Abstract
PAD4 has been strongly implicated in the pathogenesis of autoimmune, cardiovascular and oncological diseases, through clinical genetics and gene disruption in mice. Novel, selective PAD4 inhibitors binding to a calcium-deficient form of the PAD4 enzyme have, for the first time, validated the critical enzymatic role of human and mouse PAD4 in both histone citrullination and neutrophil extracellular trap formation. The therapeutic potential of PAD4 inhibitors can now be explored.
Citrullination of arginine residues, catalysed by five closely-related protein arginine deiminases (PADs 1–4 and PAD6), affects numerous physiological and pathological processes1. PAD4, predominantly expressed in granulocytes, is strongly linked to diverse diseases. In rheumatoid arthritis (RA), autoantibodies against citrullinated joint proteins manifest years before symptoms, whilst antibodies against PAD4 occur during advanced disease. PAD4 is over-expressed in multiple tumours, affecting p53 function and downstream clearance pathways. PAD4 is also linked to diseases characterised by aberrant levels of neutrophil extracellular traps (NETs). Although considered a host defence mechanism2, excessive NET load is a hallmark of vasculitis3, lupus4, 5, thrombosis6 and sepsis7. Neutrophils from PAD4-deficient mice lack NETs8 and the mice show increased susceptibility to infection, suggesting that PAD4 and NETs are critical in innate immunity.
Calcium binding to PAD4 promotes the bioactive conformation, increasing PAD4 activity by 10,000-fold9. The first characterised PAD inhibitors (e.g. F- and Cl-amidine) were irreversible, with preference for the calcium-bound enzyme10. Based on the PAD4 substrate benzoyl-arginine amide (BAA), these peptido-mimetics contain a halo-acetamidine group which covalently binds to Cys645 in the active site. These important tool molecules have aided characterisation of the wider deiminase family, and spawned more potent, second-generation inhibitors11–13. However, since these all inhibit PAD family members with similar potencies11, the precise role of PAD4 in cellular processes such as NET formation, remains poorly understood. Herein, we report the first highly potent PAD4-specific reversible inhibitors, define their novel inhibitory mechanism and confirm the enzymatic role of PAD4 in NETosis.
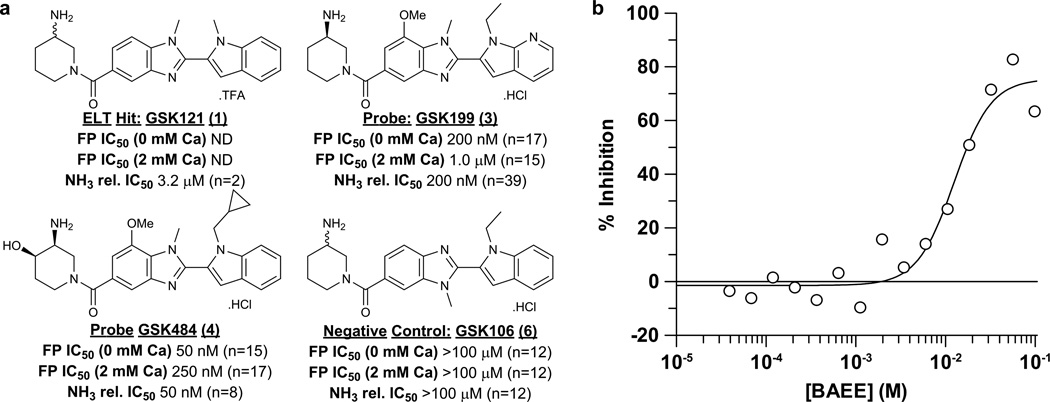
Full-length PAD4 was screened against GSK’s DNA-encoded small-molecule libraries14 with and without added calcium, leading to the identification of GSK121 (1, Fig. 1a, Supplementary Results, Supplementary Table 1). A fluorescently labelled exemplar from this series (GSK215 (2), Supplementary Fig. 1a) facilitated fluorescence polarisation (FP) ligand binding studies, conducted with and without calcium. GSK215 demonstrated high affinity binding to the low-calcium form of PAD4 (Supplementary Fig. 1b). Optimisation of GSK121 led to compounds GSK199 (3) and GSK484 (4) with IC50 potencies, in the absence of calcium, of 200 nM and 50 nM, respectively, (Fig. 1a, Supplementary Fig. 1c-d). In the presence of 2 mM calcium, we observed notably lower potencies (1 µM and 250 nM respectively). GSK199 and GSK484 also inhibited PAD4 citrullination (at 0.2 mM calcium) of benzoyl-arginine ethyl ester (BAEE) substrate in a concentration-dependent manner, as detected using an NH3 release assay. Additionally, both mass spectrometry and dialysis (Supplementary Figs 2–3) confirmed reversible binding, contrasting with the irreversible mechanism reported for the halo-acetamidine inhibitors10, 15 and their preference for the high-calcium form of PAD4.
Figure 1.
Structure and biochemical characterisation of PAD4 inhibitors. a) Summary of biochemical potency data from binding and functional assays for PAD4 inhibitors and control compound GSK106. The FP binding assay was run at a range of calcium concentrations to assess dependency. Replicate numbers are indicated in parentheses (ND = not determined). b) BAEE displacement of 10 nM GSK215 binding from PAD4 in the absence of calcium; the IC50 for BAEE was calculated as 12.5 mM. Analogous experiments in the presence of added calcium were difficult to configure and interpret due to enhanced catalytic activity and hence turnover of BAEE.
Competition studies, utilising the GSK215 FP binding assay at varying concentrations of BAEE without calcium, inferred direct competition between BAEE and GSK215 within the low-calcium form of PAD4 (Fig. 1b). Functional kinetic analysis (measuring citrullination directly) in the presence of calcium, (Supplementary Table 2), demonstrated a mixed mode of inhibition.
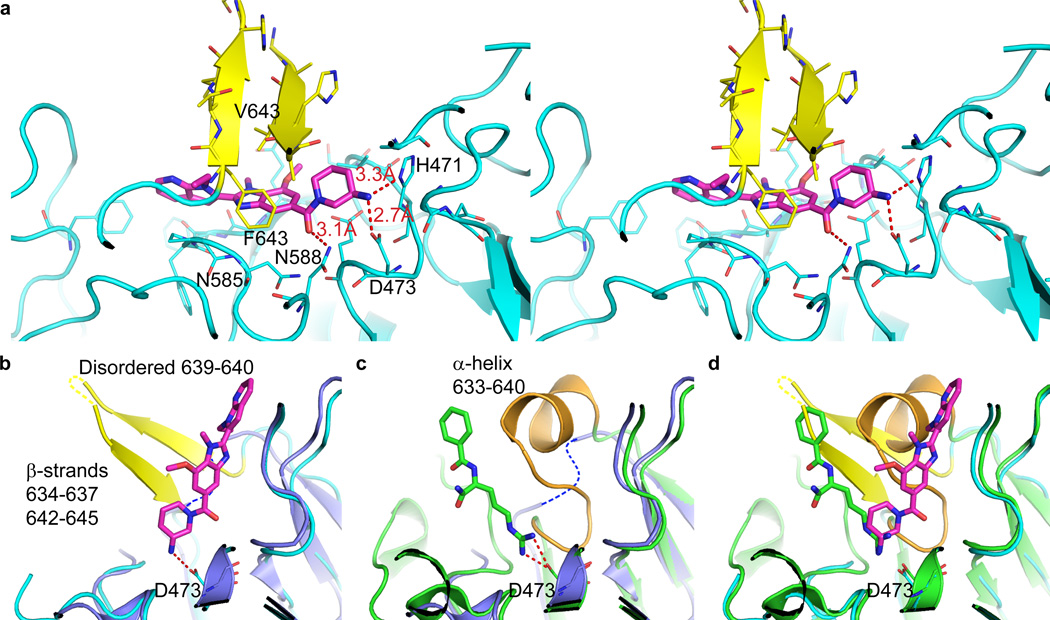
To better understand the mechanism of these molecules, we solved crystal structures of human PAD4 C645A complexed with either GSK199 at 3.3 Å or the closely related inhibitor GSK147 (5) at 3.1 Å (Supplementary Table 3). Both compounds bound in the same manner (Supplementary Figs. 4–5). However, neither structure had all five calcium sites occupied. The crystal structure of GSK199 (Fig. 2a) rationalised key SAR observed for this series. The primary amine interacted with Asp473, preserving a critical salt bridge also seen with arginine-containing ligands such as BAA (Fig. 2b-c). The proximity of the main chain NH of Asn585 to a central ring nitrogen, distance 3.6 Å, (Fig. 2a) explains why GSK106 (6, which is methylated at this position – Fig. 1a) was inactive. The ethyl group of GSK199 bound in a small hydrophobic pocket. Groups with increased complementarity to this pocket, such as the cyclopropyl of GSK484, enhanced affinity. In the absence of calcium, residues 633–645 were disordered16 (Fig. 2c). The binding of GSK199 to PAD4 induced a newly observed β-hairpin structure for these residues which allowed the hydrophobic residues Phe634 and Val643 to pack over the central part of the inhibitor (Fig. 2a-b, Supplementary Fig. 6). The close packing of Phe634 against the benzimidazole moiety of GSK199 (3.8 Å between Phe634 ring centroid and closest carbon of benzimidazole) likely accounted for the high specificity of GSK199 for PAD4 over PADs 1–3, 6, as this residue is not conserved in the other human PADs (Supplementary Fig. 7). This selectivity was found both with recombinant enzymes (Supplementary Table 4) and within cell lysates (Supplementary Fig. 8). Other pentein-fold family members, to which PAD4 belongs, also exhibit a similar induced fit of loops upon ligand binding17, 18.
Figure 2.
GSK199 binds to a re-ordered PAD4 active site. a) 3.29 Å PAD4/GSK199 complex showing hydrogen bonding interactions as dotted red lines. PAD4 is shown in cyan except for residues 633–645 (yellow), which form a hydrophobic lid with F634 and V643 packing ‘on top’ of GSK199 in this view. b) Overlay of calcium-free PAD4 structure (1WD8 – dark blue, with disordered residues 633–645 indicated by dashed blue line) with the structure of PAD4/GSK199. c) Overlay of calcium-free PAD4 structure (1WD8 – dark blue, with disordered residues 633–645 indicated by dashed blue line) with PAD4/benzoyl-L-arginine amide (BAA – green, PDB:1WDA) which has the substrate analogue inhibitor (BAA – green stick format) bound at the active site (C645A mutant). (d) Overlay of the PAD4/GSK199 and PAD4/BAA structures.
The ordering of residues 633–645, partial overlap of GSK199 with substrate (Fig. 2d; Supplementary Figs. 9–10), and lack of full occupation of calcium sites gave a structural explanation for the mixed mode of inhibition observed in kinetic inhibition experiments and the binding preference for calcium-deficient enzyme.
Both GSK199 and GSK484 showed negligible off-target activity against a panel of 50 unrelated proteins (Supplementary Table 5). Importantly, this extended to a selection of cysteine-utilising enzymes and chromatin-modifying enzymes (e.g. histone deacetylases 1–11 < 50% inhibition at 100 µM). Whilst GSK199 showed partial activation of HDACs 1 and 8 at high concentrations, the more potent GSK484 showed no activation across HDACs 1–11 even at 100 µM. A related inhibitor, immobilised on beads, clearly interacted with endogenous PAD4 in lysates from differentiated NB4 cells (Supplementary Fig. 11).
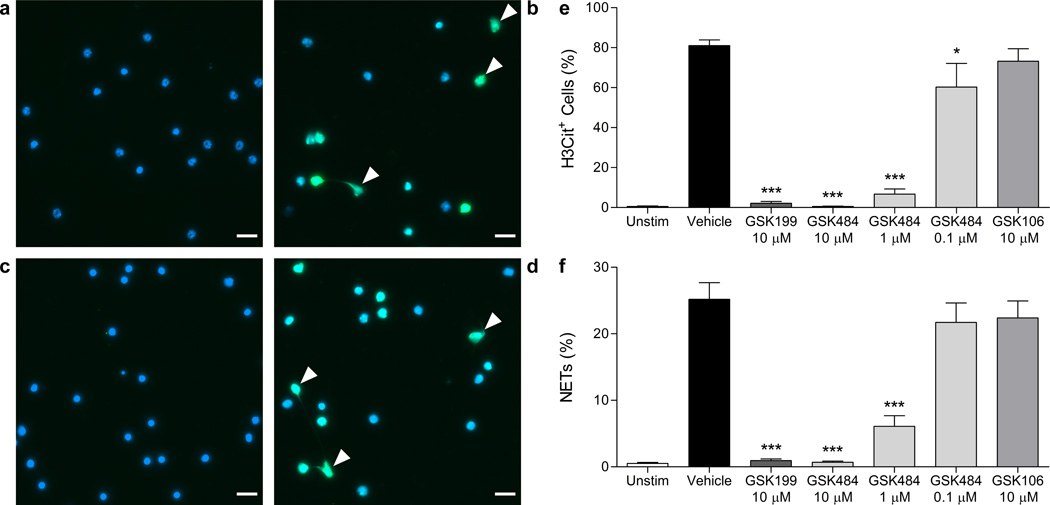
We next demonstrated inhibition of citrullination in neutrophils stimulated with calcium ionophore or bacteria by both Western blotting and a novel imaging assay (Supplementary Fig. 12) and progressed into investigating effects on in vitro NET formation in mouse and human neutrophils. In mice, ionomycin provoked histone H3 hypercitrullination, nuclear swelling and NET formation (Fig. 3a-b). Pre-treatment with 10 µM GSK484 dramatically diminished citrullination and NET formation (Fig. 3c). GSK199 produced the same effect (Supplementary Fig. 13) whilst control compound GSK106 was ineffective (Fig. 3d). GSK484 also showed a dose-dependent reduction of these endpoints (Fig. 3e-f). These data correlated with the biochemical potencies, albeit with a typical ca. ten-fold drop in cellular potency, possibly due to the biochemical assay utilising a BAEE substrate at Km, whilst H3 citrullination potency involves native substrate with unknown Km. Despite good physicochemical properties, this could also reflect cellular permeability and/or intracellular protein binding. The neutrophils exhibited no signs of either nuclear swelling or chromatin decondensation with PAD4 inhibitors (like neutrophils from PAD4−/− mice8). We also evaluated the inhibitors in human neutrophils stimulated with S. aureus (Supplementary Fig. 14), using a custom-designed computer algorithm to quantify distinct NETotic morphologies19. GSK484 caused a clear, statistically-significant reduction in diffused NETs compared with vehicle, whilst GSK199 showed less marked inhibition and GSK106 was ineffective. Parallel co-stained images (Supplementary Fig. 15) demonstrated that the diffused NETs remaining lacked citrullination. Importantly, we observed no effect on viability of neutrophils (human or mouse) or S. aureus cultures with these molecules (Supplementary Figure 16).
Figure 3.
Inhibition of mouse NETs. a) Image of control unstimulated neutrophils. b) Image of calcium ionophore-stimulated neutrophils. c) and d) Representative images of stimulated neutrophils in the presence of 10 µM GSK484 or 10 µM GSK106, respectively. All images are representative of n=5–10. Immunostaining for citrullinated histone H3 (green) was performed with DNA counterstained with Hoechst 33342 (blue). White arrows indicate NETs identified as H3Cit+ cells with spread nuclear morphology. Scale bar = 20 µm. e) Quantification of hypercitrullinated H3-stained neutrophils following ionomycin stimulation. Results are expressed as mean ± SEM and are representative of n=11 (unstimulated), n=13 (ionomycin) or n=5–9 for treatment with GSK199, GSK484 or GSK106. f) Quantification of NET-forming neutrophils following ionomycin stimulation. NETs were identified as H3Cit+ cells with spread nuclear morphology as visualised by Hoechst 33342 stain. Results are representative of n=10 (unstimulated and ionomycin treatments) or n=5–9 for treatment with GSK199, GSK484 or GSK106. *p<0.05, *** p<0.001. Effects of the inhibitors on neutrophil viability were ruled out (Supplementary Fig 16a).
Taken together, this manuscript highlights three important advances. Firstly, we describe the characterisation of the first highly-selective and reversible PAD4 inhibitors. Although second generation irreversible inhibitors such as TDFA12 demonstrate improvements in PAD4 selectivity, our data highlight GSK199 and GSK484 as the first inhibitors with a strong preference for PAD4 over other family members. We confirmed this selectivity with both recombinant enzyme and lysates from cells separately overexpressing four PAD isozymes, and via chemoproteomic mass spectrometry analysis of differentiated NB4 lysates (Supplementary Table 6), where inhibitor-conjugated beads demonstrated specific association with the PAD4 protein alone. Interestingly, mass spectrometry detected no other PAD family members specifically and, although PAD4 binds a number of proteins20–22, we identified no additional binding partners, showing that PAD4 is not captured as part of a higher order complex, at least in this configuration. Previous suggestions that intermolecular interactions with endogenous binding partners may modulate PAD4 activity in cells23, precedented by the altered calcium sensitivity recently reported with anti-PAD3/4 antibodies from RA patients24, are not evident in these data. However, this does not preclude the enzyme existing in multimeric complexes in its activated form.
Secondly, we show that inhibitors can be developed against a low calcium PAD enzyme conformation, consistent with a recent report25 but in marked contrast with inactivation of ‘active’ PAD4 by Cl-amidine10. Crystal structures of our inhibitor series show that they bind to a novel conformation of the PAD4 active site where part of the site is re-ordered to form a β-hairpin. The structures explain the mixed mode of inhibition with respect to the substrate analogue BAEE, as inhibitor binding to a low-calcium form of the enzyme would hinder the transition to the ‘active’ conformation, which binds substrates such as BAEE. The plasticity within the active site revealed by this new conformation raises the question of whether calcium may alter the substrate specificity, as well as the activity of PAD4, perhaps providing an additional mechanism of regulation.
Finally, phenotypic data from primary immune cells provide the first evidence that PAD4-selective small-molecule inhibitors can affect cellular citrullination and mimic the deficiency in NET production previously reported in PAD4-deficient mice8 and, crucially, also extend these findings to inhibition of human NETs. Since our compounds still allow partial formation of DNA-stained, diffused, NET-like structures in human neutrophils whilst fully inhibiting mouse NETosis, this process may be differentially regulated between mouse and human neutrophils. Multiple factors may account for these differences, e.g. variations in the temporal dynamics of NET formation between species, although future research could investigate whether the human NETs which remain but lack citrullination are still functional. Nevertheless, these highly selective, novel, tool molecules are now available to unlock the complex biology of NETosis further and represent potential therapeutic interventions against devastating NET diseases.
Online Methods
FP binding affinity studies
PAD4 was serially diluted in the presence of 10 nM GSK215 in Assay Buffer (100 mM HEPES, pH 8, 50 mM NaCl, 5% glycerol, 1 mM CHAPS, 1 mM DTT) at varying concentrations of calcium (0, 0.2, 2 and 10 mM). Following incubation for 50 min, apparent Kds for each calcium concentration were determined using a single site saturation curve. For IC50 determination, test compounds were serially diluted in DMSO (1% final assay concentration) and tested at the same range of calcium concentrations in the presence of PAD4 (at the calculated Kd for each calcium condition) and 10 nM GSK215 in the same assay buffer and volume. Reactions were incubated for 50 min after which IC50 values were calculated using a four-parameter logistic equation.
PAD4 functional assay
Citrullination was detected via ammonia release based on published methodology26. PAD4 was diluted to 30 nM in Assay Buffer (100 mM HEPES, 50 mM NaCl, 2 mM DTT, 0.6 mg/mL BSA, pH 8), and added to wells containing various concentrations of compound or DMSO vehicle (0.8% final) in a high volume black 384-well plate (Greiner). Following a 30 min pre-incubation at RT, the reaction was initiated by the addition of substrate (3 mM N-α-benzoyl-L-arginine ethyl ester (BAEE) in 100 mM HEPES, 50 mM NaCl, 600 µM CaCl2, 2 mM DTT, pH 8). The reaction was stopped after 60 min by the addition of stop/detection buffer containing 50 mM EDTA, 2.6 mM o-phthalaldehyde and 2.6 mM DTT. Assays were incubated at RT for 90 min before measuring fluorescence (λex 405/λem 460) on an Envision plate reader (Perkin Elmer).
Calculation and measurement of physicochemical properties
CHROMlogD and solubility (by chemiluminescent nitrogen detection) were measured using previously-described methodology27. cLogP values were calculated using Daylight software v4.9: http://www.daylight.com/dayhtml/doc/clogp/index.html (accessed 27th June, 2013).
FP substrate competition studies
BAEE competition was determined using the PAD4 FP ligand assay with no added calcium. 10 nM GSK215 was added to mixtures of BAEE and PAD4 (60 nM, ~1 × Kd) in Assay Buffer (100 mM HEPES, pH 8, 50 mM NaCl, 5% glycerol, 1 mM CHAPS, 1 mM DTT). Assay plates were read immediately and the data processed using a four-parameter logistic equation to determine the IC50 of BAEE.
Protein expression and purification
The inactive mutant, PAD4 C645A, with N-terminal 6His-FLAG tag and PreScission Protease cleavage site downstream of the tag was expressed in BL21 (DE3) E. coli cells in Magic Media (Life Technologies). Cell pellet was lysed at RT in BugBuster Protein Extraction Reagent (Novagen, 70584-4), containing 1 mM TCEP, protease inhibitor cocktail (Sigma, P8340), Lysozyme (200 µg/mL) and Benzonase (25 U/mL). The lysate, clarified by centrifugation, was purified by Ni-NTA affinity chromatography using a HisTrap column. Protein was eluted from the column with Imidazole in 50 mM Tris-HCl, pH 8, 400 mM NaCl, 1 mM EDTA buffer solution and further purified on anti-FLAG M2 affinity resin. The protein solution was batch incubated with the anti-FLAG resin for 1h and then the resin was washed with 50 mM Tris-HCl, pH 8, 400 mM NaCl buffer and packed into the column. In order to cleave the 6His-FLAG tag from the resin-bound protein, the wash buffer was removed and replaced with 1 column volume of buffer containing 50 mM Tris-HCl, pH 8, 400 mM NaCl, 1 mM EDTA and PreSission protease (25 U/mL) and left overnight at 4 °C. The cleaved protein was eluted with 50 mM Tris-HCl, pH 8, 400 mM NaCl buffer. The PreSission protease was removed from the cleaved protein by glutathione affinity chromatography utilising the GST tag.
Crystallisation
Native PAD4 crystals were grown by vapour diffusion at 293K by mixing protein (5 mg/mL in 50 mM Tris-HCl, pH 8, 400 mM NaCl, 0.5 mM TCEP) with precipitant (100 mM imidazole, pH 7.8, 200 mM Li2SO4, 2 mM TCEP, 6–7.5% PEG 2K MME) at a protein concentration of 5 mg/mL. Prior to ligand soaking, a pre-incubation step was used to stabilise the crystals; this was achieved by transfer of a crystal to a solution containing 100 mM imidazole pH 7.8, 2 mM TCEP, 50 mM CaCl2 (60 min) and then to the same solution with an additional 25 mM ligand (GSK147 or GSK199) for 14 h. Cryogenic protection was achieved from a solution containing 100 mM imidazole pH 7.8, 35% Ethylene Glycol, 2 mM TCEP, 50 mM CaCl2 and 25 mM ligand, which was added stepwise to the ethylene glycol free drop until a four-fold increase in the crystal soaking drop size was achieved. Crystals were then transferred to the full concentration cryogenic solution prior to freezing in liquid nitrogen. Attempts to soak compounds into PAD4 crystals in the absence of CaCl2 failed, causing degrading crystals which diffracted very poorly.
Data collection
Diffraction data were collected to resolutions of 3.1 Å (GSK147) and 3.29 Å (GSK199) on I02 at Diamond Light Source using a pilatus 6M detector at a wavelength of 0.9795 Å. Data were processed using Xia2. Merging data with Aimless gave estimated resolution limits along a*, b* and c* of 3.03, 3.8, and 3.03 Å for the GSK147 dataset and 3.29, 3.52, and 3.29 Å for the GSK199 dataset, showing that the data were anisotropic.
Structure solution and refinement
The two datasets (Supplementary Table 3) were in a C2 cell with similar dimensions to those of all other human PAD4 structures in the PDB. The 3.1 Å structure of the PAD4 complex with GSK147 was refined first. The calcium-free human PAD4 structure (PDB code 3APN) was used as the starting model for refinement, and after rigid body refinement (three domains) the R-factor (R-free) was 31.6% (32.4%) respectively. Cycles of rebuilding with Coot28, and refinement with Refmac29, Buster30 and Phenix31, produced the final refined PAD4/GSK147 structure. Waters were placed in peaks >3.5sigma in Fo-Fc maps when there were waters at equivalent positions in the 2.1 Å PAD4 structure (2DEX); in addition three waters were modelled into density adjacent to Cys144 (this density likely represents an unknown covalent modification of Cys144), and one water was placed at a crystal contact. The structure of the GSK199 complex with PAD4 was solved from the GSK147 complex and refined in a similar manner (the R-free set was transferred from the GSK147 dataset to the GSK199 dataset to reduce bias in the refinement of the 3.29 Å PAD4/GSK199 complex). The refinement statistics for the two complexes are summarised in Supplementary Table 3; both final structures have 96% of residues in favoured Ramachandran regions and no residues as outliers. The final 2Fo-Fc electron density map for the PAD4/GSK147 complex and difference maps (Fo-Fc) for both ligands are shown in Supplementary Fig. 5, whilst the final electron density map for the two new β-strands (634–637 and 624–647) in the PAD4/GSK199 complex is shown in Supplementary Fig. 6. In both structures, the aminopiperidine and benzimidazole moieties are well defined, forming directional interactions with residues in the PAD4 binding site (Figure 2a) and are enclosed in the binding site by the newly formed β-strands, (Supplementary Figures 4, 5, 9, 10). The pyrrolopyridine moiety is less well defined. It is in a much more solvent-exposed environment than the other parts of the compound and does not form any directional interactions with the site which would anchor it in a single position. It is, therefore, likely to have greater mobility. The higher temperature factors observed for the pyrrolopyridine (Supplementary Table 2) are consistent with static and/or dynamic disorder for this group.
Mouse NET assays
Peripheral blood neutrophils were isolated from mouse blood by density centrifugation followed by hypotonic lysis. Neutrophil purity was assessed (by Wright-Giemsa staining of cytospins) to be routinely >90%. Cells were incubated at 37 °C in RPMI with 10 mM HEPES and allowed to settle and adhere to non-coated ‘No. 1.5’ glass-bottom wells for 15 min. Compounds or vehicle controls were added to each well for 20 min, prior to stimulation with 4 µM ionomycin (Invitrogen). Following incubation at 37 °C for 2 h, cells were fixed in 2% PFA overnight at 4 °C. After permeabilisation with PBS/0.1% Triton X-100, and blocking with PBS/3% BSA, cells were incubated with an anti-citrullinated histone H3 antibody (Abcam, ab5103, 0.3 µg/mL) overnight at 4 °C, washed with PBS and incubated with 1 µg/mL Alexa Fluor 488-conjugated donkey anti-rabbit IgG (Life Technologies, A21206). Nuclei were counterstained with Hoechst 33342 (1 µg/mL, Invitrogen). Fluorescent images were acquired using an Axiovert 200 inverted fluorescence microscope (Zeiss) in conjunction with a Zeiss Axiocam MRm monochromatic CCD camera and analysed with ImageJ software. Cells were identified as H3Cit+ or as spread NETs based on nuclear morphology by Hoechst 33342 stain as previously described32. All quantification was performed in a blinded fashion.
Steady state citrulline production assays
The kinetic parameters were determined, using a standard citrulline production assay9 in the presence and absence of inhibitor. Briefly, varying concentrations of BAEE in reaction buffer (50 mM HEPES, 50 mM NaCl, 2 mM DTT, 10 mM CaCl2) were pre-incubated at 37 °C for 10 min prior to the addition of a PAD isozyme (0.2–0.5 µM). After incubation at 37 °C for 6 min, samples were frozen in liquid nitrogen and citrulline levels quantified using the COLDER assay23. The initial rates thus obtained were fitted to eq 1,
| eq 1 |
using GraFit version 5.0.1.1. For the assays performed in the presence of inhibitor, the data were globally fitted to equations representative of competitive (eq 2), non-competitive (eq 3; Kis = Kii), and mixed inhibition (eq 3; Kis ≠ Kii),
| eq 2 |
| eq 3 |
where v is velocity, Vmax is the maximum rate, Km is the Michaelis-Menten constant, [S] is the substrate concentration, [I] is the inhibitor concentration, Kis is the dissociation constant for the EI complex, and Kii is the dissociation constant for the ESI complex. The most accurate fits of the data were determined by comparison of the reduced chi2 values, as well as by visually inspecting the global data fits and the Lineweaver-Burk Plots.
Mass spectrometry compound reversibility studies
10 µM test compounds (1% DMSO final assay concentration) were incubated with 2 µM PAD4 in the presence and absence of 10 mM calcium in assay buffer (100 mM HEPES, pH 8, 50 mM NaCl, 5% glycerol and 1 mM CHAPS) overnight. Samples were run through an Agilent 6224 Time of Flight mass spectrometer attached to a 1200 LC system to determine if covalent binding to PAD4 was observed with any of the compounds. Two controls were run in the assay: a DMSO (no compound) control and a known irreversible compound (Cl-amidine).
Dialysis compound reversibility studies experiments
PAD4 (2 µM final) was incubated with 100 µM GSK199 for 1 h at 37 °C. A 6 µL aliquot of this mixture was assayed for residual activity by incubating the enzyme in reaction buffer (50 mM HEPES, 50 mM NaCl, 2 mM DTT, 10 mM CaCl2) containing 10 mM BAEE which had been pre-incubated at 37 °C for 10 min prior to the addition of enzyme. Following incubation at 37 °C for 6 min, samples were frozen in liquid nitrogen and citrulline levels quantified using the COLDER assay23. The remaining enzyme was transferred to a 3.5 kDa Slide-A-Lyzer dialysis cassette and dialysed against long-term storage buffer (20 mM Tris-HCl pH 8, 1 mM EDTA, 2 mM DTT, 500 mM NaCl, 10% glycerol) for 18 h at 4 °C. The residual activity present in this sample was then analysed using the methodology described above. Enzymatic activity was normalised by setting the samples without inhibitor to 100% activity.
PAD4 chemoproteomic binding assay
All cultures were performed at 37 °C in a humid atmosphere of 5% CO2. NB4 cells (DSMZ, ACC-207) were grown in RPMI medium supplemented with 10% Foetal Bovine Serum and differentiated with 1.25% DMSO for 24 h followed by 1 µM all-trans retinoic acid (Tocris Bioscience, 0695) for another 24 h. CGB208 (7) was immobilised on Sepharose beads as previously described33. CGB208 beads were incubated at 4 °C for 1 h with NB4 cell extract (5 mg/mL), which had been pre-incubated with inhibitor or vehicle alone. Beads were transferred to disposable columns, washed with lysis buffer and eluted with 2× SDS sample buffer. Proteins were separated by SDS-PAGE (NuPAGE 4–12% Bis-Tris, Life Technologies) and transferred to PVDF membranes. For concentration-dependent inhibition curves, 5 µL of CGB208 beads and 100 µL (5 mg/mL) of lysate were used per well (96-well filter plate, Millipore, MSHVN4550). Eluted samples were loaded directly on to nitrocellulose membrane. PAD4 was detected using an anti-PAD4 antibody (Abcam, ab50332).
Citrullination assay in HEK293 cells
HEK293 cells stably expressing N-terminal FLAG-tagged PAD1, PAD2, PAD3 or PAD4 were engineered by retroviral transduction as described34. Cells were grown in 15 cm diameter plates to subconfluency in DMEM supplemented with 10% Foetal Bovine Serum, harvested by centrifugation and washed once in PBS/2 mM EGTA. Cells were lysed in 50 mM Tris-Cl, pH 7.4, 1.5 mM MgCl2, 5% glycerol, 150 mM NaCl, 25 mM NaF, 1 mM Na3VO4, 0.4% NP40, 1 mM DTT with protease inhibitors. Lysates were pre-incubated for 20 min at 4 °C with DMSO alone (2%), 100 µM of GSK199, GSK484, GSK106 or 200 µM Cl-amidine (J&W Pharmalab, 15-0242S). Citrullination reactions were performed for 30 min at 37 °C in the presence of 2 mM calcium. Extracts were loaded on to gels, proteins separated by SDS-PAGE and transferred to PVDF membranes. Citrullinated proteins were then chemically modified following manufacturer’s instructions (Millipore, 17-347) and detected using anti-modified citrulline antibody (Millipore, 07-2168). FLAG-PAD constructs were detected using anti-FLAG antibody (Sigma, F-1804).
Neutrophil citrullination blots
Human biological samples were sourced ethically and their research use was in accordance with the terms of written informed consents. Neutrophils were isolated from human peripheral blood using 3% dextran density gradient, followed by centrifugation in Ficoll and erythrocytes lysed in 0.2% w/v saline. Neutrophils were >90–95% pure. Cells were resuspended at a density of 2×105/mL and pre-treated with compounds for 30 min. Cells were then stimulated with either 1 µM calcium ionophore (final concentration) for 45 min or S. aureus (ATCC 13709) at 10× MOI (multiplicity of infection) for 90 min, or left unstimulated as controls. During treatment, the cells were incubated at 37 °C, 5% CO2. Following stimulation, cells were centrifuged and the pellets treated on ice with lysis buffer (62.5 mM Tris-HCl pH 6.8, 1% SDS, 5% 2-mercaptoethanol) and frozen overnight. The following day, the lysates were thawed on ice and sonicated for 6× 30 sec bursts at 4 °C in a Bioruptor (Diagenode) connected to a refrigerated bath circulator (Thermo Scientific, NESLAB RTE7). Lysates were separated by SDS-PAGE. Western immunoblotting was performed to detect citrullination using the polyclonal H3Cit antibody (Abcam, ab5103), with MAPK being used as a loading control (Cell Signalling #9102). Pan-citrullination was detected using the AMC kit (Millipore, 17-347). All blots were probed with an HRP-linked secondary antibody and exposed on Image Station 4000 MM PRO (Carestream), generating digital images of the blots.
H3Cit imaging assays
Human neutrophils were isolated as above and diluted to a density of 3.5×105/mL. Cells were incubated with compounds in 384-well micro-titre plates for 45 min at 37 °C, 5% CO2. Cells were then stimulated with 2 µM calcium ionophore (final concentration) for 60 min at 37 °C. The cells were fixed with 1.3% PFA for 45 min at RT, then permeabilised in PBS/2% Triton X-100 for 10 min. Cells were washed with PBST (PBS/0.1% Tween 20) and blocked in PBST/2% BSA for 16 h at 4 °C. H3 citrullination was measured using a mouse anti-H3Cit monoclonal antibody (BioCat, 133483, generated at GSK, 6 µg/mL) and a secondary Alexa Fluor 488 goat anti-mouse IgG (Life Technologies, A11001, 2 µg/mL) with parallel Hoechst 33342 staining (Life Technologies). Plates were imaged on an IN Cell Analyzer 2000 (GE Healthcare) and the FITC intensity in neutrophil nuclei was used as a measure of H3 citrullination as determined using the IN Cell Investigator software (GE Healthcare).
Human NET assays
Pathophysiological stimulation with S. aureus (ATCC, 13709) was precedented by previous work35. Isolated neutrophils were allowed to adhere to 96-well culture plates (Costar) in RPMI (2.5×104 cells/well) for 20 min at 37 °C, 5% CO2. Neutrophils were pre-incubated for 30 min with PAD4 inhibitors or vehicle controls (0.1% DMSO final concentration). S. aureus from colonies grown on agar, was shaken in LB at 37 °C for 20 h. The bacteria were washed in ice-cold PBS prior to addition to the neutrophils at 5× MOI (5 bacteria per neutrophil). Following stimulation for 3 h at 37 °C, cells were fixed with 2% PFA. Cells were stained with Hoechst 33342. Plates were imaged on an Opera confocal imaging system (Perkin Elmer) and cells were classified as being 1) lobulated, 2) delobulated or 3) diffused NETS via an algorithm developed using the Acapella software (Perkin Elmer). Images were captured and analysed from 60 fields across 3 wells for each treatment.
Viability was assessed using Cell Titer Glo (Promega) following incubation of human (and mouse) neutrophils with compound alone for identical durations as used in parallel NET studies.
Co-staining of DNA and citrullinated histone H3 protein in stimulated human neutrophils
Neutrophils were stimulated with bacteria (5× MOI) for 3 h at 37 °C, 5% CO2, fixed with 2% PFA and stained with Hoechst 33342. Following a PBS wash the cells were permeabilised with PBS/0.1% Triton X-100 for 15 min on ice. The plates were blocked with PBS/0.1 % Triton X-100/3% BSA for 45 min at 37 °C and then stained with rabbit anti-H3Cit antibody (Abcam, ab5103), for 6 h at RT. The plates were washed in PBS and stained with anti-rabbit IgG Alexafluor 488 for 2 h at RT to allow co-imaging of DNA and H3Cit. The plates were stored in PBS at 4 °C until they were imaged using a Nikon inverted microscope attached to a Leica RGB camera. Images captured for Hoechst 33342 and Alexafluor 488 staining were merged using Adobe Photoshop CS.
Supplementary Material
Acknowledgements
This work was supported in part by NIH grants GM079357 (to P.R.T.) and R01 HL102101 (to D.D.W.). We thank the following colleagues for their medicinal and synthetic chemistry contributions to this manuscript: Dominique Amans, Hawa Diallo, Clément Douault, Neil Garton, Katherine Jones, Jessica Renaux, Tracy Shipley, Ann Walker, Bob Watson and Chris Wellaway. We also gratefully acknowledge Sarah Ready for diligently preparing and quantifying S. aureus for neutrophil studies. For the chemoproteomics data, we would like to thank Markus Boesche and Cellzome’s tissue culture and target validation teams for technical expertise, and Marcus Bantscheff for stimulating discussion. We thank Margarete Neu and Paul Homes for assistance for structural studies, and Bob Nolte for advice on crystallography.
Footnotes
Author contributions
H.D.L., J.L. and J.E.C. led the project, designed the research, interpreted data and drafted the manuscript with input from all authors. S.J.A., M.D.B., M.C. R.J.S. and D.M.W. synthesised or designed key compounds. K.E.L., C.E.R. and G.Y. performed and analysed the screening which identified the PAD4 inhibitors. K.L.B., R.P.B., P.D.C., K.L. and D.J.S. designed the biochemical assays and deduced the MoA of compounds. D.E., G.J. and G.D. generated chemproteomic data. Y.H.C., R.P.D., D.E., C.M., K.M., C.P. and M.R. performed cellular assays. For structures, O.P purified protein, J.T. crystallised protein, soaked crystals and collected data, B.D.B. refined structures and P.T. performed sequence and structural analysis. C.W.C., D.D.W., P.R.T., R.K.P. and D.M.W. guided aspects of this work. C.M, S.J.A, C.W.C and H.D.L. also contributed invaluably to the revision and formatting of the final manuscript.
Competing financial interests
The authors declare competing financial interests. P.R.T. is a co-founder and consultant to Padlock Therapeutics. The majority of the other authors, as indicated, are pharmaceutical industry employees and shareholders.
Reference List
- 1.Jones JE, Causey CP, Knuckley B, Slack-Noyes JL, Thompson PR. Protein arginine deiminase 4 (PAD4): Current understanding and future therapeutic potential. Curr. Opin. Drug Discov. Devel. 2009;12:616–627. [PMC free article] [PubMed] [Google Scholar]
- 2.Brinkmann V, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 3.Ohlsson SM, et al. Neutrophils from vasculitis patients exhibit an increased propensity for activation by anti-neutrophil cytoplasmic antibodies. Clin. Exp. Immunol. 2014 doi: 10.1111/cei.12301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hakkim A, et al. Impairment of neutrophil extracellular trap degradation is associated with lupus nephritis. Proc. Natl. Acad. Sci. U. S. A. 2010;107:9813–9818. doi: 10.1073/pnas.0909927107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Villanueva E, et al. Netting neutrophils induce endothelial damage, infiltrate tissues, and expose immunostimulatory molecules in systemic lupus erythematosus. J. Immunol. 2011;187:538–552. doi: 10.4049/jimmunol.1100450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinod K, et al. Neutrophil histone modification by peptidylarginine deiminase 4 is critical for deep vein thrombosis in mice. Proc. Natl. Acad. Sci. U. S. A. 2013;110:8674–8679. doi: 10.1073/pnas.1301059110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark SR, et al. Platelet TLR4 activates neutrophil extracellular traps to ensnare bacteria in septic blood. Nat. Med. 2007;13:463–469. doi: 10.1038/nm1565. [DOI] [PubMed] [Google Scholar]
- 8.Li P, et al. PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J. Exp. Med. 2010;207:1853–1862. doi: 10.1084/jem.20100239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knuckley B, et al. Substrate specificity and kinetic studies of PADs 1, 3, and 4 identify potent and selective inhibitors of protein arginine deiminase 3. Biochemistry. 2010;49:4852–4863. doi: 10.1021/bi100363t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo Y, et al. Inhibitors and inactivators of protein arginine deiminase 4: functional and structural characterization. Biochemistry. 2006;45:11727–11736. doi: 10.1021/bi061180d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Causey CP, et al. The development of N-alpha-(2-carboxyl)benzoyl-N(5)-(2-fluoro-1-iminoethyl)-l-ornithine amide (o-F-amidine) and N-alpha-(2-carboxyl)benzoyl-N(5)-(2-chloro-1-iminoethyl)-l-ornithine amide (o-Cl-amidine) as second generation protein arginine deiminase (PAD) inhibitors. J. Med. Chem. 2011;54:6919–6935. doi: 10.1021/jm2008985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones JE, et al. Synthesis and screening of a haloacetamidine containing library to identify PAD4 selective inhibitors. ACS Chem. Biol. 2012;7:160–165. doi: 10.1021/cb200258q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, et al. Anticancer peptidylarginine deiminase (PAD) inhibitors regulate the autophagy flux and the mammalian target of rapamycin complex 1 activity. J. Biol. Chem. 2012;287:25941–25953. doi: 10.1074/jbc.M112.375725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark MA, et al. Design, synthesis and selection of DNA-encoded small-molecule libraries. Nat. Chem. Biol. 2009;5:647–654. doi: 10.1038/nchembio.211. [DOI] [PubMed] [Google Scholar]
- 15.Luo Y, Knuckley B, Lee YH, Stallcup MR, Thompson PR. A fluoroacetamidine-based inactivator of protein arginine deiminase 4: design, synthesis, and in vitro and in vivo evaluation. J. Am. Chem. Soc. 2006;128:1092–1093. doi: 10.1021/ja0576233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arita K, et al. Structural basis for Ca(2+)-induced activation of human PAD4. Nat. Struct. Mol. Biol. 2004;11:777–783. doi: 10.1038/nsmb799. [DOI] [PubMed] [Google Scholar]
- 17.Linsky T, Fast W. Mechanistic similarity and diversity among the guanidine-modifying members of the pentein superfamily. Biochim. Biophys. Acta. 2010;1804:1943–1953. doi: 10.1016/j.bbapap.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Humm A, Fritsche E, Steinbacher S, Huber R. Crystal structure and mechanism of human L-arginine:glycine amidinotransferase: a mitochondrial enzyme involved in creatine biosynthesis. EMBO J. 1997;16:3373–3385. doi: 10.1093/emboj/16.12.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hakkim A, et al. Activation of the Raf-MEK-ERK pathway is required for neutrophil extracellular trap formation. Nat. Chem. Biol. 2011;7:75–77. doi: 10.1038/nchembio.496. [DOI] [PubMed] [Google Scholar]
- 20.Li P, et al. Regulation of p53 target gene expression by peptidylarginine deiminase 4. Mol. Cell Biol. 2008;28:4745–4758. doi: 10.1128/MCB.01747-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li P, et al. Coordination of PAD4 and HDAC2 in the regulation of p53-target gene expression. Oncogene. 2010;29:3153–3162. doi: 10.1038/onc.2010.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slack JL, Jones LE, Jr, Bhatia MM, Thompson PR. Autodeimination of protein arginine deiminase 4 alters protein-protein interactions but not activity. Biochemistry. 2011;50:3997–4010. doi: 10.1021/bi200309e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kearney PL, et al. Kinetic characterization of protein arginine deiminase 4: a transcriptional corepressor implicated in the onset and progression of rheumatoid arthritis. Biochemistry. 2005;44:10570–10582. doi: 10.1021/bi050292m. [DOI] [PubMed] [Google Scholar]
- 24.Darrah E, et al. Erosive Rheumatoid Arthritis Is Associated with Antibodies That Activate PAD4 by Increasing Calcium Sensitivity. Sci. Transl. Med. 2013;5:186ra65. doi: 10.1126/scitranslmed.3005370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewallen DM, et al. A FluoPol-ABPP PAD2 high-throughput screen identifies the first calcium site inhibitor targeting the PADs. ACS Chem. Biol. 2014;9:913–921. doi: 10.1021/cb400841k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sugawara K, Oyama F. Fluorogenic reaction and specific microdetermination of ammonia. J. Biochem. 1981;89:771–774. doi: 10.1093/oxfordjournals.jbchem.a133257. [DOI] [PubMed] [Google Scholar]
- 27.Young RJ, Green DV, Luscombe CN, Hill AP. Getting physical in drug discovery II: the impact of chromatographic hydrophobicity measurements and aromaticity. Drug Discov. Today. 2011;16:822–830. doi: 10.1016/j.drudis.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 28.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D. Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smart OS, et al. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. D. Biol. Crystallogr. 2012;68:368–380. doi: 10.1107/S0907444911056058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Demers M, et al. Cancers predispose neutrophils to release extracellular DNA traps that contribute to cancer-associated thrombosis. Proc. Natl. Acad. Sci. U. S. A. 2012;109:13076–13081. doi: 10.1073/pnas.1200419109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bantscheff M, et al. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat. Biotechnol. 2007;25:1035–1044. doi: 10.1038/nbt1328. [DOI] [PubMed] [Google Scholar]
- 34.Bouwmeester T, et al. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat. Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 35.Fuchs TA, et al. Novel cell death program leads to neutrophil extracellular traps. J. Cell Biol. 2007;176:231–241. doi: 10.1083/jcb.200606027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.