Abstract
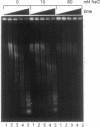
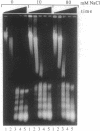
New studies on chromatin fiber morphology, using the technique of scanning force microscopy (SFM), have caused us to reexamine recent analysis of nuclease digestion of chromatin. Chicken erythrocyte chromatin fibers, glutaraldehyde-fixed at 0, 10, and 80 mM NaCl, were imaged with the help of SFM. The chromatin fibers possessed a loose three-dimensional 30-nm structure even in the absence of added salt. This structure slightly condensed upon addition of 10 mM NaCl, and highly compacted, irregularly segmented fibers were observed at 80 mM NaCl. This sheds new light upon our previously reported analysis of the kinetics of digestion by soluble and membrane-immobilized micrococcal nuclease [Leuba, S. H., Zlatanova, J. & van Holde, K. (1994) J. Mol. Biol. 235, 871-880]. While the low-ionic-strength fibers were readily digested, the highly compacted structure formed at 80 mM NaCl was refractory to nuclease attack, implying that the linkers were fully accessible in the low-ionic-strength conformation but not in the condensed fibers. We now find that cleavage of the linker DNA by a small molecule, methidiumpropyl-EDTA-Fe(II), proceeds for all types of conformations at similar rates. Thus, steric hindrance is responsible for the lack of accessibility to micrococcal nuclease in the condensed fiber. Taken in total the data suggest that reexamination of existing models of chromatin conformation is warranted.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cartwright I. L., Elgin S. C. Nonenzymatic cleavage of chromatin. Methods Enzymol. 1989;170:359–369. doi: 10.1016/0076-6879(89)70056-2. [DOI] [PubMed] [Google Scholar]
- Giannasca P. J., Horowitz R. A., Woodcock C. L. Transitions between in situ and isolated chromatin. J Cell Sci. 1993 Jun;105(Pt 2):551–561. doi: 10.1242/jcs.105.2.551. [DOI] [PubMed] [Google Scholar]
- Hertzberg R. P., Dervan P. B. Cleavage of DNA with methidiumpropyl-EDTA-iron(II): reaction conditions and product analyses. Biochemistry. 1984 Aug 14;23(17):3934–3945. doi: 10.1021/bi00312a022. [DOI] [PubMed] [Google Scholar]
- Leuba S. H., Zlatanova J., van Holde K. On the location of linker DNA in the chromatin fiber. Studies with immobilized and soluble micrococcal nuclease. J Mol Biol. 1994 Jan 21;235(3):871–880. doi: 10.1006/jmbi.1994.1045. [DOI] [PubMed] [Google Scholar]
- Libertini L. J., Small E. W. Salt induced transitions of chromatin core particles studied by tyrosine fluorescence anisotropy. Nucleic Acids Res. 1980 Aug 25;8(16):3517–3534. doi: 10.1093/nar/8.16.3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka Y., Leder P. The complete sequence of a chromosomal mouse alpha--globin gene reveals elements conserved throughout vertebrate evolution. Cell. 1979 Nov;18(3):875–882. doi: 10.1016/0092-8674(79)90139-9. [DOI] [PubMed] [Google Scholar]
- Rattner J. B., Hamkalo B. A. Higher order structure in metaphase chromosomes. I. The 250 A fiber. Chromosoma. 1978 Dec 6;69(3):363–372. doi: 10.1007/BF00332139. [DOI] [PubMed] [Google Scholar]
- Rattner J. B., Hamkalo B. A. Nucleosome packing in interphase chromatin. J Cell Biol. 1979 May;81(2):453–457. doi: 10.1083/jcb.81.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russanova V. R., Dimitrov S. I., Makarov V. L., Pashev I. G. Accessibility of the globular domain of histones H1 and H5 to antibodies upon folding of chromatin. Eur J Biochem. 1987 Sep 1;167(2):321–326. doi: 10.1111/j.1432-1033.1987.tb13339.x. [DOI] [PubMed] [Google Scholar]
- Taniuchi H., Anfinsen C. B., Sodja A. The amino acid sequence of an extracellular nuclease of Staphylococcus aureus. 3. Complete amino acid sequence. J Biol Chem. 1967 Oct 25;242(20):4752–4758. [PubMed] [Google Scholar]
- Thoma F., Koller T., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979 Nov;83(2 Pt 1):403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock C. L., Grigoryev S. A., Horowitz R. A., Whitaker N. A chromatin folding model that incorporates linker variability generates fibers resembling the native structures. Proc Natl Acad Sci U S A. 1993 Oct 1;90(19):9021–9025. doi: 10.1073/pnas.90.19.9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock C. L., McEwen B. F., Frank J. Ultrastructure of chromatin. II. Three-dimensional reconstruction of isolated fibers. J Cell Sci. 1991 May;99(Pt 1):107–114. doi: 10.1242/jcs.99.1.107. [DOI] [PubMed] [Google Scholar]
- Yager T. D., McMurray C. T., van Holde K. E. Salt-induced release of DNA from nucleosome core particles. Biochemistry. 1989 Mar 7;28(5):2271–2281. doi: 10.1021/bi00431a045. [DOI] [PubMed] [Google Scholar]