Abstract
Post-translational histone modifications are believed to allow the epigenetic transmission of distinct chromatin states, independently of associated DNA sequences. H3K9 methylation is essential for heterochromatin formation, however, a demonstration of its epigenetic heritability is lacking. Fission yeast has a single H3K9 methyltransferase, Clr4, that directs all H3K9 methylation and heterochromatin. Utilizing releasable tethered Clr4 reveals that an active process rapidly erases H3K9 methylation from tethering sites in wild-type cells. However, inactivation of the putative histone demethylase Epe1 allows H3K9 methylation and silent chromatin maintenance at the tethering site through many mitotic divisions, and transgenerationally through meiosis, after release of tethered Clr4. Thus, H3K9 methylation is a heritable epigenetic mark whose transmission is usually countered by its active removal, which prevents the unauthorised inheritance of heterochromatin.
In most eukaryotes the methylation of nucleosomal histone H3 on lysine 9 (H3K9me) is required for the assembly of constitutive heterochromatin (1). H3K9me2/3 is bound by HP1/Swi6 proteins and Suv39/Clr4 H3K9 methyltransferases to form heterochromatic regions (2-6). Since Suv39/Clr4 can bind the H3K9me2/3 mark that they generate, and HP1 proteins may also facilitate recruitment of these methyltransferases (7), it is thought that H3K9 methylation, and heterochromatin, can be maintained by self-propagation, even when the initiator is withdrawn (8, 9). However, in eukaryotic systems that exhibit overtly heritable chromatin states there is often a tight relationship between DNA methylation, H3K9 methylation and heterochromatin, confounding analyses of the heritability of H3K9 methylation (10, 11). Fission yeast lacks DNA methylation and a single non-essential methyltransferase, Clr4 (Suv39 ortholog), is responsible for all H3K9me-dependent heterochromatin (12). Thus fission yeast is an ideal system in which to determine whether H3K9me-dependent heterochromatin is truly heritable. Clr4 normally requires sequence-directed targeting to particular chromosomal regions via RNAi in a process involving heterochromatin nucleation, spreading and maintenance (13-15). The inheritance of heterochromatin on centromere repeat DNA inserted at ectopic locations also requires RNAi and mating-type locus heterochromatin is dependent on DNA binding factors in the absence of RNAi (13-17). However, constitutive tethering of Clr4 to a euchromatic locus via the Gal4 DNA binding domain (GBD) allows the assembly of an extensive domain of H3K9me-heterochromatin, independently of RNAi (18). Here we tether a regulatable TetRoff-Clr4 fusion protein to determine if H3K9me is a persistent histone modification that can be stably copied through mitotic cell divisions and meiosis following release or loss of the TetRoff-Clr4 initiator.
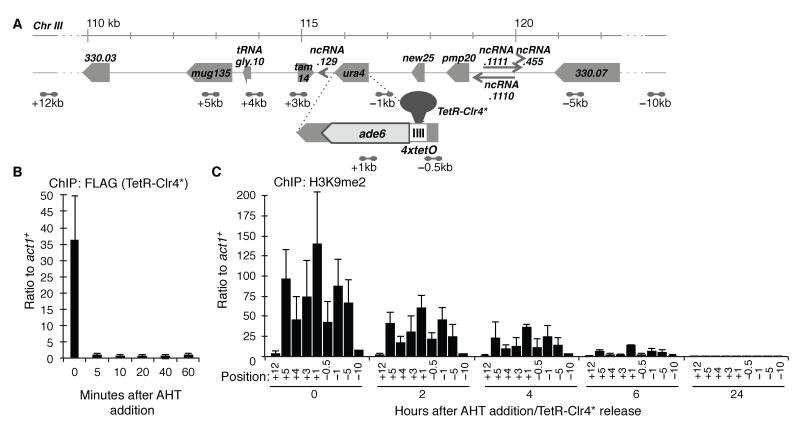
TetRoff-2xFLAG-Clr4-cdd fusion protein (abbreviated TetR-Clr4*), lacking the Clr4 chromodomain, was stably expressed in cells with an ade6+ gene downstream of 4xtetO binding sites at the ura4 locus (4xtetO-ade6+; Fig. 1A) (19). TetR-Clr4* silences 4xtetO-ade6+ independently of RNAi (ago1Δ, dcr1Δ), similar to GBD-Clr4 (18), resulting in reduced RNAPII association, and high H3K9me2 levels and silencing over a broad region (Fig. 1C, fig. S1, and fig. S9C-E). TetR-Clr4* is released within 5 minutes from tetO sites by addition of anhydrotetracycline (AHT; Fig. 1B). All strains utilised also express wild-type Clr4 which can interact via its chromodomain with TetR-Clr4-directed H3K9me and thus potentially use its read-write capabilities to methylate newly incorporated H3 and allow heterochromatin transmission to daughter cells following TetR-Clr4* release. However, in a time course, H3K9me2 rapidly declines over 4xtetO-ade6+ following AHT addition; >90% is lost within six hours (Fig. 1C, and fig. S2A). AHT itself does not affect endogenous heterochromatin integrity (fig. S2B). H3 levels do not decline on 4xtetO-ade6+ over this period (fig. S2A). Swi6HP1 is also lost from 4xtetO-ade6+ when cells are grown with AHT (fig. S1F).
Fig. 1. H3K9 methylation is rapidly lost upon release of tethered TetR-Clr4*.
(A) Positions of 4xtetO, tethered TetR-Clr4* beside ade6+ at ura4, and surrounding S. pombe chromosome III genes. Dumbbells indicate primer pairs.
(B and C) qChIP time course of FLAG-TetR-Clr4* (B) and H3K9me2 (C) levels on 4xtetO-ade6+ following AHT addition using indicated primers. Data are mean ±SD (n=3), P<0.05 (t-test).
We also tethered TetR-Clr4* within two non-essential genes with long open reading frames, which are less likely to contain unannotated features that might interfere with heterochromatin integrity. Moreover, both sib1+ (15,005 bp) and vps1302+ (9,200 bp) exhibit expression levels and rates of H3 turnover that are ~3-fold lower than those of ade6+ (Fig. 2A-C, and fig. S3). 4xtetO and 1xtetO sites were placed within sib1 and vps1203, respectively (Fig. 2D and E). sib1:4xtetO and vps1302:1xtetO were also placed under the control of low, medium (med), and high versions of the constitutive adh1 promoter (20). We also generated sib1:4xtetO and vps1302:1xtetO without promoters (no). All strains expressed wild-type Clr4 and TetR-Clr4*. Both sib1+ and vps1302+ were expressed at low levels when their promoters were removed and at much higher levels from med-adh1 or high-adh1 compared to their own, or low-adh1, promoters (fig. S4). TetR-Clr4* was unable to establish significant levels of H3K9me2 when tethered to sib1:4xtetO or vps1302:1xtetO expressed from hi-adh1 and relatively low levels when expressed from med-adh1, but substantial H3K9me2 occurred when either gene had no, its own or, the low-adh1 promoter (fig. S4). However, as with 4xtetO-ade6+, rapid loss of H3K9me2 followed TetR-Clr4* release from even no and own promoter constructs; again >90% was lost within 6 hours (Fig. 2, D and E). Although high levels of transcription across tethering sites prevents the establishment of H3K9me by TetR-Clr4*, neither low promoter strength nor low H3 turnover renders H3K9me more persistent upon methyltransferase release. Thus the inability to maintain H3K9 methylation upon removal of the initiating tethered Clr4 methyltransferase is likely a general feature of euchromatic loci.
Fig. 2. Tethering TetR-Clr4* at loci with low expression and histone turnover does not stablize H3K9 methylation.
(A) Read distribution (log2RPKM) from S. pombe polyA RNA-seq relative to gene length. ade6+, sib1+ and vps1302+ indicated.
(B) qRT-PCR of ade6+, sib1+ and vps1302+ RNA levels. Data are mean ±SD (n=3) P<0.005 (t-test)
(C) Recombination-induced tag exchange monitoring incorporation of new H3-T7 on act1+, ade6+, sib1+ vps1302+ and cen-dg repeats. Data are mean ±SD (n=3). H3 turnover on sib1+ and vps1302+ was significantly lower than on act1+ and ade6+ P<0.05 (t-test).
(D and E) sib1+ and vps1302+ lose H3K9me2 after TetR-Clr4* release. Position of tetO sites within sib1 and vps1302. own promoters were replaced with ura4+ (no) or swapped to low, medium (med), or high adh1 promoter versions (20). Dumbbells indicate primers. qChIP of H3K9me2 levels, at time points relative to AHT addition, on sib1:4xtetO (D) and vps1302:1xtetO (E) with no or indicted promoters. Data are mean ±SD (n=3), P<0.05 (t-test). H3K9me2 level within sib1 carrying its own promoter is decreased with a probability of respectively P=0.068 and P=0.051, 3h and 6h following TetR-Clr4* release.
To determine if the loss of H3K9 methylation from the tethering site is coupled to replication or passage through the cell cycle we released TetR-Clr4* from 4xtetO-ade6+ in cdc25-22 synchronized cultures (Fig. 3A). H3K9me2 levels on 4xtetO-ade6+ dropped by 70% within one cell cycle following the addition of AHT to these synchronized cultures and no accelerated H3K9me2 loss was evident during S phase which is coincident with septation (21). We also released TetR-Clr4* from 4xtetO-ade6+ in non-cycling G2 blocked cdc25-22 cells (Fig. 3B). TetR-Clr4* was lost from 4xtetO-ade6 within an hour and H3K9me declined to less than 25% of initial levels within 4 hours. Thus, after release of the initiating methyltransferase, rather than being passively diluted through chromatin replication, H3K9 methylation must be removed by an active process.
Fig. 3. H3K9 methylation rapidly declines through the cell cycle and in non-cycling cells.
Regimes for release of TetR-Clr4* from 4xtetO-ade6+ following AHT addition to cdc25-22 G2 synchronised cultures (A) or double-blocked cdc25-22 G2 cells (B). Synchrony was assessed by septation index. qChIP time course of H3K9me2 or FLAG-TetR-Clr4* levels on 4xtetO-ade6+ using indicated primers. Data are mean ±SD (n=3).
Known and putative histone demethylases might act to remove H3K9me and thus disassemble heterochromatin from TetR-Clr4* tethering sites. We therefore tested if mutation of genes for six JmjC domain (Epe1, Jmj1, Jmj2, Jmj4, Lid2, Msc1) (22) or two SWIRM/Amino-oxidase domain proteins (Lsd1 or Lsd2; 23) allowed long-term 4xtetO-ade6+ silencing after tethered TetR-Clr4* release. Wild-type 4xtetO-ade6+ TetR-Clr4* cells form red/ade6-repressed colonies on indicator plates lacking AHT, white/ade6-expressing colonies appear on +AHT plates due to loss of H3K9me-dependent heterochromatin over 4xtetO-ade6+. Of the eight tested mutants only epe1Δ consistently formed red/pink colonies on +AHT plates, indicating that 4xtetO-ade6+ can remain repressed without bound TetR-Clr4* (Fig. 4A, fig. S5 and fig. S6). Catalytically inactivating mutations in the Fe(II) or 2-oxyglutarate binding sites of the Epe1 putative demethylase (epe1-H297A and epe1-K314A) had a similar phenotype (Fig. 4A, fig. S6 and Table S3). The variable silencing and colony colour most likely reflects stochastic events at the 4xtetO-ade6+ locus in epe1Δ cells in which H3K9me domains are known to expand and additional heterochromatin islands also appear, potentially titrating and redistributing heterochromatin proteins between various loci in individual cells (24-27). Maintenance of the silenced state in epe1Δ cells is not dependent on the RNAi component Ago1 as ago1Δepe1Δ cells form red/ade6-silent colonies on +AHT plates (fig. S7A) but it does require untethered wild-type Clr4 with an intact Clr4 chromodomain and Swi6 (fig. S8). This reliance on untethered, intact Clr4 and Swi6 is consistent with a simple read-write propagation mechanism (fig. S10).
Fig. 4. epe1 mutants retain heterochromatin without tethered Clr4 methyltransferase through multiple cell divisions and meiosis.
(A) wild-type, epe1Δ, epe1-K314A and epe1-H297A cells carrying 4xtetO-ade6+ and expressing TetR-Clr4*, were grown −/+ AHT. Colony colour assay to assess 4xtetO-ade6+ silencing (red-pink colonies; % of total indicated) and H3K9me2 qChIP on 4xtetO-ade6+ with (−AHT) or without (+AHT) tethered TetR-Clr4* Data are mean ±SD (n=3), P<0.05 (t-test).
(B) TetR-Clr4* was completely removed from F0 epe1Δ 4xtetO-ade6+ tetR-Clr4* cells by crossing to epe1Δ lacking TetR-Clr4* and 4xtetO-ade6+. F1 progeny were crossed to epe1Δ cells, generating epe1Δ F2 progeny. epe1+ F2xwt progeny were produced by crossing epe1+ into epe1Δ 4xtetO-ade6+ F2 cells. Naïve epe1Δ 4xtetO-ade6+ cells never expressed TetR-Clr4*. Colony colour, qRT-PCR and qChIP assays to assess silencing and transcription of 4xtetO-ade6+, and H3K9me2 levels on 4xtetO-ade6+ in indicated cell types. Data are mean ±SD (n=3). 4xtetO-ade6+ RNA levels are significantly reduced in F0, F1 and F2 compared to wild-type cells without TetR-Clr4*; P<0.05 (t-test).
Silencing of 4xtetO-ade6+ can be propagated through multiple cell divisions in epe1 mutants (lost in 4% of cells/division), and a high proportion of descendant cells retain silencing of, and 30-70% of H3K9me2 on, 4xtetO-ade6+ after TetR-Clr4* release by AHT. In contrast, 4xtetO-ade6+ silencing and H3K9me2 are completely lost in wild-type cells (Fig. 4A, and fig. S7B-E). The relative levels of H3K9me2 and H3K9me3 detected on 4xtetO-ade6+ are similar in wild-type and epe1Δ cells and surrounding genes are silenced by H3K9me2 in both wild-type and epe1Δ. (fig. S9). To determine if H3K9me on 4xtetO-ade6+ in epe1Δ cells is maintained through meiosis in the absence of TetR-Clr4*, epe1Δ 4xtetO-ade6+ tetR-clr4* cells (F0) were crossed to epe1Δ cells devoid of both 4xtetO-ade6+ and TetR-clr4* and then F1 epe1Δ 4xtetO-ade6+ progeny lacking TetR-Clr4* were again crossed to epe1Δ cells. A high proportion of resulting F2 epe1Δ 4xtetO-ade6+ progeny formed red-pink/ade6-repressed colonies and H3K9me2 was retained (Fig. 4B, and fig. S6B). Thus, epe1Δ allows silencing and H3K9me to persist through multiple mitotic divisions, and meiosis, in the complete absence of the tethered TetR-Clr4* that initiated H3K9me-dependent heterochromatin on 4xtetO-ade6+. Crossing of red F2 epe1Δ 4xtetO-ade6+ cells to wild-type epe1+ cells resulted in loss of silencing (white colonies only) and H3K9me2 from the 4xtetO-ade6+ locus. Thus, provision of epe1+ results in removal of persistent H3K9me and loss of silencing (Fig. 4B). Genetically identical naïve epe1Δ 4xtetO-ade6+ cells, that were never exposed to the TetR-Clr4* initiator, formed only white/ade6-expressing colonies and H3K9me2 was absent (Fig. 4B). We conclude that the transient tethering of TetR-Clr4* adjacent to 4xtetO-ade6+ allows establishment of H3K9me-dependent heterochromatin which can be propagated epigenetically through mitotic cell divisions and meiosis using endogenous read-write copying mechanisms, provided Epe1 is rendered non-functional (Model: fig. S10).
Propagation of heterochromatin on 4xtetO-ade6+ in epe1 mutants requires recognition of TeR-Clr4*-mediated H3K9me by the chromodomain of Clr4, and also Swi6 (fig. S8). Epe1 associates with Swi6HP1 and clearly opposes heterochromatin formation (24-28). Indeed Epe1 associates with TetR-Clr4*-mediated heterochromatin (fig. S7C). Although Epe1 contains a JmjC domain, its Fe(II) binding site is unusual and histone demethylase activity has not been detected (22). However, the human PHF2 JmjC domain bears a similar anomaly but phosphorylation activates its latent H3K9 demethylase activitity (29). The analyses presented here are consistent with Epe1 normally acting as an H3K9 demethylase that removes H3K9 methylation from ectopic sites of heterochromatin formation. Moreover, additional heterochromatin islands and domain expansion in epe1 mutants are best explained by loss of an H3K9 demethylase that prevents excessive H3K9me-dependent heterochromatin formation. Epe1-dependent removal of H3K9me ensures regulation of centromeric heterochromatin and makes the RNAi pathway essential for the systematic replenishment of H3K9me every cell cycle (30, 31). Epe1 itself may be regulated in response to environmental cues in order to retain or eliminate H3K9 methylation at specific locations (26). Indeed Epe1 levels are regulated and this may aid the persistence of centromeric H3K9me-dependent heterochromatin (28). Thus opposing H3K9 methyltransferase and demethylase activities must be finely tuned to allow controlled heterochromatin formation and prevent its inappropriate mitotic and transgenerational inheritance. It seems counterintuitive for heterochromatin to carry a means of self-destruction, however, such an inbuilt safety mechanism averts the inappropriate, and potentially deleterious, silencing of genes by removing repressive heterochromatin and preventing its propagation.
Supplementary Material
Acknowledgments
We thank I. Stancheva and the Allshire Lab for valuable discussions and E.S. Choi for RNA-seq data. We are grateful to S. Grewal, F. van Leeuwen, L. Bayne, Y. Shi, H.D. Madhani, T. Urano and H. Watanabe for providing strains and materials. PNCBA was supported by the Wellcome Trust 4 Year PhD programme in Cell Biology (093852). RCA is supported a Wellcome Trust Principal Research Fellowship (095021) and the EC-NOE-EpiGeneSys (HEALTH-F4-2010-257082) and core funding to the Wellcome Trust Centre for Cell Biology (092076). RNA-seq data NCBI-GEO accession: SRX689922.
Footnotes
References and Notes
- 1.Almouzni G, Probst AV. Heterochromatin maintenance and establishment: Lessons from the mouse pericentromere. nucleus. 2011;2:332–338. doi: 10.4161/nucl.2.5.17707. [DOI] [PubMed] [Google Scholar]
- 2.Bannister AJ, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 3.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 4.Zhang K, Mosch K, Fischle W, Grewal S. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol. 2008;15:381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- 5.Wang T, et al. Crystal structure of the human SUV39H1 chromodomain and its recognition of histone H3K9me2/3. PLoS ONE. 2012;7:e52977. doi: 10.1371/journal.pone.0052977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacobs SA, Fischle W, Khorasanizadeh S. Assays for the determination of structure and dynamics of the interaction of the chromodomain with histone peptides. Meth Enzymol. 2004;376:131–148. doi: 10.1016/S0076-6879(03)76009-1. [DOI] [PubMed] [Google Scholar]
- 7.Aagaard L, et al. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 1999;18:1923–1938. doi: 10.1093/emboj/18.7.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rivera C, Gurard-Levin ZA, Almouzni G, Loyola A. Histone lysine methylation and chromatin replication. Biochimica et Biophysica Acta (BBA) 2014 doi: 10.1016/j.bbagrm.2014.03.009. doi:10.1016/j.bbagrm.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 9.Zhu B, Reinberg D. Epigenetic inheritance: uncontested? Cell Res. 2011;21:435–441. doi: 10.1038/cr.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng X. Structural and Functional Coordination of DNA and histone methylation. Cold Spring Harb Pespect Biol. 2014;6 doi: 10.1101/cshperspect.a018747. pii:a018747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hathaway NA, et al. Dynamics and memory of heterochromatin in living cells. Cell. 2012;149:1447–1460. doi: 10.1016/j.cell.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 13.Reyes-Turcu FE, Grewal SI. Different means, same end—heterochromatin formation by RNAi and RNAi-independent RNA processing factors in fission yeast. Curr Opin Genet Dev. 2012;22:156–163. doi: 10.1016/j.gde.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet. 2013;14:100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buscaino A, et al. Distinct roles for Sir2 and RNAi in centromeric heterochromatin nucleation, spreading and maintenance. EMBO J. 2013;32:1250–64. doi: 10.1038/emboj.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wheeler BS, Ruderman BT, Willard HF, Scott KC. Uncoupling of genomic and epigenetic signals in the maintenance and inheritance of heterochromatin domains in fission yeast. Genetics. 2012;190:549–557. doi: 10.1534/genetics.111.137083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia S, Noma K-I, Grewal SIS. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science. 2004;304:1971–1976. doi: 10.1126/science.1099035. [DOI] [PubMed] [Google Scholar]
- 18.Kagansky A, et al. Synthetic heterochromatin bypasses RNAi and centromeric repeats to establish functional centromeres. Science. 2009;324:1716–1719. doi: 10.1126/science.1172026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Materials and methods are available as supporting material online.
- 20.Sakuno T, Tada K, Watanabe Y. Kinetochore geometry defined by cohesion within the centromere. Nature. 2009;458:852–858. doi: 10.1038/nature07876. [DOI] [PubMed] [Google Scholar]
- 21.Kim SM, Huberman JA. Regulation of replication timing in fission yeast. EMBO J. 2001;20:6115–6126. doi: 10.1093/emboj/20.21.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsukada Y-I, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2005;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 23.Shi Y, et al. Histone Demethylation Mediated by the Nuclear Amine Oxidase Homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 24.Zofall M, Grewal SIS. Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Mol Cell. 2006;22:681–692. doi: 10.1016/j.molcel.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 25.Ayoub N, et al. A novel jmjC domain protein modulates heterochromatization in fission yeast. Mol Cell Biol. 2003;23:4356–4370. doi: 10.1128/MCB.23.12.4356-4370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zofall M, et al. RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation. Science. 2012;335:96–100. doi: 10.1126/science.1211651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trewick SC, Minc E, Antonelli R, Urano T, Allshire RC. The JmjC domain protein Epe1 prevents unregulated assembly and disassembly of heterochromatin. EMBO J. 2007;26:4670–4682. doi: 10.1038/sj.emboj.7601892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Braun S, et al. The Cul4-Ddb1(Cdt)2 ubiquitin ligase inhibits invasion of a boundary-associated antisilencing factor into heterochromatin. Cell. 2011;144:41–54. doi: 10.1016/j.cell.2010.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baba A, et al. PKA-dependent regulation of the histone lysine demethylase complex PHF2-ARID5B. Nat Cell Biol. 2011;13:669–676. doi: 10.1038/ncb2228. [DOI] [PubMed] [Google Scholar]
- 30.Chen ES, et al. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- 31.Kloc A, Zaratiegui M, Nora E, Martienssen R. RNA Interference Guides Histone Modification during the S Phase of Chromosomal Replication. Curr Biol. 2008;18:490–495. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verzijlbergen KF, et al. Recombination-induced tag exchange to track old and new proteins. Proc Natl Acad Sci USA. 2010;107:64–68. doi: 10.1073/pnas.0911164107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.