Abstract
The pathogenesis of Alzheimer's disease (AD) is a critical unsolved question; and although recent studies have demonstrated a strong association between altered brain immune responses and disease progression, the mechanistic cause of neuronal dysfunction and death is unknown. We have previously described the unique CVN-AD mouse model of AD, in which immune-mediated nitric oxide is lowered to mimic human levels, resulting in a mouse model that demonstrates the cardinal features of AD, including amyloid deposition, hyperphosphorylated and aggregated tau, behavioral changes, and age-dependent hippocampal neuronal loss. Using this mouse model, we studied longitudinal changes in brain immunity in relation to neuronal loss and, contrary to the predominant view that AD pathology is driven by proinflammatory factors, we find that the pathology in CVN-AD mice is driven by local immune suppression. Areas of hippocampal neuronal death are associated with the presence of immunosuppressive CD11c+ microglia and extracellular arginase, resulting in arginine catabolism and reduced levels of total brain arginine. Pharmacologic disruption of the arginine utilization pathway by an inhibitor of arginase and ornithine decarboxylase protected the mice from AD-like pathology and significantly decreased CD11c expression. Our findings strongly implicate local immune-mediated amino acid catabolism as a novel and potentially critical mechanism mediating the age-dependent and regional loss of neurons in humans with AD.
Keywords: Alzheimer's disease, amino acid deprivation, arginase, arginine, immune suppression, microglia
Introduction
Alzheimer's disease (AD) is a disease of aging associated with the accumulation of β-amyloid (Aβ) peptides within the brain parenchyma and cerebral blood vessels and hyperphosphorylated and aggregated tau within neurons. Although the mechanisms causing brain atrophy and neuronal loss are unknown, increasing evidence suggests that immunity plays a critical role in AD pathogenesis. Genome-wide association studies have linked immune genes to increased risk for AD (Kamboh et al., 2012). Recently, an integrated analysis of AD-associated genes demonstrated that AD neuropathology has the strongest association with changes in immune and microglial gene networks (Zhang et al., 2013a). Despite this, we have not yet identified the specific mechanisms by which immunity contributes to AD pathogenesis. Although some studies have demonstrated increased expression of classical proinflammatory mediators in human AD, the inflammatory milieu also includes immunosuppressive components (Colton et al., 2006). This immunosuppressive bias is consistent with the brain's status as an immune privileged site (Carson et al., 2006; Mellor and Munn, 2008; Ransohoff and Cardona, 2010), and the contribution of immunosuppression to neuronal cell loss and other AD pathology remains largely unexplored.
In mice, transgenic expression of mutated human Aβ precursor protein (APP) leads to substantial Aβ deposition and behavioral deficits but does not commonly lead to tau pathology or neuronal death. As a result, mouse models of AD have been called “incomplete” models of human AD (Irizarry et al., 1997; Radde et al., 2008a). In contrast, we have previously reported that CVN-AD mice, which are mNos2-deficient and transgenic for the Swedish K670N/M671L vasculotropic Dutch/Iowa E693Q/D694N mutant (APPSwDI) APP, display the cardinal characteristics of AD progression, including Aβ plaques, phosphorylated tau protein, spatial memory impairments, and significant hippocampal neuronal death (Colton et al., 2008, 2014; Wilcock et al., 2008). The prevention of inducible nitric oxide synthase (iNOS) protein expression in this model recapitulates the biology of human myeloid cells, which produce relatively little iNOS and nitric oxide (NO) (Weinberg et al., 1995; Geller and Billiar, 1998; Colton et al., 2008; Wink et al., 2011; Guo et al., 2012).
Here, we present a detailed time course analysis of brain immune responses in CVN-AD mice and describe a novel hypothesis for AD pathophysiology. We find that CVN-AD mice develop neuropathology in an age-dependent fashion that mimics the progression of human AD. This neuropathology includes the accumulation of a specific population of activated microglia in the hippocampus and cortex and the predominant expression of immunosuppressive genes that alter utilization of key amino acids. Specifically, we find that arginase-1 is highly expressed in regions of Aβ accumulation, suggesting that arginine depletion and nutrient deprivation are responsible for neuronal cell death. In support of this hypothesis, we find that CVN-AD mice have decreased total brain arginine, and pharmacologic inhibition of arginine utilization enzymes reverses amyloid production, immunosuppression, and memory impairments. Together, these findings suggest that AD may be a disease of local immune suppression, rather than proinflammatory activation, and that the cause of neuronal death in AD is chronic immune-mediated amino acid catabolism resulting in nutrient deprivation.
Materials and Methods
Animals.
All animal experiments were performed in accordance with protocols approved by the Institutional Animal Care and Use committee at Duke University Medical Center under the National Institutes of Health Guide for the Utilization and Care of Vertebrate Animals Used in Testing, Research and Training.
CVN-AD mice.
Homozygous APPSwDI/mNos2−/− (CVN; CVN-AD) mice were produced by crossing mice expressing the Swedish K760N/M671L, Dutch E693Q, and Iowa D694N human APP mutations under control of the Thy-1 promoter (Davis et al., 2004) with mNos2−/− (B6 129P2NOS2tau1Lau/J) mice (Laubach et al., 1998) (The Jackson Laboratory). APPSwDI mice were generously provided by Dr. Bill Van Nostrand and Judianne Davis at Stony Brook University Medical Center. Mice were genotyped using standard PCR methods. To determine whether mice used in the experiments expressed the Rd1 mutation in the Pde68 gene that causes blindness in mice (Carter-Dawson and LaVail, 1979), CVN-AD mice were genotyped and found to be negative for Rd1. All mice were also genotyped for the Rd8 mutation of the Crb1 gene that, when expressed, causes retinal pathology but is not associated with blindness (Mattapallil et al., 2012). CVN-AD mice were found to be heterozygous for this gene. Animals were fed standard mouse chow and housed under 12 h light/12 h dark cycles at 21 ± 3°C in an Institutional Animal Care and Use Committee approved barrier facility under Institutional Animal Care and Use Committee-approved animal protocols. Mice were aged to 6, 12, 24, 36, or 52 weeks of age before behavioral assays and harvest of tissue. Mixed genders were used in the experiments, and gender-specific effects were not examined due to the complexity of the experimental protocol and the significantly increased number of mice required. General pathological features of CVN-AD mice include severe cerebral vascular amyloid deposition, tau pathology, and neuronal loss associated with a decline in learning and memory as previously described (Wilcock et al., 2008).
Immunohistochemistry.
After injection with a lethal mixture of ketamine/xylazine, mice were intracardially perfused with 25 ml of PBS to remove intravascular circulating blood cells. Perfused brains were then rapidly removed and bisected in the mid-sagittal plane. The left hemisphere was frozen in liquid nitrogen for use in ELISA and gene expression analysis, and the right hemisphere was immersion fixed in 4% PFA. For immunohistochemistry, brains were cryoprotected by sequential passage through 10%, 20%, and 30% sucrose for 24 h. Frozen sagittal or coronal sections (25 μm) were then cut. Sections equally spaced at 600 μm apart were immunostained with standard techniques using the following antibodies: anti- Aβ (H31L21; Invitrogen), anti-phospho tau (AT8; Thermo Scientific), anti-CD45 (YW62.3, AbD Serotec), anti-CD11b (5C6, AbD Serotec), anti-CD11c (N418; AbD Serotec), anti-Iba-1 (polyclonal 01-1941; Wako Pure Chemicals), anti-CD206 (AF2535; R&D Systems), and anti-arginase-1 (gift from Dr. Sidney Morris, University of Pittsburgh, and commercially available as ABS535; Millipore). Secondary antibodies, ABC kit, and DAB kit were purchased from Vector Laboratories.
Quantitative RT-PCR.
Frozen cryo-pulverized brain samples from CVN-AD, mNos2−/−, WT, and APPSwDI mice at 6, 12, 24, 36, and 52 weeks of age were used to prepare RNA for analysis of immune gene expression. mRNA was extracted from frozen tissue using the RNeasy Tissue Kit (QIAGEN) according to the manufacturer's instructions. RNA was quantified using a NanoDrop spectrophotometer, and cDNA was generated using the cDNA high-capacity kit (Applied Biosystems). Real-time PCR was performed using the TaqMan Gene Expression kit (Applied Biosystems), according to the manufacturer's instructions and as previously described (Wilcock et al., 2011b). All PCR probe sets were purchased from Applied Biosystems. Beta-actin served as the internal standard, and fold change in gene expression levels was calculated using the 2(−delta delta C(T)) method (Livak and Schmittgen, 2001). WT mice at the same age were used as the comparator except where delineated.
Flow cytometry.
Anesthetized mice were intracardially perfused with PBS, and brains were then rapidly harvested, manually dissociated, and digested for 1 h at 37°C with 1.5 mg/ml collagenase A (Roche Applied Science) and 0.4 mg/ml DNase I (Roche Applied Science) in 5% FBS with 10 mm HEPES. Cells from the digested tissue were then strained through a 70 μm filter and washed with PBS. Cells were centrifuged in a 30% over 70% Percoll (Invitrogen) in PBS density gradient. Cells from the interface were isolated, and red blood cells were lysed with ammonium/chloride/potassium lysis buffer (Invitrogen). Cells were counted and then stained with Live/Dead Aqua (Invitrogen), and the following antibodies (all from eBioscience) were used: CD11b FITC, CD11c PE-Cy5.5, CD45 PE-Cy7, CD3ε APC, Ly6G AF700, CD11b APC-Cy7, Ly6C V450, and IA-IE Qdot655. Flow cytometry was run on a BD LSR-II Flow Cytometer (BD Biosciences) in the Duke Human Vaccine Institute Flow Research Facility and analyzed with FlowJo (TreeStar).
Cell isolation by FACS and RNA isolation and amplification.
CVN-AD, mNos2−/−, and WT (C57BL/6) mice (n = 4 per genotype), all at 48 weeks of age, were anesthetized and intracardially perfused with PBS. Cells were isolated for flow cytometry as described above. Cells were counted and stained with the following antibodies (all from eBioscience): CD11c PE-Cy5.5, CD45 PE-Cy7, CD11b APC-Cy7, and Ly6G V450. FACS sorting was run on a BD FACS Aria II Special Order Research Product Flow Cytometer (BD Biosciences) in the Duke Human Vaccine Institute Flow Research Facility. Cells were gated on Ly6G− CD11b+ CD45low microglia for mNos2−/−, C57BL/6, and CVN-AD samples and further subgated into CD11chigh and CD11clow populations in CVN-AD samples. A total of 1000 microglia were collected from each mNos2−/− and C57BL/6 brain, and 1000 CD11chigh and CD11clow microglia were collected per CVN-AD brain and sorted in a volume of 1.0 μl directly into 6.4 μl of SuperAmp Lysis Buffer (Miltenyi Biotec). Samples were then incubated at 45°C for 10 min and frozen at −20°C before shipment to Miltenyi Biotec on dry ice.
Microarray analysis.
SuperAmp RNA amplification was performed by Miltenyi Biotec based on a global PCR protocol using mRNA-derived cDNA. cDNA integrity was checked via Agilent 2100 Bioanalyzer platform (Agilent Technologies). cDNA was labeled with Cy3 and hybridized to an Agilent Whole Mouse Genome Oligo Microarray 8×60K. After washing and staining, fluorescence signals were detected with Agilent's Microarray Scanner System (Agilent Technologies). The Agilent Feature Extraction Software was used to read out and process microarray image files (Agilent Technologies). Partek Genomics Suite (Partek) was used for data analysis, where we ran an ANOVA with the four experimental groups to identify significantly differentially expressed genes. Significance threshold for the effect size was set at twofold change and false discovery rate at 5% (q = 0.05).
Measurement of brain amino acids.
Amino acids were extracted from cryo-pulverized 20- to 24-week-old mouse brain tissue (n = 3–12 mice/strain) using 4% CHAPS/50 mm EDTA solution with probe sonication. Stable-isotope-labeled internal standards for l-arginine, l-ornithine, and l-citrulline (Cambridge Isotope) were added to the extraction mixture following sonication and used for MS-based quantitation. Samples were clarified with centrifugation, subjected to solid phase extraction purification on Strata-X-C columns (Phenomenex), dried under air, and resuspended in isopropanol/0.2% formic acid. Analytes and their internal standards were then quantified in positive-ion liquid chromatography-tandem mass spectrometry (LC-MS/MS) mode, using previously described methodology (Brown et al., 2011). Briefly, separations were performed on an Atlantic hydrophilic-interaction chromatography (HILIC) column (Waters) with mobile phases of 75% acetonitrile/25% methanol/0.2% formic acid (mobile phase A) or 0.2% formic acid in water (mobile phase B) at 0.2 ml/min flow rate. Samples were introduced to a 4000 QTrap LC-MS/MS (AB SCIEX) via electrospray ionization, and analytes and internal standards quantified using multiple-reaction monitoring. Analyte concentrations were calculated from the slope of a calibration curve generated by serial dilutions of internal standards.
Difluoromethylornithine (DFMO) treatment.
CVN-AD mice at 6–8 weeks of age were treated with 10 mg/kg DFMO plus 1 mg/kg putrescine in saline by oral gavage 3 times a week (Mon/Wed/Fri) for 14 weeks (n = 8 mice/ group). Control CVN-AD mice were treated on the same schedule using oral gavage with 1 mg/kg putrescine in saline alone (n = 7 mice/group). DFMO (trade name Eflornithine; pharmaceutical grade) was provided by Dr. Patrick M. Woster, Department of Pharmacology, Wayne State University (Detroit) and was stored at −80°C and aliquoted at the time of use. Cotreatment with putrescine was used to reduce potential DFMO-mediated damage to the epithelial cells in the gastrointestinal tract. All mice were weighed twice per week, and no significant differences in weight were observed between DFMO-treated and saline-treated mice. After 14 weeks of treatment, mice were tested in the radial-arm water maze (RAWM) for learning and memory. On completion of behavioral testing, brains were harvested for pathological assessment and gene analysis.
Two day Radial arm water maze (RAWM) test for learning and memory.
CVN-AD, WT (C57BL/6), and mNos2−/− mice were tested at the ages indicated using the 2 day RAWM described in detail previously (Alamed et al., 2006). Briefly, a six arm maze is submerged in a black plastic pool of water, and a platform is placed at the end of one arm below the surface. The mouse receives 15 trials per day for 2 d (30 total) and is started in a different arm each trial while the goal arm containing the platform remains constant for each mouse. Using static visual cues, the mouse learns the position of the escape platform. On the first day, the first 12 trials are considered training and alternate between a visible and a hidden platform, whereas the final 3 trials use a hidden platform. All trials on day 2 use a hidden platform. The number of errors (incorrect arm entries) is counted over a 1 min period. The errors for each consecutive three trials (1–3, 4–6, 7–9, 10–12, 13–15) are averaged as one block, resulting in 5 blocks per day. After completion of all hidden maze tasks, mice were tested for sensory or motor deficits using the open pool task with visible platform as described previously (Alamed et al., 2006). Mice that did not perform successfully in the open pool task were excluded from data analysis. All behavioral tests were performed with treatment groups blinded to the investigator.
Statistical analysis and comparisons between groups.
Average values ± SEM were determined for each of the outcome measures at 6, 12, 24, 36, and/or 52 weeks of age in the CVN-AD and control mice. Mixed genders were used in the analyses, and gender based-differences were not investigated. Significant differences across age within strain were determined using one-way ANOVA, and statistical significance between genotypes and age was determined by two-way ANOVA using the PRISM statistical program (GraphPad Software). Significance was set at p ≤ 0.05. The number of mice analyzed ranged from 3 to 14 mice per group depending on the outcome assay.
Results
Immune suppression is an early feature of disease progression
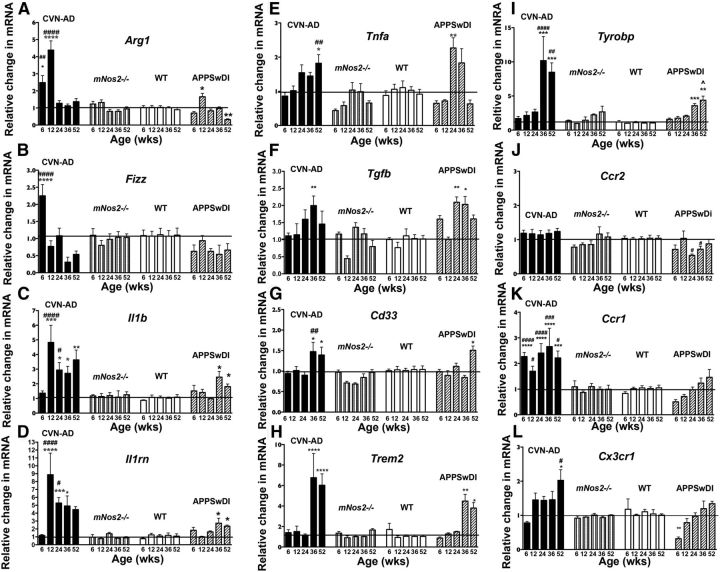
CVN-AD mice display characteristic neuropathology resembling human AD as they age, which include the accumulation of Aβ deposits starting at 6 weeks of age, the presence of hyperphosphorylated and aggregated tau beginning at 12 weeks of age, spatial memory deficits beginning ∼24 weeks of age, and neuronal loss at ∼36 weeks of age (Colton et al., 2014). We thus reasoned that these mice could be used to identify immune abnormalities that arise early in the course of disease as well as with disease progression during aging. We used qRT-PCR of total brain lysate to measure gene expression for a variety of proinflammatory and anti-inflammatory genes implicated in AD in CVN-AD, mNos2−/−, and WT (C57BL/6) mice at 6, 12, 24, 36, and 52 weeks of age. We also examined mRNA expression levels in the parent APPSwDI strain to determine whether Aβ accumulation alone was a factor in the immune changes, as levels of soluble and insoluble Aβ are not significantly different in APPSwDI versus CVN-AD mice (Wilcock et al., 2008). Because significant neuronal death begins ∼36 weeks in CVN-AD mice (Colton et al., 2014), we grouped the genes into two major patterns of expression: genes that were upregulated before 36 weeks (Fig. 1A–D) and genes upregulated at or after 36 weeks (Fig. 1E–I). We also examined gene expression levels for chemokine receptors and ligands from 6 to 52 weeks of age (Fig. 1J–L).
Figure 1.
Expression of immune-related genes from whole-brain lysates. Average values and SEM for the relative change in mRNA levels are shown for immune-related genes from whole-brain lysate samples from CVN-AD, mNos2−/−, WT (C57BL/6), and APPSwDI mice. A–D, Genes that were upregulated in CVN-AD mice before 36 weeks of age. E–I, Genes that were upregulated in CVN-AD mice at or after 36 weeks of age. J–L, Gene expression of chemokine receptors. mRNA expression was measured using quantitative RT-PCR and represent the 2(−delta delta C(T)) compared with WT mice. Samples were analyzed by two-way ANOVA and post hoc multiple comparison test with Bonferonni's correction. *Comparisons between CVN-AD or APPSwDI and WT. #Comparisons between CVN-AD and APPSwDI mice. *p < 0.05. **p < 0.01. ***p < 0.001. ****p < 0.0001. #p < 0.05. ##p < 0.01. ###p < 0.001. ####p < 0.0001. n > 5 mice per group.
Genes that were upregulated at the earliest time points in CVN-AD mice were predominately those that have been linked to immune suppression in macrophages (Fig. 1). This included arginase-1 (Arg1), Found in Inflammatory Zone 1 (Fizz1), and interleukin 1 receptor antagonist (Il1rn). These genes tended to display the highest expression level only transiently, returning toward WT control levels by 24 weeks. Expression of these anti-inflammatory genes was significantly greater in CVN-AD mice compared with the parent APPSwDI strain, with the exception of Arg1 expression at 12 weeks of age, where a small increase in mRNA level was observed in APPSwDI brain lysates. Interleukin-1β (Il1b), the classic proinflammatory mediator, was also expressed early in CVN-AD mice, beginning at 12 weeks of age. After this initial peak, IL-1β gene expression levels remained significantly elevated throughout the 52 weeks of the study. Compared with APPSwDI mice, which exhibited only slight increases in Il1b and Il1rn at 24 and 52 weeks, CVN-AD mice had greater expression levels for Il1b and Il1rn at all time points.
Specific genes that were increased in CVN-AD mice at or after 36 weeks of age included the canonical proinflammatory gene Tnfa, which was significantly increased only at the latest time point studied (52 weeks), and Tgfb, an anti-inflammatory gene that was increased transiently beginning at 36 weeks of age. Also increased was Cd33 (Siglec-3), an inhibitory lectin that has been implicated in genome-wide association studies of AD and that is believed to alter microglial phagocytosis of Aβ (Salminen et al., 2009; Bradshaw et al., 2013). Other immunosuppressive genes known to be associated with AD, namely, Trem2 and Tryobp (DAP12), were also increased late in disease progression in CVN-AD mice. Data from APPSwDI mice brain showed a similar age-specific pattern of expression for these late genes, but no significant changes were found in either mNos2−/− or wild-type control mice at any age.
Expression of mRNAs for the chemokine receptor Ccr2 (Fig. 1J) and its ligand Ccl2 (MCP1) (data not shown) was unchanged at all time points in CVN-AD brains. Ccr2 is expressed on inflammatory monocytes and has been shown to mediate the migration of these cells from the periphery into the brain in mice expressing mutated human APP (El Khoury et al., 2007; Naert and Rivest, 2011), although others have observed that circulating monocytes from humans with AD have depressed CCR2 expression (Zhang et al., 2013b). Expression of Cxcl1 (KC, GROa), which can mediate neutrophil recruitment and may have a role in neuroprotection, and Ccl11 (eotaxin 1), a chemoattractive factor for eosinophils, also remained unchanged (data not shown). However, Ccr1 and Cx3cr1 expression was significantly increased in CVN-AD mice but was not observed in any of the control mice, including APPSwDI mice. Increased Ccr1 expression in CVN-AD mice was observed early and elevated at all ages. CCR1 has been shown to associate with plaques in the entorhinal cortex and hippocampus, is an early marker for clinical dementia in humans with AD, and has been shown to directly correlate with cognitive decline (Halks-Miller et al., 2003). Expression of Cx3cr1, which is expressed primarily by microglia in the brain, increased progressively in CVN-AD brains (Fig. 1L), consistent with the increasing number of microglia (see Fig. 3). However, gene expression for Cx3cr1 receptor ligand, Cx3cl1 (fractalkine), was not increased at any time (data not shown).
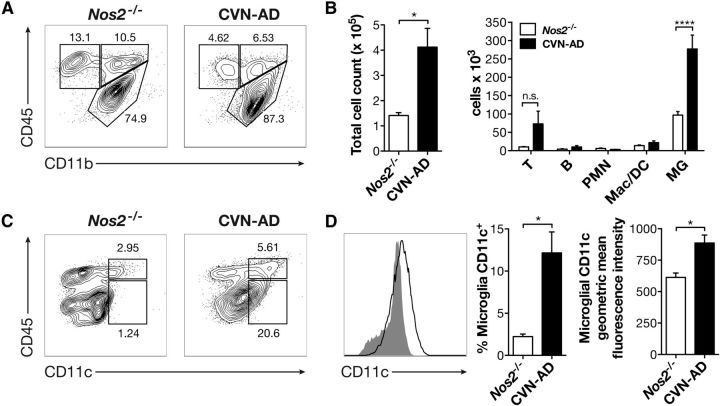
Figure 3.
CD11c+ cells from CVN-AD brains have a microglial phenotype. A, Representative flow cytometry plots from aged 48-week-old mNos2−/− and CVN-AD brains after gating on CD45+ cells. B, Quantification of total CD45+ cells and individual cell types distinguished by FACS in 48-week-old mNos2−/− and CVN-AD brains, including T lymphocytes (T), B lymphocytes (B), neutrophils (PMN), monocytes/macrophages/DCs (Mac/DC), and microglia (MG) from aged 48-week-old mNos2−/− and CVN-AD brains. *p < 0.05; ****p < 0.0001. C, Representative flow plots of CD45 and CD11c expression from aged 48-week-old mNos2−/− and CVN-AD brains after gating on all CD11b+ cells. D, Representative histogram of CD11c expression on mNos2−/− (closed gray) and CVN-AD (open black) CD11b+ CD45low cells, as well as quantitative summaries of the percentages and geometric mean fluorescence intensities of CD11c from mNos2−/− and CVN-AD CD11b+ CD45low microglia. *p < 0.01. n = 4 mice per group.
CVN-AD mice show progressive cellular inflammation with CD11c+ microglia in areas of Aβ deposition
To better understand the cellular basis of the altered immune responses found in whole-brain lysates of CVN-AD mice compared with control mice, we examined the presence and characteristics of typical immune cells using immunocytochemistry on brain sections through one year of the disease process. Age-matched mNos2−/− and C57BL/6 mice were used as controls. Immune markers that are characteristically upregulated in tissue sections from autopsied human AD brain include CD45, MHC-II, CD11b, and CD11c (Rozemuller et al., 1986; McGeer et al., 1987; Rogers et al., 2002; Mrak and Griffin, 2005; Eikelenboom et al., 2010). Changes in these markers are used as a pathological indicator of the immune response in humans with AD and are also observed in mouse models of AD (Tan et al., 2000; Butovsky et al., 2006; Nichol et al., 2008; Tambuyzer et al., 2009; Mildner et al., 2011). In CVN-AD mice, tissue expression of CD45, Iba-1, and CD11c is correlated with amyloid deposition with aging (Fig. 2). Aβ plaques appeared initially in the subiculum of the hippocampus and increased in staining density throughout the cortex and hippocampus with age (Fig. 2A–D). Aβ deposition was accompanied in time and spatial location by increased immunoreactivity for CD45 (Fig. 2E–H). CD45+ cells had the morphological features of reactive microglia with bushy, shortened processes (data not shown). To confirm that CD45 cells in CVN-AD brains represent microglia, sections from the same CVN-AD mice were stained with Iba-1, a calcium binding protein that is expressed specifically on microglia in the CNS (Fig. 2I–L). Iba-1 stained microglia throughout the brain, as well as cells within the hippocampus and cortex that correlated with the pattern of CD45 staining in these mice.
Figure 2.
CVN-AD amyloid deposition is associated with cells expressing characteristic markers of microglia. Representative sagittal sections from a CVN-AD mouse at 6, 12, 24, and 52 weeks of age immunostained for Aβ (A–D), CD45 (E–H), Iba-1 (I–L), and CD11c (M–P). For each age, panels represent sister sections from the same mouse. For example, A, E, I, and M are sections from the same 6-week-old mouse. Scale bar, 500 μm. Q, Representative micrographs of CD11c and Iba-1 costaining in the subiculum of the hippocampus from a 24-week-old CVN-AD mouse. Scale bar, 25 μm. R, Increased CD11c immunostaining is associated with increased total brain gene expression of CD11c (Itgax). Gene expression levels (mean ± SEM) were measured using quantitative RT-PCR and represent the 2(−delta delta C(T)) compared with WT mice. Samples were analyzed by two-way ANOVA and post hoc multiple comparison test with Bonferonni's correction. *Comparisons between CVN-AD or APPSwDI and WT. #Comparisons between CVN-AD and APPSwDI mice. *p < 0.05. **p < 0.01. ***p < 0.001. ****p < 0.0001. #p < 0.05. ##p < 0.01. ###p < 0.001. ####p < 0.0001. n > 5 mice per group.
We also examined the expression pattern of CD11c, which is not commonly found on either resting or reactive microglia in mouse brain, but is found on resting and activated human microglia (Akiyama and McGeer, 1990). An increasing pattern of expression was similar to that of CD45 was observed for CD11c (Fig. 2M–P), and CD11c expression found to overlap with Iba-1+ microglia using double labeling techniques (Fig. 2Q). To further confirm the change in levels of CD11c with age, we also performed qRT-PCR on whole-brain lysates. Gene expression levels for Itgax (CD11c) increased dramatically from 6 weeks of age, leading to an ∼100-fold increase at 52 weeks of age (Fig. 2R). In contrast, the expression levels of Itgax mRNA in the APPSwDI parent mouse strain showed a relatively delayed increase in gene expression levels and were significantly less than the fold changes in mRNA found in CVN-AD mice brain. Similarly, we observed less and delayed CD11c immunostaining at all ages when comparing APPSwDI with CVN-AD mice (data not shown). No changes in CD11c mRNA levels were found for mNos2−/− or WT mice brain lysates at any age studied.
The above findings suggest that cellular inflammation in CVN-AD mice is primarily due to an increased number of reactive microglial cells that coexpress CD45 and CD11c. To confirm the identity of the CD11c+ cells, we performed 12-color flow cytometric analysis of homogenized whole brains from 42-week-old mNos2−/−, which lack CD11c+ microglia, and CVN-AD mice (Fig. 3). Using this technique, we found increased total cell numbers in the brains of CVN-AD mice that was caused by a significant increase in the numbers of CD45low CD11b+ microglia, and secondarily, by a trend toward increased numbers of T cells (Fig. 3B). The total numbers of B cells, neutrophils, eosinophils, macrophages, Ly6Clow resident monocytes, and Ly6Chigh inflammatory monocytes, however, were unchanged in CVN-AD brains (Fig. 3B). CD11c was predominantly expressed by CD45low microglia (Fig. 3C). At 42 weeks of age, ∼12.2 ± 2.5% of all microglia were CD11c+, compared with 2.24 ± 0.3% in mNos2−/− controls, and there was a significant increase in the mean fluorescence intensity of microglial expression of CD11c (Fig. 3D). Thus, both immunocytochemical and flow cytometric analyses demonstrated that CVN-AD mice develop substantial cellular inflammation that is caused primarily by the expansion of microglial cells. These microglial cells display a reactive morphology, accumulate specifically in areas of Aβ deposition, and can be distinguished from microglia in control mice by their increased expression of CD11c. This pattern of cellular inflammation is consistent with that seen in humans with AD (Dickson et al., 1988; Akiyama and McGeer, 1990).
CD11c+ microglia show an immunosuppressive phenotype
To better understand the potential functional capabilities of the CD11c-immunopositive microglia in CVN-AD mice, we used flow sorting to isolate this population from whole-brain lysates. Mice were used at 48 weeks of age to ensure a large population of cells for the study. Cells were gated on Ly6G− CD11b+ CD45low microglia for mNos2−/−, WT, and CVN-AD brain samples and further subgated into CD11chigh and CD11clow populations in CVN-AD mice. Messenger RNA from each collected cell sample was then globally amplified using SuperAmp to produce cDNA for use in an Agilent Whole Mouse Genome Oligo microarray. Gene expression from the CVN-AD CD11c+ cells was compared with that of the Ly6G− CD11b+ CD45low CD11c− microglia from CVN-AD, C57BL/6, and mNos2−/− brains. ANOVA with the four experimental groups was used to identify significantly differentially expressed genes. We set the significance threshold for the effect size at twofold change and false discovery rate at 5% (q = 0.05).
Approximately 375 genes were significantly upregulated and 85 genes downregulated in CVN-AD CD11c+ microglia compared with CD11c− microglia from CVN-AD, C57BL/6, and mNos2−/− brains (Table 1). Upregulated genes included those genes that are used to identify microglia in humans with AD, such as Itgax (Cd11c/LeuM5; CR4) and Cd200r (Cell surface glycoprotein CD200 receptor 2; OX2R). The 124-fold increase in Itgax gene expression verified our previous observations and our FACS isolation process. Strikingly, many of the genes that were upregulated in CVN-AD CD11c+ microglia, such as Spp1, Wfdc17, Gp49a, Apoe, and Pdcd1, were genes associated with immune suppression and increased arginase activity. Similarly, many of the genes that were downregulated were from proinflammatory pathways, such as Apobec3, Ifngr1, and Siglech, or represent negative regulators of immune suppression, such as Klf6 (Table 1). Together, along with additional genes that were changed significantly (Table 1), we observed that CD11c+ microglia from CVN-AD mice had a predominantly immunosuppressive phenotype.
Table 1.
Gene expression in CVN-AD CD11c+ microglia compared with CVN-AD CD11c− microglia, C57BL/6 microglia, and mNos2−/− microgliaa
| Gene | Name | Fold change | p value | Reported actions | References |
|---|---|---|---|---|---|
| Mamdc | MAM domain containing protein 2 | 515 | 9.7 × 10−7 | Glycosaminoglycan binding, linked to negative regulation of synapses | Pettem et al., 2013 |
| Spp1 | Secreted phosphoprotein 1; osteopontin | 515 | 5.9 × 10−6 | Increased in CSF of AD patients; enhances immunosuppression | Comi et al., 2010; Sangaletti et al., 2014 |
| Gpnmb | Glycoprotein-transmembrane nmb; osteoactivin, Fe65-like1 | 442 | 1.5 × 10−6 | Tissue repair, M2 state; phagocytic vesicle processing | Duffield, 2010 |
| Wfdc17 | Whey acidic protein four disulfide core domain 17; AMWAP | 136 | 4.1 × 10−5 | Overexpression increases Arg-1, reduces IL-6, IL-1β | Karlstetter et al., 2010 |
| Itgax | Integrin alpha X (complement component 3 receptor 4 subunit), CD11c | 124 | 5.8 × 10−6 | Leukocyte-specific integrin; associated with dendritic cells; phagocytosis of complement-coated particles; found on microglia in AD | Akiyama and McGeer, 1990; Tooyama et al., 1990; Becher and Antel, 1996; Butovsky et al., 2007 |
| Gp49a | Glycoprotein 49a; Lirb4 | 107 | 2.8 × 10−4 | Member of inhibitory Ig superfamily; increased IL-4; suppresses LPS; inhibits Fc-gamma-mediated phagocytosis | Arm et al., 1997; McCormick et al., 1999 |
| Gng12 | Guanine nucleotide binding protein 12 | 64 | 1.7 × 10−4 | Inhibits LPS-mediated pro-inflammation | Larson et al., 2010 |
| Pdcd1 | Programmed cell death 1; PD-L1, CD279 | 51 | 4.1 × 10−6 | Immunoglobin superfamily; shifts microglia to M2 phenotype; regulates Arg-1 activity | Yao et al., 2014 |
| Apbb2 | Amyloid precursor protein binding protein 2 | 25.8 | 2.4 × 10−4 | Adaptor protein binds to cytoplasmic domain of APP; polymorphisms associated with dementia in aged population; involved in ECM synthesis by Macs | Wright et al., 2005; Grupe et al., 2006; Golanska et al., 2013 |
| TIMP2 | Tissue inhibitor of metalloprotease-2 | 20.7 | 3.2 × 10−4 | Blocks metalloprotease activity; associated with M2 phenotype | Ridnour et al., 2007; Wilcock et al., 2011a |
| Igf1 | Insulin-like growth factor 1; 2; somatomedin c | 27.4 | 2.0 × 10−5 | Found in human microglia, protects from IL-1- and IFN-γ-mediated damage; promotes Aβ clearance | Trueba-Sáiz et al., 2013 |
| Igf2 | 8.0 | 2.1 × 10−3 | |||
| Apoe | Apolipoprotein E | 20.1 | 3.6 × 10−4 | Strongest single gene risk factor for AD, suppresses proinflammatory cytokines | Saunders et al., 1996 |
| CD200r | Cell surface glycoprotein CD200 receptor 2, OX2R | 13.8 | 3.3 × 10−3 | Inhibitory immune receptor found on microglia, marker for M2 activation in human MG, less so in mice | Denieffe et al., 2013; Walker and Lue, 2013 |
| Klf6 | Kruppel-like factor 6 | 0.12 | 4.9 × 10−6 | Suppression is linked to M2 phenotype | Date et al., 2014 |
| Apobec3 | Apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 | 0.18 | 4.1 × 10−4 | Promotes antiviral immunity through production of neutralizing antibody | Santiago et al., 2008 |
| Ifngr1 | Interferon-γ receptor 1; CD119 | 0.19 | 9.1 × 10−6 | Encodes ligand binding domain for IFN-γ; downregulated by TLR2; IFN-β | Curry et al., 2004; Kearney et al., 2013 |
| Siglech | Siglec-H | 0.42 | 2.6 × 10−4 | DAP12 signaling molecule, microglial “sensome,” decreased with aging | Hickman et al., 2013 |
aSelect genes that were significantly upregulated or downregulated in CVN-AD CD11c+ microglia compared with CVN-AD CD11c− microglia, C57BL/6 microglia, and mNos2−/− microglia, including fold change in expression, p value, and reported actions with references.
CVN-AD pathology is associated with increased arginine utilization and decreased brain arginine bioavailability
The immunosuppressive phenotype observed in CVN-AD mice brain suggested a potential causative role for this immune process in disease progression. In particular, Arg1, a critical anti-inflammatory gene that codes for the enzyme arginase-1, was expressed predominantly in young mice in brain areas associated with Aβ deposits but before neuronal loss in this model of AD. Arginase-1 regulates the microenvironmental level of arginine, a semiessential amino acid, by altering arginine catabolism (Bronte and Zanovello, 2005; Gabrilovich and Nagaraj, 2009; Pesce et al., 2009; Tang et al., 2009). Increased usage of arginine results in a drop in tissue levels of arginine (Reeds, 2000), which if not replaced, initiates amino acid deprivation responses in susceptible cells. Sustained arginine deprivation leads to cell death (Kuma and Mizushima, 2010). To determine whether induction of Arg1 may play a role in AD-like pathology in CVN-AD mice, we examined arginase-1 protein expression and its spatial and temporal relationship with Aβ deposition and CD11c+ microglial at different ages using immunocytochemistry. Arginase-1 protein accumulated in the subiculum and CA1 regions of the hippocampus, areas of primary neuronal loss, starting between 6 and 12 weeks and peaked at 24 weeks (Fig. 4A), consistent with the onset of behavioral deficits at 24 weeks. In contrast, no arginase-1 staining was observed at any time point in Nos2−/− brains (Fig. 4B; and data not shown). Of note, arginase-1 staining in CVN-AD brains did not primarily localize to cells but instead diffusely stained the hippocampus and subiculum, suggesting that arginase was distributed in the extracellular space surrounding the cells (Fig. 4B). Additionally, arginase-1 staining in the hippocampus displayed a spatial correlation with that of Aβ, Iba-1, and CD11c (Fig. 4C), highly suggesting that CD11c+ microglia are a likely source of arginase-1 production and that expression is associated with Aβ.
Figure 4.
CVN-AD pathology is associated with arginase-1. A, Representative sagittal sections from CVN-AD mice at 6, 12, 24, and 52 weeks of age stained for arginase-1 in sister sections from the same mice as in Figure 1. Scale bar, 500 μm. B, Magnified view of arginase immunoreactivity in the subiculum. Representative sagittal sections from 24-week-old mNos2−/− and CVN-AD stained for arginase-1. Scale bars: top, 500 μm; bottom, 50 μm. C, Sister coronal sections from the same 52-week-old CVN-AD brain stained for Aβ, CD11c, Iba-1, and arginase-1 to show regional associations.
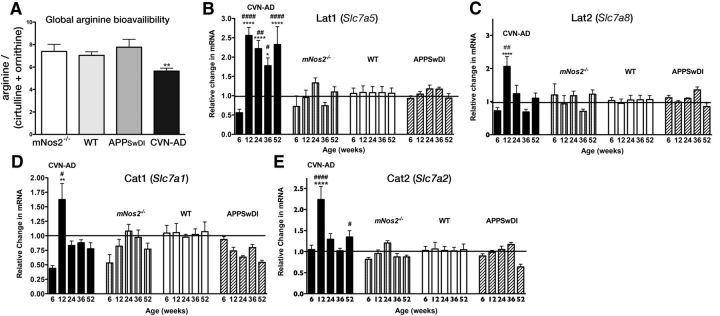
To determine whether arginase-1 expression alters brain arginine levels in CVN-AD mice, we examined brain amino acid levels and the compensatory expression of amino acid transporters in these animals. Total brain arginine and arginine metabolites, including ornithine and citrulline, were measured using HILIC and LC-MS/MS (Brown et al., 2011). In disease contexts, the global arginine bioavailability ratio (GABR), which is the ratio of arginine to its metabolites ornithine and citrulline (arginine/(ornithine + citrulline)), is often a better indicator of dysregulated arginine metabolism than arginine concentration alone (Tang et al., 2009; Morris, 2012). Calculating the GABR revealed that, compared with WT, mNos2−/−, and APPSwDI mice, CVN-AD mice had significantly reduced GABR (Fig. 5A), indicating that CVN-AD brains exhibited increased arginine catabolism.
Figure 5.
CVN-AD brains have decreased total l-arginine bioavailability and increased expression of arginine transporters. A, Global arginine bioavailability (arginine/(ornithine + citrulline)) for CVN-AD, mNos2−/−, WT, and APPSwDI mice. Average values (± SEM) per genotype were calculated for individual mice (n = 3–12 mice per group). Amino acid levels were measured using HILIC LC-MS/MS. **p < 0.01 (one-way ANOVA). B–E, Relative gene expression (mean ± SEM) was measured in total brain homogenates from CVN-AD, mNos2−/−, WT, and APPSwDI mice for the neutral arginine transporters Slc7a5 (LAT1) and Slc7a8 (LAT2) (B, C) and for the cationic amino acid transporters Slc7a1 (CAT1) and Slc7a2 (CAT2) (D, E). Samples were analyzed by two-way ANOVA and post hoc multiple comparison test with Bonferonni's correction. *Comparisons between CVN-AD or APPSwDI and WT. #Comparisons between CVN-AD and APPSwDI mice. *p < 0.05. **p < 0.01. ***p < 0.001. ****p < 0.0001. #p < 0.05. ##p < 0.01. ###p < 0.001. ####p < 0.0001. n = 4–8 mice per group.
Cells deprived of arginine attempt to compensate by increasing expression of amino acid transporters that regulate the cellular uptake of arginine from the local environment (Hyatt et al., 1997; Bröer, 2002; Closs et al., 2006). To determine whether such mechanisms were active in CVN-AD brains, we measured whole-brain lysate mRNA levels of cell membrane-bound amino acid transporters that mediate arginine uptake (Fig. 5B–E). These included cationic amino acid transporter 1 (CAT1; Slc7A1), cationic amino acid transporter 2 (CAT2; Slc7A2), cationic amino acid transporter 3 (CAT3; Scl7A3), and the large neutral amino acid transporters LAT1 (Slc7A5) and LAT2 (Slc7A8). We observed no significant changes for all of these genes in Nos2−/−, C57BL/6, and APPSwDI mice (Fig. 5B–E). In contrast, in CVN-AD brain, we found that LAT1 mRNA was consistently increased from 12 weeks through 52 weeks (Fig. 5B). We also observed transient increased gene expression for LAT2, CAT1, and CAT2 at 12 weeks of age and returning to baseline expression levels by 24 weeks (Fig. 5C–E). CAT3 showed no significant change at any age (data not shown), consistent with its restricted expression on a small number of neuronal populations (Hosokawa et al., 1999). These changes in GABR and arginine transporters indicate that, compared with control strains, CVN-AD mice have dysregulated arginine metabolism.
Blockade of arginine utilization reverses memory loss
Our data on arginase expression and arginine depletion in the brains of CVN-AD mice raised the possibility that chronic brain arginine deprivation promotes neurodegeneration. To determine whether there is a causal relationship between arginine depletion and the development of pathology and cognitive defects in CVN-AD mice, we reduced the biological effects of arginine catabolism by blocking key enzymes in the arginine utilization pathway (Fig. 6). We chose to use eflornithine (DFMO), an irreversible inhibitor of ornithine decarboxylase (ODC) (Abeloff et al., 1984; Pepin et al., 1987; Seiler, 2003) and a partial inhibitor of arginase that has been previously used to block arginine utilization in vivo (Selamnia et al., 1998). Because the polyamine pathway, which is downstream of ODC, is important for cell proliferation particularly in the gastrointestinal system (Buts et al., 1993), we supplemented CVN-AD mice with putrescine to reduce gastrointestinal damage created by decreased polyamine levels (Kameji et al., 1979; Löser et al., 1999).
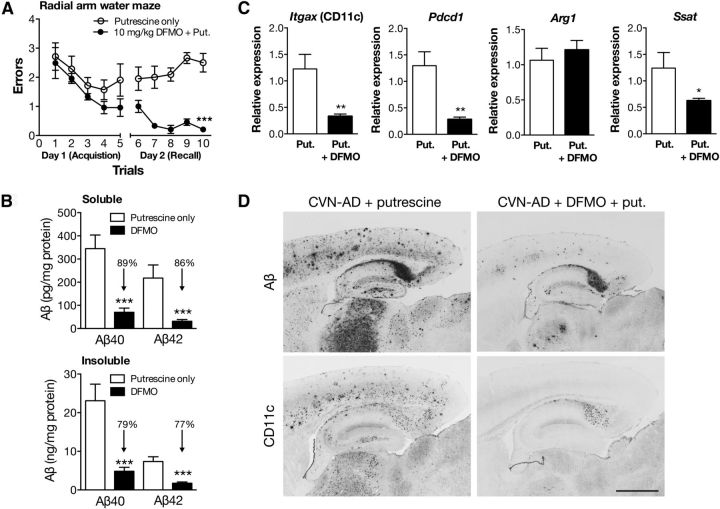
Figure 6.
CVN-AD memory deficits and pathology are reversed by an inhibitor of arginine utilization. A, RAWM assessment of spatial memory acquisition and recall in CVN-AD mice treated with putrescine (put.) alone or putrescine and DFMO (10 mg/kg) by oral gavage for 3 d/week for 14 weeks. All mice were naive to the behavioral procedure and tested on the final 2 d of treatment. Day 1 depicts 5 trial groups for the acquisition phase (learning), and day 2 depicts successive trials for memory recall. Data represent the average number of errors (± SEM) made finding the escape platform for each group of trials. Open circles represent CVN-AD mice treated with putrescine and vehicle. Closed circles represent CVN-AD mice treated with DFMO and putrescine. Data were analyzed by two-way ANOVA and post hoc multiple comparison test with Bonferonni's correction. ***p < 0.001. n = 7 or 8 mice per group. B, Soluble and insoluble Aβ40 and Aβ42 peptides in total brain homogenates from CVN-AD mice treated with vehicle containing putrescine only or putrescine plus DFMO as measured by ELISA. Data represent average levels (± SEM) of Aβ40 or Aβ42 peptides. ***p < 0.001 for DFMO-treated compared with putrescine/vehicle-treated using an unpaired Student's t test. n = 7 or 8 mice/group. C, DFMO treatment alters mRNA levels of immune genes. Average mRNA expression levels (± SEM) for Itgax, Pdcd1 (programmed death receptor 1), Arg1, and Ssat were measured in total brain lysates from CVN-AD mice treated with putrescine in vehicle or putrescine and DFMO. Gene expression levels were measured using quantitative RT-PCR and represent the 2(−delta delta C(T)) with untreated mice as the comparator. Significance between untreated and treated was determined using the unpaired Student's t test. *p < 0.05. **p < 0.01. n = 7 or 8 mice per group. D, Representative sagittal sections from CVN-AD mice treated with either putrescine alone or putrescine and DFMO stained for Aβ or CD11c. For each treatment group, panels represent sister sections from the same mouse. Scale bar, 500 μm.
Two treatment groups were established: CVN-AD mice treated with oral gavage of putrescine alone and CVN-AD mice treated with oral gavage of putrescine plus DFMO. Because increased arginase-1 mRNA and protein levels were observed at the earliest time points in the disease process, treatment was started at 6–8 weeks of age with oral gavage 3 times a week for 14 weeks. Immediately on conclusion of treatment, we tested learning and memory behavior in the putrescine only-treated and DFMO plus putrescine-treated mice. We have previously demonstrated that CVN-AD mice have deficits in spatial memory as measured by RAWM and make significantly more errors than either mNos2−/− or C57BL/6 controls at 24 weeks of age (Colton et al., 2014). CVN-AD mice treated with DFMO plus putrescine demonstrated significantly improved acquisition and recall compared with vehicle (putrescine only)-treated CVN-AD mice (Fig. 6A), suggesting that blockade of arginine catabolism reverses the behavioral phenotype of memory loss found in CVN-AD mice.
To determine whether this reversal was associated with previously identified AD-like pathologies, we measured levels of both soluble and insoluble Aβ40 and Aβ42 in whole-brain lysates. We found that soluble and insoluble Aβ40 and Aβ42 were significantly reduced by DFMO plus putrescine treatment (Fig. 6B). To determine whether neuronal numbers were altered, the number of neurons in the CA3 region of the hippocampus was counted using unbiased stereology. As we expected, we found no difference in the number of hippocampal neurons between treatment groups of 20- to 24-week-old mice, as CVN-AD mice do not develop significant neuronal loss until ∼36 weeks of age (data not shown). We measured mRNA levels for Itgax (CD11c) in whole-brain lysates from these mice and found that Itgax expression was significantly reduced in the DFMO-treated group (Fig. 6C). We also measured Pdcd1, a principal immunosuppressive gene that regulates arginase activity and T-cell activity (Liu et al., 2009; Krempski et al., 2011) and was highly increased in our gene screen. DFMO treatment significantly reduced Pdcd1 expression (Fig. 6C). Genes associated with arginine utilization were also measured. As expected, Arg1 mRNA levels did not change, whereas gene expression for spermine synthase s-acetyltransferase, which is induced by polyamine activity (Colton et al., 2004; Krempski et al., 2011), was significantly decreased by DFMO treatment (Fig. 6C). Immunohistochemical staining for Aβ revealed that DFMO treatment in CVN-AD mice resulted in less Aβ plaques, particularly in the cortex and thalamus, and to a lesser extent, in the hippocampus. Similarly, we found that DFMO-treated CVN-AD mice had less CD11c+ cells in the cortex, thalamus, and hippocampus. Interestingly, treatment with DFMO was less efficacious for reducing Aβ and CD11c in the subiculum of the hippocampus (Fig. 6D). Together, these observations indicate that treatment with DFMO reversed arginine utilization and prevented AD pathology.
Discussion
Our studies using CVN-AD mice show that: (1) CD11c+ microglia accumulate at sites of Aβ deposition, (2) these microglia show an immunosuppressive phenotype, (3) extracellular arginase accumulates in these same regions, (4) global arginine bioavailability significantly decreases, and (5) inhibition of abnormal arginine utilization results in a marked improvement in pathology and cognitive function. Importantly, CD11c+ microglial and arginase accumulation in the hippocampus and subiculum in CVN-AD mice corresponds to brain areas previously associated with neuronal damage and loss (Wilcock et al., 2008; Colton et al., 2014).
The role of immunity in AD pathogenesis is a critical and unresolved question. One current view proposes that extracellular Aβ peptide activates CNS immune cells, including microglia and perivascular macrophages, to produce inflammatory cytokines, such as TNFα, IL-1β, and reactive nitrogen or oxygen species that initiate neuronal death. Although this type of response is clearly observed in acute brain diseases, such as bacterial or viral infection and trauma, our data suggest that proinflammatory toxicity is not a primary factor in AD pathogenesis and associated neuronal cell loss. Although increased proinflammatory genes are observed in the CVN-AD mice with increasing age, these changes are late in the course of the disease and counteracted by increased expression of immunosuppressive genes (IL-1Ra, TGFβ, CD33, TREM2, and DAP12). We propose, instead, that a different type of immune pathology that involves immune suppression leads to neuronal death. In association with Aβ deposition, a subset of resident brain microglia assume a specific immunosuppressive phenotype that includes expression of CD11c and production of extracellular arginase, which acts to deplete brain extracellular arginine levels.
In contrast with other tissues, the brain is particularly susceptible to arginine deprivation as arginine is actively transported into the brain via rate-limiting amino acid transporters (Boado et al., 2004; Hawkins et al., 2005, 2006; Closs et al., 2006). Whereas baseline blood arginine levels are ∼115 μm, whole-brain arginine is ∼50 μm and even lower in the hippocampus (Dawson et al., 2004; Erdely et al., 2010). Furthermore, under inflammatory conditions, endogenous arginine synthesis and transport at the blood–brain barrier are decreased. Thus, prolonged and abnormal immune-mediated activation of arginine utilization pathways in susceptible brain regions, such as the hippocampus, may further reduce already low arginine levels, facilitating neuronal damage. When arginine utilization is interrupted by treatment with the arginase and ODC inhibitor DFMO, we show that AD-like pathology is prevented. Further studies will be required to confirm that DFMO improves arginine bioavailability and to determine whether arginase blockade can protect against neuronal death and memory deficit in a therapeutic fashion.
Our findings strikingly reflect previous observations from human AD. CD11c+ microglia are a well-known feature of human AD pathology, although their functional role has not been explicitly defined (Akiyama and McGeer, 1990; Tooyama et al., 1990; Dick et al., 1997; Walker and Lue, 2005). Direct evidence also supports that dysregulated arginine utilization may contribute to human AD. We and others have demonstrated that arginase mRNA expression and enzymatic activity are increased in human AD brain samples (Colton et al., 2006; Hansmannel et al., 2010; Liu et al., 2014). Consistent with increased arginase activity, the frontal cortex of AD patients has significantly decreased levels of l-arginine and l-ornithine (Gueli and Taibi, 2013). Furthermore, others have found increased polyamines (putresecine, spermidine, and spermine), the downstream products of arginase activity, in human AD brain (Inoue et al., 2013). Amino acid starvation leads to GCN2 kinase-mediated phosphorylation of eIF2α, leading to cell autophagy or apoptosis (Young et al., 2009; Altman and Rathmell, 2012). Indeed, increased phosphorylation of GCN2 kinase has been observed in human AD brain (Ma et al., 2013), suggesting that the integrated stress response pathway is activated as a result of amino acid deprivation. Collectively, these data support that arginine utilization, which is immune regulated, can be dysfunctional in AD and impact neurodegeneration.
Arginine is the sole substrate of two opposing enzyme systems: the iNOS pathway and the arginase pathway (Fig. 7). Both iNOS and arginase are immune regulated; and, in general, iNOS is associated with the proinflammatory phase of an immune response and arginase is associated with the anti-inflammatory phase. iNOS and arginase are often coexpressed, but competition between these two opposing enzymes favors arginase because of its higher expression level and greater Vmax (Morris, 2007; Wu et al., 2009). In humans, in contrast to C57BL/6 mice, the competitive advantage is further skewed in favor of arginase. hNOS2 expression is tightly regulated through promoter differences and post-transcriptional repressors, resulting in less iNOS-mediated NO production in humans compared with mice (Colton et al., 1996; Ganster et al., 2001; Mestas and Hughes, 2004; Guo et al., 2012). iNOS transcript, protein, and protein activity are unchanged or even decreased in human AD (Colton et al., 2006; Liu et al., 2014). Low iNOS enzymatic activity also increases arginase enzyme activity due to the loss of N-(ω)-hydroxy-l-arginine, an intermediate in the l-arginine to NO metabolic pathway and inhibitor of arginase (Boucher et al., 1999; Tenu et al., 1999).
Figure 7.
Simplified schematic of arginine catabolism and the actions of DFMO to block arginine utilization. NOHA, N-hydroxyarginine. Gray text indicates a reduction of this protein/product in CVN-AD mice.
Our findings offer one explanation for why most mouse models of AD show abundant Aβ deposition but, despite large increases in proinflammatory cytokines, do not show significant neuronal cell loss (Irizarry et al., 1997; Radde et al., 2008b). By crossing mutant APPSwDI mice onto an mNos2-deficient background, we developed a mouse model of AD that more closely mimics the reduced iNOS activity found in humans (Vitek et al., 1997). In contrast to CVN-AD mice, the parent APPSwDI mice had much reduced expression of arginase at all ages and showed no changes in arginine transporter mRNA or global arginine bioavailability. Furthermore, reconstituting the human NOS2 gene into the CVN-AD mouse strain resulted in equivalent AD-like pathology as found in CVN-AD mice (Colton et al., 2014), demonstrating that reducing NOS2 expression to more human-like levels favors arginase in the competition for arginine and highlights the species difference in NOS2 regulation of arginine utilization. Interestingly, this shift in immune-mediated redox conditions likely accounts for the accelerated and more prominent expression of CD11c, although the mechanisms governing this difference are unknown.
The exact cell type associated with increased arginine consumption remains unclear. Neurons, astrocytes, microglia, and circulating monocytes are all capable of expressing arginase-1; thus, each (or all) may participate in depletion of extracellular arginine. However, few neurons demonstrate arginase immunoreactivity, and the neuronal expression of arginase appears to be limited to specific tracts in the visual cortex and cerebellum (Yu et al., 2001). Astrocytes express a similar profile of arginine transporters to microglia and may actively reduce environmental arginine levels via robust uptake. Future studies are required to understand the kinetics and cellular origin of arginase expression, particularly in human AD pathology. However, we hypothesize that microglia are the primary source for arginine consumption. First, Aβ itself may initiate induction of immunosuppression in microglia (Kurnellas et al., 2013). Second, similarly to our observations, human myeloid cells commonly release extracellular arginase under pathologic conditions, as has been observed in glioblastoma and other tumors (Raychaudhuri et al., 2011). The appearance of CD11c+ microglia very early in pathology and their tight correlation with AD pathology and extracellular arginase support a role for this subtype of microglia in immune-mediated arginase release. Importantly, when we blocked arginase utilization in CVN-AD mice, we found significantly decreased expression of CD11c.
Although it cannot be firmly established with the present data, we anticipate that the CD11c+ cells arise from the brain's endogenous microglial population and not from infiltrating monocytes. Influx of Ly6C+ inflammatory monocytes into the mouse brain depends largely on expression of CCR2 and its ligands (El Khoury et al., 2007; Mildner et al., 2007), and no change was observed in total brain expression of CCR2 or CCL2 in CVN-AD mice at any age. Although infiltrating Ly6C+ monocytes could comprise a rare cell population, and thus total brain Ccr2 would be unchanged, we also did not observe any significant increase in brain monocytes at multiple ages by flow cytometry. Supporting these findings, Zhang et al. (2013b) previously demonstrated that monocytes from humans with AD have decreased expression of CCR2, suggesting decreased capacity for chemotaxis. In concordance with data from human AD, we found increased CCR1 expression in CVN-AD brain, although how CCR1 regulates chemotaxis in AD remains unknown (Halks-Miller et al., 2003). Thus, although our data highly implicate endogenous hippocampal microglia and not peripheral cells, further studies are required to prove this point.
In conclusion, based on our analysis of CNS immunity during aging in a mouse model of AD, we conclude that, contrary to the predominant paradigm that AD pathology is driven by proinflammatory factors, our data support an alternative mechanism for neuronal death in AD. We suggest that immune suppression and arginine catabolism lead to a loss of arginine, a critical semiessential amino acid, and this nutrient deprivation is followed by cell death. This is a novel and potentially critical mechanism that may explain the temporal and spatial induction of the slow and persistent loss of neurons in humans with AD.
Footnotes
This work was supported by the National Institutes of Health Grants NIA AG031124 and AG031845 to C.A.C. and National Institute of General Medical Sciences Medical Scientist Training Program Grant T32-GM007171 to M.J.K.
M.P.V. is a founder of Cognosci, Inc. and Oncotide Pharmaceuticals, Inc. Matters of conflict of interest for C.A.C. and M.P.V. are approved and managed by the Duke Conflict of Interest Committee. The remaining authors declare no competing financial interests.
References
- Abeloff MD, Slavik M, Luk GD, Griffin CA, Hermann J, Blanc O, Sjoerdsma A, Baylin SB. Phase I trial and pharmacokinetic studies of alpha-difluoromethylornithine-an inhibitor of polyamine biosynthesis. J Clin Oncol. 1984;2:124–130. doi: 10.1200/JCO.1984.2.2.124. [DOI] [PubMed] [Google Scholar]
- Akiyama H, McGeer PL. Brain microglia constitutively express beta-2 integrins. J Neuroimmunol. 1990;30:81–93. doi: 10.1016/0165-5728(90)90055-R. [DOI] [PubMed] [Google Scholar]
- Alamed J, Wilcock DM, Diamond DM, Gordon MN, Morgan D. Two-day radial-arm water maze learning and memory task: robust resolution of amyloid-related memory deficits in transgenic mice. Nat Protoc. 2006;1:1671–1679. doi: 10.1038/nprot.2006.275. [DOI] [PubMed] [Google Scholar]
- Altman BJ, Rathmell JC. Metabolic stress in autophagy and cell death pathways. Cold Spring Harbor Perspect Biol. 2012;4:a008763. doi: 10.1101/cshperspect.a008763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arm JP, Nwankwo C, Austen KF. Molecular identification of a novel family of human Ig superfamily members that possess immunoreceptor tyrosine-based inhibition motifs and homology to the mouse gp49B1 inhibitory receptor. J Immunol. 1997;159:2342–2349. [PubMed] [Google Scholar]
- Becher B, Antel JP. Comparison of phenotypic and functional properties of immediately ex vivo and cultured human adult microglia. Glia. 1996;18:1–10. doi: 10.1002/(SICI)1098-1136(199609)18:1<1::AID-GLIA1>3.3.CO%3B2-I. [DOI] [PubMed] [Google Scholar]
- Boado RJ, Li JY, Wise P, Pardridge WM. Human LAT1 single nucleotide polymorphism N230K does not alter phenylalanine transport. Mol Genet Metab. 2004;83:306–311. doi: 10.1016/j.ymgme.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Boucher JL, Moali C, Tenu JP. Nitric oxide biosynthesis, nitric oxide synthase inhibitors and arginase competition for l-arginine utilization. Cell Mol Life Sci. 1999;55:1015–1028. doi: 10.1007/s000180050352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw EM, Chibnik LB, Keenan BT, Ottoboni L, Raj T, Tang A, Rosenkrantz LL, Imboywa S, Lee M, Von Korff A, Morris MC, Evans DA, Johnson K, Sperling RA, Schneider JA, Bennett DA, De Jager PL. CD33 Alzheimer's disease locus: altered monocyte function and amyloid biology. Nat Neurosci. 2013;16:848–850. doi: 10.1038/nn.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröer S. Adaptation of plasma membrane amino acid transport mechanisms to physiological demands. Pflugers Arch. 2002;444:457–466. doi: 10.1007/s00424-002-0840-y. [DOI] [PubMed] [Google Scholar]
- Bronte V, Zanovello P. Regulation of immune responses by l-arginine metabolism. Nat Rev Immunol. 2005;5:641–654. doi: 10.1038/nri1668. [DOI] [PubMed] [Google Scholar]
- Brown CM, Becker JO, Wise PM, Hoofnagle AN. Simultaneous determination of 6 l-arginine metabolites in human and mouse plasma by using hydrophilic-interaction chromatography and electrospray tandem mass spectrometry. Clin Chem. 2011;57:701–709. doi: 10.1373/clinchem.2010.155895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovsky O, Ziv Y, Schwartz A, Landa G, Talpalar AE, Pluchino S, Martino G, Schwartz M. Microglia activated by IL-4 or IFN-gamma differentially induce neurogenesis and oligodendrogenesis from adult stem/progenitor cells. Mol Cell Neurosci. 2006;31:149–160. doi: 10.1016/j.mcn.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Butovsky O, Bukshpan S, Kunis G, Jung S, Schwartz M. Microglia can be induced by IFN-gamma or IL-4 to express neural or dendritic-like markers. Mol Cell Neurosci. 2007;35:490–500. doi: 10.1016/j.mcn.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Buts JP, De Keyser N, Kolanowski J, Sokal E, Van Hoof F. Maturation of villus and crypt cell functions in rat small intestine: role of dietary polyamines. Dig Dis Sci. 1993;38:1091–1098. doi: 10.1007/BF01295726. [DOI] [PubMed] [Google Scholar]
- Carson MJ, Doose JM, Melchior B, Schmid CD, Ploix CC. CNS immune privilege: hiding in plain sight. Immunol Rev. 2006;213:48–65. doi: 10.1111/j.1600-065X.2006.00441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter-Dawson LD, LaVail MM. Rods and cones in the mouse retina: II. Autoradiographic analysis of cell generation using tritiated thymidine. J Comp Neurol. 1979;188:263–272. doi: 10.1002/cne.901880205. [DOI] [PubMed] [Google Scholar]
- Closs EI, Boissel JP, Habermeier A, Rotmann A. Structure and function of cationic amino acid transporters (CATs) J Membr Biol. 2006;213:67–77. doi: 10.1007/s00232-006-0875-7. [DOI] [PubMed] [Google Scholar]
- Colton C, Wilt S, Gilbert D, Chernyshev O, Snell J, Dubois-Dalcq M. Species differences in the generation of reactive oxygen species by microglia. Mol Chem Neuropathol. 1996;28:15–20. doi: 10.1007/BF02815200. [DOI] [PubMed] [Google Scholar]
- Colton CA, Xu Q, Burke JR, Bae SY, Wakefield JK, Nair A, Strittmatter WJ, Vitek MP. Disrupted spermine homeostasis: a novel mechanism in polyglutamine-mediated aggregation and cell death. J Neurosci. 2004;24:7118–7127. doi: 10.1523/JNEUROSCI.1233-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colton CA, Mott RT, Sharpe H, Xu Q, Van Nostrand WE, Vitek MP. Expression profiles for macrophage alternative activation genes in AD and in mouse models of AD. J Neuroinflammation. 2006;3:27. doi: 10.1186/1742-2094-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colton CA, Wilcock DM, Wink DA, Davis J, Van Nostrand WE, Vitek MP. The effects of NOS2 gene deletion on mice expressing mutated human AbetaPP. J Alzheimers Dis. 2008;15:571–587. doi: 10.3233/jad-2008-15405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colton CA, Wilson JG, Everhart A, Wilcock DM, Puolivali J, Heikkinen T, Oksman J, Jääskeläinen O, Lehtimäki K, Laitinen T, Vartiainen N, Vitek MP. mNos2 deletion and human NOS2 replacement in Alzheimer disease models. J Neuropathol Exp Neurol. 2014;73:752–769. doi: 10.1097/NEN.0000000000000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comi C, Carecchio M, Chiocchetti A, Nicola S, Galimberti D, Fenoglio C, Cappellano G, Monaco F, Scarpini E, Dianzani U. Osteopontin is increased in the cerebrospinal fluid of patients with Alzheimer's disease and its levels correlate with cognitive decline. J Alzheimers Dis. 2010;19:1143–1148. doi: 10.3233/JAD-2010-1309. [DOI] [PubMed] [Google Scholar]
- Curry H, Alvarez GR, Zwilling BS, Lafuse WP. Toll-like receptor 2 stimulation decreases IFN-gamma receptor expression in mouse RAW264.7 macrophages. J Interferon Cytokine Res. 2004;24:699–710. doi: 10.1089/jir.2004.24.699. [DOI] [PubMed] [Google Scholar]
- Date D, Das R, Narla G, Simon DI, Jain MK, Mahabeleshwar GH. Kruppel-like transcription factor 6 regulates inflammatory macrophage polarization. J Biol Chem. 2014;289:10318–10329. doi: 10.1074/jbc.M113.526749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J, Xu F, Deane R, Romanov G, Previti ML, Zeigler K, Zlokovic BV, Van Nostrand WE. Early-onset and robust cerebral microvascular accumulation of amyloid beta-protein in transgenic mice expressing low levels of a vasculotropic Dutch/Iowa mutant form of amyloid beta-protein precursor. J Biol Chem. 2004;279:20296–20306. doi: 10.1074/jbc.M312946200. [DOI] [PubMed] [Google Scholar]
- Dawson LA, Organ AJ, Winter P, Lacroix LP, Shilliam CS, Heidbreder C, Shah AJ. Rapid high-throughput assay for the measurement of amino acids from microdialysates and brain tissue using monolithic C18-bonded reversed-phase columns. J Chromatogr B Analyt Technol Biomed Life Sci. 2004;807:235–241. doi: 10.1016/j.jchromb.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Denieffe S, Kelly RJ, McDonald C, Lyons A, Lynch MA. Classical activation of microglia in CD200-deficient mice is a consequence of blood brain barrier permeability and infiltration of peripheral cells. Brain Behav Immun. 2013;34:86–97. doi: 10.1016/j.bbi.2013.07.174. [DOI] [PubMed] [Google Scholar]
- Dick AD, Pell M, Brew BJ, Foulcher E, Sedgwick JD. Direct ex vivo flow cytometric analysis of human microglial cell CD4 expression: examination of central nervous system biopsy specimens from HIV-seropositive patients and patients with other neurological disease. AIDS. 1997;11:1699–1708. doi: 10.1097/00002030-199714000-00006. [DOI] [PubMed] [Google Scholar]
- Dickson DW, Farlo J, Davies P, Crystal H, Fuld P, Yen SH. Alzheimer's disease: a double-labeling immunohistochemical study of senile plaques. Am J Pathol. 1988;132:86–101. [PMC free article] [PubMed] [Google Scholar]
- Duffield JS. Macrophages and immunologic inflammation of the kidney. Semin Nephrol. 2010;30:234–254. doi: 10.1016/j.semnephrol.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eikelenboom P, van Exel E, Hoozemans JJ, Veerhuis R, Rozemuller AJ, van Gool WA. Neuroinflammation: an early event in both the history and pathogenesis of Alzheimer's disease. Neurodegener Dis. 2010;7:38–41. doi: 10.1159/000283480. [DOI] [PubMed] [Google Scholar]
- El Khoury J, Toft M, Hickman SE, Means TK, Terada K, Geula C, Luster AD. Ccr2 deficiency impairs microglial accumulation and accelerates progression of Alzheimer-like disease. Nat Med. 2007;13:432–438. doi: 10.1038/nm1555. [DOI] [PubMed] [Google Scholar]
- Erdely A, Kepka-Lenhart D, Salmen-Muniz R, Chapman R, Hulderman T, Kashon M, Simeonova PP, Morris SM., Jr Arginase activities and global arginine bioavailability in wild-type and ApoE-deficient mice: responses to high fat and high cholesterol diets. PLoS One. 2010;5:e15253. doi: 10.1371/journal.pone.0015253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganster RW, Taylor BS, Shao L, Geller DA. Complex regulation of human inducible nitric oxide synthase gene transcription by Stat 1 and NF-kappa B. Proc Natl Acad Sci U S A. 2001;98:8638–8643. doi: 10.1073/pnas.151239498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller DA, Billiar TR. Molecular biology of nitric oxide synthases. Cancer Metastasis Rev. 1998;17:7–23. doi: 10.1023/A:1005940202801. [DOI] [PubMed] [Google Scholar]
- Golanska E, Sieruta M, Gresner SM, Pfeffer A, Chodakowska-Zebrowska M, Sobow TM, Klich I, Mossakowska M, Szybinska A, Barcikowska M, Liberski PP. APBB2 genetic polymorphisms are associated with severe cognitive impairment in centenarians. Exp Gerontol. 2013;48:391–394. doi: 10.1016/j.exger.2013.01.013. [DOI] [PubMed] [Google Scholar]
- Grupe A, Li Y, Rowland C, Nowotny P, Hinrichs AL, Smemo S, Kauwe JS, Maxwell TJ, Cherny S, Doil L, Tacey K, van Luchene R, Myers A, Wavrant-De Vrièze F, Kaleem M, Hollingworth P, Jehu L, Foy C, Archer N, Hamilton G, et al. A scan of chromosome 10 identifies a novel locus showing strong association with late-onset Alzheimer disease. Am J Hum Genet. 2006;78:78–88. doi: 10.1086/498851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueli MC, Taibi G. Alzheimer's disease: amino acid levels and brain metabolic status. Neurol Sci. 2013;34:1575–1579. doi: 10.1007/s10072-013-1289-9. [DOI] [PubMed] [Google Scholar]
- Guo Z, Shao L, Zheng L, Du Q, Li P, John B, Geller DA. miRNA-939 regulates human inducible nitric oxide synthase posttranscriptional gene expression in human hepatocytes. Proc Natl Acad Sci U S A. 2012;109:5826–5831. doi: 10.1073/pnas.1118118109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halks-Miller M, Schroeder ML, Haroutunian V, Moenning U, Rossi M, Achim C, Purohit D, Mahmoudi M, Horuk R. CCR1 is an early and specific marker of Alzheimer's disease. Ann Neurol. 2003;54:638–646. doi: 10.1002/ana.10733. [DOI] [PubMed] [Google Scholar]
- Hansmannel F, Sillaire A, Kamboh MI, Lendon C, Pasquier F, Hannequin D, Laumet G, Mounier A, Ayral AM, DeKosky ST, Hauw JJ, Berr C, Mann D, Amouyel P, Campion D, Lambert JC. Is the urea cycle involved in Alzheimer's disease? J Alzheimers Dis. 2010;21:1013–1021. doi: 10.3233/JAD-2010-100630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RA, Mokashi A, Simpson IA. An active transport system in the blood–brain barrier may reduce levodopa availability. Exp Neurol. 2005;195:267–271. doi: 10.1016/j.expneurol.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Hawkins RA, O'Kane RL, Simpson IA, Viña JR. Structure of the blood–brain barrier and its role in the transport of amino acids. J Nutr. 2006;136:218S–226S. doi: 10.1093/jn/136.1.218S. [DOI] [PubMed] [Google Scholar]
- Hickman SE, Kingery ND, Ohsumi TK, Borowsky ML, Wang LC, Means TK, El Khoury J. The microglial sensome revealed by direct RNA sequencing. Nat Neurosci. 2013;16:1896–1905. doi: 10.1038/nn.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa H, Ninomiya H, Sawamura T, Sugimoto Y, Ichikawa A, Fujiwara K, Masaki T. Neuron-specific expression of cationic amino acid transporter 3 in the adult rat brain. Brain Res. 1999;838:158–165. doi: 10.1016/S0006-8993(99)01686-8. [DOI] [PubMed] [Google Scholar]
- Hyatt SL, Aulak KS, Malandro M, Kilberg MS, Hatzoglou M. Adaptive regulation of the cationic amino acid transporter-1 (Cat-1) in Fao cells. J Biol Chem. 1997;272:19951–19957. doi: 10.1074/jbc.272.32.19951. [DOI] [PubMed] [Google Scholar]
- Inoue K, Tsutsui H, Akatsu H, Hashizume Y, Matsukawa N, Yamamoto T, Toyo'oka T. Metabolic profiling of Alzheimer's disease brains. Sci Rep. 2013;3:2364. doi: 10.1038/srep02364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry MC, McNamara M, Fedorchak K, Hsiao K, Hyman BT. APPSw transgenic mice develop age-related A beta deposits and neuropil abnormalities, but no neuronal loss in CA1. J Neuropathol Exp Neurol. 1997;56:965–973. doi: 10.1097/00005072-199709000-00002. [DOI] [PubMed] [Google Scholar]
- Kamboh MI, Demirci FY, Wang X, Minster RL, Carrasquillo MM, Pankratz VS, Younkin SG, Saykin AJ, Saykin AJ, Jun G, Baldwin C, Logue MW, Buros J, Farrer L, Pericak-Vance MA, Haines JL, Sweet RA, Ganguli M, Feingold E, Dekosky ST, et al. Genome-wide association study of Alzheimer's disease. Transl Psychiatry. 2012;2:e117. doi: 10.1038/tp.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameji T, Murakami Y, Hayashi S. Effect of diaminobutene and diaminopropane on diet-stimulated polyamine synthesis and cell proliferation in rat liver. J Biochem. 1979;86:191–197. [PubMed] [Google Scholar]
- Karlstetter M, Walczak Y, Weigelt K, Ebert S, Van den Brulle J, Schwer H, Fuchshofer R, Langmann T. The novel activated microglia/macrophage WAP domain protein, AMWAP, acts as a counter-regulator of proinflammatory response. J Immunol. 2010;185:3379–3390. doi: 10.4049/jimmunol.0903300. [DOI] [PubMed] [Google Scholar]
- Kearney SJ, Delgado C, Eshleman EM, Hill KK, O'Connor BP, Lenz LL. Type I IFNs downregulate myeloid cell IFN-gamma receptor by inducing recruitment of an early growth response 3/NGFI-A binding protein 1 complex that silences ifngr1 transcription. J Immunol. 2013;191:3384–3392. doi: 10.4049/jimmunol.1203510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krempski J, Karyampudi L, Behrens MD, Erskine CL, Hartmann L, Dong H, Goode EL, Kalli KR, Knutson KL. Tumor-infiltrating programmed death receptor-1+ dendritic cells mediate immune suppression in ovarian cancer. J Immunol. 2011;186:6905–6913. doi: 10.4049/jimmunol.1100274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Mizushima N. Physiological role of autophagy as an intracellular recycling system: with an emphasis on nutrient metabolism. Semin Cell Dev Biol. 2010;21:683–690. doi: 10.1016/j.semcdb.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Kurnellas MP, Adams CM, Sobel RA, Steinman L, Rothbard JB. Amyloid fibrils composed of hexameric peptides attenuate neuroinflammation. Sci Transl Med. 2013;5:179ra142. doi: 10.1126/scitranslmed.3005681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson KC, Lipko M, Dabrowski M, Draper MP. Gng12 is a novel negative regulator of LPS-induced inflammation in the microglial cell line BV-2. Inflamm Res. 2010;59:15–22. doi: 10.1007/s00011-009-0062-2. [DOI] [PubMed] [Google Scholar]
- Laubach VE, Foley PL, Shockey KS, Tribble CG, Kron IL. Protective roles of nitric oxide and testosterone in endotoxemia: evidence from NOS-2-deficient mice. Am J Physiol. 1998;275:H2211–H2218. doi: 10.1152/ajpheart.1998.275.6.H2211. [DOI] [PubMed] [Google Scholar]
- Liu P, Fleete MS, Jing Y, Collie ND, Curtis MA, Waldvogel HJ, Faull RL, Abraham WC, Zhang H. Altered arginine metabolism in Alzheimer's disease brains. Neurobiol Aging. 2014;35:1992–2003. doi: 10.1016/j.neurobiolaging.2014.03.013. [DOI] [PubMed] [Google Scholar]
- Liu Y, Yu Y, Yang S, Zeng B, Zhang Z, Jiao G, Zhang Y, Cai L, Yang R. Regulation of arginase I activity and expression by both PD-1 and CTLA-4 on the myeloid-derived suppressor cells. Cancer Immunol Immunother. 2009;58:687–697. doi: 10.1007/s00262-008-0591-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Löser C, Eisel A, Harms D, Fölsch UR. Dietary polyamines are essential luminal growth factors for small intestinal and colonic mucosal growth and development. Gut. 1999;44:12–16. doi: 10.1136/gut.44.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, Trinh MA, Wexler AJ, Bourbon C, Gatti E, Pierre P, Cavener DR, Klann E. Suppression of eIF2alpha kinases alleviates Alzheimer's disease-related plasticity and memory deficits. Nat Neurosci. 2013;16:1299–1305. doi: 10.1038/nn.3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattapallil MJ, Wawrousek EF, Chan CC, Zhao H, Roychoudhury J, Ferguson TA, Caspi RR. The Rd8 mutation of the Crb1 gene is present in vendor lines of C57BL/6N mice and embryonic stem cells, and confounds ocular induced mutant phenotypes. Invest Ophthalmol Vis Sci. 2012;53:2921–2927. doi: 10.1167/iovs.12-9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick MJ, Castells MC, Austen KF, Katz HR. The gp49A gene has extensive sequence conservation with the gp49B gene and provides gp49A protein, a unique member of a large family of activating and inhibitory receptors of the immunoglobulin superfamily. Immunogenetics. 1999;50:286–294. doi: 10.1007/s002510050604. [DOI] [PubMed] [Google Scholar]
- McGeer PL, Itagaki S, Tago H, McGeer EG. Reactive microglia in patients with senile dementia of the Alzheimer type are positive for the histocompatibility glycoprotein HLA-DR. Neurosci Lett. 1987;79:195–200. doi: 10.1016/0304-3940(87)90696-3. [DOI] [PubMed] [Google Scholar]
- Mellor AL, Munn DH. Creating immune privilege: active local suppression that benefits friends, but protects foes. Nat Rev Immunol. 2008;8:74–80. doi: 10.1038/nri2233. [DOI] [PubMed] [Google Scholar]
- Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- Mildner A, Schmidt H, Nitsche M, Merkler D, Hanisch UK, Mack M, Heikenwalder M, Brück W, Priller J, Prinz M. Microglia in the adult brain arise from Ly-6ChiCCR2+ monocytes only under defined host conditions. Nat Neurosci. 2007;10:1544–1553. doi: 10.1038/nn2015. [DOI] [PubMed] [Google Scholar]
- Mildner A, Schlevogt B, Kierdorf K, Böttcher C, Erny D, Kummer MP, Quinn M, Brück W, Bechmann I, Heneka MT, Priller J, Prinz M. Distinct and non-redundant roles of microglia and myeloid subsets in mouse models of Alzheimer's disease. J Neurosci. 2011;31:11159–11171. doi: 10.1523/JNEUROSCI.6209-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SM., Jr Arginine metabolism: boundaries of our knowledge. J Nutr. 2007;137:1602S–1609S. doi: 10.1093/jn/137.6.1602S. [DOI] [PubMed] [Google Scholar]
- Morris SM., Jr Arginases and arginine deficiency syndromes. Curr Opin Clin Nutr Metab Care. 2012;15:64–70. doi: 10.1097/MCO.0b013e32834d1a08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrak RE, Griffin WS. Glia and their cytokines in progression of neurodegeneration. Neurobiol Aging. 2005;26:349–354. doi: 10.1016/j.neurobiolaging.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Naert G, Rivest S. CC chemokine receptor 2 deficiency aggravates cognitive impairments and amyloid pathology in a transgenic mouse model of Alzheimer's disease. J Neurosci. 2011;31:6208–6220. doi: 10.1523/JNEUROSCI.0299-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichol KE, Poon WW, Parachikova AI, Cribbs DH, Glabe CG, Cotman CW. Exercise alters the immune profile in Tg2576 Alzheimer mice toward a response coincident with improved cognitive performance and decreased amyloid. J Neuroinflammation. 2008;5:13. doi: 10.1186/1742-2094-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepin J, Milord F, Guern C, Schechter PJ. Difluoromethylornithine for arseno-resistant Trypanosoma brucei gambiense sleeping sickness. Lancet. 1987;2:1431–1433. doi: 10.1016/S0140-6736(87)91131-7. [DOI] [PubMed] [Google Scholar]
- Pesce JT, Ramalingam TR, Mentink-Kane MM, Wilson MS, El Kasmi KC, Smith AM, Thompson RW, Cheever AW, Murray PJ, Wynn TA. Arginase-1-expressing macrophages suppress Th2 cytokine-driven inflammation and fibrosis. PLoS Pathog. 2009;5:e1000371. doi: 10.1371/journal.ppat.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettem KL, Yokomaku D, Takahashi H, Ge Y, Craig AM. Interaction between autism-linked MDGAs and neuroligins suppresses inhibitory synapse development. J Cell Biol. 2013;200:321–336. doi: 10.1083/jcb.201206028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radde R, Duma C, Goedert M, Jucker M. The value of incomplete mouse models of Alzheimer's disease. Eur J Nucl Med Mol Imaging. 2008a;35(Suppl 1):S70–S74. doi: 10.1007/s00259-007-0704-y. [DOI] [PubMed] [Google Scholar]
- Radde R, Duma C, Goedert M, Jucker M. The value of incomplete mouse models of Alzheimer's disease. Eur J Nucl Med Mol Imaging. 2008b;35(Suppl 1):S70–S74. doi: 10.1007/s00259-007-0704-y. [DOI] [PubMed] [Google Scholar]
- Ransohoff RM, Cardona AE. The myeloid cells of the central nervous system parenchyma. Nature. 2010;468:253–262. doi: 10.1038/nature09615. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri B, Rayman P, Ireland J, Ko J, Rini B, Borden EC, Garcia J, Vogelbaum MA, Finke J. Myeloid-derived suppressor cell accumulation and function in patients with newly diagnosed glioblastoma. Neuro-oncology. 2011;13:591–599. doi: 10.1093/neuonc/nor042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeds PJ. Dispensable and indispensable amino acids for humans. J Nutr. 2000;130:1835S–1840S. doi: 10.1093/jn/130.7.1835S. [DOI] [PubMed] [Google Scholar]
- Ridnour LA, Windhausen AN, Isenberg JS, Yeung N, Thomas DD, Vitek MP, Roberts DD, Wink DA. Nitric oxide regulates matrix metalloproteinase-9 activity by guanylyl-cyclase-dependent and -independent pathways. Proc Natl Acad Sci U S A. 2007;104:16898–16903. doi: 10.1073/pnas.0702761104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J, Strohmeyer R, Kovelowski CJ, Li R. Microglia and inflammatory mechanisms in the clearance of amyloid beta peptide. Glia. 2002;40:260–269. doi: 10.1002/glia.10153. [DOI] [PubMed] [Google Scholar]
- Rozemuller JM, Eikelenboom P, Stam FC. Role of microglia in plaque formation in senile dementia of the Alzheimer type: an immunohistochemical study. Virchows Arch B Cell Pathol Incl Mol Pathol. 1986;51:247–254. doi: 10.1007/BF02899034. [DOI] [PubMed] [Google Scholar]
- Salminen A, Ojala J, Kauppinen A, Kaarniranta K, Suuronen T. Inflammation in Alzheimer's disease: amyloid-beta oligomers trigger innate immunity defence via pattern recognition receptors. Prog Neurobiol. 2009;87:181–194. doi: 10.1016/j.pneurobio.2009.01.001. [DOI] [PubMed] [Google Scholar]
- Sangaletti S, Tripodo C, Sandri S, Torselli I, Vitali C, Ratti C, Botti L, Burocchi A, Porcasi R, Tomirotti A, Colombo MP, Chiodoni C. Osteopontin shapes immunosuppression in the metastatic niche. Cancer Res. 2014;74:4706–4719. doi: 10.1158/0008-5472.CAN-13-3334. [DOI] [PubMed] [Google Scholar]
- Santiago ML, Montano M, Benitez R, Messer RJ, Yonemoto W, Chesebro B, Hasenkrug KJ, Greene WC. Apobec3 encodes Rfv3, a gene influencing neutralizing antibody control of retrovirus infection. Science. 2008;321:1343–1346. doi: 10.1126/science.1161121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders AM, Hulette O, Welsh-Bohmer KA, Schmechel DE, Crain B, Burke JR, Alberts MJ, Strittmatter WJ, Breitner JC, Rosenberg C. Specificity, sensitivity, and predictive value of apolipoprotein-E genotyping for sporadic Alzheimer's disease. Lancet. 1996;348:90–93. doi: 10.1016/S0140-6736(96)01251-2. [DOI] [PubMed] [Google Scholar]
- Seiler N. Thirty years of polyamine-related approaches to cancer therapy: retrospect and prospect: 2. Structural analogues and derivatives. Curr Drug Targets. 2003;4:565–585. doi: 10.2174/1389450033490876. [DOI] [PubMed] [Google Scholar]
- Selamnia M, Mayeur C, Robert V, Blachier F. Alpha-difluoromethylornithine (DFMO) as a potent arginase activity inhibitor in human colon carcinoma cells. Biochem Pharmacol. 1998;55:1241–1245. doi: 10.1016/S0006-2952(97)00572-8. [DOI] [PubMed] [Google Scholar]
- Tambuyzer BR, Ponsaerts P, Nouwen EJ. Microglia: gatekeepers of central nervous system immunology. J Leukoc Biol. 2009;85:352–370. doi: 10.1189/jlb.0608385. [DOI] [PubMed] [Google Scholar]
- Tan J, Town T, Mori T, Wu Y, Saxe M, Crawford F, Mullan M. CD45 opposes beta-amyloid peptide-induced microglial activation via inhibition of p44/42 mitogen-activated protein kinase. J Neurosci. 2000;20:7587–7594. doi: 10.1523/JNEUROSCI.20-20-07587.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WH, Wang Z, Cho L, Brennan DM, Hazen SL. Diminished global arginine bioavailability and increased arginine catabolism as metabolic profile of increased cardiovascular risk. J Am Coll Cardiol. 2009;53:2061–2067. doi: 10.1016/j.jacc.2009.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenu JP, Lepoivre M, Moali C, Brollo M, Mansuy D, Boucher JL. Effects of the new arginase inhibitor N(omega)-hydroxy-nor-l-arginine on NO synthase activity in murine macrophages. Nitric Oxide. 1999;3:427–438. doi: 10.1006/niox.1999.0255. [DOI] [PubMed] [Google Scholar]
- Tooyama I, Kimura H, Akiyama H, McGeer PL. Reactive microglia express class I and class II major histocompatibility complex antigens in Alzheimer's disease. Brain Res. 1990;523:273–280. doi: 10.1016/0006-8993(90)91496-4. [DOI] [PubMed] [Google Scholar]
- Trueba-Sáiz A, Cavada C, Fernandez AM, Leon T, González DA, Fortea Ormaechea J, Lleó A, Del Ser T, Nuñez A, Torres-Aleman I. Loss of serum IGF-I input to the brain as an early biomarker of disease onset in Alzheimer mice. Transl Psychiatry. 2013;3:e330. doi: 10.1038/tp.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitek MP, Snell J, Dawson H, Colton CA. Modulation of nitric oxide production in human macrophages by apolipoprotein-E and amyloid-beta peptide. Biochem Biophys Res Commun. 1997;240:391–394. doi: 10.1006/bbrc.1997.7408. [DOI] [PubMed] [Google Scholar]
- Walker DG, Lue LF. Investigations with cultured human microglia on pathogenic mechanisms of Alzheimer's disease and other neurodegenerative diseases. J Neurosci Res. 2005;81:412–425. doi: 10.1002/jnr.20484. [DOI] [PubMed] [Google Scholar]
- Walker DG, Lue LF. Understanding the neurobiology of CD200 and the CD200 receptor: a therapeutic target for controlling inflammation in human brains? Future Neurol. 2013;8:3. doi: 10.2217/fnl.13.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg JB, Misukonis MA, Shami PJ, Mason SN, Sauls DL, Dittman WA, Wood ER, Smith GK, McDonald B, Bachus KE. Human mononuclear phagocyte inducible nitric oxide synthase (iNOS): analysis of iNOS mRNA, iNOS protein, biopterin, and nitric oxide production by blood monocytes and peritoneal macrophages. Blood. 1995;86:1184–1195. [PubMed] [Google Scholar]
- Wilcock DM, Lewis MR, Van Nostrand WE, Davis J, Previti ML, Gharkholonarehe N, Vitek MP, Colton CA. Progression of amyloid pathology to Alzheimer's disease pathology in an amyloid precursor protein transgenic mouse model by removal of nitric oxide synthase 2. J Neurosci. 2008;28:1537–1545. doi: 10.1523/JNEUROSCI.5066-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcock DM, Morgan D, Gordon MN, Taylor TL, Ridnour LA, Wink DA, Colton CA. Activation of matrix metalloproteinases following anti-Abeta immunotherapy: implications for microhemorrhage occurrence. J Neuroinflammation. 2011a;8:115. doi: 10.1186/1742-2094-8-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcock DM, Zhao Q, Morgan D, Gordon MN, Everhart A, Wilson JG, Lee JE, Colton CA. Diverse inflammatory responses in transgenic mouse models of Alzheimer's disease and the effect of immunotherapy on these responses. ASN Neuro. 2011b;3:249–258. doi: 10.1042/AN20110018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wink DA, Hines HB, Cheng RY, Switzer CH, Flores-Santana W, Vitek MP, Ridnour LA, Colton CA. Nitric oxide and redox mechanisms in the immune response. J Leukoc Biol. 2011;89:873–891. doi: 10.1189/jlb.1010550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright KO, Murray DA, Crispe NI, Pierce RH. Quantitative PCR for detection of the OT-1 transgene. BMC Immunol. 2005;6:20. doi: 10.1186/1471-2172-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Bazer FW, Davis TA, Kim SW, Li P, Rhoads MJ, Satterfield CM, Smith SB, Spencer TE, Yin Y. Arginine metabolism and nutrition in growth, health and disease. Amino Acids. 2009;37:153–168. doi: 10.1007/s00726-008-0210-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao A, Liu F, Chen K, Tang L, Liu L, Zhang K, Yu C, Bian G, Guo H, Zheng J, Cheng P, Ju G, Wang J. Programmed death 1 deficiency induces the polarization of macrophages/microglia to the M1 phenotype after spinal cord injury in mice. Neurotherapeutics. 2014;11:636–650. doi: 10.1007/s13311-013-0254-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JE, Martinez RA, La Spada AR. Nutrient deprivation induces neuronal autophagy and implicates reduced insulin signaling in neuroprotective autophagy activation. J Biol Chem. 2009;284:2363–2373. doi: 10.1074/jbc.M806088200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Iyer RK, Kern RM, Rodriguez WI, Grody WW, Cederbaum SD. Expression of arginase isozymes in mouse brain. J Neurosci Res. 2001;66:406–422. doi: 10.1002/jnr.1233. [DOI] [PubMed] [Google Scholar]
- Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA, Zhang C, Xie T, Tran L, Dobrin R, Fluder E, Clurman B, Melquist S, Narayanan M, Suver C, Shah H, Mahajan M, Gillis T, Mysore J, MacDonald ME, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer's disease. Cell. 2013a;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Miller RG, Madison C, Jin X, Honrada R, Harris W, Katz J, Forshew DA, McGrath MS. Systemic immune system alterations in early stages of Alzheimer's disease. J Neuroimmunol. 2013b;256:38–42. doi: 10.1016/j.jneuroim.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]