Abstract
We have identified and characterized a spontaneous Brown Norway from Janvier rat strain (BN-J) presenting a progressive retinal degeneration associated with early retinal telangiectasia, neuronal alterations, and loss of retinal Müller glial cells resembling human macular telangiectasia type 2 (MacTel 2), which is a retinal disease of unknown cause. Genetic analyses showed that the BN-J phenotype results from an autosomal recessive indel novel mutation in the Crb1 gene, causing dislocalization of the protein from the retinal Müller glia (RMG)/photoreceptor cell junction. The transcriptomic analyses of primary RMG cultures allowed identification of the dysregulated pathways in BN-J rats compared with wild-type BN rats. Among those pathways, TGF-β and Kit Receptor Signaling, MAPK Cascade, Growth Factors and Inflammatory Pathways, G-Protein Signaling Pathways, Regulation of Actin Cytoskeleton, and Cardiovascular Signaling were found. Potential molecular targets linking RMG/photoreceptor interaction with the development of retinal telangiectasia are identified. This model can help us to better understand the physiopathologic mechanisms of MacTel 2 and other retinal diseases associated with telangiectasia.
Keywords: adherens junction, disease model, genetics, microcirculation, retinal blood vessels, retinal degeneration
Introduction
Retinal Müller glial (RMG) cells span the entire thickness of the retina and establish links between retinal blood vessels and photoreceptors, providing nutritional support, removing metabolic waste, and maintaining homeostasis of extracellular medium (Bringmann et al., 2006). RMG cells intervene in the formation and maintenance of the inner blood–retinal barrier (Tout et al., 1993; Tretiach et al., 2005) and connect to photoreceptors with adherens and tight-like junctions at the outer limiting membrane (OLM) (Omri et al., 2010). It was recently suggested that RMG cells may play a role in the development of diabetic retinopathy (Fletcher et al., 2005; Bringmann et al., 2006) and macular telangiectasia type 2 (MacTel 2; Powner et al., 2010).
MacTel 2 is a progressive retinal disease characterized by vascular abnormalities, depletion of macular luteal pigment, and cystic cavities with focal disorganization of retinal lamination (Yannuzzi et al., 2006). Photoreceptor degeneration is associated with visual impairment (Ooto et al., 2011). In vivo optical coherence tomography further showed OLM defects associated with photoreceptor disruption (Zhu et al., 2013). Loss of RMG markers and reduction of RMG-associated proteins in the macula have been revealed on MacTel 2 retinas, providing evidences on the role of RMG in the disease pathogenesis (Powner et al., 2010; Len et al., 2012).
During retinal development, RMG cells are required for photoreceptor outer segment assembly (Jablonski and Iannaccone, 2000; Wang et al., 2005) and, in the postnatal period, genetic RMG destruction led to retinal dysplasia and retinal degeneration (Dubois-Dauphin et al., 2000). Conversely, RMG proliferation in mice lacking the cell cycle inhibitor protein p27Kip1 also induced retinal dysplasia, OLM disruption, and leaky vascular dilation (Dyer and Cepko, 2000).
The Crumbs (CRB) proteins, particularly CRB1, located in the subapical region above the OLM, form a molecular scaffold with Pals1 and Patj and interact with the Par6/Par3/aPKC complex and with β-catenin (Alves et al., 2014). CRB1, expressed in mammalian RMG cells, is essential for OLM formation and for photoreceptor morphogenesis (Mehalow et al., 2003; van de Pavert et al., 2004). Interestingly, crb1 mutations lead to retinal degenerations that are potentially associated with coats-like vascular telangiectasia (den Holländer et al., 2004; Henderson et al., 2011).
This report describes a Brown Norway from Janvier rat strain (BN-J) that spontaneously develops progressive focal retinal layer disorganization, loss of photoreceptors, cystic cavitation, and RMG abnormalities associated with early retinal vascular telangiectasia and late stage subretinal neovascularization. This phenotype bears marked resemblance to the telangiectasia-like model obtained by specific RMG depletion (Shen et al., 2012) and reminiscent of human MacTel 2 (Charbel Issa et al., 2013). A new mutation in exon 6 of the rat crb1 was identified to be responsible for this retinal phenotype. In addition, the full profile of genes differentially expressed in RMG cells extracted from the Crb1-mutated BN rat retina compared with two wild-type strains allowed identification of possible molecular targets. These data link CRB1-associated functions with rat retinal telangiectasia and possibly with human MacTel 2.
Materials and Methods
Animals.
All experiments were performed in accordance with the European Communities Council Directive 86/609/EEC and approved by local ethical committees. BN rats obtained from Janvier Breeding Center (pathological BN-J rat) or Harlan Laboratories (wild-type BN-H rat) and Lewis rats from Janvier Breeding Center were used. Rats of either sex were used. Animals were kept in pathogen-free conditions with food, water, and litter and housed in a 12 h light/12 h dark cycle. For genetic analyses, four couples of pure parental strains (BN-H × BN-J) were cross-bred, which resulted in an F1. Four F1 couples were then cross-bred to produce an F2. Anesthesia was induced by intramuscular ketamine (40 mg/kg) and xylazine (4 mg/kg). Animals were killed by carbon dioxide inhalation.
Fluorescein angiography.
BN-H and BN-J rats of different ages (8 weeks and 6 months, n = 6 rats per time point) were used. Fluorescein (0.1 ml of 10% fluorescein in saline) was injected in the tail vein of anesthetized rats. In vivo angiography was performed with a confocal scanning laser ophthalmoscope (cSLO, HRA; Heidelberg Engineering). Images were collected at early and late time points.
Electroretinogram.
Electroretinographic (ERG) analyses were performed on 3-week-old BN-H and BN-J rats (n = 4–5 per strain) using a VisioSystem device (Siem Biomedicale). Animals were dark adapted overnight. Scotopic ERG was performed in the dark with light intensities of flashes ranging from 0.0003 to 10 cd · s/m2. For each intensity, the average response to 5 flashes at a frequency of 0.5 Hz was recorded. Basic overall retinal responses were recorded after flashes at 0 dB intensity for 40 ms at a frequency of 0.5 Hz. Five responses were averaged. For photopic recordings, animals were light adapted for 10 min with a background light of 25 cd/m2 and then the response after a single light flash of 10 cd · s/m2 was recorded.
Histology.
BN-J and BN-H rats were killed [adults at 8 weeks and 6 months of age, n = 4 rats per time point per strain, and postnatal day 1 (P1), P8, and P15, n = 3 per time point and per strain], and eyes enucleated for histological analyses using historesine sections (5 μm) stained with toluidine blue as described previously (Zhao et al., 2012).
Semithin and ultrathin sections.
Eyes from BN rats (8 weeks and 6 months of age, n = 4 rats per time point and per strain) were fixed in 2.5% glutaraldehyde in cacodylate buffer (0.1 mol/L, pH7.4) and then dissected, postfixed in 1% osmium tetroxide in cacodylate buffer, and dehydrated in a graded series of alcohol before being included in epoxy resin. Semithin sections (1 μm) were stained with toluidine blue. Ultrathin sections (80 nm) were contrasted by uranyl acetate and lead citrate and observed with a transmission electron microscope (TEM) and photographed.
Retinal flat-mounts.
BN-H and BN-J rats at 8 weeks were killed (n = 10 rats per strain). Rat flat-mounted retinas were prepared as described previously (Zhao et al., 2010). The following primary antibodies were used: rabbit anti-glial fibrillary acidic protein (GFAP, 1:100; Dako), rabbit anti-glutamine synthetase (GS, 1:100; Sigma-Aldrich), and secondary antibody Alexa Fluor 594-conjugated goat anti-rabbit IgG (1:100; Invitrogen). Blood vessels were stained with FITC-labeled lectin from Bandeiraea simplicifolia (1:100; Sigma-Aldrich). Images were taken using a confocal laser scanning microscope Zeiss LSM 710 and analyzed using ImageJ.
Immunohistochemistry on cryosections.
Eyes of 8-week-old BN rats (n = 4 rats per strain) were used for cryosections. Cryostat sections were incubated with the following primary antibodies: mouse anti-CD31 (1:100; BD Pharmingen), rabbit anti-GFAP (1:200), rabbit anti-GS (1:200), rabbit anti-cone arrestin (1:100; Millipore), mouse anti-rhodopsin (Rho4D2, 1:100; Abcam), mouse anti-protein kinase C-α (PKC-α, 1:400; Santa Cruz Biotechnology), rabbit anti-synaptophysin (1:200; Abcam), rabbit anti-CRB1 (AK2, 1:150; van de Pavert et al., 2004), and secondary antibodies: Alexa Fluor 488- or 594-conjugated goat anti-mouse IgG (1:200; Invitrogen) and Alexa Fluor 488- or 594-conjugated goat anti-rabbit IgG (1:200; Invitrogen). Cone photoreceptor segments were labeled with FITC-conjugated peanut agglutinin (PNA, 1:100; Sigma-Aldrich). Cell nuclei were stained with DAPI (1:3000; Sigma-Aldrich). Negative controls were performed without primary antibodies. Images were taken using a fluorescence microscope (BX51; Olympus).
In a separate experiment using five animals per strain, retinal sections at the level of the optic nerve head were obtained. RMG cells were stained using rabbit anti-GS and rabbit anti-cellular retinaldehyde-binding protein (CRALBP, 1:250, kind gift from John Saari, University of Washington, Seattle, WA), both RMG markers. RMG processes in the inner plexiform layer were counted on the entire retinal section. In addition, RMG cells were also counted using p27kip1 (1:100; Abcam), an RMG nuclear marker (Dyer and Cepko, 2000). Cone-arrestin positive cells (cones) were also counted. Using ImageJ, rhodopsin-positive areas of rod outer segments were analyzed.
Statistics.
Experimental results were analyzed by Mann–Whitney U test or two-way ANOVA using the GraphPad Prism5 program. A p-value of 0.05 or less was considered statistically significant. Data are presented graphically in figures as mean ± SE.
RMG cell primary culture.
RMG primary cultures were obtained from 3 consecutive P17 litters for each BN-H, BN-J, and Lewis rat strain. Animals were killed and eyes were enucleated. RMG cells were isolated as described previously (Zhao et al., 2010).
RNA-sequencing and data analysis.
Total RNA was extracted from primary RMG cells of the 3 rat strains (n = 3 samples per strain) using an RNeasy Mini Kit (Qiagen) including DNase I (Qiagen) treatment. RNA integrity was checked on the Agilent 2100 Bioanalyzer. RNA sequencing was performed on Illumina HiSeq 2000 platform according to the manufacturer's instructions. The average number of reads per sample was 27 M. Reads from each sample were processed as follows. First, reads were trimmed using an in-house Perl script with a minimum phred quality of 20 per base and a minimum read length of 30 bp. On average, 24% of reads per sample were discarded. The resulting reads were later aligned to the Rattus norvegicus genome assembly 3.4 (from Ensembl) using Tophat (Trapnell et al., 2009). Differential expression between BN-J and Lewis, BN-J and BN-H, and BN-H and Lewis was calculated using the Cuffdiff program from the Cufflinks suite (Trapnell et al., 2010). Fold changes >1.5 and FDR-corrected p-values <0.05 were used as filters. The corresponding comparisons will be further reported as JL, JH, and HL, respectively.
Signaling pathways for Rattus norvegicus were retrieved from WikiPathways (Kelder et al., 2012). Pathways with FDR-corrected p-values < 0.05 were selected as enriched. Pathvisio 2 (van Iersel et al., 2008) was used to visualize the pathways and map the values from each protein set.
Genetic analyses.
DNA was extracted from rats' tails by proteinase K (0.5 mg/ml; catalog #P-2308; Sigma) digestion overnight at 56°C in lysis buffer (50 mm Tris-HCl, pH 8.0, 100 mm EDTA, 100 mm NaCl, 1% SDS) and then purified with the DNAzol kit (catalog #DN127; MRC) according to the manufacturer's protocol. Genomic DNA was then used as a template for PCRs targeting coding exons of the crb1 gene using 35 cycles (94°C 2 min; 59°C 30 s; 72°C 30 s). PCR products were subsequently cleaned using the ExoSAP-IT kit (catalog #78201; Affymetrix), sequenced by the Sanger method (BigDye Terminator v1.1, catalog #4337450; Applied Biosystems) according to standard procedures and finally purified on EDGE gel filtration cartridges (catalog #42453; EdgeBio) before injection into an ABI Prism 3100 Sequencer.
Results
Vascular abnormalities in BN-J rat
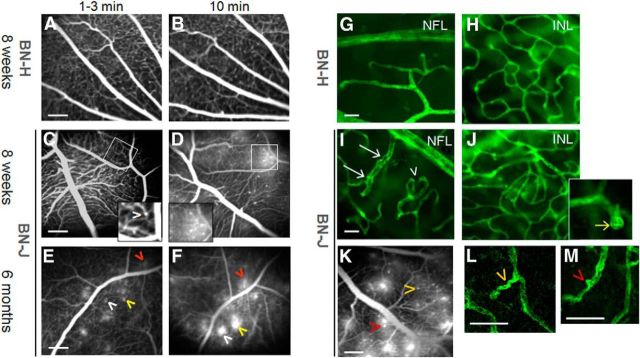
Retinal vessels were visualized in vivo by fluorescein angiography performed both on wild-type BN-H and pathological BN-J rats at young adult and older age (respectively, 8 weeks and 6 months of age). Digital images were taken at early (1–3 min) and later (10 min) time points after fluorescein injection. BN-H rats showed normal vascular aspect and circulatory filling (Fig. 1A,B). In 8-week-old BN-J rats at early time points, very subtle capillary dilations could be observed (Fig. 1C, inset) that became more visible at later time points as fluorescein leaked from the vascular telangiectasia (Fig. 1D, inset). Eyes of older BN-J rats (6 months) presented similar but leaky capillary ectasia (Fig. 1E,F, arrowheads of the same color indicate the same spot). Fluorescein angiography performed on younger BN-J rats (15 d of life) showed that sparse capillary ectasia was already present at this early age (data not shown).
Figure 1.
Vascular abnormalities in BN-J rats. Shown is in vivo fluorescein angiography of retinal vessels of BN-H and BN-J rats (A–F). Normal retinal vessels of BN-H rat at early (1–3 min, A) and late phase (10 min, B) of the angiographic sequence. Eight-week-old BN-J rat exhibits subtle capillary dilation hardly detected in the early phase (1–3 min) of angiography (C, arrowhead in the inset) that becomes more visible with leakage at later time point of 10 min (D and inset). At 6 months of age, similar but leakier capillary ectasia are observed (E, F, arrowheads, the same color indicates the same spot). Hyperfluorent leaking dots are observed at 1–3 min and their size increases at 10 min. Scale bar, 200 μm. Confocal imaging of lectin-stained retinal vessels on flat-mounted retinas from BN-H and BN-J rats is shown (G–J, L, M). Normal retinal vascular network (green) at the nerve fiber layer (NFL) and in the deep plexus at the INL from BN-H rat (G, H). In the BN-J rat retina, irregular vascular diameter (white arrows) and increased tortuosity (arrowhead) are observed at the NFL level (I). In the INL, disorganized capillary plexus is observed (J), together with multiple capillary telangiectasia (inset, yellow arrow). Images of a lectin-labeled flat-mounted retina of BN-J rat are linked to their corresponding angiographic pattern (K). Higher magnifications of the lectin-labeled vessels show that leaky telangiectasia (K) correspond to capillary tortuousness (L) and focal capillary ectasia (M). Arrowheads of the same color indicate the same spot. Scale bars: G–J, 20 μm; K, 200 μm; L, M, 50 μm.
On flat-mounted retinas stained with lectin, compared with BN-H rats (Fig. 1G,H), BN-J rats exhibited nonhomogenous vascular diameter (Fig. 1I, arrows), tortuous capillaries (Fig. 1I, arrowheads), and a global deep capillary network disorganization noted at different depth (some capillaries are seen underneath) (Fig. 1J). Vascular telangiectasia were clearly observed in the inner nuclear layer (INL) deep plexus (inset in Fig. 1J, yellow arrow).
Images of lectin-labeled flat-mounted retinas were correlated with the corresponding angiographic images showing that telangiectasia and leaky capillaries on angiography (Fig. 1K) correspond with capillary tortuousness (Fig. 1L) and focal capillaries ectasia on flat-mounted retinas (Fig. 1M, arrowheads of the same color indicate the same spot). Using CD-31 immunohistochemistry as an endothelial marker on flat-mounted retinas, endothelial cell discontinuity was found to be associated with nonhomogenous capillary diameter in BN-J rats, which may partially explain the leakage of fluorescein on angiography (data not shown).
Morphological retinal lesions in BN-J rats
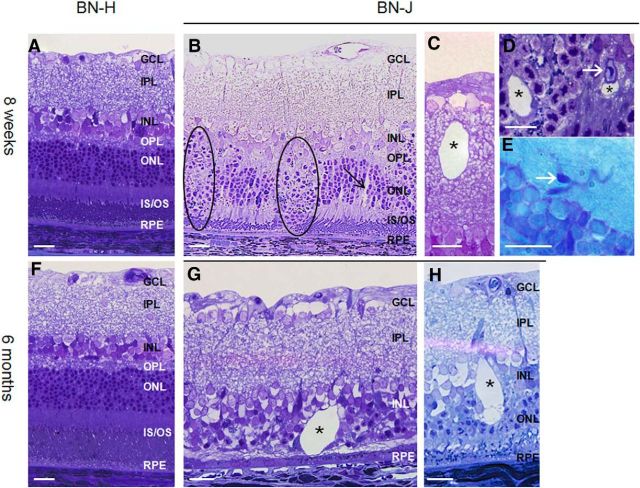
On semithin sections of the BN-J retina, at 8 weeks of age, focal disorganization of both the outer nuclear layer (ONL) and INL with loss of the outer segments of photoreceptors (Fig. 2B, zones in dark circles) were observed. Intraretinal cysts (asterisks) formed in both the inner (Fig. 2C) and the outer retina (Fig. 2D). Interestingly, around these focal areas of retinal lamination loss, the gross retina structure appeared preserved, but vascular tortuosity and capillary telangiectasia (white arrows) were noticeable in disorganized (Fig. 2D) and in normal areas (Fig. 2E). Small cysts were also observed surrounding retinal vessels (Fig. 2D). At 6 months, BN-J rats presented focal disappearance of the ONL containing photoreceptor cells in numerous areas that were spread across the entire retina (example in Fig. 2G). Large intraretinal cysts (Fig. 2G,H, asterisk) were specifically observed in BN-J animals compared with BN-H rats of the same age (Fig. 2F), showing the progression of retinal degeneration.
Figure 2.
Retinal morphology of BN-H and BN-J rats at 8 weeks and 6 months. Compared with the normally developed retina of BN-H rat at 8 weeks (A), the retina of the BN-J rat shows focal disorganization of the outer retinal layers (B, dark circles), where segments are not formed and nuclei of photoreceptors dive toward the retinal pigment epithelium (RPE). In areas where segments are present, swollen RMG cells can be observed (B, black arrow). Cysts (asterisks) can be found in both the inner (C) and the outer (D) retina. Telangiectasia are also identified on histological sections (D, E, white arrow). At 6 months, the BN-H rat retina is unchanged (F), whereas the retina of the BN-J rat shows variable degrees of degeneration. Photoreceptors have totally disappeared in some areas (G) and cysts are more abundant with irregular shapes (G, H, asterisks). GCL, Ganglion cell layer; IPL, inner plexiform layer; OPL, outer plexiform layer; IS/OS; inner and outer segments of photoreceptors. Scale bar, 20 μm.
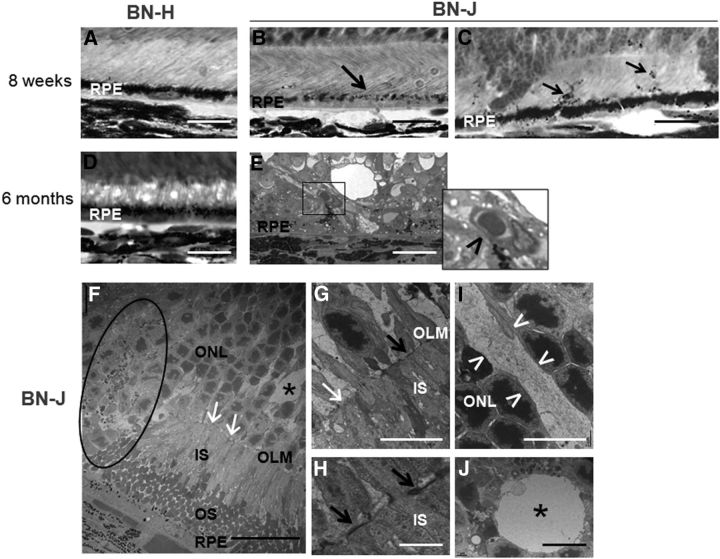
Closer observation showed other abnormalities in the outer retina of 8-week-old BN-J rats such as focal loss of pigment in retinal pigment epithelial (RPE) cells (Fig. 3B, arrow) and pigment migration (Fig. 3C, arrows) compared with BN-H rats (Fig. 3A). In the outer retina of 6-month-old BN-J rats, abnormal neovessels could be observed above RPE (Fig. 3E, inset and arrowhead).
Figure 3.
Outer retinal alterations in BN-J rats. A–E, Histological sections of the outer retina. F–J, TEM images. Contrasting with the heavy pigments located in the apical side of retinal pigment epithelium (RPE) in BN-H retina (A), melanosomes are poorly formed in BN-J rat at 8 weeks, even in areas where the segments have formed (B, black arrow) and pigments migrate in the photoreceptor segment layer (C, black arrows). At 6 months, the outer retina of BN-H rat does not change (D), whereas abnormal vessels are observed between RPE cells and the degenerated retina of BN-J rat (E, inset and black arrowhead), potentially corresponding to neovascularization. TEM analysis allows detection of more subtle changes in BN-J retina such as focal decrease in junction structures (F, G, white arrows) at the outer limiting membrane (OLM), alternating with normal OLM structures (G, H, black arrows). Abrupt disorganization of retinal layers is observed (F, dark circle). Swollen RMG cells (I, in between the white arrowheads) are identified in the ONL and cysts (F, J, asterisks) are surrounded by a membrane-like structure, suggesting intracellular swollen. IS, Inner segments of photoreceptors; OS, outer segments of photoreceptors. Scale bars: A–E, 20 μm; F, 25 μm; G, I, J, 10 μm; H, 2 μm.
TEM (Fig. 3, bottom) observation confirmed the abrupt transition between normal and abnormal retinal areas (Fig. 3F, circled area). However, even in areas where photoreceptor structure was maintained, focal disruption of junction structures (appearing black in TEM) was identified at the OLM (Fig. 3F,G, white arrows show loss of junctions, G,H, black arrows show maintained junctions). Swollen RMG processes were present between photoreceptor nuclei (Fig. 3I, arrowheads) and cysts (Fig. 3J, asterisk) appeared surrounded by membrane-like structures (Fig. 3J).
To determine at what age retinal abnormalities start, eyes from P1, P8, and P15 BN rats were examined. Although no difference could be observed in BN-J and BN-H retinas at P1 and P8 (Fig. 4A,B,D,E), at P15, sparse zones of irregular and/or without photoreceptor segment elongation were observed in BH-J rat retina (Fig. 4F, circled areas), suggesting RMG/photoreceptor interaction abnormalities (Rapaport et al., 2004). BN-J rats raised in the dark from birth until 3 weeks exhibit similar retinal abnormalities as the rats raised in normal light-dark cycles (data not shown), suggesting that retinal degeneration is not light dependent.
Figure 4.
Postnatal retinal development morphology of BN-H and BN-J rats. From P1 to P8, the neuronal layers are segmented into inner neuroblastic (INbL) and outer neuroblastic layers (ONbL) both in BN-H (A, B) and in BN-J (D, E) rats. However, from P8 to P15, whereas inner and outer segments (IS and OS) elongate normally in the BN-H retina (C), focal areas without segment elongation and persistent neuroblastic nuclei (circled areas) are observed in the BN-J retina (F). Dilated capillaries can be observed in the INL of BN-J retina (F, arrow). GCL, Ganglion cell layer. Scale bar, 20 μm.
Retinal neuron alterations in BN-J rat
Because the outer retina of BN-J rat is focally disorganized, we investigated photoreceptors (cones and rods), bipolar cells, and their synapses using specific immunohistochemistry staining. Cone photoreceptors were labeled in adult BN-H and BN-J rats using a cone arrestin antibody staining the entire cone cells, including outer segments and synaptic bodies (Fig. 5A–D). In BN-J rats, segments and axonal connections were completely absent in some areas of the outer plexiform layer (Fig. 5C,D, asterisk). Some cone cells were displaced toward the INL (Fig. 5C,D, arrow). Cell count on the entire retinal section showed a reduction of cones in BN-J rats compared with BN-H rats (Fig. 5E). Immunostaining of rhodopsin exhibited disappearance of outer segments of rod photoreceptors in the focal disorganized areas of the BN-J retina (Fig. 5G, asterisk), whereas the remaining outer segments (Fig. 5G, arrows) appeared shorter than those in the retina of BN-H rat (Fig. 5F). The rhodopsin-positive surface in the BN-J rat was significantly reduced compared with the BN-H rat (Fig. 5H), suggesting that rods may be more widely altered than primarily suspected.
Figure 5.
Immunohistochemistry of retinal neurons of BN-J rats. Different neuronal types are immunostained with specific markers in the BN-H retina: Cone arrestin stains the entire cone photoreceptors, including outer segments and synaptic bodies (A), rhodopsin stains the outer segments (OS) of rod photoreceptors (F), PKC-α labels the bipolar cells (I), and synaptophysin labels synaptic connections between retinal neurons (M). B, F, J and N are merged images with DAPI (blue). In the BN-J retina, cone segments are shorter or even absent (C, arrowhead) and cones are missing in cystic formations (C, asterisk) and nuclei of some cones without segments are displaced (C, arrow). Cell count shows significant reduction of cone cells in BN-J rats (E). Rods are absent in disorganized area (G, asterisk) and their segments are shorter in other regions (G, arrows), suggesting segment elongation disruption. Quantification of rhodopsin-positive surface shows significant decrease in rod outer segment areas in BN-J rats (H). In disorganized areas, nuclei of bipolar cell are internally displaced (K, circle) and neuronal synapses are disrupted in the outer plexiform layer (OPL, O, arrows). D, G, L and P are merged images with DAPI. GCL, Ganglion cell layer; IPL, inner plexiform layer. Scale bars: A–D, F, G, M–P, 50 μm; I–L, 20 μm. For E and H, n = 5 rats per strain; **p < 0.01.
PKC-α labels the bipolar cells. In BN-H rats, PKC-α-expressing cells were located in the INL extending their processes to the innermost part of the inner plexiform layer (Fig. 5I,J), whereas in some areas of the BN-J rat retina, the nuclei of bipolar cells were internally displaced by the nuclei of photoreceptors invading in the INL (Fig. 5K,L, circled area).
Synaptophysin immune labeling showed focal disruption of synapses in the outer plexiform layer of the BN-J retina (Fig. 5O,P) compared with intact synapses in the BN-H retina (Fig. 5M,N).
Glial abnormalities in BN-J rat
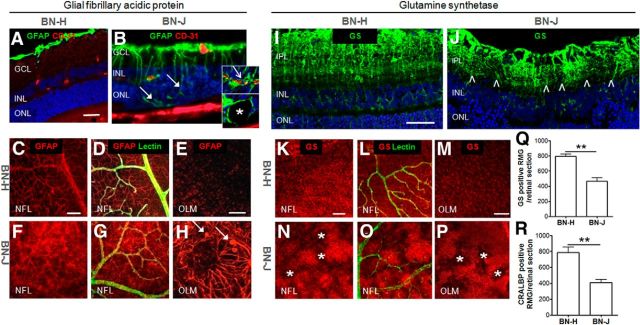
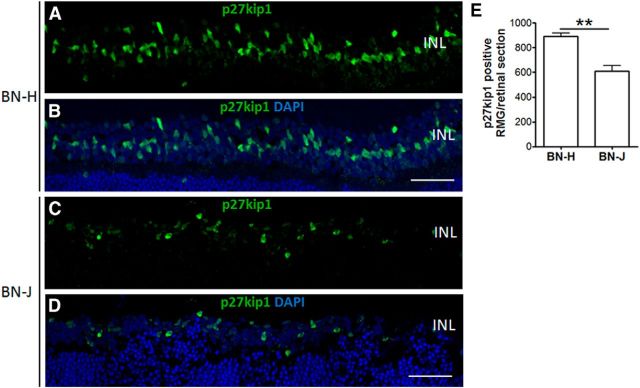
GFAP labels astrocytes and activated RMG cells. GFAP staining in BN-H rats was restricted, as expected, to RMG end feet and astrocytes (Fig. 6A). In BN-J rats, enhanced GFAP fluorescence was observed in Müller end feet and in activated swollen RMG cells (Fig. 6B, filled arrows) extending up to the vascular processes, as demonstrated by the colabeling of GFAP with CD31 (Fig. 6B and top inset, open arrow). Some cysts appeared surrounded by GFAP labeling (Fig. 6B and bottom inset, asterisk). On flat-mounted retinas of BN-J rats, RMG cells were highly activated because GFAP staining was spread all along their end feet and processes with hypertrophic apices at the OLM (Fig. 6F–H) compared with BN-H retina (Fig. 6C–E). GS is also a Müller cell marker that labels from their end feet to their apical processes, as observed in BN-H rats (Fig. 6I,K–M). In BN-J retinas, GS immunoreactivity was decreased in focal areas in between hypertrophic RMG cells (Fig. 6J,N–P). GS and CRALBP (another RMG marker) positive RMG cells were significantly reduced in BN-J rat compared with BN-H rat (Fig. 6Q,R). Additional immunostaining experiments using the RMG nuclear marker p27kip1 confirmed the loss of RMG cells in BN-J rat retina (Fig. 7).
Figure 6.
RMG morphologic alterations in sectioned and flat-mounted BN-J retinas. GFAP stains RMG end feet and astrocytes on BN-H retinal cross-section (A). On flat-mounted retinas, GFAP staining is surrounding vessels (labeled with lectin) in the nerve fiber layer (NFL, C, D) and, in RMG apices, in the outer limiting membrane (OLM, E). In the BN-J retina, activated RMG cells extend up to the subretinal space in disorganized areas (B, filled arrows). Activated RMG cells surround vessels in the INL (B, top inset, open arrow) and form the border of cysts (B, bottom inset, asterisk). On flat-mounted BN-J retinas, GFAP is enhanced in the RMG end feet in the NFL (F, G) and extends up to the OLM, where swollen apices (H, filled arrows) and disorganization of RMG (H) are observed. In the BN-H retina, GS stains the RMG from their end feet to processes around the vessels (labeled with lectin) and to their apices (retinal section, I, and flat-mounted retina, K–M). In the BN-J retina, GS immunoreactivity is enhanced in hypertrophic RMG and reduced in the surrounding areas, as observed in both retinal section (J, between the arrowheads) and flat-mounted retina (N–P, asterisks), suggesting focal loss of RMG cells. GS-positive and CRALBP (another RMG marker)-positive RMG cells are significantly decreased in BN-J rat retinas (Q, R). GCL, Ganglion cell layer; IPL, inner plexiform layer. Scale bars: A, B, I, J, 50 μm; C, D, F, G, K, L, N, O, 100 μm; E, H, M, P, 50 μm. For Q and R, n = 5 rats per strain; **p < 0.01.
Figure 7.
Loss of Müller glial cells in BN-J rat retinas. RMG cells were immunostained with p27kip1, an RMG nuclear marker. P27kip1-positive nuclei in the INL are reduced in the BN-J rat retina (C) compared with BN-H rat retina (A). B and D are merged images with DAPI. Scale bar, 50 μm. Cell count of p27kip1-positive RMG shows significant decrease in BN-J rat retinas (E). n = 7 rats for BN-H and 6 rats for BN-J; **p < 0.01.
Early retinal functional abnormalities in BN-J rat
To evaluate retinal functional changes in BN-J rats, ERG was performed as early as 3 weeks, when retinal focal abnormalities were already identified on histology. Overall ERG responses showed a significant reduction in b-wave amplitude and a trend, but not a significant reduction, in the a-wave amplitude, translating the postreceptoral disturbance of the visual signal particularly at the bipolar and RMG cells (Fig. 8A,B). Scotopic ERGs showed significant reduced a- and b-wave amplitudes even at low intensities (0.1 cd · s/m2; Fig. 8C,D), whereas no significant difference was observed in photopic ERGs (data not shown), suggesting that rod function is affected earlier than cone function, a finding also observed in MacTel 2 patients (Schmitz-Valckenberg et al., 2008).
Figure 8.
Early retinal function alterations in BN-J rats. ERG was performed on 3-week-old BN-H and BN-J rats. Although global ERG responses show a trend but not significant reduction in a-wave amplitude (A), the b-wave amplitude is significantly decreased (B), suggesting postreceptoral disturbance of the visual signal in the inner retina. Scotopic ERG shows a significant reduction in a-wave (C) and b-wave (D) amplitudes from 0.1 to 3 cd · s/m2, suggesting intense rod visual pathway dysfunction. n = 5 rats for BN-H and n = 4 for BN-J. *p < 0.05.
Genetic analyses
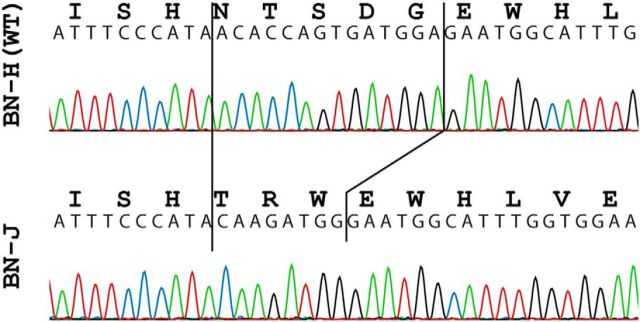
In humans, mutations in the crb1 gene usually cause recessively inherited retinitis pigmentosa with preserved para-arteriolar RPE and Leber congenital amaurosis (congenital retinal blindness; den Holländer et al., 2004). However, less frequently, they cause retinitis pigmentosa with retinal cysts and peripheral (nonmacular) telangiectasia, potentially associated at late stages with vascular coats-like telangiectasia (den Holländer et al., 2004; Henderson et al., 2011). We therefore screened for mutations in the entire coding region of crb1 in BN-H and BN-J rats. In these latter animals, we identified a homozygous insertion-deletion (indel) in exon 6. This small DNA rearrangement (c.1685_1698delinsCAAGATGG; reference: NM_001107182.1) involved the ablation of 14 nt of the wild-type rat DNA sequence and the insertion of 8 new ones while preserving at the same time the canonical open reading of the gene (Fig. 9). At the protein level, this change would translate into the replacement of amino acid residues 562 to 566 (NTSDG) with 3 new ones: TRW. Residues 562 to 566 of CRB1 are identical in human and rat and are well conserved across vertebrates (the last three of which, SDG, being invariant from man to zebrafish; data not shown), indicating that this portion of the protein may be rather important for its function. Finally, as expected, we did not detect any DNA variation within the crb1 coding sequence in BN-H animals or in the karyotype of BN-J rats.
Figure 9.
Electropherograms of part of crb1 exon 6 in BN-H and BN-J rats. The insertion–deletion in the BN-J sequence is indicated by solid black lines.
To ascertain whether this change in crb1 represented a true mutation responsible for the retinal phenotype of BN-J rats, we analyzed the cosegregation of the indel with the aberrant retinal phenotype over an extended set of animals that were the offspring of targeted mating. Crosses of pure parental strains (BN-H × BN-J) resulted in an F1 composed of 18 phenotypically normal rats, as ascertained by retinal histology, which were verified to be heterozygous for the BN-J indel. Four F1 couples were then cross-bred, to produce an F2 composed of a total of 30 pups, which as adults were all phenotyped and genotyped by investigators who were reciprocally masked. Of these 30 animals, 24 had normal retinas and 6 presented with defects that were indistinguishable from those displayed by the parental BN-J strain. Genotyping showed that all specimens with abnormal retinas were homozygotes for the BN-J indel, whereas those with normal retinal morphology were either wild-type or heterozygotes (5 and 19 animals, respectively). Together, these results indicate that the crb1 indels detected in BN-J rats acts as a recessive allele to determine the observed retinal phenotype in homozygous animals, with an associated p-value < 2.5 × 10−7 (likelihood of phenotypes and genotypes cooccurring by chance; that is, the retinal BN-J phenotype not being associated with the detected indel mutation = 0.256 × 0.7524).
Mislocalization of CRB1 protein in BN-J rat
We further studied CRB1 expression by immunofluorescence in adult BN-J and BN-H rats. In BN-H rat retina, CRB1 was observed particularly in the apical region above the OLM labeled by GS (Fig. 10A–C, left inset and arrows). CRB1 was colocalized with GS in the microvilli of RMG cells (Fig. 10C, arrowheads in the right inset). CRB1 was also diffusely distributed in the inner segments of photoreceptors (Fig. 10A,D). Double staining of CRB1 and PNA showed colocalization in the inner segments of cone cells (Fig. 10F, arrowheads in the inset). In BN-J rat retina, CRB1 was still expressed in photoreceptor inner segments (Fig. 10G,J–L, inset), but its localization in the subapical region was missing and CRB1 did not colocalize with GS (Fig. 10G–I, inset). The CRB1 phenotype of BN-J rats is therefore more pronounced in RMG cells and results in a mislocalization of the protein in the RMG/photoreceptor cell junction.
Figure 10.
CRB1 immunolocalization in BN-H and BN-J retinas. In the BN-H retina, CRB1 localizes in the subapical region above the level of outer limiting membrane (OLM) labeled with GS (A–C, open arrows in the left inset). Higher magnification shows colocalization of CRB1 with GS in the microvilli of RMG cells (arrowheads in the right inset). CRB1 stains also the inner segments (IS) of photoreceptors (A, D). Double staining with PNA (E) shows colocalization of CRB1 with cone IS (F, arrowheads in the inset). In the BN-J retina, CRB1 loses its localization in the subapical region (G–I, inset), OLM is even disrupted in disorganized areas (H, filled arrow). In organized areas, CRB1 remains in the photoreceptor IS (J). Double staining with PNA (K) shows colocalization with cone IS (L, arrowhead in the inset). Scale bar, 20 μm.
Transcriptome analysis and deregulated signaling pathways in BN-J RMG cells
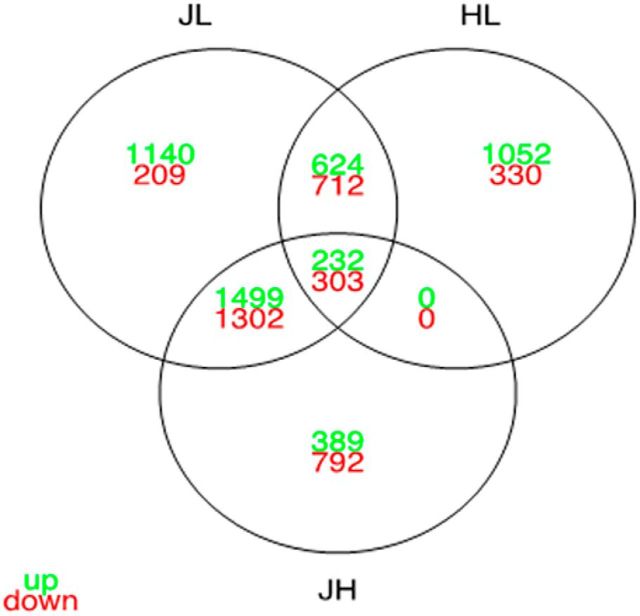
To identify the molecular mechanisms linking CRB-1 to the BN-J retinal phenotype, we analyzed the differential transcriptome of primary RMG cells extracted at P17, a time when RMG cells have acquired polarization and differentiation markers (Wurm et al., 2006), from BN-J, BN-H, and control wild-type Lewis rats. The results showed respectively 11808, 11358, and 11873 expressed genes (FPKM values ≥ 2). Differential expression analyses were performed between the three strains. A total of 6021 differentially expressed genes resulted from the JL comparison (between BN-J and Lewis RMGs), 4517 differentially expressed genes from the JH comparison (between BN-J and BN-H RMGs), and 3253 differentially expressed genes from the HL comparison (between BN-H and Lewis RMGs) (Fig. 11). The data showed an expression profile in BN-J RMG that is further away from the ones of BN-H or Lewis RMG than between these two last strains together. The common differentially expressed genes between both JL and JH are of importance because they may contain the “BN-J specific set of genes,” the misregulation of which plays a predominant role in the early development of the pathological process (observed in the BN-J strain). The corresponding intersection contains a total of 3336 genes (corresponding to, respectively, 73% and 53% of the JH and JL differentially expressed genes). As expected, BN-J and BN-H are closer together than each of them separately with the Lewis strain. Moreover, the majority (73%) of the JH differentially expressed genes are also differentially expressed between the pathological BN-J and a further distant wild-type strain, such as the Lewis strain. The BN-J specific set of genes represent candidate genes potentially involved in the pathological development of BN-J rats. The results showed a significant reduction in crb1 expression in RMG cells (crb1 gene name referred as D3ZZL8_RAT) from BN-J and BN-H compared with the Lewis control (log2FC JL = −3.76; log2FC HL = −3,05) and a reduction in BN-J compared with BN-H (log2FC JH = −0.72).
Figure 11.
Crossing of the differential expression analyses of BN-J, BN-H, and Lewis P17 primary RMG cells. Venn diagram shows the number of overexpressed (in green) and underexpressed genes (in red) that are shared and unique for each comparison.
To decipher the essential functions of the RMG cells at this early stage, we performed a pathway enrichment analysis on the three studied strains (Table 1). With the aim of distinguishing between the potentially pathological pathways and the strain-related ones, we conducted further pathway enrichment analyses on the JL, JH, and HL sets of genes (Table 2). In agreement with the previous results on gene-based distances between strains (number of misregulated genes for log2FC JL>log2FC JH>log2FC HL>0.6 or log2FC JL<log2FC JH<log2FC HL<0.6), we found 48, 28, and 10 enriched signaling pathways in JL, JH, and HL, respectively. After enriched pathways have been grouped by similar functions, we focused on the pathways that were enriched in both JL and JH sets of genes (i.e., the “BN-J specific set of genes”). Among those pathways, TGF-β signaling, matrix metalloproteinases, kit receptor signaling, type II interferon signaling, MAPK cascade, growth factor signaling pathways, inflammatory pathways, G-protein signaling pathways, regulation of actin cytoskeleton, cardiovascular signaling, calcium regulation in the cardiac cell, and EGFR1 signaling pathway were found.
Table 1.
Signaling pathways enrichment in primary RMG cells from BN-J, Harlan, and Lewis rat strains
| Pathways | Enriched in |
adj. p-value |
||||
|---|---|---|---|---|---|---|
| BNJ | BNH | Lewis | BNJ | BNH | Lewis | |
| D-GLOCUSE-INS1-RXRA | ✓ | 4,0E-02 | 5,5E-02 | 6,5E-02 | ||
| Pentose phosphate pathway | ✓ | 5,0E-02 | 5,8E-02 | 6,3E-02 | ||
| Type II interferon signaling (IFNG) | ✓ | 9,3E-03 | 6,1E-02 | 1,4E-01 | ||
| Urea cycle and metabolism of amino groups | ✓ | ✓ | 1,5E-02 | 2,0E-02 | 7,2E-02 | |
| ATM | ✓ | ✓ | ✓ | 4,7E-05 | 4,9E-04 | 1,0E-04 |
| Adipogenesis | ✓ | ✓ | ✓ | 1,8E-03 | 6,8E-06 | 2,9E-06 |
| Alpha6-Beta4 integrin signaling pathway | ✓ | ✓ | ✓ | 2,5E-03 | 1,0E-02 | 3,2E-03 |
| Androgen receptor signaling pathway | ✓ | ✓ | ✓ | 8,0E-16 | 6,6E-16 | 2,5E-14 |
| Apoptosis | ✓ | ✓ | ✓ | 2,7E-09 | 1,0E-08 | 5,8E-09 |
| Apoptosis modulation by HSP70 | ✓ | ✓ | ✓ | 4,8E-04 | 4,3E-03 | 6,6E-05 |
| B cell receptor signaling pathway | ✓ | ✓ | ✓ | 1,3E-19 | 2,9E-19 | 1,5E-17 |
| Beta oxidation meta pathway | ✓ | ✓ | ✓ | 4,1E-03 | 6,2E-03 | 6,8E-04 |
| CDKN1A-EGF-CREB | ✓ | ✓ | ✓ | 1,0E-08 | 2,7E-09 | 7,8E-09 |
| Calcium regulation in the cardiac cell | ✓ | ✓ | ✓ | 1,6E-02 | 1,8E-03 | 7,5E-05 |
| Cardiovascular signaling | ✓ | ✓ | ✓ | 6,7E-03 | 3,4E-03 | 1,4E-03 |
| Cell cycle | ✓ | ✓ | ✓ | 3,8E-14 | 1,5E-14 | 6,6E-13 |
| Cholesterol biosynthesis | ✓ | ✓ | ✓ | 9,6E-04 | 1,3E-03 | 1,5E-03 |
| Cholesterol metabolism | ✓ | ✓ | ✓ | 9,3E-05 | 1,6E-04 | 1,1E-03 |
| Cytoplasmic ribosomal proteins | ✓ | ✓ | ✓ | 1,7E-26 | 2,6E-25 | 2,3E-24 |
| DNA replication | ✓ | ✓ | ✓ | 2,6E-07 | 2,1E-09 | 8,3E-08 |
| Delta-Notch signaling pathway | ✓ | ✓ | ✓ | 1,0E-09 | 1,4E-11 | 2,0E-09 |
| Diurnally regulated genes with circadian orthologs | ✓ | ✓ | ✓ | 8,1E-07 | 2,0E-06 | 5,2E-07 |
| EBV LMP1 signaling | ✓ | ✓ | ✓ | 4,5E-03 | 1,3E-03 | 1,7E-03 |
| EGFR1 signaling pathway | ✓ | ✓ | ✓ | 1,3E-20 | 3,7E-20 | 6,4E-20 |
| EPO receptor signaling | ✓ | ✓ | ✓ | 1,4E-02 | 5,7E-03 | 2,4E-02 |
| Electron transport chain | ✓ | ✓ | ✓ | 2,6E-25 | 3,3E-24 | 1,3E-24 |
| Endochondral ossification | ✓ | ✓ | ✓ | 8,8E-03 | 1,3E-03 | 2,0E-03 |
| ErbB signaling pathway | ✓ | ✓ | ✓ | 9,8E-03 | 2,6E-03 | 9,7E-03 |
| Eukaryotic transcription initiation | ✓ | ✓ | ✓ | 6,9E-10 | 1,7E-09 | 3,1E-09 |
| FAS pathway and stress induction of HSP regulation | ✓ | ✓ | ✓ | 1,4E-08 | 3,9E-07 | 6,7E-07 |
| Fatty acid beta oxidation | ✓ | ✓ | ✓ | 2,1E-03 | 3,3E-03 | 3,3E-04 |
| Fatty acid beta oxidation 1 | ✓ | ✓ | ✓ | 2,3E-02 | 3,2E-02 | 4,3E-03 |
| Fatty acid beta oxidation 3 | ✓ | ✓ | ✓ | 2,0E-02 | 2,4E-02 | 2,7E-02 |
| G-protein signaling pathways | ✓ | ✓ | ✓ | 9,4E-06 | 1,8E-08 | 4,7E-08 |
| G1 to S cell cycle control | ✓ | ✓ | ✓ | 5,7E-08 | 4,7E-09 | 6,8E-08 |
| G13 signaling pathway | ✓ | ✓ | ✓ | 2,2E-03 | 2,8E-04 | 8,5E-05 |
| Glycogen metabolism | ✓ | ✓ | ✓ | 2,0E-03 | 7,0E-04 | 9,5E-04 |
| Glycolysis and Gluconeogenesis | ✓ | ✓ | ✓ | 1,7E-02 | 1,1E-02 | 1,5E-02 |
| Heme biosynthesis | ✓ | ✓ | ✓ | 2,3E-02 | 2,8E-02 | 3,1E-02 |
| Homologous recombination | ✓ | ✓ | ✓ | 1,0E-02 | 1,3E-02 | 1,6E-02 |
| IL-1 signaling pathway | ✓ | ✓ | ✓ | 1,2E-03 | 1,6E-03 | 1,1E-02 |
| IL-2 signaling pathway | ✓ | ✓ | ✓ | 4,9E-09 | 2,9E-09 | 4,2E-08 |
| IL-3 signaling pathway | ✓ | ✓ | ✓ | 2,3E-13 | 2,4E-14 | 1,1E-13 |
| IL-4 signaling pathway | ✓ | ✓ | ✓ | 8,6E-11 | 2,6E-10 | 7,0E-10 |
| IL-5 signaling pathway | ✓ | ✓ | ✓ | 2,4E-10 | 8,2E-11 | 2,1E-09 |
| IL-6 signaling pathway | ✓ | ✓ | ✓ | 1,9E-15 | 1,2E-16 | 4,9E-16 |
| IL-7 signaling pathway | ✓ | ✓ | ✓ | 4,1E-08 | 9,5E-08 | 1,9E-07 |
| IL-9 signaling pathway | ✓ | ✓ | ✓ | 3,9E-04 | 5,9E-04 | 7,6E-04 |
| Id signaling pathway | ✓ | ✓ | ✓ | 2,6E-04 | 1,5E-04 | 6,5E-05 |
| Insulin signaling | ✓ | ✓ | ✓ | 1,2E-15 | 6,0E-16 | 4,9E-19 |
| Integrin-mediated cell adhesion | ✓ | ✓ | ✓ | 3,2E-06 | 3,4E-06 | 6,8E-07 |
| Keap1-Nrf2 | ✓ | ✓ | ✓ | 3,4E-02 | 4,3E-02 | 1,0E-02 |
| Kit receptor signaling pathway | ✓ | ✓ | ✓ | 3,6E-08 | 1,9E-08 | 2,3E-07 |
| MAPK cascade | ✓ | ✓ | ✓ | 2,1E-04 | 5,3E-05 | 4,2E-07 |
| MAPK signaling pathway | ✓ | ✓ | ✓ | 9,2E-16 | 2,2E-15 | 3,3E-15 |
| Mitochondrial gene expression | ✓ | ✓ | ✓ | 3,1E-03 | 4,4E-03 | 5,5E-03 |
| Mitochondrial LC-fatty acid beta-oxidation | ✓ | ✓ | ✓ | 1,9E-03 | 2,6E-03 | 3,2E-03 |
| Myometrial relaxation and contraction pathways | ✓ | ✓ | ✓ | 1,2E-07 | 3,3E-08 | 2,0E-09 |
| NR3C1-PKL1 | ✓ | ✓ | ✓ | 4,2E-06 | 9,9E-06 | 1,7E-05 |
| Non-homologous end joining | ✓ | ✓ | ✓ | 3,6E-02 | 4,3E-02 | 4,7E-02 |
| Notch signaling pathway | ✓ | ✓ | ✓ | 1,4E-03 | 2,3E-04 | 3,4E-03 |
| Nucleotide metabolism | ✓ | ✓ | ✓ | 8,9E-05 | 1,3E-04 | 1,5E-04 |
| One carbon metabolism | ✓ | ✓ | ✓ | 2,4E-02 | 3,4E-02 | 4,1E-02 |
| Oxidative stress | ✓ | ✓ | ✓ | 1,5E-03 | 2,1E-03 | 6,6E-05 |
| Oxidative phosphorylation | ✓ | ✓ | ✓ | 2,2E-17 | 1,1E-16 | 3,3E-16 |
| PI3K_AKT_NFKB pathway | ✓ | ✓ | ✓ | 5,4E-06 | 1,2E-05 | 3,4E-06 |
| PKC-SCP2 | ✓ | ✓ | ✓ | 3,2E-04 | 2,5E-04 | 4,1E-04 |
| Proteasome degradation | ✓ | ✓ | ✓ | 1,5E-11 | 4,8E-11 | 1,3E-10 |
| Regulation of actin cytoskeleton | ✓ | ✓ | ✓ | 2,0E-06 | 2,0E-07 | 2,4E-07 |
| Renin–angiotensin system | ✓ | ✓ | ✓ | 6,0E-04 | 1,1E-04 | 4,8E-05 |
| Selenium metabolism selenoproteins | ✓ | ✓ | ✓ | 2,3E-03 | 3,3E-03 | 4,0E-03 |
| Senescence and autophagy | ✓ | ✓ | ✓ | 6,5E-08 | 2,9E-08 | 6,6E-08 |
| Signal transduction of S1P | ✓ | ✓ | ✓ | 7,1E-03 | 9,9E-03 | 3,2E-03 |
| Signaling of hepatocyte growth factor receptor | ✓ | ✓ | ✓ | 1,0E-08 | 2,1E-08 | 4,2E-08 |
| T cell receptor signaling pathway | ✓ | ✓ | ✓ | 1,3E-09 | 7,3E-09 | 2,2E-07 |
| TCA cycle | ✓ | ✓ | ✓ | 2,9E-05 | 5,0E-05 | 6,6E-05 |
| TGF beta signaling pathway | ✓ | ✓ | ✓ | 9,3E-05 | 2,7E-06 | 4,6E-06 |
| TGF-beta receptor signaling pathway | ✓ | ✓ | ✓ | 1,1E-19 | 2,2E-21 | 1,6E-19 |
| TNF-alpha NF-kB signaling pathway | ✓ | ✓ | ✓ | 6,4E-32 | 1,1E-32 | 2,2E-30 |
| TNF-alpha and mucus production in lung epythelium | ✓ | ✓ | ✓ | 3,2E-06 | 5,7E-06 | 7,7E-06 |
| The effect of glucocorticoids on target gene expression | ✓ | ✓ | ✓ | 2,7E-03 | 3,6E-03 | 4,3E-03 |
| Toll-like receptor signaling pathway | ✓ | ✓ | ✓ | 3,1E-08 | 1,0E-07 | 5,3E-09 |
| Translation factors | ✓ | ✓ | ✓ | 9,3E-11 | 2,5E-10 | 5,7E-10 |
| VEGF-receptor signal transduction | ✓ | ✓ | ✓ | 9,3E-04 | 5,4E-03 | 3,4E-04 |
| Wnt signaling pathway net path | ✓ | ✓ | ✓ | 3,6E-08 | 4,1E-08 | 5,7E-10 |
| Wnt signaling pathway and pluripotency | ✓ | ✓ | ✓ | 2,2E-03 | 1,3E-03 | 1,2E-05 |
| Estrogen signaling | ✓ | ✓ | ✓ | 4,5E-13 | 1,2E-13 | 4,0E-13 |
| Genetic alternations of lung cancer | ✓ | ✓ | ✓ | 3,5E-05 | 5,6E-05 | 7,5E-05 |
| mRNA processing | ✓ | ✓ | ✓ | 0,0E + 00 | 2,4E-25 | 0,0E + 00 |
| p38 MAPK signaling pathway (BioCarta) | ✓ | ✓ | ✓ | 2,5E-07 | 5,9E-06 | 4,3E-08 |
| p53 pathway | ✓ | ✓ | ✓ | 7,1E-07 | 4,3E-05 | 2,9E-06 |
| p53 signal pathway | ✓ | ✓ | ✓ | 2,1E-04 | 3,3E-04 | 4,6E-04 |
| Alanine and aspartate metabolism | ✓ | 5,6E-01 | 5,9E-01 | 4,1E-02 | ||
| Fatty acid biosynthesis | ✓ | 7,2E-02 | 8,9E-02 | 9,9E-03 | ||
| NLR proteins | ✓ | 1,3E-01 | 1,4E-01 | 2,9E-02 | ||
| Wnt signaling pathway | ✓ | 2,1E-01 | 1,2E-01 | 3,2E-02 | ||
List of enriched Wikipathway signaling pathways from genes expressed in BNJ, BNH, and Lewis rat RMG cells and their corresponding significance (adjusted P-values).
Table 2.
Signaling pathways enrichment in BN-J versus Lewis or Harlan rat primary RMG cells
| Signaling pathways | JL | JH | HL |
|---|---|---|---|
| TGF beta signaling pathway | ✓ | ✓ | |
| Matrix metalloproteinases | ✓ | ✓ | |
| Kit receptor signaling pathway | ✓ | ✓ | |
| Type II interferon signaling (IFNG) | ✓ | ✓ | |
| MAPK cascade | ✓ | ✓ | |
| p38 MAPK signaling pathway | ✓ | ||
| Signal transduction of S1P | ✓ | ||
| Adipogenesis | ✓ | ✓ | |
| Endochondral ossification | ✓ | ✓ | |
| Signaling of hepatocyte growth factor receptor | ✓ | ||
| Apoptosis | ✓ | ✓ | |
| Senescence and autophagy | ✓ | ✓ | |
| p53 signal pathway | ✓ | ||
| Apoptosis modulation by HSP70 | ✓ | ||
| FAS pathway and stress induction of HSP regulation | ✓ | ||
| Toll-like receptor signaling pathway | ✓ | ||
| B cell receptor signaling pathway | ✓ | ✓ | |
| IL-3 signaling pathway | ✓ | ✓ | |
| IL-4 signaling pathway | ✓ | ✓ | |
| IL-5 signaling pathway | ✓ | ✓ | |
| IL-2 signaling pathway | ✓ | ||
| T cell receptor signaling pathway | ✓ | ||
| IL-6 signaling pathway | ✓ | ||
| IL-7 signaling pathway | ✓ | ||
| IL-9 signaling pathway | ✓ | ✓ | |
| Cytokines and inflammatory response | ✓ | ||
| G-protein signaling pathways | ✓ | ✓ | |
| Myometrial relaxation and contraction pathways | ✓ | ✓ | ✓ |
| Small ligand GPCRs | ✓ | ✓ | |
| GPCRs | ✓ | ||
| Regulation of actin cytoskeleton | ✓ | ✓ | |
| Striated muscle contraction | ✓ | ✓ | |
| G13 signaling pathway | ✓ | ||
| Cardiovascular signaling | ✓ | ✓ | |
| Integrin-mediated cell adhesion | ✓ | ✓ | ✓ |
| Focal adhesion | ✓ | ||
| EGFR1 signaling pathway | ✓ | ✓ | |
| CDKN1A-EGF-CREB | ✓ | ||
| Hypertrophy model | ✓ | ||
| Calcium regulation in the cardiac cell | ✓ | ✓ | ✓ |
| Insulin signaling | ✓ | ✓ | ✓ |
| Osteoclast | ✓ | ✓ | |
| Delta-Notch signaling pathway | ✓ | ||
| Wnt signaling pathway | ✓ | ||
| Oxidative stress | ✓ | ||
| Id signaling pathway | ✓ | ||
| PI3K_AKT_NF-κB pathway | ✓ | ||
| EBV LMP1 signaling | ✓ | ||
| TNF-alpha NF-κB signaling pathway | ✓ | ||
| EPO receptor signaling | ✓ | ||
| Renin–angiotensin system | ✓ | ||
| Complement and coagulation cascades | ✓ | ✓ | |
| Glutathione metabolism | ✓ | ✓ | |
| Urea cycle and metabolism of amino groups | ✓ | ||
| Eicosanoid synthesis | ✓ |
The presented signaling pathways were selected based on significance (p ≤ 0.05) unless stated otherwise. JL, Signaling pathways enriched for genes differentially expressed between Janvier Brown Norway and Lewis rat RMGs; JH, signaling pathways enriched for genes differentially expressed between Janvier and Harlan Brown Norway rat RMGs; HL, signaling pathways enriched for genes differentially expressed between Harlan Brown Norway and Lewis rat RMGs.
In addition, we have compared our rat data with previously described classical Müller glial markers and Müller glial markers derived from the mouse transcriptome study of Roesch et al. (2008) and found that 64.7% (11 of 17) of those Müller glia markers are downregulated in BN-J compared with BN-H or Lewis rats, supporting the fact that the number of mature Müller glia is reduced in BN-J retina. The downregulated markers are Aqp4, Clu, Kir4.1/Kcnj10, S100a16, CRALBP-1/Rlbp1, GS/Glul, Dkk3, Chx-10/Vsx2, Spbc25/Spc25, GPR37, and Car2.
Discussion
This report describes a recessively inherited retinal phenotype of a rat strain carrying abnormalities as observed in the human MacTel 2 disease: focal loss of retinal lamination, OLM disruptions, retinal cysts, RMG, photoreceptor and RPE alterations associated with retinal telangiectasia, and late-stage intraretinal neovascularization. It was found that this phenotype is caused by a new mutation in exon 6 of rat crb1. This model was then used to decipher the molecular pathways deregulated in RMG cells and thus provided a full spectrum of targets with which to study the pathogenesis of MacTel 2 and of other retinal diseases associated with telangiectasia.
A specific focus directed toward RMG cells in the histology of a MacTel 2 patient retina showed prominent loss of RMG cell markers in the central retina and RMG metabolic disorders (Powner et al., 2010). The link between RMG cell depletion and retinal vessel telangiectasia was further highlighted by the group of Mark Gillies, who generated a transgenic mouse model with conditional RMG cell ablation by using a portion of the regulatory region of the retinaldehyde binding protein 1 gene. The selective killing of RMG cells in adult mice led to photoreceptor apoptosis, vascular telangiectasia, blood–retinal barrier breakdown, and late intraretinal neovascularization (Shen et al., 2012). Interestingly, the retinal pathology of this animal model is very similar to the one displayed by the BN-J rat reported here, for which retinal abnormalities development coincides with RMG cell maturation in rats (Wurm et al., 2006).
In this context, the differential transcriptomic analysis of mature RMG cells from BN-J rats and two control strains (BN-H and Lewis) was performed. A restricted list of pathways was identified, most of which were found also in the whole retina transcriptomic analysis of the transgenic conditional mice model described above (Chung et al., 2013). These similarities support the hypothesis that early Müller glia dysregulation could induce the retinal vascular pathology observed in the BN-J rat, although it is still unclear whether a focal loss of RMG cells is required or if prior RMG dysfunction alone could induce retinal alterations observed in MacTel 2.
RMG cell processes surround retinal capillaries and the basement membrane of the perivascular Müller cells merge with the self-propagating vessels wall, demonstrating the very close interaction of RMG cells with the retinal vasculature. Conversely, RMG cells communicate with photoreceptor cells through adherens junctions and serve as sensors for any environmental changes. In the healthy retina, RMG cells contribute to control retinal angiogenesis through the production of the anti-angiogenic PAI-1 factor (Abukawa et al., 2009) and meteorin, which, interestingly, also controls GFAP expression (Lee et al., 2010). Indirect evidence originating from in vitro studies suggest that RMG cells could also participate in the blood–retinal barrier through TGF-β and MMP9 expression (Behzadian et al., 2001), a pathway and a factor, respectively, that were identified in the transcriptomic differential analysis of the BN-J rat. RMG cells are therefore now viewed as a component of the neurovascular unit of the retina (Reichenbach and Bringmann, 2013).
The genetic analysis of the BN-J rat showed that the retinal phenotype is transmitted as a Mendelian recessive trait, apparently in contrast to current knowledge on inheritance of human macular telangiectasia (Parmalee et al., 2012). The disease allele is an indel mutation in exon-6 of the crb1 gene, a DNA change that has not been described previously in other crb1-related retinal degenerations in humans or in animal models. The recessive inheritance of the rat phenotype seems to suggest that the indel leads to some crb1 loss of function that is completely tolerated in heterozygotes. However, it is still unclear whether the mutation completely abrogates protein functionality or if it represents a hypomorphic allele that still allows some residual activity. The preservation of crb1 canonical reading frame, despite five seemingly important amino acids being replaced, is compatible with this latter hypothesis and may support the notion that the rat phenotype similar to the human MacTel 2 phenotype could be considered as a milder manifestation of more severe crb1-linked retinal degenerations. Despite RNA sequencing showing a significant reduction in crb1 (ENSRNOG00000010903) expression in RMG cells from BN-J and BN-H compared with the Lewis control, a reduction in BN-J compared with BN-H, the variant CRB1 protein remains present in the retina of BN-J rats. However, the CRB1 protein is mislocalized and loses its concentration in the subapical regions above the adherens junctions between RMG and photoreceptors of BN-J retina. Among proteins involved in the correct localization of CRB, the small GTPase Cdc42 that belongs to the Ras superfamily is of particular interest because it plays a major and unique role in epithelium permeability (Citalán-Madrid et al., 2013); retina-specific Cdc42-knock-down mice showed not only retinal degeneration but also important vascular abnormalities (Heynen et al., 2013).
In contrast to phenotypes resulting from other crb1 mutations, the BN-J rat presents early retinal vascular leaky telangiectasia and late intraretinal neovascularization and this degeneration is not light dependent. These differences can result from different type of mutations or from different genetic setups displayed by different animal species. Interestingly, using exome sequencing analysis, crb1 defect was recently found to be associated with an unusual form of macular dystrophy, suggesting that some CRB1 dysfunction could be specifically expressed in the macula (Tsang et al., 2014).
CRB proteins interact with β-catenin, N-cadherin, and with the PAR3/PAR6/atypical PKC pathway and with PALS-1/MPP3/MPP5, which belongs to the MAGUK proteins (Alves et al., 2014). Complex interactions maintain this molecular scaffold and alterations of different partners may induce variable retinal phenotypes. For example, progressive retinal degeneration and vascular abnormalities have been recently described in a conditional knock-out mouse for MPP3 that is normally localized in apices of RMG and regulates the levels of PALS1 (Dudok et al., 2013). This suggests that mutations in genes encoding different proteins interacting with CRB could induce retinal degeneration and vascular phenotypes. Interestingly, retinal vessel development was recently shown to be dynamically regulated by VEGF receptor endocytosis and the activity of cell polarity proteins, particularly PAR3/atypical PKC (Nakayama et al., 2013). In addition, we recently found that the activity of atypical PKC-ζ in the retina is deregulated early by hyperglycemia and contributes to OLM disruptions (Omri et al., 2013), which could be a link between increased susceptibility to MacTel 2 in diabetic patients (Clemons et al., 2013). So far, attempts to find the gene(s) responsible for MacTel 2 by candidate-gene screening have been unsuccessful (Parmalee et al., 2010). Whether CRB1 and/or other proteins associated with adherens junctions between cone photoreceptors and RMG cells in the macula are associated with MacTel 2 phenotype in humans should be evaluated.
The exact mechanisms linking CRB1 mislocalization to the BN-J retina phenotype are yet to be determined. To identify potential pathways, we studied the molecular imbalances of primary BN-J developing RMG cells mutated for crb1 using transcriptome analysis. Pathways such as TGF-β signaling, matrix metalloproteinases, kit receptor signaling, type II interferon signaling, MAPK cascade, growth factor signaling pathways, inflammatory pathways, G-protein signaling pathways, regulation of actin cytoskeleton, cardiovascular signaling, and EGFR1 signaling pathway were found to be deregulated in the rat model. Among these, known cellular process and pathways associated with MacTel 2 disease were found, such as (cardio)vasculogenesis, apoptosis, or oxidative stress. In addition, regulation of actin cytoskeleton and calcium regulation in the cardiac cell contained genes associated with adherens junctions, where CRB1 appears to be mislocalized in BN-J rat. Focal adhesion and integrin-mediated cell adhesion pathways regulating the blood–retinal barrier were also found to be affected in BN-J rats. TGF-β signaling and matrix metalloproteinases were strongly dysregulated. A direct correlation of TGF-β effects on MMP9 as a potential cause of the blood–retinal barrier breakdown was already hypothesized (Behzadian et al., 2001). Small G-proteins (such as RAP1,which was identified in the transcriptomic differential analysis of BN-J rat) have also been reported to play a critical role in the stabilization of endothelial junctions (Wilson and Ye, 2014). Several studies have also established the role of growth factors (in particular VEGF) and inflammation in the pathophysiology of the MacTel 2 disease. Moreover, different clinical trials with anti-VEGF compounds (Kovach and Rosenfeld, 2009; Charbel Issa et al., 2011; Narayanan et al., 2012) and a retrospective interventional case description on the positive effect of topical anti-inflammatory agents on a phakic cystoid macular edema secondary to idiopathic macular telangiectasia have been reported (Dunn et al., 2013).
In conclusion, we have identified and characterized spontaneous retinal abnormalities in a strain of BN rats that are very close to other models of MacTel 2 created by depletion of RMG cells and strongly reminiscent of the human phenotype. We have identified the genetic mutation responsible for this phenotype in the rat crb1 gene and have studied the transcriptome of RMG cells from these animals, highlighting the involvement of numerous cellular pathways and potential regulatory targets. This rat model could be used to evaluate potential new therapeutic options for retinal telangiectasia.
Footnotes
This work was supported by the European People Marie Curie Actions Program, Marie Curie European Reintegration (Grant FP7-PEOPLE-2010-RG to C.A.-S.), the Swiss National Science Foundation (Grant 310030_138346 to C. R.), INSERM, Union National des Aveugles et Deficients Visuels, and the University of Lausanne. In vivo morphological and functional explorations were performed on rat eyes at the Centre d'Explorations Fonctionnelles of Centre de Recherche des Cordeliers. We thank Christophe Klein of Centre de Recherche des Cordeliers for his help in confocal microscopy and Iharilalao Dubail of Faculté de Pharmacie of Paris Descartes University for providing animal facility.
The authors declare no competing financial interests.
References
- Abukawa H, Tomi M, Kiyokawa J, Hori S, Kondo T, Terasaki T, Hosoya K. Modulation of retinal capillary endothelial cells by Muller glial cell-derived factors. Mol Vis. 2009;15:451–457. [PMC free article] [PubMed] [Google Scholar]
- Alves CH, Pellissier LP, Wijnholds J. The CRB1 and adherens junction complex protiens in retinal development and maintenance. Prog Retin Eye Res. 2014;40:35–52. doi: 10.1016/j.preteyeres.2014.01.001. [DOI] [PubMed] [Google Scholar]
- Behzadian MA, Wang XL, Windsor LJ, Ghaly N, Caldwell RB. TGF-beta increases retinal endothelial cell permeability by increasing MMP-9: possible role of glial cells in endothelial barrier function. Invest Ophthalmol Vis Sci. 2001;42:853–859. [PubMed] [Google Scholar]
- Bringmann A, Pannicke T, Grosche J, Francke M, Wiedemann P, Skatchkov SN, Osborne NN, Reichenbach A. Muller cells in the healthy and diseased retina. Prog Retin Eye Res. 2006;25:397–424. doi: 10.1016/j.preteyeres.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Charbel Issa P, Finger RP, Kruse K, Baumüller S, Scholl HP, Holz FG. Monthly ranibizumab for nonproliferative macular telangiectasia type 2: a 12-month prospective study. Am J Ophthalmol. 2011;151:876–886.e1. doi: 10.1016/j.ajo.2010.11.019. [DOI] [PubMed] [Google Scholar]
- Charbel Issa P, Gillies MC, Chew EY, Bird AC, Heeren TF, Peto T, Holz FG, Scholl HP. Macular telangiectasia type 2. Prog Retin Eye Res. 2013;34:49–77. doi: 10.1016/j.preteyeres.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung SH, Shen W, Jayawardana K, Wang P, Yang J, Shackel N, Gillies MC. Differential gene expression profiling after conditional Muller-cell ablation in a novel transgenic model. Invest Ophthalmol Vis Sci. 2013;54:2142–2152. doi: 10.1167/iovs.12-11559. [DOI] [PubMed] [Google Scholar]
- Citalán-Madrid AF, García-Ponce A, Vargas-Robles H, Betanzos A, Schnoor M. Small GTPases of the Ras superfamily regulate intestinal epithelial homeostasis and barrier function via common and unique mechanisms. Tissue Barriers. 2013;1:e26938. doi: 10.4161/tisb.26938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemons TE, Gillies MC, Chew EY, Bird AC, Peto T, Wang JJ, Mitchell P, Ramdas WD, Vingerling JR, Macular Telangiectasia Project Research Group Medical characteristics of patients with macular telangiectasia type 2 (MacTel Type 2) MacTel project report no. 3. Ophthalmic Epidemiol. 2013;20:109–113. doi: 10.3109/09286586.2013.766757. [DOI] [PubMed] [Google Scholar]
- den Holländer AI, Davis J, van der Velde-Visser SD, Zonneveld MN, Pierrottet CO, Koenekoop RK, Kellner U, van den Born LI, Heckenlively JR, Hoyng CB, Handford PA, Roepman R, Cremers FP. CRB1 mutation spectrum in inherited retinal dystrophies. Hum Mutat. 2004;24:355–369. doi: 10.1002/humu.20093. [DOI] [PubMed] [Google Scholar]
- Dubois-Dauphin M, Poitry-Yamate C, de Bilbao F, Julliard AK, Jourdan F, Donati G. Early postnatal Muller cell death leads to retinal but not optic nerve degeneration in NSE-Hu-Bcl-2 transgenic mice. Neuroscience. 2000;95:9–21. doi: 10.1016/s0306-4522(99)00313-9. [DOI] [PubMed] [Google Scholar]
- Dudok JJ, Sanz AS, Lundvig DM, Sothilingam V, Garrido MG, Klooster J, Seeliger MW, Wijnholds J. MPP3 regulates levels of PALS1 and adhesion between photoreceptors and Muller cells. Glia. 2013;61:1629–1644. doi: 10.1002/glia.22545. [DOI] [PubMed] [Google Scholar]
- Dunn EN, Gregori NZ, Goldhardt R. Phakic cystoid macular edema secondary to idiopathic macular telangiectasia type 1 responsive to topical anti-inflammatory agents. Semin Ophthalmol. 2013;28:84–87. doi: 10.3109/08820538.2012.760615. [DOI] [PubMed] [Google Scholar]
- Dyer MA, Cepko CL. Control of Muller glial cell proliferation and activation following retinal injury. Nat Neurosci. 2000;3:873–880. doi: 10.1038/78774. [DOI] [PubMed] [Google Scholar]
- Fletcher EL, Phipps JA, Wilkinson-Berka JL. Dysfunction of retinal neurons and glia during diabetes. Clin Exp Optom. 2005;88:132–145. doi: 10.1111/j.1444-0938.2005.tb06686.x. [DOI] [PubMed] [Google Scholar]
- Henderson RH, Mackay DS, Li Z, Moradi P, Sergouniotis P, Russell-Eggitt I, Thompson DA, Robson AG, Holder GE, Webster AR, Moore AT. Phenotypic variability in patients with retinal dystrophies due to mutations in CRB1. Br J Ophthalmol. 2011;95:811–817. doi: 10.1136/bjo.2010.186882. [DOI] [PubMed] [Google Scholar]
- Heynen SR, Meneau I, Caprara C, Samardzija M, Imsand C, Levine EM, Grimm C. CDC42 is required for tissue lamination and cell survival in the mouse retina. PLoS One. 2013;8:e53806. doi: 10.1371/journal.pone.0053806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski MM, Iannaccone A. Targeted disruption of Muller cell metabolism induces photoreceptor dysmorphogenesis. Glia. 2000;32:192–204. doi: 10.1002/1098-1136(200011)32:2<192::AID-GLIA80>3.0.CO%3B2-6. [DOI] [PubMed] [Google Scholar]
- Kelder T, van Iersel MP, Hanspers K, Kutmon M, Conklin BR, Evelo CT, Pico AR. WikiPathways: building research communities on biological pathways. Nucleic Acids Res. 2012;40:D1301–1307. doi: 10.1093/nar/gkr1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach JL, Rosenfeld PJ. Bevacizumab (avastin) therapy for idiopathic macular telangiectasia type II. Retina. 2009;29:27–32. doi: 10.1097/IAE.0b013e31818ba9de. [DOI] [PubMed] [Google Scholar]
- Lee HS, Han J, Lee SH, Park JA, Kim KW. Meteorin promotes the formation of GFAP-positive glia via activation of the Jak-STAT3 pathway. J Cell Sci. 2010;123:1959–1968. doi: 10.1242/jcs.063784. [DOI] [PubMed] [Google Scholar]
- Len AC, Powner MB, Zhu L, Hageman GS, Song X, Fruttiger M, Gillies MC. Pilot application of iTRAQ to the retinal disease macular telangiectasia. J Proteome Res. 2012;11:537–553. doi: 10.1021/pr200889t. [DOI] [PubMed] [Google Scholar]
- Mehalow AK, Kameya S, Smith RS, Hawes NL, Denegre JM, Young JA, Bechtold L, Haider NB, Tepass U, Heckenlively JR, Chang B, Naggert JK, Nishina PM. CRB1 is essential for external limiting membrane integrity and photoreceptor morphogenesis in the mammalian retina. Hum Mol Genet. 2003;12:2179–2189. doi: 10.1093/hmg/ddg232. [DOI] [PubMed] [Google Scholar]
- Nakayama M, Nakayama A, van Lessen M, Yamamoto H, Hoffmann S, Drexler HC, Itoh N, Hirose T, Breier G, Vestweber D, Cooper JA, Ohno S, Kaibuchi K, Adams RH. Spatial regulation of VEGF receptor endocytosis in angiogenesis. Nat Cell Biol. 2013;15:249–260. doi: 10.1038/ncb2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan R, Chhablani J, Sinha M, Dave V, Tyagi M, Pappuru RR, Kuppermann BD. Efficacy of anti-vascular endothelial growth factor therapy in subretinal neovascularization secondary to macular telangiectasia type 2. Retina. 2012;32:2001–2005. doi: 10.1097/IAE.0b013e3182625c1d. [DOI] [PubMed] [Google Scholar]
- Omri S, Omri B, Savoldelli M, Jonet L, Thillaye-Goldenberg B, Thuret G, Gain P, Jeanny JC, Crisanti P, Behar-Cohen F. The outer limiting membrane (OLM) revisited: clinical implications. Clin Ophthalmol. 2010;4:183–195. doi: 10.2147/opth.s5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omri S, Behar-Cohen F, Rothschild PR, Gélizé E, Jonet L, Jeanny JC, Omri B, Crisanti P. PKCzeta mediates breakdown of outer blood-retinal barriers in diabetic retinopathy. PLoS One. 2013;8:e81600. doi: 10.1371/journal.pone.0081600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooto S, Hangai M, Takayama K, Arakawa N, Tsujikawa A, Koizumi H, Oshima S, Yoshimura N. High-resolution photoreceptor imaging in idiopathic macular telangiectasia type 2 using adaptive optics scanning laser ophthalmoscopy. Invest Ophthalmol Vis Sci. 2011;52:5541–5550. doi: 10.1167/iovs.11-7251. [DOI] [PubMed] [Google Scholar]
- Parmalee NL, Schubert C, Merriam JE, Allikmets K, Bird AC, Gillies MC, Peto T, Figueroa M, Friedlander M, Fruttiger M, Greenwood J, Moss SE, Smith LE, Toomes C, Inglehearn CF, Allikmets R. Analysis of candidate genes for macular telangiectasia type 2. Mol Vis. 2010;16:2718–2726. [PMC free article] [PubMed] [Google Scholar]
- Parmalee NL, Schubert C, Figueroa M, Bird AC, Peto T, Gillies MC, Bernstein PS, Kiryluk K, Terwilliger JD, Allikmets R, MacTel Project Identification of a potential susceptibility locus for macular telangiectasia type 2. PLoS One. 2012;7:e24268. doi: 10.1371/journal.pone.0024268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powner MB, Gillies MC, Tretiach M, Scott A, Guymer RH, Hageman GS, Fruttiger M. Perifoveal muller cell depletion in a case of macular telangiectasia type 2. Ophthalmology. 2010;117:2407–2416. doi: 10.1016/j.ophtha.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport DH, Wong LL, Wood ED, Yasumura D, LaVail MM. Timing and topography of cell genesis in the rat retina. J Comp Neurol. 2004;474:304–324. doi: 10.1002/cne.20134. [DOI] [PubMed] [Google Scholar]
- Reichenbach A, Bringmann A. New functions of Muller cells. Glia. 2013;61:651–678. doi: 10.1002/glia.22477. [DOI] [PubMed] [Google Scholar]
- Roesch K, Jadhav AP, Trimarchi JM, Stadler MB, Roska B, Sun BB, Cepko CL. The transcriptome of retinal Muller glial cells. J Comp Neurol. 2008;509:225–238. doi: 10.1002/cne.21730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Valckenberg S, Fan K, Nugent A, Rubin GS, Peto T, Tufail A, Egan C, Bird AC, Fitzke FW. Correlation of functional impairment and morphological alterations in patients with group 2A idiopathic juxtafoveal retinal telangiectasia. Arch Ophthalmol. 2008;126:330–335. doi: 10.1001/archopht.126.3.330. [DOI] [PubMed] [Google Scholar]
- Shen W, Fruttiger M, Zhu L, Chung SH, Barnett NL, Kirk JK, Lee S, Coorey NJ, Killingsworth M, Sherman LS, Gillies MC. Conditional Muller cell ablation causes independent neuronal and vascular pathologies in a novel transgenic model. J Neurosci. 2012;32:15715–15727. doi: 10.1523/JNEUROSCI.2841-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tout S, Chan-Ling T, Holländer H, Stone J. The role of Muller cells in the formation of the blood-retinal barrier. Neuroscience. 1993;55:291–301. doi: 10.1016/0306-4522(93)90473-S. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretiach M, Madigan MC, Wen L, Gillies MC. Effect of Muller cell co-culture on in vitro permeability of bovine retinal vascular endothelium in normoxic and hypoxic conditions. Neurosci Lett. 2005;378:160–165. doi: 10.1016/j.neulet.2004.12.026. [DOI] [PubMed] [Google Scholar]
- Tsang SH, Burke T, Oll M, Yzer S, Lee W, Xie YA, Allikmets R. Whole exome sequencing identifies CRB1 defect in an unusual maculopathy phenotype. Ophthalmology. 2014;121:1773–1782. doi: 10.1016/j.ophtha.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Pavert SA, Kantardzhieva A, Malysheva A, Meuleman J, Versteeg I, Levelt C, Klooster J, Geiger S, Seeliger MW, Rashbass P, Le Bivic A, Wijnholds J. Crumbs homologue 1 is required for maintenance of photoreceptor cell polarization and adhesion during light exposure. J Cell Sci. 2004;117:4169–4177. doi: 10.1242/jcs.01301. [DOI] [PubMed] [Google Scholar]
- van Iersel MP, Kelder T, Pico AR, Hanspers K, Coort S, Conklin BR, Evelo C. Presenting and exploring biological pathways with PathVisio. BMC Bioinformatics. 2008;9:399. doi: 10.1186/1471-2105-9-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Iannaccone A, Jablonski MM. Contribution of Muller cells toward the regulation of photoreceptor outer segment assembly. Neuron Glia Biol. 2005;1:1–6. doi: 10.1017/S1740925X05000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CW, Ye W. Regulation of vascular endothelial junction stability and remodeling through Rap1-Rasip1 signaling. Cell Adh Migr. 2014:8. doi: 10.4161/cam.28115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurm A, Pannicke T, Iandiev I, Wiedemann P, Reichenbach A, Bringmann A. The developmental expression of K+ channels in retinal glial cells is associated with a decrease of osmotic cell swelling. Glia. 2006;54:411–423. doi: 10.1002/glia.20391. [DOI] [PubMed] [Google Scholar]
- Yannuzzi LA, Bardal AM, Freund KB, Chen KJ, Eandi CM, Blodi B. Idiopathic macular telangiectasia. Arch Ophthalmol. 2006;124:450–460. doi: 10.1001/archopht.124.4.450. [DOI] [PubMed] [Google Scholar]
- Zhao M, Valamanesh F, Célérier I, Savoldelli M, Jonet L, Jeanny JC, Jaisser F, Farman N, Behar-Cohen F. The neuroretina is a novel mineralocorticoid target: aldosterone upregulates ion and water channels in Muller glial cells. FASEB J. 2010;24:3405–3415. doi: 10.1096/fj.09-154344. [DOI] [PubMed] [Google Scholar]
- Zhao M, Célérier I, Bousquet E, Jeanny JC, Jonet L, Savoldelli M, Offret O, Curan A, Farman N, Jaisser F, Behar-Cohen F. Mineralocorticoid receptor is involved in rat and human ocular chorioretinopathy. J Clin Invest. 2012;122:2672–2679. doi: 10.1172/JCI61427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Krilis M, Gillies MC. The relationship between inner retinal cavitation, photoreceptor disruption, and the integrity of the outer limiting membrane in macular telangiectasia type 2. Retina. 2013;33:1547–1550. doi: 10.1097/IAE.0b013e318285cb9c. [DOI] [PubMed] [Google Scholar]