted CGCTCACCCCGCCCC; tvar3/r4 = heterozygote of deleted ACCCCGCCCC and ACCCC; tvar3/r5 = heterozygote of deleted ACCCCGCCCC and CGCTCACCCC; tvar3/r6 = heterozygote of deleted ACCCCGCCCC and CGCTCACCCCGCCCC; tvar4/r5 = heterozygote of deleted ACCCC and CGCTCACCCC; tvar4/r6 = heterozygote of deleted ACCCC and CGCTCACCCCGCCCC; and tvar5/r6 = heterozygote of deleted CGCTCACCCC and CGCTCACCCCGCCCC.
Identification of novel tva receptor gene splicing variants
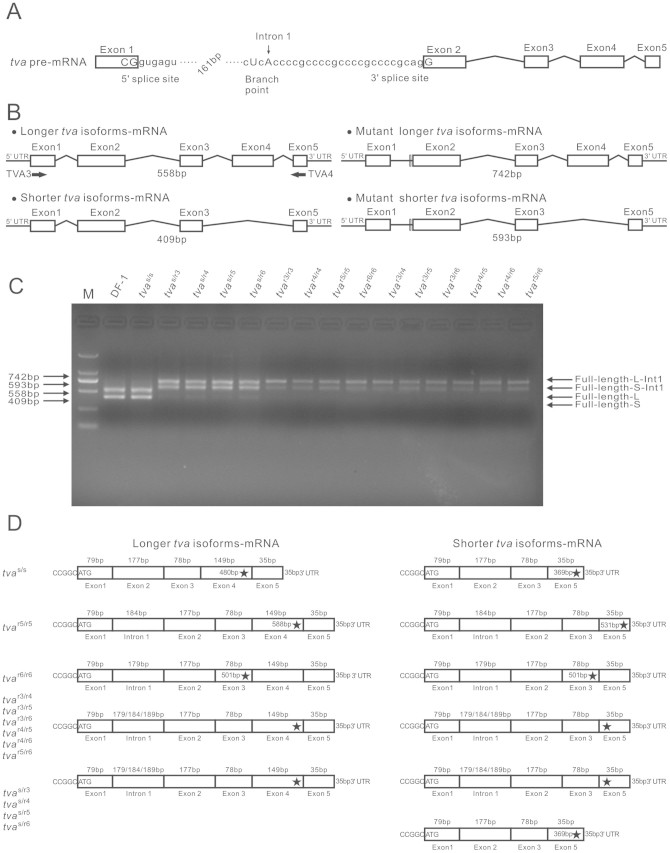
Sequencing revealed that both the tvar5 and tvar6 alleles delete the CTCAC consensus sequence of the branch point signal17 (Fig. 1). These deletions include the A nucleotide, which is required for the first cleavage-ligation step of the splicing reaction18. Therefore, we hypothesized that the deletion mutations may disrupt splicing of the tva precursor mRNA (pre-mRNA) (Fig. 2A).
Figure 2. Deletion of internal intron 1 sequences affects splicing of tva receptor gene.
(A) Schematic diagram of intron 1 in tva pre-mRNA showing the 5′ splice site, 3′ splice site and the branch point sequence (corresponding bases are indicated by dots). The adenosine residue which is required for the first cleavage-ligation step of the splicing reaction18 is marked by an arrow. (B) Schematic diagram of RT-PCR strategy. The use of PCR primers TVA3 and TVA4 generated easily discernible whole cDNA fragments that were amplified from the longer and shorter tva forms, respectively, as well as the longer and shorter tva forms with intron 1 retention. Sizes of diagnostic PCR products are indicated. Exons are drawn as boxes, retained introns are shown as black lines and spliced introns as diagonal lines. The vertical white bar indicates the position of the intronic deletion. (C) RT-PCR of RNA isolated from DF-I cells and samples from lives of defined origin. Lane 1–16 indicated the RT-PCR products from DF-I cells and the tvas/s, tvas/r3, tvas/r4, tvas/r5, tvas/r6, tvar3/r3, tvar4/r4, tvar5/r5, tvar6/r6, tvar3/r4, tvar3/r5, tvar3/r6, tvar4/r5, tvar4/r6 and tvar5/r6 samples from lives, respectively. Positions of full-length and intron 1 retention PCR products are indicated on the right, and sizes of diagnostic PCR products are indicated on the left. Spliced products from the full-length longer and shorter tva forms (Full-length-L and Full-length-S) migrated slightly faster than the corresponding RNAs of the mutants (Full-length-L-Int1 and Full-length-S-Int1). The gels have been run under the same experimental conditions, and the cropped gels are used. The full-length gel images are presented in the supplementary information. (D) Separate sequence analysis of PCR products revealed normal splicing of longer and shorter tva forms of tvas/s homozygotes, while splicing of longer and shorter tva forms of tvar5/r5 and tvar6/r6 homozygotes containing intron 1 with the corresponding deletion mutation, however, the tvas/r3, tvas/r4, tvas/r5 and tvas/r6 heterozygotes generate both the normal splicing of shorter tva forms and abnormal splicing of longer and shorter tva forms. Stars represent premature TGA stop codons identified in the alternative transcript.
To determine whether these specific mutations would interfere with the process of tva pre-mRNA splicing, the full-length tva coding sequence from lives samples of defined origin and from DF-1 cells as a control were amplified by RT-PCR using primers crossing the entire coding sequence of the tva receptor gene (Fig. 2B). The cDNA products from the DF-1 cells and tvas/s samples from lives were of the expected sizes corresponding to the longer and shorter tva isoforms.13 However, the cDNA products from the tvar5/r5 and tvar6/r6 homozygous samples from lives, as well as those from the tvar3/r4, tvar3/r5, tvar3/r6, tvar4/r5, tvar4/r6 and tvar5/r6 heterozygous samples, were longer by approximately 200 bp (Fig. 2C), similar to those of the tvar3/r3 and tvar4/r4 homozygous samples14. These longer sequences suggest that the first intron is retained (i.e., nucleotides 337 to 530, based on the published tvas allele genomic sequence, GenBank accession number: AY531262.1).Interestingly, the products from tvas/r3, tvas/r4, tvas/r5 and tvas/r6 heterozygous samples from lives by RT-PCR have three transcripts. Except for one transcript was the same with the normal shorter tva isoforms, the another two transcripts were longer by approximately 200 bp compared with those of the longer and shorter tva isoforms, respectively (Fig. 2C).
In order to verify the retention of intron 1 in the transcripts of the tvar5 and tvar6 alleles, as well as to identify the heterozygotes, their RT-PCR products were then cloned into the T-vector for sequencing. Separate sequence analysis of PCR products revealed that the retention of the first intron in the RT-PCR products and a deletion of the corresponding sequence (CGCTCACCCC or CGCTCACCCCGCCCC) within the first intron in both the longer and shorter tva isoforms of tvar5/r5 and tvar6/r6 homozygotes, as well as the group of heterozygotes included tvar3/r4, tvar3/r5, tvar3/r6, tvar4/r5, tvar4/r6 and tvar5/r6 (Fig. 2D). Although these similar sequencing results were found in the tvas/r3, tvas/r4, tvas/r5 and tvas/r6 heterozygotes, the splicing of the shorter tva isoforms in these heterozygotes were normal (Fig. 2D). Therefore, the retention of intron 1 in both the longer and shorter tva isoforms of the tvar5 and tvar6 alleles were confirmed via alternative splicing, as well as identified in the heterozygotes. The position of the consensus sequence (CTCAC) of the putative branch point signal positioned at 23 nucleotides upstream of the 3′ splice signal, which is within the optimal distance from the intron-exon boundary19,20, suggesting that it is used in splicing out the first intron from the tva pre-mRNA and its deletion would abrogate the splicing event. Retention of intron 1 in the tvar5 allele changes the translational reading frame of both forms of the tva mRNAs immediately after the end of exon 1 and introduces a premature stop codons TGA in exon 4 or 5 of the longer or shorter mRNA, respectively (Fig. 2D). These two nonspliced mRNAs should be translated into proteins of 196 and 177 amino acids, respectively. However, the retention of the first intron in the tvar6 allele changes the translational reading frame of both forms of the tva mRNAs immediately after the end of exon 1 and introduces premature a stop codon TGA in exon 3 of both the longer and shorter mRNA, causing both nonspliced mRNAs to be translated into proteins of 167 amino acids (Fig. 2D). Therefore, we have demonstrated that intronic deletions comprising the deduced branch point signal within the first intron lead to inefficient splicing of tva mRNA.
Construction of ASLV subgroup A reporter virus
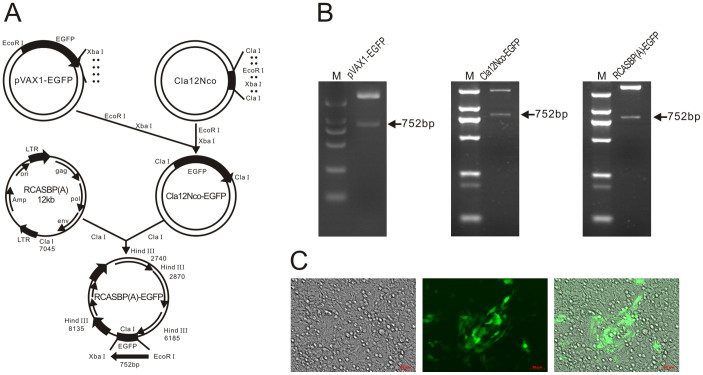
To simplify the procedure and improve the quantitative assessment of ASLV-A infection, we constructed an expression vector RCASBP(A)-EGFP to produce the green fluorescent protein (GFP) reporter virus of subgroup A specificity based on the RCAS retrovirus vector21 (Fig. 3A). The EGFP gene was successfully cloned into the RCASBP(A) vector (Fig. 3B). The resulting replication-competent virus, RCASBP(A)-EGFP, was propagated in DF-1 cells and reached a titer of 106 IU/mL. We then checked the subgroup specificity of the RCASBP(A)-EGFP virus by infection of chicken DF-1 cells and human 293 T cells. The proportion of GFP-positive cells in virus-inoculated DF-1 cells gradually increased over time, providing evidence for the replication and spread of virus (Fig. 3C). By comparison, less than 0.1% of inoculated as well as non-inoculated 293 T cells were GFP-positive, which may be attributed to the background auto-fluorescence (data not shown).
Figure 3. Construction of RCASBP(A)-EGFP.
(A) Schematic representation of strategy for construction of RCASBP(A)-EGFP. Restriction sites used for construction of RCASBP(A)-EGFP are indicated. EGFP, with the size, is indicated. (B) Restriction enzyme digestion analysis of pVAX1-EGFP, Cla12Nco-EGFP and RCASBP(A)-EGFP vectors. (C) Fluorescence microscopy of transduced DF-I cells confirming EGFP marker gene expression.
Reduced susceptibility of chinese chickens to subgroup A ASLV
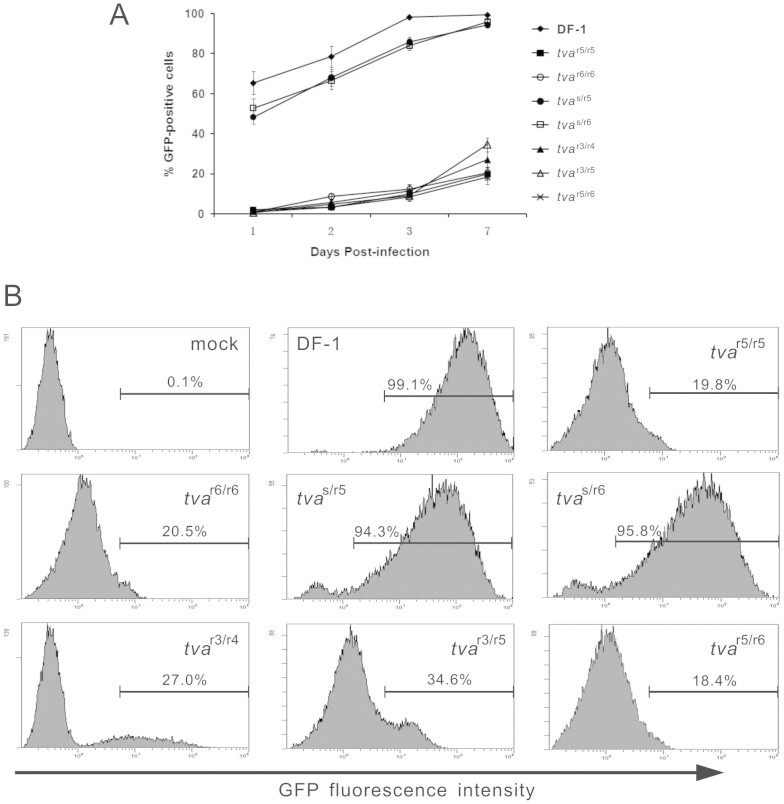
Since two independent intronic deletions that resulted in defective splicing of tva mRNA were identified, we hypothesized that these deletion mutations may confer their functional significance. To determine the effects of tvar5 and tvar6 alleles, as well as their heterozygous variants, on subgroup A ASLV susceptibility, CEFs of defined origin were infected with the RCASBP(A)-EGFP reporter virus, a replication-competent ASLV vector encoding EGFP, and the time course of infection was followed by quantitating the percentage of green fluorescent cells by flow cytometry over 7 subsequent days. DF-1 cells (tvas/s) are susceptible to subgroup A ASLV and thus were used as a positive control. As expected, the DF-1 cells was efficiently infected by RCASBP(A)-EGFP, with more than 60% of the cells being infected on day 1 post-infection, and virtually complete infection of cells was achieved by day 3 (Fig. 4A). However, a very different result was observed when the tvar5/r5 and tvar6/r6 CEFs were infected with RCASBP(A)-EGFP. Both the tvar5/r5 and tvar6/r6 CEFs were much less efficiently infected with RCASBP(A)-EGFP, with only 2.0% and 1.0% of the cells infected on day 1, respectively, and the virus spread very slowly, with only 19.8% and 20.5% of the cells infected by day 7, respectively (Fig. 4C). We also determined the susceptibility of tvar3/r3 and tvar4/r4 CEFs to subgroup A ASLV, and the results were consistent with those reported by Reinišová et al.14 (date not shown). In a separate experiment, quantitative differences in host susceptibility to ASLV-A were observed between heterozygotes which could be classified into two groups. The tvas/r5 and tvas/r6 CEFs were efficiently infected with RCASBP(A)-EGFP at a similar rate to that of DF-1 cells (tvas/s) (Fig. 4B), with 94.3% and 95.8% of the cells infected by day 7, respectively (Fig. 4C). However, the tvar3/r4, tvar3/r5 and tvar5/r6 CEFs were inefficiently infected by RCASBP(A)-EGFP (Fig. 3B), with only 27.0%, 34.6% and 18.4% of the cells being infected at 7 days post-infection, respectively (Fig. 4C). These results clearly indicated the inefficient infection and slow spread of the subgroup A ASLV in the tvar5/r5 and tvar6/r6 homozygous CEFs and the tvar3/r4, tvar3/r5 and tvar5/r6 heterozygous CEFs.
Figure 4. Time course of ASLV-A infection of DF-I cells and CEFs of defined origin.
DF-I cells (tvas/s) and CEFs of defined origin were inoculated at a the multiplicity of infection of 10 with replication-competent ASLV encoding the EGFP reporter protein, RCASBP(A)-EGFP. (A) Proportions of GFP-positive cells were determined by FACS on indicated days post-infection. Results are means of data from three parallel dishes. (B) Representative FACS histograms of CEFs infected with RCASBP(A)-GFP at 7 days post-infection. The relative GFP fluorescence is plotted against the cell count, and the percentage of GFP-positive cells is indicated.
Inefficient infection of chinese chickens by subgroup A ALV
In order to further determine the in vivo effects of the tvar5 and tvar6 alleles, as well as their heterozygous variants, commercial broilers randomly collected from lines 208, 419, 502 and 603 were inoculated with subgroup A ALV, and blood samples of chicks were tested for ALV-A infection. One-day-old chicks were inoculated with 0.2 mL of subgroup A ALV (2.4 S/P) into the abdominal cavity, and inoculated once again when they were 5 days old. At one month post-infection, the status of ALV-A infection was determined by RT-PCR of RNA extracted from whole blood samples. As expected, the tvas/s birds were all positive for ALV-A. While the tvas/r3, tvas/r4, tvas/r5 and tvas/r6 birds were nearly all positive for ALV-A, in the cohort of 9, 5, 9 and 5 of the tvas/r3, tvas/r4, tvas/r5 and tvas/r6 chicks, respectively, all birds were positive for ALV-A, except for one tvas/r5 bird (Table 2). However, the susceptibility to subgroup A ALV of the homozygous tvar3/r3, tvar4/r4, tvar5/r5 and tvar6/r6 chicks and the heterozygous tvar3/r4, tvar3/r5, tvar3/r6, tvar4/r5, tvar4/r6 and tvar5/r6 chicks decreased. In the cohort of 32, 18, 15, 11, 6, 9, 5, 8, 6 and 17 of the tvar3/r3, tvar4/r4, tvar5/r5, tvar6/r6, tvar3/r4, tvar3/r5, tvar3/r6, tvar4/r5, tvar4/r6 and tvar5/r6 chicks, only 4, 1, 1 and 1 of the tvar3/r3, tvar4/r4, tvar6/r6, and tvar5/r6 chicks were positive for ALV-A (Table 2). These results were consistent with those of the in vitro experiment.
Table 2. Incidence of RT-PCR positivity for avian leukosis virus subgroup A at one month post-infection in chicks with different genotypes of the tvamut502–516 multiple allele1.
| tvas/s2 | tvas/r3 | tvas/r4 | tvas/r5 | tvas/r6 | tvar3/r3 | tvar4/r4 | tvar5/r5 | tvar6/r6 | tvar3/r4 | tvar3/r5 | tvar3/r6 | tvar4/r5 | tvar4/r6 | tvar5/r6 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 45/45 | 9/9 | 5/5 | 8/9 | 5/5 | 4/32 | 1/18 | 0/15 | 1/11 | 0/6 | 0/9 | 0/5 | 0/8 | 0/6 | 1/17 |
1tvamut502–516 = multiple allele of tvar3, tvar4, tvar5 and tvar6 at the tva locus.
2tvas/s = susceptible homozygote of tva receptor gene; tvas/r3 = heterozygote of deleted ACCCCGCCCC; tvas/r4 = heterozygote of deleted ACCCC; tvas/r5 = heterozygote of deleted CGCTCACCCC; tvas/r6 = heterozygote of deleted CGCTCACCCCGCCCC; tvar3/r3 = homozygote of deleted ACCCCGCCCC; tvar4/r4 = homozygote of deleted ACCCC; tvar5/r5 = homozygote of deleted CGCTCACCCC; tvar6/r6 = homozygote of deleted CGCTCACCCCGCCCC; tvar3/r4 = heterozygote of deleted ACCCCGCCCC and ACCCC; tvar3/r5 = heterozygote of deleted ACCCCGCCCC and CGCTCACCCC; tvar3/r6 = heterozygote of deleted ACCCCGCCCC and CGCTCACCCCGCCCC; tvar4/r5 = heterozygote of deleted ACCCC and CGCTCACCCC; tvar4/r6 = heterozygote of deleted ACCCC and CGCTCACCCCGCCCC; and tvar5/r6 = heterozygote of deleted CGCTCACCCC and CGCTCACCCCGCCCC.
Taken together, these results clearly demonstrated that the deletion of the branch point signal within the first tva intron disrupt mRNA splicing of the tva receptor gene, which explains the decreased susceptibility of the homozygous tvar5/r5 and tvar6/r6 birds and the heterozygous tvar3/r4, tvar3/r5, tvar3/r6, tvar4/r5, tvar4/r6 and tvar5/r6 birds to infection by subgroup A ASLV.
Discussion
In the present study, we described the identification of two similar intronic deletions encompassing the deduced branch point signal in the tva receptor gene, which were shown to significantly decrease the susceptibility of chickens to infection by ASLV-A. Furthermore, we identified two groups of heterozygotes based on their quantitative differences in host susceptibility to ASLV-A. This study is the first to report genetic defects in the tva receptor gene that account for a quantitative effect on ASLV-A susceptibility and pathogenesis in Chinese chickens. The altered susceptibility to infection by ASLV-A was observed both in cultured cells and in chicks challenged with subgroup A ALV.
Alternative splicing allows individual genes to produce two or more variant mRNAs, which in many cases encode functionally distinct proteins19,22,23. Indeed, two tva receptor gene splicing variants created by alternative splicing were identified previously7,24. Binding of more than one receptor, probably two, is needed for entry of virions via the Tva800 receptor that is encoded by the shorter tva forms, whereas binding of just one Tva950 receptor encoded by the longer tva forms is sufficient for successful entry25. Alternative splicing can be modulated by variation both in the cis genomic splicing signals and in the cellular pathways that regulate splicing22,26,27. In this study, we identified the alternative splicing of the tvar5 and tvar6 resistant alleles resulting from intronic deletions encompassing the deduced branch point signal, leading to the retention of the first intron and introduction of premature TGA stop codons in the longer and shorter tva forms (Fig. 2). Alternative splicing events generally create a premature termination codon that would cause the resulting mRNA to be degraded by nonsense-mediated mRNA decay28,29. Therefore, alternative splicing of mRNAs that changes the encoded proteins has profound functional effects23,30,31. Experimental analysis of distinct protein isoforms showed that alternative splicing regulates binding between proteins, between proteins and nucleic acids, as well as between proteins and membranes23,31,32,33. In most cases, the binding affinity is modulated, but the binding is not abolished completely34,35,36,37. The tvar3 and tvar4 alleles harbor an intronic deletion of the branch point and disrupt splicing of the tva receptor gene, leading to decreasing binding affinity between the Tva receptor and envelope glycoproteins of ASLV-A and display of the Tva receptor on the cell surface, and consequently reduced host susceptibility to subgroup A ASLV14. Given that the tvar3, tvar4, tvar5 and tvar6 alleles all contain a deletion of the same branch point signal, the same effects are also likely exerted by the tvar5 and tvar6 alleles in homozygotes, as well as the tvar3/r4, tvar3/r5, tvar3/r6, tvar4/r5, tvar4/r6 and tvar5/r6 resistant heterozygotes, as evidenced by their significantly decreased susceptibility to infection by ASLV-A. In contrast to resistant heterozygotes, the tvas/r3, tvas/r4, tvas/r5 and tvas/r6 susceptible heterozygotes may retain a receptor conformation that is at least partially suitable for binding of ASLV-A envelope glycoproteins and subsequent viral infection. A previously similar study were reported by Kučerová16, the deletion of W38 completely abrogates receptor activity and explains the resistance of chukar to the J subgroup of ALV; however, alleles with W38 replaced by G or E conferred susceptibility to the virus when overexpressed in the virus entry assay. Alternative splicing is not only important for normal cellular functions but also frequently is involved in disease pathogenesis27,38,39,40,41. Interestingly, published studies have identified intronic deletions causing exon skipping or intron retention in the human LDLR gene of patients with familial hypercholesterolemia42,43,44,45,46. Because the human LDLR gene is homologous to the tva receptor gene, such deletions as described above may be speculated as a common mechanism of mutagenesis within this gene family.
From the point of view of virus-host coevolution, it is tempting to speculate that the tvar5 and tvar6 alleles with decreased susceptibility to ASLV-A in Chinese chickens have been selected by pressure from subgroup A ASLV. Although eradication management strategies at the breeder level and high biosecurity level of flock production are used to control avian leukosis47,48, these conventional methods are not a practical means of completely eliminating the occurrence and spread of infection in developing countries49,50,51. The prevalence of ASLV in chicken populations may have imposed a strong selection pressure toward resistance or at least decreased susceptibility to ASLV infection. ASLV infection is known to cause a variety of neoplastic disease conditions and other production problems in affected flocks52. Under these conditions, a wide prevalence of resistance alleles in the ASLV receptor genes can be expected. To date, resistance alleles in the tva, tvb and tvc genes have been found in particular inbred lines of White Leghorns11,12,13,14,15. The resistance conferred by some alleles are caused by premature termination codons or frame shift mutations, such as tvar2, tvbr and tvcr11–13, which do not encode any product that can carry out normal cellular functions, but their potentially detrimental impact on resistant birds is unknown. Given the presence of such counter-selection and rapid evolution of ASLV envelope glycoproteins, complete resistance to ASLV entry can appear but cannot prevail and be fixed in the chicken population. Hence, we suggest that genetic variations with modest effects on both ASLV entry and natural receptor functions provided a positive selective advantage in the chicken population. Selection of mutant Tva proteins that have lost the ability to function as an ASLV-A receptor while possibly retaining the“normal” Tva function may have provided the greatest selective advantage, resulting in fixing of the tvar5 and tvar6 loci in the germ lines of certain lines of chickens.
The tvar3, tvar4, tvar5 and tvar6 resistant alleles are prevalent in Chinese commercial broiler lines (Table 1), indicating that the potential for genetic improvement of resistance to ASLV-A is great and selective breeding for chickens genetically resistant to ASLV-A is feasible. Despite the potential benefits to be derived from breeding for enhanced resistance to ASLV-A, evaluating whether trade-offs between disease resistance and other economically important traits of chickens exist would be prudent. In fact, in a separate study we have evaluated the effects of genetic resistance to subgroup A ASLV on growth, feed efficiency and carcass traits using an F2 resource population by reciprocally crossing Huiyang Bearded chickens and fast-growing Chinese yellow broilers. The results demonstrated that genetic selection for tvar3, tvar4, tvar5 and tvar6 resistant loci can improve genetic resistance to subgroup A ASLV, but does not compromise production performance (data not shown). Selection for genetic resistance to subgroup B ASLV has been found to have no negative effect on laying performance in White Leghorn hens53. Furthermore, genetic selection for tvar and tvbr alleles can improve laying performance and growth rate, as well as greatly reduce lymphoid leukosis in White Leghorn hens54,55.
In conclusion, our study identified two novel genetic resistant alleles, tvar5 and tvar6, in Chinese commercial broilers. These two resistant alleles both harbor an internal intronic deletions comprising the deduced branch point signal within the first intron and leading to alternative splicing of the tva receptor gene, resulting in significantly decreased the susceptibility to subgroups A ASLV. We also demonstrated a difference in the susceptibility to subgroup A ASLV between tvar3/r4, tvar3/r5, tvar3/r6, tvar4/r5, tvar4/r6 and tvar5/r6 heterozygotes and tvas/r3, tvas/r4, tvas/r5 and tvas/r6 heterozygotes. This study provides valuable insight into mechanisms of genetic resistance to ASLV and retrovirus-host coevolution.
Methods
Ethics statement
The animal experiments were conducted in accordance with the guidelines of the Guangdong province on the review of welfare and ethics of laboratory animals approved by the Guangdong province administration office of laboratory animals (GPAOLA). All the animal procedures were approved by the Animal Care Committee of the College of Animal Science, South China Agricultural University, Guangzhou, China (approval ID: 201004152).
Amplification and analysis of tva alleles from commercial broiler lines
Genomic DNA was prepared from blood samples of 22 commercial broiler lines using phenol-chloroform extraction. These commercial broiler lines were maintained at Guangdong Wen’s Food Group Co., Ltd. The sequence of the genomic region including exon 1, intron 1 and exon 2 of the tva gene was amplified using forward TVA1 primer 5′-GTTCAGCAGATCCTCATCTCCCG-3′ and reverse TVA2 primer 5′-GGCCATTGTGCGATCTAAGAGGG-3′. The PCR procedure was as follows: an initial denaturation at 95°C for 5 min, followed by 35 cycles of 94°C for 30 s, 67°C for 45 s and 72°C for 90 s, and a final extension of 72°C for 10 min with KOD-Plus Neo (Toyobo, Tokyo, Japan). In total, 712 birds from different commercial broiler lines were genotyped. The final PCR product with an expected length of 1308 bp was directly sequenced using an ABI 3730 sequencer (Applied Biosystems, Foster City, CA, USA).
Splicing analysis by RT-PCR
Total RNAs from lives of commercial broiler lines 208, 419, 502 and 603 were isolated using TRIZOL reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. cDNA was obtained by reverse transcription of 1 μg of total RNA with ReverTra Ace® qPCR RT Master Mix with gDNA Remover (Toyobo, Tokyo, Japan). The whole tva coding sequence was amplified using the forward TVA3 primer 5′- CCGGCATGGTGCGGTTGTTG-3′ containing the tva initiation codon and the reverse TVA4 primer 5′-AGCCAGGTTCCACGGTCAGC-3′, which is complementary to the 3′ untranslated region. The PCR conditions were as follows: an initial denaturation at 95°C for 5 min, followed by 30 cycles of 94°C for 30 s, 58°C for 30 s and 72°C for 50 s, and a final extension of 72°C for 5 min with KOD FX (Toyobo, Tokyo, Japan). To identify the transcripts of the tva alleles and their heterozygotes, the RT-PCR products were visualized by electrophoresis on 2% agarose gels. The purified RT-PCR products were ligated into the pMD19-T vector (TaKaRa, Dalian, China) and sequenced using an ABI 3730 sequencer (Applied Biosystems).
Construction of ASLV subgroup A reporter vector and virus propagation
The EGFP gene was isolated from the pVAX1-EGFP vector (stored at our laboratory) as an EcoRI-XbaI fragment and cloned into the Cla12Nco adapter plasmid at the same restriction sites21,56. The resulting clones with the proper orientation of EGFP were isolated as ClaI fragments and subcloned into the corresponding site of the RCASBP(A) vector21, which was kindly obtained from Stephen H. Hughes (HIV Drug Resistance Program, National Cancer Institute, USA). The resulting expression construct was designated RCASBP(A)-EGFP. The RCASBP(A)-EGFP virus, transducing the EGFP reporter gene, was propagated by transfection of plasmid DNA containing the reporter vector into DF-1 cells, which are free of closely related endogenous retrovirus loci. Transfection was performed by using the X-tremeGENE 9 transfection reagent (Roche, Basel, Switzerland) according to the manufacturer’s instructions. Infection and virus spread were observed as an increasing proportion of GFP-positive cells, and virus stocks were harvested on day 9 post-transfection. The cell supernatants were cleared of debris by centrifugation at 2,000 × g for 10 min at 10°C, and aliquoted viral stocks were stored at −80°C. The virus titer was determined by terminal dilution and subsequent infection of DF-1 cells, reached 106 IU/mL.
Preparation of CEFs and cell culture
Primary CEFs were prepared from 9 to 11-day-old embryos from commercial broilers lines 208, 419, 502 and 603. The procedure was described previously57. The genotypes of CEFs were determined by direct sequencing as described above. All embryo fibroblasts, as well as the chicken permanent cell line DF-158, were propagated in growth medium containing Dulbecco’s modified Eagle’s medium (DMEM) (Gibco/Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum (Gibco/Invitrogen, Carlsbad, CA, USA), and penicillin/streptomycin (100 mg/ml each) at 37°C and 5% CO2.
RCASBP(A)-EGFP virus spread assayed by fluorescence-activated cell sorting (FACS)
CEFs of defined origin were mock infected or infected by incubation with diluted RCASBP(A)-EGFP virus stock. Briefly, the CEFs were seeded in triplicate wells at a density of 5 × 104 per well in a 24-well plate and inoculated with 5 × 105 IU of RCASBP(A)-EGFP virus 24 h after seeding. The virus was applied in 0.25 mL medium for 1 h. The percentage of GFP-positive cells was quantitated by FACS using a Cytomics FC 500 analyzer (Beckman Coulter, Brea, CA, USA) on days 1, 2, 3 and 7 post-infection. For FACS analysis, the cells of three wells were trypsinized, washed in phosphate-buffered saline (PBS) and then analyzed.
Animal experiment
The animal experiment design included two independent experiments with 100 birds each. Chicks randomly collected from commercial broilers lines 208, 419, 502 and 603 were randomly divided into four groups with 25 birds each. Chicks were maintained in four negatively-pressured biosecurity isolators under quarantine conditions and provided with water and commercial feed ad libitum. One-day-old chicks were inoculated with 2.4 S/P of avian leukosis virus subgroup A (ALV-A) strain GD0849, which was kindly provided by Weisheng Cao at South China Agricultural University, P. R. China, in 0.2 mL into the abdominal cavity. These chicks were inoculated once again at five days of age. A whole blood sample from each one-day-old chick was drawn from the wing vein and used for genomic DNA isolation by the phenol-chloroform method for genotyping as above described. To determine whether blood samples of chicks were positive for ASLV-A at one month post-infection, a whole blood sample from each bird was drawn for preparing total RNA using the TRIZOL reagent (Invitrogen, Carlsbad, CA, USA). The coding sequence of the env gene of ALV-A was amplified using the forward H5 primer 5′- GGATGAGGTGACTAAGAAAG-3′ and reverse H6 primer 5′- AGAGAAAGAGGGGTGTCTAAGGAGA-3′59. The PCR conditions were as follows: a reverse translation at 50°C for 30 min, then an initial denaturation at 94°C for 3 min, followed by 30 cycles of 94°C for 30 s, 56°C for 30 s and 72°C for 60 s, and a final extension of 72°C for 5 min with PrimeScript® One Step RT-PCR Kit Ver. 2 (TaKaRa, Dalian, China). To identify the infection status of ALV-A, the PCR products were visualized by electrophoresis on 2% agarose gels.
Author Contributions
W.G.C., Y.L. and Q.M.X. conceived and designed the project. W.G.C., Y.L. and H.X.L. performed the experiments. W.G.C., H.X.L. and S.C. analyzed the data. W.G.C. and H.M.Z. wrote the paper. D.M.S., H.M.Z. and F.C. contributed the materials. H.M.Z. and Q.M.X. carried out the principal investigator. W.G.C. and Y.L. contributed equally to the work. All of the authors read and approved the final manuscript.
Acknowledgments
We thank Guangdong Wen’s Food Group Co., Ltd., Guangdong, China, for providing the commercial broiler lines 202, 203, 204, 205, 207, 208, 209, 301, 306, 406, 407, 408, 411, 413, 417, 418, 419, 501, 502, 505, 511, 603 samples, and Stephen H. Hughes from HIV Drug Resistance Program, National Cancer Institute, USA, for providing the Cla12Nco adapter plasmid and RCASBP(A) vector, as well as Weisheng Cao at South China Agricultural University, P R. China, for providing the avian leukosis virus subgroup A strain GD08. This study was supported by the Natural Science Foundation of Guangdong Province (Grant No. S2013030013313), National Modern Agricultural Industry Technology System Project of China (CARS-41) and Guangdong Province Science and technology plan project (Grant No. 2012B020306002 and Grant No. 2012B091100078).
References
- Weiss R. A. Cellular receptors and viral glycoproteins involved in retrovirus entry. Levy J. A., ed. (ed.), pp. 1–108 (Plenum Press, NY, 1993). [Google Scholar]
- Hunter E. Viral entry and receptors. Coffin J. M., , Hughes S. H., & Varmus H. E., eds. (ed.), pp. 71–120. (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 1997). [PubMed] [Google Scholar]
- Earp L. J., Delos S. E., Park H. E. & White J. M. The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol. 285, 25–66 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard R. J. O., Elleder D. & Young J. A. T. Avian sarcoma and leucosis virus-receptor interactions: from classical genetics to novel insights into virus-cell membrane vision. Virology 344, 25–29 (2006). [DOI] [PubMed] [Google Scholar]
- Barnard R. J. & Young J. A. T. Alpharetrovirus envelope-receptor interactions. Curr. Top. Microbiol. Immunol. 281, 107–136 (2003). [DOI] [PubMed] [Google Scholar]
- Chai N. & Bates P. . Na/H exchanger type 1 is a receptor for pathogenic subgroup J avian leukosis virus. Proc. Natl. Acad. Sci. 103, 5531–5536 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates P., Young J. A. & Varmus H. E. A receptor for subgroup A Rous sarcoma virus is relatedto the low density lipoprotein receptor. Cell 74, 1043–1051 (1993). [DOI] [PubMed] [Google Scholar]
- Brojatsch J. et al. CAR1, a TNFR-related protein, is a cellular receptor for cytopathic avian leukosis-sarcoma viruses and mediates apoptosis. Cell 87, 845–855 (1996). [DOI] [PubMed] [Google Scholar]
- Adkins H. B. et al. Identification of a cellular receptor for subgroup E avian leukosis virus. Proc. Natl. Acad. Sci. 94, 11617–11622 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adkins H. B., Blacklow S. C. & Young J. A. T. Two functionally distinct forms of a retroviral receptor explain the nonreciprocal receptor interference among subgroup B, D, and E avian leukosis viruses. J. Virol. 75, 3520–3526 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elleder D. et al. The receptor for the subgroup C avian sarcoma and leukosis viruses, Tvc, is related to mammalian butyrophilins, members of the immunoglobulin superfamily. J. Virol. 79, 10408–10419 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klucking S., Adkins H. B. & Young J. A. T. Resistance to infection by subgroups B, D, and E avian sarcoma and leukosis viruses is explained by a premature stop codon within a resistance allele of the tvb receptor gene. J. Virol. 76, 7918–7921 (2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elleder D. et al. Two different molecular defects in the Tva receptor gene explain the resistance of two tvar lines of chickens to infection by subgroup A avian sarcoma and leukosis viruses. J. Virol. 78, 13489–13500 (2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinišová M. et al. Intronic deletions that disrupt mRNA splicing of the tva receptor gene result in decreased susceptibility to infection by avian sarcoma and leukosis virus subgroup A. J. Virol. 86, 2021–2030 (2012). [DOI] [PMC free article] [PubMed]
- Reinišová M. et al. A single-amino-acid substitution in the TvbS1 receptor results in decreased susceptibility to infection by avian sarcoma and leukosis virus subgroups B and D and resistance to infection by subgroup E in vitro and in vivo. J. Virol. 82, 2097–2105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kučerová D. et al. Nonconserved tryptophan 38 of the cell surface receptor for subgroup J avian leukosis virus discriminates sensitive from resistant avian species. J. Virol. 87, 8399–8407 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao K., Masuda A., Matsuura T. & Ohno K. Human branch point consensus sequence is yUnAy. Nucleic Acids Res. 36, 2257–2267 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Query C. C. et al. Branch nucleophile selection in pre-mRNA splicing: evidence for the bulged duplex model. Genes & Dev. 8, 587–597 (1994). [DOI] [PubMed] [Google Scholar]
- Matlin A. J., Clark F. & Smith C. W. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell. Bio. 6, 386–398 (2005). [DOI] [PubMed] [Google Scholar]
- Blencowe B. J. Alternative splicing: new insights from global analyses. Cell 126, 37–47 (2006). [DOI] [PubMed] [Google Scholar]
- Hughes S. H. The RCAS vector system. Folia Biol. 50, 107–119 (2004). [PubMed] [Google Scholar]
- Black D. L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. biochem. 72, 291–336 (2003). [DOI] [PubMed] [Google Scholar]
- Kelemen O. et al. Function of alternative splicing. Gene 514, 1–30 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J. A. T., Bates P. & Varmus H. E. Isolation of a chicken gene that confers susceptibility to infection by subgroup A avian leukosis and sarcoma viruses. J. Virol. 67, 1811–1816 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray E. R., Illingworth C. J., Coffin J. M. & Stoye J. P. Binding of more than one Tva800 molecule is required for ASLV-A entry. Retrovirology 8, 1–11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres J. F. & Kornblihtt A. R. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet. 18, 186–193 (2002). [DOI] [PubMed] [Google Scholar]
- Liu S. & Cheng C. Alternative RNA splicing and cancer. Wiley Interdisciplinary Reviews: RNA 4, 547–566 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlincy N. J. & Smith C. W. Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends. Biochem. Sci. 33, 385–393 (2008). [DOI] [PubMed] [Google Scholar]
- Nicholson P. & Mühlemann O. Cutting the nonsense: the degradation of PTC-containing mRNAs. Biochem. Soc. Trans. 36, 1615–1620 (2010). [DOI] [PubMed] [Google Scholar]
- Ward A. J. & Cooper T. A. The pathobiology of splicing. J. Pathol. 220, 152–163 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalea A. Z., Schmidt A. M. & Hudson B. I. Alternative splicing of RAGE: roles in biology and disease. Front Biosci. 17, 2756–2770 (2011). [DOI] [PubMed] [Google Scholar]
- Bechtel J. M. et al. The Alternative Splicing Mutation Database: a hub for investigations of alternative splicing using mutational evidence. BMC research notes 1, 3 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhy A. et al. Genetic variants affecting alternative splicing of human cholesteryl ester transfer protein. Biochem Biophys Res Commun. 443, 1270–4 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S. F. et al. Three- and four-repeat TAU regulate the dynamic instability of two distinct microtubule subpopulations in qualitatively different manners. J. Biol. Chem. 280, 13520–13528 (2005). [DOI] [PubMed] [Google Scholar]
- Itoh M., Nagatomo K., Kubo Y. & Saitoh O. Alternative splicing of RGS8 gene changes the binding property to the M1 muscarinic receptor to confer receptor type-specific Gq regulation. J. Neurochem. 99, 1505–1516 (2006). [DOI] [PubMed] [Google Scholar]
- Dubielecka P. M. et al. Differential regulation of macropinocytosis by Abi1/Hssh3bp1 isoforms. PLoS One 5, e10430 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushiyama M. et al. Alternative splicing of the pituitary adenylate cyclaseactivating polypeptide (PACAP) receptor contributes to function of PACAP-27. J. Mol. Neurosci. 42, 341–348 (2010). [DOI] [PubMed] [Google Scholar]
- Tazi J., Bakkour N. & Stamm S. Alternative splicing and disease. BBA-Mol. Basis. Dis. 1792, 14–26 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Pezzi E., Gómez-Salinero J., Gatto A., García-Pavía P. The alternative heart: impact of alternative splicing in heart disease. J Cardiovasc Transl Res. 6, 945–55 (2013). [DOI] [PubMed] [Google Scholar]
- Fan X. & Tang L. Aberrant and alternative splicing in skeletal system disease. Gene 528, 21–6 (2013). [DOI] [PubMed] [Google Scholar]
- Gamazon E. R. & Stranger B. E. Genomics of alternative splicing: evolution, development and pathophysiology. Hum Genet. 133, 679–687 (2014). [DOI] [PubMed] [Google Scholar]
- Webb J. C. et al. Genetic variation at a splicing branch point in intron 9 of the low density lipoprotein (LDL) receptor gene: a rare mutation that disrupts mRNA splicing in a patient with familial hypercholesterolaemia and a common polymorphism. Hum. Mol. Genet. 5, 1325–1331 (1996). [DOI] [PubMed] [Google Scholar]
- Amsellem S. et al. Intronic mutations outside of Alu-repeat-rich domains of the LDL receptor gene are a cause of familial hypercholesterolemia. Hum. Genet. 111, 501–510 (2002). [DOI] [PubMed] [Google Scholar]
- Defesche J. C. et al. Silent exonic mutations in the low-density lipoprotein receptor gene that cause familial hypercholesterolaemia by affecting mRNA splicing. Clin. Genet. 73, 573–578 (2008). [DOI] [PubMed] [Google Scholar]
- Jelassi A. et al. A novel splice site mutation of the LDL receptor gene in a Tunisian hypercholesterolemic family. Clin. Chim. Acta. 392, 25–29 (2008). [DOI] [PubMed] [Google Scholar]
- Cameron J. et al. Splicesite mutation c.3131, GA in intron 3 of the LDL receptor gene results in transcripts with skipping of exon 3 and inclusion of intron 3. Clin. Chim. Acta. 403, 131–135 (2009). [DOI] [PubMed] [Google Scholar]
- Kreager K. S. Chicken industry strategies for control of tumor virus infections. Poult Sci. 77, 1213–6 (1998). [DOI] [PubMed] [Google Scholar]
- Fadly, A. M. & Payne, L. N., Leukosis-sarcoma group. Saif, Y. M. et al. (ed.), pp. 465–516 (Kennett Square, PA: American Association of Avian Pathologists, 2003).
- Zhang Q. C., Zhao D. M., Guo H. J. & Cui Z. Z. Isolation and identification of a subgroup A avian leukosis virus from imported meat-type grand-parent chickens. Virol Sin. 25, 130–136 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai H. et al. Isolation and characterization of emerging subgroup J avian leukosis virus associated with hemangioma in egg-type chickens. Vet. Microbiol. 151, 275–283 (2011). [DOI] [PubMed] [Google Scholar]
- Gao Y. et al. Molecular epidemiology of avian leukosis virus subgroup J in layer flocks in China. J. Clin. Microbiol. 50, 953–960 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadly, A. M. & Nair, V., Leukosis/sarcoma group. Saif, Y. M. et al. (ed.), pp. 514–568 (Ames, IA: Blackwell Publishing, 2008).
- Bao M., Yu J., Wang D., Li C. & Ning Z. Association Analysis of TVB Genotype and Laying Performance in Fast-feathering and Slow-feathering White Leghorns. China Poultry 35, 6–10 (2013). [Google Scholar]
- Hartmann W., Hagen D. & Heil G. Genetic resistance to Leukosis virus infection: effects on mortality and egg production in Leghorn hens. Brit Poult Sci. 25, 43–52 (1984). [DOI] [PubMed] [Google Scholar]
- Smith E. J. & Nelsen T. C. Effect of tumor virus susceptibility alleles on the late-feathering gene K on early growth of White Leghorns. Poult Sci. 72, 1400–1404 (1993). [DOI] [PubMed] [Google Scholar]
- Federspiel M. J. & Hughes S. H. Retroviral gene delivery. Methods Cell Biol. 52, 179–214 (1997). [PubMed] [Google Scholar]
- Maas R., van Zoelen D., Oei H. & Claassen I. Replacement of primary chicken embryonic fibroblasts (CEF) by the DF-1 cell line for detection of avian leucosis viruses. Biologicals 34, 177–181 (2006). [DOI] [PubMed] [Google Scholar]
- Himly M. et al. The DF-1 chicken fibroblast cell line: transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology 248, 295–304 (1998). [DOI] [PubMed] [Google Scholar]
- Bieth E. I. & Darlix J. L. Complete nucleotide sequence of a highly infectious avian leukosis virus. Nucleic Acids Res. 20, 367 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]