Abstract
Background
In captive breed turtles and tortoises conjunctival disease is common. Our aim was to investigate the bacterial and fungal flora present in the eyes of healthy and pathological chelonians and to compare findings in turtles with those in tortoises.
Results
Samples were taken from the conjunctival sacs of 34, diseased and healthy, chelonians (18 tortoises and 16 turtles) and submitted to bacterial and fungal investigation. All samples showed bacterial growth. Thirteen animals (38%), harboured a single bacterial species as sole isolate and twenty-one animals (62%) harboured more than one species. Detection of multiple bacterial infection was clearly greater in tortoises compared to turtles. Most frequently isolated bacterial species were Bacillus spp. (13 isolates), Staphylococcus xylosus (10 isolates), Sphingomonas paucimobilis (6 isolates), Staphylococcus sciuri and Aeromonas hydrophila/caviae (each 5 isolates), Ochrobactrum anthropi (3 isolates), Citrobacter freundii, Enterobacter cloacae and Pseudomonas luteola (each 2 isolates). Only one isolate of Kocuria varians/rosea, Staphylococcus aureus, Staphylococcus auricularis, Staphylococcus haemolyticus, Staphylococcus lentus, Morganella morganii, Pasteurella multocida, Pasteurella pneumotropica/haemolytica, Proteus spp., Pseudomonas putida, Salmonella enterica ssp. arizonae, Stenotrophomonas maltophilia and Vibrio parahaemolyticus was evidenced. The presence in 8 animals of Mycoplasma spp. and in 1 animal with severe conjunctivitis of Chlamydia spp. was detected by PCR. Candida spp. was also isolated from two healthy animals.
Conclusions
A clear predominance of Gram positive isolates in tortoises and Gram negative isolates in turtles was found. However, we cannot ascribe the observed difference to the diversity of animal species, as other factors, including especially different characteristics of the living environments, may play a role. Almost all bacterial species isolated may have clinical significance, mostly as opportunistic pathogens, both for humans and animals. That chelonians are often carrier of bacteria with zoonotic potential is a well-known fact, in particular with regard to Salmonella spp. Therefore, it is not surprising the detection of a strain of Salmonella enterica ssp. arizonae in the eye of one of the animals tested. Worthy of note is the finding of Chlamydia spp. in a severe case of conjunctivitis, though we cannot epidemiologically assess a cause-effect relationship between presence of chlamydia and disease.
Electronic supplementary material
The online version of this article (doi:10.1186/s12917-015-0405-x) contains supplementary material, which is available to authorized users.
Keywords: Turtles, Conjunctiva, Microbial flora, Zoonosis
Background
Chelonians are kept in captivity as pets or for commercial purpose. However, the progressive increase in the number of these animals which are in close contact with humans has not been matched with an appropriate increase in medical knowledge. Chelonians may have different common names, depending on their habitat. Turtles, as those who belong to the genera Trachemys, Emydura and Pseudemys, spends most of their life in the water, while tortoises, which includes the genus Testudo, are land-dwellers. Finally, Pelomedusa subrufa can be considered a terrapin and spends its time both on land and in water. Chelonians belong to the class of reptilian, order testudines. The suborder Cryptodira includes tortoises as Testudo spp., which belong to the family of Testudinidae, as well as turtles as Trachemys scripta and Pseudemys spp., which belong to the family of Emydidae. Pelomedusa subrufa and Emydura subglobosa belong to the Pleurodira suborder, Pelomedusidae and Chelidae families, respectively.
Chelonians are ectothermic animals and they need direct ultraviolet B (UV-B) irradiation to maintain optimal body temperature and vitamin D3 activation. The bactericidal property of UV radiation contribute to the control of cutaneous bacterial flora. Furthermore, inadeguated UV irradiation often results in a reduction of immune system activity [1]. In chelonians immune depression and vitamin deficiencies (hypovitaminosis A and D) are the main causes of metabolic disease that often occur with conjunctivitis [2]. Conjunctivitis is also a common symptom of upper respiratory tract disease (URTD), a widespread disease among chelonians, characterized by mild to severe rhinitis, nasal and ocular discharge, conjunctivitis, and periocular edema [3].
Knowledge of the conjunctival microbical flora, particularly those of pathological cases, is essential for treatment of eye diseases. The aim of this study was to investigate the bacterial and fungal flora present in the eyes of healthy and pathological chelonians and to compare findings in turtles with those in tortoises.
Methods
Animals
Thirty-four animals, belonging to the Cryptodira suborder (8 Testudo graeca, 8 Testudo hermanni, 1 Testudo hosfieldi, 1 Testudo marginata, 1 Pseudemys spp., and 13 Trachemys scripta) and to the Pleurodira suborder (1 Pelomedusa subrufa and 1 Emydura subglobosa), brought to the veterinary hospital of the Department of Veterinary-Medical Sciences of the University of Parma, underwent routine ophthalmic examination as part of a general health check and were subjected to microbiological examination. All the animals were captive animals and came from private collections of northern Italy, with the exception of an healthy wild Trachemys scripta elegans (n. 27) caught in the river Po. As reported in Table 1, four of the 34 animals showed bilateral conjunctivitis, but they were not affected by URTD or any other symptom. Moreover, another animal (n. 18) was weakened, with no specific symptoms. In all the diseased animals hypovitaminosis was excluded. The research comply with the current law of the European Union and Italy regarding the protection of animals used for experimental and other scientific purposes (Directive 86/609/EEC - D. L.vo 116/92) and was approved by the Ethics Committee on animal experimentation of the University of Parma.
Table 1.
Results of Nested-PCRs and cultures on eye swabs from healthy and diseased tortoises and turtles
| Sample | Species | Tortoise | Turtle | Clinical signs | Chlamydia PCR | Mycoplasma PCR | Gram positive isolates* | Gram negative isolates § | Candida spp. | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | ||||||||
| 1 | T. graeca | X | None | + | + | ||||||||||||||||||||||||
| 10 | T. graeca | X | Conjunctivitis | + | + | + | |||||||||||||||||||||||
| 11 | T. hosfieldi | X | Conjunctivitis | + | + | ||||||||||||||||||||||||
| 13 | T. graeca | X | None | + | + | ||||||||||||||||||||||||
| 14 | T. hermanni | X | Conjunctivitis | + | + | ||||||||||||||||||||||||
| 15 | T. graeca | X | None | + | + | + | |||||||||||||||||||||||
| 16 | T. graeca | X | None | + | + | + | |||||||||||||||||||||||
| 17 | T. graeca | X | None | + | + | ||||||||||||||||||||||||
| 18 | T. graeca | X | Weakeness | + | + | + | + | ||||||||||||||||||||||
| 19 | T. hermanni | X | None | + | + | + | |||||||||||||||||||||||
| 20 | T. hermanni | X | None | + | + | + | + | ||||||||||||||||||||||
| 21 | T. hermanni | X | None | + | + | ||||||||||||||||||||||||
| 22 | T. hermanni | X | None | + | + | + | + | ||||||||||||||||||||||
| 24 | T. graeca | X | None | + | + | ||||||||||||||||||||||||
| 25 | T. marginata | X | None | + | + | ||||||||||||||||||||||||
| 26 | T. hermanni | X | None | + | + | ||||||||||||||||||||||||
| 33 | T. hermanni | X | None | + | + | ||||||||||||||||||||||||
| 34 | T. hermanni | X | None | + | |||||||||||||||||||||||||
| 2 | T. scripta | X | None | + | + | ||||||||||||||||||||||||
| 3 | T. scripta | X | None | + | |||||||||||||||||||||||||
| 4 | T. scripta | X | None | + | + | ||||||||||||||||||||||||
| 5 | P. subrufa | X | None | + | + | ||||||||||||||||||||||||
| 6 | E. subglobosa | X | None | + | + | + | |||||||||||||||||||||||
| 7 | T. scripta | X | None | + | |||||||||||||||||||||||||
| 8 | T. scripta | X | None | + | + | ||||||||||||||||||||||||
| 9 | T. scripta | X | None | + | + | ||||||||||||||||||||||||
| 12 | T. scripta | X | None | + | |||||||||||||||||||||||||
| 23 | T. scripta | X | Conjunctivitis | + | + | ||||||||||||||||||||||||
| 27 | T. scripta | X | None | + | + | ||||||||||||||||||||||||
| 28 | T. scripta | X | None | + | |||||||||||||||||||||||||
| 29 | T. scripta | X | None | + | |||||||||||||||||||||||||
| 30 | T. scripta | X | None | + | + | + | |||||||||||||||||||||||
| 31 | T. scripta | X | None | + | |||||||||||||||||||||||||
| 32 | Pseudemys spp. | X | None | + | |||||||||||||||||||||||||
*Gram positive isolates: A=Bacillus spp.; B=Kocuria varians/rosea; C=Staphylococcus aureus; D=Staphylococcus auricularis; E=Staphylococcus haemolyticus; F=Staphylococcus lentus; G=Staphylococcus sciuri; H=Staphylococcus xylosus.
§Gram negative isolates: I=Aeromonas hydrophila/caviae; J=Citrobacter freundii; K=Enterobacter cloacae; L=Morganella morganii; M=Ochrobactrum anthropic; N=Pasteurella multocida; O=Pasteurella pneumotropica/ haemolytica; P=Proteus spp.; Q=Pseudomonas luteola; R=Pseudomonas putida; S=Salmonella enterica ssp. arizonae; T=Sphingomonas paucimobilis; U=Stenotrophomonas maltophilia; V=Vibrio parahaemolyticus.
Culture
Samples were taken from the conjunctival sac of each eye with sterile swabs and bacterial and fungal investigation were immediately performed. Tryptose agar (Beckton Dickinson, Sparks, Maryland, USA) containing 5% of bovine erythrocytes, MacConkey agar (Beckton Dickinson), Sabouraud agar (Beckton Dickinson), Brain Heart Infusion broth (BHI) (Beckton Dickinson) and Mycoplasma agar (Beckton Dickinson) were inoculated and incubated in aerobic and/or microaerophilic (air with 5% CO2) atmosphere. Plates were inoculated and incubated for 24 hours at 37°C or for 48 hours at 35°C for bacteriological and micological investigation, respectively. The incubation of Mycoplasma agar plates in 5% CO2 air was extended to 7 days and plates were examined daily with a plate microscope. BHI broth is an enrichment medium and was inoculated to be subcultured in case of total absence of growth on agar plates. Bacterial isolates were identified using standard microbiological procedures, as growth and colonial characteristics, Gram staining, cellular morphology, catalase and oxidase reactions, coagulase test (Slidex Staph Plus, bioMérieux, Marcy-l’Etoile, France), haemolysin production. Species identification was carried out using the API biochemical test systems (bioMérieux, Marcy-l’Etoile, France), as well as conventional biochemical tests [4].
DNA extraction
To detect the presence of Mycoplasma spp. or Chlamydia spp., one swab from each eye was placed in a 2 ml tube containing 400 μl of PBS and 1 mM EDTA, thoroughly vortexed and stored at −80°C until DNA extraction. For DNA extraction, each sample was thawed and thoroughly vortexed, swab was removed and the solution was transferred to a 1.5 ml tube containing Phase Lock Gel Light (Eppendorf, Hamburg, Germany). Four-hundred μl of phenol/chlorophorm (Invitrogen) were added and mixed by repeated inversion. After 10 min of incubation at RT, the tube was centrifuged (Microfuge 18 Centrifuge, Beckman Coulter) at 14000 rpm for 10 min at room temperature (RT). The acqueous phase was removed and transferred to a new eppendorf tube. Four-hundred μl of isopropanol and 40 μl of 3 M, pH 4.8, potassium acetate were added and mixed by repeated inversion. After 10 min of incubation at RT, the tube was centrifuged at 14000 rpm for 10 min, the surnatant was removed and the pellet gently washed with 500 μl of 70% ethanol. After centrifugation at 14000 rpm for 5 min the ethanol was completely removed and the pellet was resuspended in 50 μl of sterile deionized water. A 5 μl aliquot of this DNA suspension was used for PCR amplification.
Mycoplasma genus-specific PCR
A nested PCR that target a conserved intergenic spacer region between the 16S and 23S rRNA gene of Mycoplasma spp. was used [5]. External primers for primary PCR were F1-sense (5′-ACACCATGGGAGCTGGTAAT-3′) and R1-antisense (5′-CTTCATCGACTTTCAGACCCAAGGCAT-3′), whilst internal primers for secondary PCR were F2-sense (5′-GTTCTTTGAAAACTGAAT-3′) and R2-antisense (5′-GCATCCACCAAAAACTCT-3′). Depending on the different mycoplasma species, the expected amplicons lengths ranged from 369 to 681 bp for the first PCR and from 145 to 237 bp for the second PCR. Five microliter of the extracted DNA were added to a reaction mixture (total volume of 50 μl) containing 2.5 mM MgCl2, PCR buffer (20 mM Tris–HCl [pH 8.4], 50 mM KCl), 0.2 mM deoxynucleoside triphosphates (Invitrogen) 0.5 μM of external primers, and 1.5 U of Taq DNA polymerase (Invitrogen). Amplification was performed using a PTC 150 thermal cycler (MJ research, Waltham, MA, USA) with the following settings: one cycle of 10 min at 94°C, followed by 30 cycles of denaturation at 94°C for 30 sec, annealing at 55°C for 2 min, extension at 72°C for 2 min, and by one cycle of final extension at 72°C for 5 min. The product (2 μl) was used in secondary PCR, performed in the same manner as the primary PCR, but with internal primers instead of external ones. The amplified products were electrophoresed on a 2.5% agarose gel and visualized by ethidium bromide fluorescence. From positive specimens a fragment of around 200 bp, or alternatively of around 150 bp, was amplified after the second round PCR (Figure 1). Mycoplasma bovis DNA was used as positive control. Deionized water was used as negative control (no template control). Positive and negative controls were included for each round of amplification. Samples analysis was repeated twice.
Figure 1.
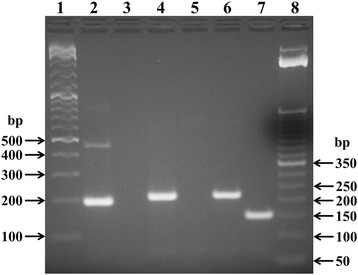
Representative electrophoresis of PCR products for the detection of Mycoplasma spp. in eye swabs samples. Lane 1: DNA size markers (GeneRuler DNA Ladder Mix, Fermentas); lane 2: positive control (Mycoplasma bovis DNA); lane 3: negative control (no template); lanes 4, 6 and 7: positive samples (animals n. 2, 4 and 5, respectively); lane 5: negative sample (animal n. 3); lane 8: DNA size markers (TrackIt 50 bp DNA Ladder, Life Technologies).
Chlamydia genus-specific PCR
A nested PCR that amplifies a portion of the MOMP gene of Chlamydia spp. was used. External primers for primary PCR were CHL2A (5′-GAAAAAACTCTTRAARTCGG-3′) and CHL2B (5′-CGNANGCTWATRGCRTCRCACCAAG-3′), whilst internal primers for secondary PCR were CHL2C (5′-TGCCTGTRGGGAAYCCWKCTGAWCCAAG-3′) and CHL2D (5′-CAAGTNCNRCAAGGATCRCAAGGATC-3′) [6]. Five microliter of the extracted DNA were added to a reaction mixture (total volume of 50 μl) containing 2.5 mM MgCl2, PCR buffer (20 mM Tris–HCl [pH 8.4], 50 mM KCl), 0.2 mM deoxynucleoside triphosphates (Invitrogen) 0.5 μM of external primers, and 1.5 U of Taq DNA polymerase (Invitrogen). Amplification was performed using a PTC 150 thermal cycler (MJ research, Waltham, MA, USA) with the following settings: one cycle of 3 min at 94°C, followed by 35 cycles of denaturation at 94°C for 20 sec, annealing at 45°C for 20 sec, extension at 72°C for 30 sec. The product (2 μl) was used in secondary PCR, performed in the same manner as the primary PCR, but with internal primers instead of external ones. The amplified products were electrophoresed on a 2.5% agarose gel and visualized by ethidium bromide fluorescence. Specimens were regarded as positive if an appropriately sized band (90 bp) was present on second round PCR (Figure 2). Chlamydia psittaci DNA was used as positive control. Deionized water was used as negative control (no template control). Positive and negative controls were included for each round of amplification.
Figure 2.
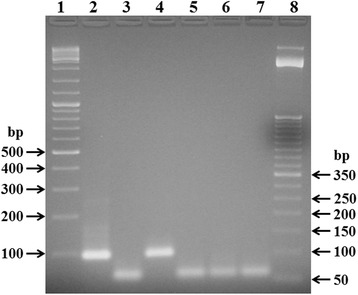
Representative electrophoresis of PCR products for the detection of Chlamydia spp. in eye swabs samples. Lane 1: DNA size markers (GeneRuler DNA Ladder Mix, Fermentas); lane 2: positive control (Chlamydia psittaci DNA); lane 3: negative control (no template); lane 4: positive sample (animal n. 10); lanes 5, 6 and 7: negative samples (animals n. 11, 12 and 13, respectively); lane 8: DNA size markers (TrackIt 50 bp DNA Ladder, Life Technologies). In the negative control and samples the lower band represent dimers of primers.
DNA sequencing
Amplicons were purified and extracted from a 2% agarose gel by a spin column technique (JETQUICK Gel Extraction Spin Kit, Genomed, Löhne, Germany). Sequences were obtained by cycle sequencing on an ABI 3730XL sequencing machine using F2 and R2 primers. GeneBank sequence alignment function (BLAST) was used for sequence analysis.
Statistical analysis
Statistical analysis was performed using the Fisher’s exact test.
Results
All the cultured samples showed bacterial growth on agarized media. Therefore, BHI broth cultures were not used for subculturing. Among fungi, only Candida spp. was detected in 2 healthy tortoises, both T. graeca (Table 1). Thirteen animals (38%), almost all turtles (10/13), harboured a single bacterial species as sole isolate. Seventeen animals (50%) harboured 2 different species of bacteria, 2 animals (6%) harboured 3 species and 2 animals (6%) harboured 4 species (Table 1). The detection of multiple bacterial infection was clearly greater (p = 0.011) in tortoises (17 out of 18 animals) compared to turtles (9 out of 16 animals). Besides Bacillus spp. (13 isolates), the most frequently isolated bacterial species were Staphylococcus xylosus (10 isolates), Sphingomonas paucimobilis (6 isolates), Staphylococcus sciuri and Aeromonas hydrophila/caviae (each 5 isolates), Ochrobactrum anthropi (3 isolates), and finally Citrobacter freundii, Enterobacter cloacae and Pseudomonas luteola (each 2 isolates). Only one isolate of Kocuria varians/rosea, Staphylococcus aureus, Staphylococcus auricularis, Staphylococcus haemolyticus, Staphylococcus lentus, Morganella morganii, Pasteurella multocida, Pasteurella pneumotropica/haemolytica, Proteus spp., Pseudomonas putida, Salmonella enterica ssp. arizonae, Stenotrophomonas maltophilia and Vibrio parahaemolyticus was evidenced (Table 1). Mycoplasma cultures were negative for all the animals. By PCR we detected eight animals, of which 1 diseased and 2 healthy tortoises and 5 healthy turtles, positive for Mycoplasma spp., whereas only one tortoise with conjunctivitis was positive for Chlamydia spp. (Table 1). Sequencing analysis, performed on Mycoplasma PCR products, confirmed that all the amplified sequences belong to the Mycoplasma genus (sequences are reported in an additional file [see Additional file 1]; BLAST analysis results are not shown).
Discussion
The number of people who raise reptiles as pets has increased, but information about microbiological is limited. Among reptiles, pet turtles and tortoises are also becoming increasingly popular. The animals involved in this survey are almost all captive chelonians, that are routinely or occasionally subjected to medical examination.
Except the presence of Bacillus spp. (13 isolates), Staphylococcus xylosus was the most frequent isolate in tortoises (10 isolates). Sphingomonas paucimobilis and Aeromonas hydrophila/caviae were likewise the most frequent isolates in turtles (6 and 5 isolates, respectively).
From Table 1 a clear predominance of Gram positive isolates in tortoises and Gram negative isolates in turtles can be noticed (p < 0.001). A preponderance of Gram positive species was already observed by the Authors in the normal conjunctival bacterial flora of green iguana, in accordance with data reported for other terrestrial animal species [7]. The prevalence of Gram negative isolates in water turtles is also a not surprising result. Captive water turtles, as those included in the present study, often live in small water volumes of aquaria and ponds, where fecal matter and other waste material tend to be concentrated. Therefore, these animals live immersed in a medium were proliferation of enteric bacteria is favoured. To date, little is known about the microbial communities or potential pathogens associated with aquarium water. However, in a survey of Smith et al. [8] the bacterial community of ornamental fish aquarium freshwater was characterized by molecular methods. In accordance with our results, they found in their samples the presence of Aeromonas and Vibrio genera, which include species that can become pathogenic for fish as for turtles in stressful conditions. On the contrary, they did not found the presence of Salmonella spp. This is not unexpected, as it is well known that reptiles are frequently carriers of Salmonella spp. [9]. However, it was reported that also bacteria belonging to Salmonella genus, as well as Aeromonas and Vibrio genera, can be present in home aquaria and probably be responsible of illness in humans due to exposure to ornamental fish [10].
The occurrence of conjunctivitis was higher in tortoises than in turtles: three out of eighteen tortoises (n. 10, 11 and 14) and one out of sixteen turtles (n. 23), showed bilateral conjunctivitis (Table 1). However, the data is not statistically significant (p = 0.282) and to be confirmed would require a greater number of samples. As already it is clear looking at the data, the statistical analysis confirms the absence of association (p = 0.398) between the presence of conjunctivitis and the presence of infection with multiple bacterial species. Bacterial flora of tortoises 11 and 14 did not differed from that of other healthy tortoises. In these animals, as well as in tortoise 10, the only common isolate was represented by Staphylococcus xylosus. However, tortoise 10 was the only one in which it was possible to detect the presence of Chlamydia spp. by PCR. Unlike in the other clinical cases, in this animal conjunctivitis was particularly severe and the animal was not able to open the eyelids themselves. This finding may indicate a role of chlamydia in inducing severe ocular disease. As reviewed by Corsaro and Venditti [11], Chlamydia or Chlamydia-like organisms infections have occasionally been reported in reptiles, including both turtles and tortoises, and almost all the reported cases concerned captive animals. Common symptoms are lethargy, anorexia and chronic respiratory disease. However, almost all Chlamydiaceae species are able to infect the conjunctiva of their vertebrate host and, with regard to reptiles, exudative conjunctivitis attributed to Chlamydia spp. infection was reported in farmed juvenile crocodiles [11,12].
On the other hand, it is noteworthy that bacterial isolates from turtle 23, which is the only acquatic turtle showing conjunctivitis, that is Staphylococcus haemolyticus and Morganella morganii, were not found in any of the other turtles. Staphylococcus haemolyticus belong to the group of coagulase-negative staphylococci (CNS). CNS are a major component of the normal microflora of the human skin and are mostly considered to be saprophytes. However, S. haemolyticus can be also an opportunistic pathogen and is the second most frequently encountered species of CNS in human clinical infections [13]. S. haemolyticus is a frequently isolated pathogen in dairy cows and small ruminants and can be sporadically involved in subclinical mastitis [14]. Less is known about the relation of this bacterium and other animal species. Instead, it is well known the ability of S. haemolyticus to acquire multi-antibiotic resistance [15]. In our case, S. haemolyticus, as well as S. lentus, was found in acquatic animals, while, on the contrary, all the other species of staphylococci were detected in animals with terrestrial habitat. One isolate of S. aureus was found in tortoise n. 1. S. aureus may be involved in case of ocular disease induced by hypovitaminosis in tortoises [16], but in our case the animal was health. M. morganii is a Gram negative facultative anaerobe, belonging to the Enterobacteriaceae family. It is commonly found in the environment and as normal flora in the intestinal tracts of humans, mammals other than humans and reptiles and is considered an opportunistic bacterium. In human, despite its wide distribution, is an uncommon cause of nosocomial infections in adults and is mostly isolated from urinary tract or wound infections, as well as can be involved in neonatal sepsis [17]. A case of postoperative bacterial endophtalmitis caused by M. morganii was also reported [18]. In animals, M. morganii was isolated from mixed infections of the marine mammal Dugong dugon [19] and was reported as one of the most common pathogens identified in human secondary wound infections following snakebites [20]. Recently, a sporadic case of fatal infection in chickens, caused by a highly pathogenic M. morganii strain, was also reported [21]. Our isolate of Morganella morganii was resistant to a wide variety of antibiotics suitable for chelonians treatment (data not showed). The presence of multiple drug resistance is significant, since the bacterium can, although rarely, cause the occurrence of zoonotic disease, particularly in weak persons.
The presence of Mycoplasma negative cultures, also from those animals that resulted positive to Mycoplasma PCR, is not a surprising result [22] as mycoplasmas have many different and often fastidious requirements for growth. The seven days incubation time may have further reduced the probability of isolating slow growing mycoplasmas. In Mycoplasma PCR we found an occasional unspecific amplification, outside the 145–237 bp range of positivity, in the negative control. This could be due to an external contamination of the water used in place of template in the negative control. Conversely, we exclude the contamination of the reaction mix, because such unspecific amplification has never occurred to any of the samples tested. However, to doubtless confirm that the amplified sequences belong to the Mycoplasma genus, we repeated twice the Mycoplasma PCR and we performed sequencing analysis on Mycoplasma PCR products. We found a higher percentage of Mycoplasma PCR positive samples in turtles compared to tortoises (Table 1). Mycoplasmas are frequently found in reptile species, particularly in case of chelonians, and some of them are part of the commensal bacterial flora of the host. However, Mycoplasma agassizii and Mycoplasma testudineum can be involved in the URTD in tortoises [23]. In particular, the most frequently reported agent of URTD in tortoises is Mycoplasma agassizii [24,25]. We found by PCR the presence of Mycoplasma spp. in 23,5% of the animals (16.7% for tortoises and 31,3% for turtles). Our result for the testudo genus is similar to those obtained by Soares et al. [25], who reported a prevalence of 15.8% for Mycoplasma agassizii in captive tortoises (Testudo spp.) in the United Kingdom. In Italy, in particular on the island of Sardinia, a higher prevalence of Mycoplasma spp. (37%) was found by Lecis et al. [24] in tortoises belonging to the Testudo genus. However, in addition to the different geographical origin of the animals, Lecis et al. [24] used wild animals, while in our case, with only one exception (animal n. 27), the animals came from private collections. Moreover, they have analyzed oral and cloacal swabs and used a different technique of DNA extraction and amplification. Considering our study, two results emerge: the fact that the detection of Mycoplasma spp. is almost double in turtles than tortoises, but with a number of observations that does not allow to assign statistical significance to the data (p = 0.196), and that the presence of Mycoplasma spp. was not statistically associated with the presence of conjunctivitis (p = 0.424). It would be interesting to assess whether, in case of URTD with simultaneous presence of conjunctivitis, such an association could occur.
Regarding the presence of Candida spp., which we have found in 2 healthy tortoises, the occurrence of yeasts in reptiles, especially in those who predominantly consuming vegetable diets, has been reported, and different Candida species have been isolated from tortoises belongin to the Testudinidae family [26]. Usually reptiles colonized with yeasts do not reveal any symptoms, as in our case, and we consider this finding not worthy of further investigation for the present study. However, representatives of the Testudinidae are frequently kept as pets, as indeed in the case of our tested animals, and could play a role in transmission of yeasts to human beings, especially in immunocompromised hosts, children and elderly or ill persons who are predisposed to candidosis.
Stenotrophomonas maltophilia causes many opportunistic infections as sepsis, pneumonia, urinary tract infection, meningitis, endocarditis, septic arthritis, and peritonitis. It has also been noted to be a pathogen in many ocular infections, including conjunctivitis, keratitis, dacryocystitis, cellulitis, infected scleral buckles, and endophthalmitis, as reviewed by Chang et al. [27]. However, in our case the bacterium was isolated from an healthy animal.
Conclusions
A clear predominance of Gram positive isolates in tortoises and Gram negative isolates in turtles was found. However, we cannot ascribe the observed difference to the diversity of animal species, as other factors, including especially the different characteristics of the living environments, may play a role. Almost all bacterial species isolated may have clinical significance, mostly as opportunistic pathogens, both for humans and animals. That chelonians are often carrier of bacteria with zoonotic potential is a well-known fact, in particular with regard to Salmonella spp. Therefore, it is not surprising the detection of a strain of Salmonella enterica ssp. arizonae in the eye of one of the animals tested. Worthy of note is the finding of chlamydia in a severe case of conjunctivitis, though we cannot epidemiologically assess a cause-effect relationship between the presence of chlamydia and disease.
Availability of supporting data
The data set supporting the results of this article is included within the article (and its additional file).
Acknowledgements
The authors wish to thank Emiliana Schiano for the preparation of culture media and technical assistance. This study was financed by internal funds of the Dipartimento di Scienze Medico-Veterinarie.
Additional file
Mycoplasma PCR products sequences. In the Additional file 1 the plain format sequences of the amplicons obtained from Mycoplasma PCR positive samples and control are reported.
Footnotes
Francesco Di Ianni and Pier Luigi Dodi contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
FDI, PLD and IP conceived the study, collected the animals and contributed to draft the manuscript. CSC and AS performed the microbiological analyses and contributed to draft the manuscript. SC, EP and FQ contributed to the revision and obtained the funding. ST conceived and designed the study, collected the samples, performed the microbiological and molecular analyses, evaluated the data and drafted the manuscript. All authors read and approved the final manuscript.
Contributor Information
Francesco Di Ianni, Email: francesco.diianni@unipr.it.
Pier Luigi Dodi, Email: pierluigi.dodi@unipr.it.
Clotilde Silvia Cabassi, Email: clotildesilvia.cabassi@unipr.it.
Igor Pelizzone, Email: visc75@yahoo.it.
Andrea Sala, Email: sala86.exovet@gmail.com.
Sandro Cavirani, Email: sandro.cavirani@unipr.it.
Enrico Parmigiani, Email: enrico.parmigiani@unipr.it.
Fausto Quintavalla, Email: fausto.quintavalla@unipr.it.
Simone Taddei, Email: simone.taddei@unipr.it.
References
- 1.Rossi JV. Biology and husbandry. In: Mader DR, editor. Reptile medicine and surgery. 2. St. Louis, Missour: Saunders Elsevier; 2006. pp. 25–41. [Google Scholar]
- 2.Williams DL. Ophthalmology of exotic pets. Chichester, West Sussex: Wiley-Blackwell; 2012. pp. 167–9. [Google Scholar]
- 3.Origgi FC, Jacobson ER. Diseases of the respiratory tract of chelonians. Vet Clin North Am Exot Anim Pract. 2000;3:537–49. doi: 10.1016/s1094-9194(17)30088-9. [DOI] [PubMed] [Google Scholar]
- 4.Quinn PJ, Carter ME, Markey BK, Carter GR. Clinical veterinary microbiology. Europe: Mosby-Year Book; 1994. Bacterial pathogens: microscopy, culture and identification; pp. 21–66. [Google Scholar]
- 5.Harasawa R, Uemori T, Asada K, Kato I. Sensitive detection of mycoplasmas in cell cultures by using two-step polymerase chain reaction. In: Kahane I, Adoni A, editors. Rapid diagnosis of mycoplasmas. New York: Plenum Publishing Corp; 1993. pp. 227–32. [Google Scholar]
- 6.Ong GM, Coyle PV, Barros D’Sa AAB, McCluggage WG, Duprex WP, O’Neill HJ, et al. Non-detection of Chlamydia species in carotid atheroma using generic primers by nested PCR in a population with a high prevalence of Chlamydia pneumoniae antibody. BMC Infect Dis. 2001;1:12. doi: 10.1186/1471-2334-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taddei S, Dodi PL, Di Ianni F, Cabassi CS, Cavirani S. Conjunctival flora of clinically normal captive green iguanas (Iguana iguana) Vet Rec. 2010;167:29–30. doi: 10.1136/vr.b4868. [DOI] [PubMed] [Google Scholar]
- 8.Smith KF, Schmidt V, Rosen GE, Amaral-Zettler L. Microbial diversity and potential pathogens in ornamental fish aquarium water. PLoS One. 2012;7 doi: 10.1371/journal.pone.0039971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warwick C, Lambiris AJ, Westwood D, Steedman C. Reptile-related salmonellosis. J R Soc Med. 2001;94:124–6. doi: 10.1177/014107680109400306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weir M, Rajić A, Dutil L, Cernicchiaro N, Uhland FC, Mercier B, et al. Zoonotic bacteria, antimicrobial use and antimicrobial resistance in ornamental fish: a systematic review of the existing research and survey of aquaculture-allied professionals. Epidemiol Infect. 2012;140:192–206. doi: 10.1017/S0950268811001798. [DOI] [PubMed] [Google Scholar]
- 11.Corsaro D, Venditti D. Emerging chlamydial infections. Crit Rev Microbiol. 2004;30:75–106. doi: 10.1080/10408410490435106. [DOI] [PubMed] [Google Scholar]
- 12.Huchzermeyer FW, Langelet E, Putterill JF. An outbreak of chlamydiosis in farmed Indopacific crocodiles (Crocodylus porosus) J S Afr Vet Assoc. 2008;79:99–100. doi: 10.4102/jsava.v79i2.253. [DOI] [PubMed] [Google Scholar]
- 13.Balows A, Hausler WJ, Herrmann KL, Isenberg HD, Shadomy HJ. Staphylococcus. In: Balows A, editor. Manual of clinical microbiology. Washington DC: American Society for Microbiology; 1991. pp. 222–37. [Google Scholar]
- 14.Leitner G, Sela S, Hammer-Muntz O, Zivotofsky D, Weisblit L, Chaffer M, et al. Outbreak of subclinical mastitis in a flock of dairy goats associated with atypical Staphylococcus haemolyticus. J Dairy Res. 2009;76:1–5. doi: 10.1017/S0022029908003646. [DOI] [PubMed] [Google Scholar]
- 15.de Allori MC, Jure MA, Romero C, de Castillo ME. Antimicrobial resistance and production of biofilms in clinical isolates of coagulase-negative Staphylococcus strains. Biol Pharm Bull. 2006;29:1592–6. doi: 10.1248/bpb.29.1592. [DOI] [PubMed] [Google Scholar]
- 16.Holt PE, Cooper JE, Needham JR. Diseases of tortoises: a review of seventy cases. J Small Anim Pract. 1979;20:269–86. doi: 10.1111/j.1748-5827.1979.tb06721.x. [DOI] [PubMed] [Google Scholar]
- 17.Chang HY, Wang SM, Chiu NC, Chung HY, Wang HK. Neonatal Morganella morganii sepsis: a case report and review of the literature. Pediatr Int. 2011;53:121–3. doi: 10.1111/j.1442-200X.2010.03241.x. [DOI] [PubMed] [Google Scholar]
- 18.Zaninetti M, Baglivo E, Safran AB. Morganella morganii endophthalmitis after vitrectomy: case report and review of the literature. Klin Monbl Augenheilkd. 2003;220:207–9. doi: 10.1055/s-2003-38176. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen KA, Owen HC, Mills PC, Flint M, Gibson JS. Bacteria isolated from dugongs (Dugong dugon) submitted for postmortem examination in Queensland, Australia, 2000–2011. J Zoo Wildl Med. 2013;44:35–41. doi: 10.1638/1042-7260-44.1.35. [DOI] [PubMed] [Google Scholar]
- 20.Chen CM, Wu KG, Chen CJ, Wang CM. Bacterial infection in association with snakebite: a 10-year experience in a Northern Taiwan medical center. J Microbiol Immunol Infect. 2011;44:456–60. doi: 10.1016/j.jmii.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 21.Zhao C, Tang N, Wu Y, Zhang Y, Wu Z, Li W, et al. First reported fatal Morganella morganii infections in chickens. Vet Microbiol. 2012;156:452–5. doi: 10.1016/j.vetmic.2011.11.021. [DOI] [PubMed] [Google Scholar]
- 22.Pinard CL, Brightman AH, Yeary TJ, Everson TD, Cox LK, Chengappa MM, et al. Normal conjunctival flora in the North American opossum (Didelphis virginiana) and raccoon (Procyon lotor) J Wildl Dis. 2002;38:851–5. doi: 10.7589/0090-3558-38.4.851. [DOI] [PubMed] [Google Scholar]
- 23.Farkas SL, Gál J. Adenovirus and mycoplasma infection in an ornate box turtle (Terrapene ornata ornata) in Hungary. Vet Microbiol. 2009;138:169–73. doi: 10.1016/j.vetmic.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 24.Lecis R, Paglietti B, Rubino S, Are BM, Muzzeddu M, Berlinguer F, et al. Detection and characterization of Mycoplasma spp. and Salmonella spp. in free-living European tortoises (Testudo hermanni, Testudo graeca, and Testudo marginata) J Wildl Dis. 2011;47:717–24. doi: 10.7589/0090-3558-47.3.717. [DOI] [PubMed] [Google Scholar]
- 25.Soares JF, Chalker VJ, Erles K, Holtby S, Waters M, McArthur S. Prevalence of Mycoplasma agassizii and Chelonian herpesvirus in captive tortoises (Testudo sp.) in the United Kingdom. J Zoo Wildl Med. 2004;35:25–33. doi: 10.1638/02-092. [DOI] [PubMed] [Google Scholar]
- 26.Milde K, Kostka V, Kaleta EF, Willems H, Jäger C. Multiplex-PCR-based differentiation and characterization of Candida-isolates derived from tortoises (Testudinidae) Vet Microbiol. 2000;76:395–402. doi: 10.1016/S0378-1135(00)00248-0. [DOI] [PubMed] [Google Scholar]
- 27.Chang JS, Flynn HW, Jr, Miller D, Smiddy WE. Stenotrophomonas maltophilia endophthalmitis following cataract surgery: clinical and microbiological results. Clin Ophthalmol. 2013;7:771–7. doi: 10.2147/OPTH.S39608. [DOI] [PMC free article] [PubMed] [Google Scholar]


