Abstract
The probiotic product Symbioflor2 (DSM 17252) is a bacterial concentrate of six different Escherichia coli genotypes, whose complete genome sequences are compared here, between each other as well as to other E. coli genomes. The genome sequences of Symbioflor2 E. coli components contained a number of virulence-associated genes. Their presence seems to be in conflict with a recorded history of safe use, and with the observed low frequency of adverse effects over a period of more than 6 years. The genome sequences were used to identify unique sequences for each component, for which strain-specific hybridization probes were designed. A colonization study was conducted whereby five volunteers were exposed to an exceptionally high single dose. The results showed that the probiotic E. coli could be detected for 3 months or longer in their stools, and this was in particular the case for those components containing higher numbers of virulence-associated genes. Adverse effects from this long-term colonization were absent. Thus, the presence of the identified virulence genes does not result in a pathogenic phenotype in the genetic background of these probiotic E. coli.
Keywords: colonization fitness, genome sequence, probiotic, virulence, Escherichia coli
Introduction
For producers and legislators alike, the safety aspects of starter cultures, probiotic products and pharmaceuticals are of utmost importance. A safety evaluation should even have priority over a functional evaluation. In addition to an array of microbiological and phenotypical assays that are available to demonstrate the absence of virulence potential in a strain earmarked for probiotic use, a genome sequence can nowadays be considered an essential part of a thorough safety evaluation. Preferably, virulence genes should be absent from the genome of probiotic and starter culture bacteria; the absence of transferable genes providing resistance to clinically relevant antibiotics is also desired. However, the interpretation of genomic data is not without difficulty. Notably, genes present in pathogens are often conserved in commensals [1] and probiotic species as well [2]. This applies in particular to Gram-negative species, where a large part of the proteobacteria for which genome sequences are available is derived from pathogens [3]. This overrepresentation of pathogens and their genes in public databases increases the chance of finding a hit for a query gene from a probiotic genome to a gene detected in a proteobacterial pathogen. Even genes that have been proven to add to the virulence potential of a given pathogen can sometimes be demonstrated in the genome of probiotic strains [2, 4].
In case a long history of safe use can be demonstrated for a given probiotic strain, the presence of genes with high similarity to virulence genes in its genome can be ignored. An example is Escherichia coli Nissle 1917, which has been used in probiotic applications for decades [5]. When its genome was analyzed, of all E. coli genomes then available, it showed most similarity to that of strain E. coli CFT073, a pathogenic E. coli causing urinary tract infections [6, 7]. Nevertheless, its documented history of safe use suggests that the presence of genes that, in a different genomic content, have been shown to contribute to virulence, can be considered as nonproblematic, when found in the context of the Nissle genome.
This argument cannot be used for strains for which a long history of safe use is not, or not yet, available. The precautionary principle dictates to mistrust the presence of virulence genes in the genome of a strain that is targeted to be consumed on purpose. Understandably, legislators have preferentially allowed novel probiotic products or starter culture strains that are closely related (for instance, belonging to the same species) to strains with a proven history of safe use. This, however, hampers the development of novel probiotic and starter culture products, as it restricts novel discoveries to a limited number of species only.
The discussion which genes, or gene combinations, can be considered “safe” and which play a role in virulence is complicated because many gene homologs can function in both benign and pathogenic host–microbe interactions. Many gut commensals use the same strategy to enhance their colonization (for instance by means of motility or attachment) that enteric pathogens also use. As a consequence, genes encoding for attachment or motility can enhance virulence of pathogens (and are often assigned a virulence function), while their equivalents in a commensal strain or species enhance their colonization without pathogenicity, and in a probiotic strain these same homologs might actually promote the beneficial effects of these bacteria.
One of the most difficult species for which to predict, from a genome sequence, virulent or commensal–probiotic interactions with the human host, is E. coli. The genomes of different E. coli strains vary considerably in size (the larger E. coli genomes are over one thousand kilobases larger than the shortest known genome of this species); consequently, genomes vary considerably in gene content [8]. Moreover, the species includes commensal as well as mild or highly pathogenic strains, whereby genome size is not necessarily correlating with pathogenicity.
We evaluated the genome content of E. coli strains that comprise a commercial probiotic product, Symbioflor2, that has been on the market for human consumption in Germany since 1954, followed in Austria, Hungary, and Switzerland (amongst other countries) a few years later. The product is composed of six live E. coli components, which are cultured separately and then mixed in equal amounts of G1/2, G3/10, G4/9, G6/7, and half the amount of G5 and G8, to produce the final product (the reason for this composition is historical). The product is marketed to regulate and improve the immune system of the gut; it is specifically recommended in case of irritable bowel disease. Consumers are advised to take 20 drops three times daily for a period up to 6 months, equivalent to a dose of 1·108 CFU per day.
The genomes of the components of Symbioflor2 have been sequenced [9], and a comparison of their predicted proteomes is presented here. The genome sequences were also used for a comparison to a subset of other E. coli genomes, both pathogenic and commensal strains. The genomes of Symbioflor2 E. coli contained a number of putative virulence genes. With the use of specific probes, we determined whether presence of these genes correlated to the colonization properties of the individual Symbioflor2 components, as we hypothesized that putative virulence genes could affect their fitness to compete with residual microflora in a human gut. The functionality of the probiotic properties of this product was not part of this investigation.
Methods
Symbioflor2 components
E. coli G1/2 (DSM 16441, serotype rough), G3/10 (DSM 16443, serotype O:35,129), G4/9 (DSM 16444, rough), G5 (DSM 16445, rough), G6/7 (DSM 16446, rough), and G8 (DSM 16447, O:169) were provided by SymbioPharm (Herborn, Germany). All strains were H−. These E. coli isolates originated from the stool of one healthy individual in Germany in 1954, and together comprise the product Symbioflor2 DSM 17252.
Genome comparison
The protein-coding genes (CDS) predicted for the Symbioflor2 genomes were compared between each other and with other E. coli strains by means of BLASTP, and visualized in a matrix [10]. Cluster analysis of these genomes and 28 additional published E. coli genomes was performed according to published methods [11], resulting in a pan-genome tree. Information on the included reference genomes can be found in Ref. [8]. A BLAST atlas was created with the CMG-Biotools system on a local computer [10]. A phylogenetic tree of concatenated fragments of housekeeping genes adk, fumC, icd, gyrB, mdh, purA, and recA was performed by ClustalW alignment and a neighbor-joining tree was constructed with NGplot.
Virulence gene identification
Two databases were used to compare the genome content of Symbioflor2 E. coli with known virulence genes: the MvirDB at LLNL (http://mvirdb.llnl.gov) [12], using genes from E. coli only, and the database of virulence factors of pathogenic bacteria (VFDB) available at http://www.mgc.ac.cn/VFs/main.htm [13]. In case of VFDB, either the virulence factors (VF) or the “VFs for comparative studies” were used. Genes identified in Symbioflor2 genomes were compared to the genes stored in these databases by BlastP, filtering for e-values <0.001, and for >98% similarity. Genes that are normally part of a locus, and only function when a complete locus is present, were checked for presence of that complete locus.
Symbioflor2-specific oligonucleotide probes and colony lift hybridization
Unique protein-coding genes for each strain were identified by BLASTP comparison [14], and checked for uniqueness in the nonredundant protein database at NCBI (February 2011). For those amino acid sequences that reported no significant hit, or only a single hit (preferentially to a species different from E. coli), the corresponding nucleotide sequences were extracted from the sequence files and these were used for a search in the nonredundant nucleotide database at NCBI (March 2011) using BLASTN. This resulted in identification of a sequence unique to the genome of interest, and hybridization probes were designed to target these presumed unique sequences (Table 1). Their specificity was confirmed by Southern blot hybridization using Symbioflor2 components, as well as against a variety of nonrelated E. coli isolates and other Enterobacteriaceae (data not shown). Probe pr G1/2 could not distinguish between strains G1/2, G6/7 and G8, whose genome sequences were very similar. Probes specific for the latter two could not be identified; thus, probe pr G1/2 was used for detection of these three strains collectively. General bacterial probe pr1037 with binding specificity for the 23S rDNA of bacteria was used as the final hybridization probe and as a reference targeting all lifted E. coli colonies. By subtracting all Symbioflor2-specific colony-counts from the pr1037 signal, the number of CFU for Symbioflor2 E. coli components was calculated.
Table 1.
Probes used in this study
| Probe name | Sequence (5ʹ → 3ʹ) | Specificity | Tm (°C) |
|---|---|---|---|
| pr G1/2 | ACAGGCAAACCAAAGGATTG | G1/2, G6/7, G8 | 58 |
| pr G3/10 | GGCTGAACTCACTGGAAAGC | G3/10 | 62 |
| pr G4/9 | CCCCTTTTGCATTTACCAAC | G4/9 | 58 |
| prG5 | AAAAATGCCCGGTTCTTCTTC | G5 | 60 |
| pr1037 | CGACAAGGAATTTCGCTAC | 23S rRNA, bacteria | 47 |
Volunteer study for colonization potential
Five healthy human volunteers participated in a colonization experiment; each person took a single high dose of Symbioflor2 after a meal on day 1, while a stool sample had been taken on day zero to provide a baseline. Persons B (male, 46 years) and E (female, 45) took 100 ml of Symbioflor2, containing 2·107 CFU/ml, persons A (male, 38) and C (male, 47) received a dose of 50 ml and person D (female, 27) took a dose of 10 ml. Stool samples were taken on days 3 to 7, days 10 to 12, and weekly for a period of 28 weeks thereafter (volunteer E was followed for 36 weeks). The volunteer study was carried out under supervision of an MD and with informed and written consent of all volunteers. The study was registered at the National Association of Statutory Health Insurance Funds (GKV Spitzenverband) Berlin, according to § 67 Abs. 6 AMG (BFarmNr. 147482). Presence of E. coli bacteria was determined by culture of serial stool dilutions on MacConkey agar. Symbioflor2 bacteria were detected by colony lift hybridization of these plates, using the Symbioflor2-specific hybridization probes of Table 1. Total E. coli counts were performed with probe pr1037. The colony lift protocol was performed according to Ref. [15]. Control experiments confirmed the specificity of the probes as shown in Table 1.
Safety evaluation following commercial use
The Periodic Safety Update Reports that had been collected in Europe according to BfArM and MedDRA pharmacovigilance regulations (www.meddra.org) were reviewed. These reports, covering a period of 6.6 years (June 2005 to December 2011) were compiled in accordance to European safety regulations and were made available by SymbioPharm GmbH.
Results
Key features of Symbioflor2 E. coli genome sequences
Some key features for the six sequenced E. coli strains comprising Symbioflor2 not available from Ref. [9] are summarized in Table 1. The variation in number of detected coding sequences (CDS) correlated with the differences in genome length (the genome of E. coli G4/9 was significantly smaller than that of the others [9]). The genome of strain G3/10 was most completely covered, although it remained in 12 contigs that could not yet be positioned relative to each other.
The presence of plasmids in these strains is noticeable, and their numbers varied between one and six. Based on the plasmid content, strains G1/2, G6/7, and G8 resembled each other, while strains G3/10, G5, and G4/9 were distinct. Four plasmids with sizes below 4 kb contained genes involved in their own replication only. One megaplasmid (pSYM1) present in strain G3/10 contained a gene coding for microcin S [9, 16], as well as conjugational protein and type 3 fimbrial proteins. Plasmid pSYM5 contained a gene for hemagglutinin, while pSYM12 contained a mob operon (which was also found in pSYM7) as well as a colicin S4 operon.
Comparison of the six Symbioflor2 strains
The protein-coding genes of the strains were deduced from the genomes, and these were binned into gene families, both within genomes and between them. Since a single gene family can contain more than one gene in a given genome, the number of gene families per genome is lower than its number of genes (Table 3). Genes not finding a homolog in any other genome result in a gene family with a single member (singletons). From this comparison, the Symbioflor-specific pan-genome was defined as comprising all genes and gene families of the six strains combined; similarly, a core genome was defined as those genes and gene families conserved in all six strains (Table 3). Combining all 28,180 genes of the six probiotic E. coli gives a pan-genome with 6486 gene families, in which there are 7281 genes extra compared to the conserved core genome of 20,899 genes; this means that only 26% of the genes found in any of the genomes is not present in all the other genomes. This is indicative of a relatively low genetic diversity between these six strains, as a set of six randomly chosen E. coli genomes would most likely produce a larger difference between pan-genome and core genome (Ref. [8] and O. Lukjancenko, personal communication). Table 3 also lists the pan- and core genome after addition of 5 nonrelated E. coli genomes (two commensal strains, two pathogenic strains, and an environmental isolate). As expected, this increased the pan-genome more than it affected the core-genome.
Table 3.
Protein genes and gene families of Symbioflor2 strains including their plasmids
| Total no. of genes |
Total no. of gene families |
Unique no. of genes |
Unique no. of gene families |
|
|---|---|---|---|---|
| G1/2 | 4836 | 4676 | 211 | 206 |
| G3/10 | 4855 | 4351 | 565 | 546 |
| G4/9 | 4138 | 3944 | 177 | 175 |
| G5 | 4442 | 4228 | 304 | 301 |
| G6/7 | 5033 | 4658 | 261 | 261 |
| G8 | 4876 | 4743 | 172 | 168 |
| Symbioflor2-specific pan-genome | 28,180 | 6486 | N.A. | N.A. |
| Symbioflor2-specific core genome | 20,899 | 3299 | N.A. | N.A. |
| Pan genome after addition of 5 nonrelated E. coli genomes* | 51,656 | 8344 | N.A. | N.A. |
| Core genome after addition of 5 nonrelated E. coli genomes* | 34.296 | 2,936 | N.A. | N.A. |
| N.A.: not applicable *For comparison, the following E. coli genomes were included: commensal strains K12 MG1655 and BL21 DE3, environmental isolate SMS-3-5, enterohemolytic strain O157:H5 EDL933, and uropathogenic CFT073. For details of these genomes see Ref. [8] | ||||
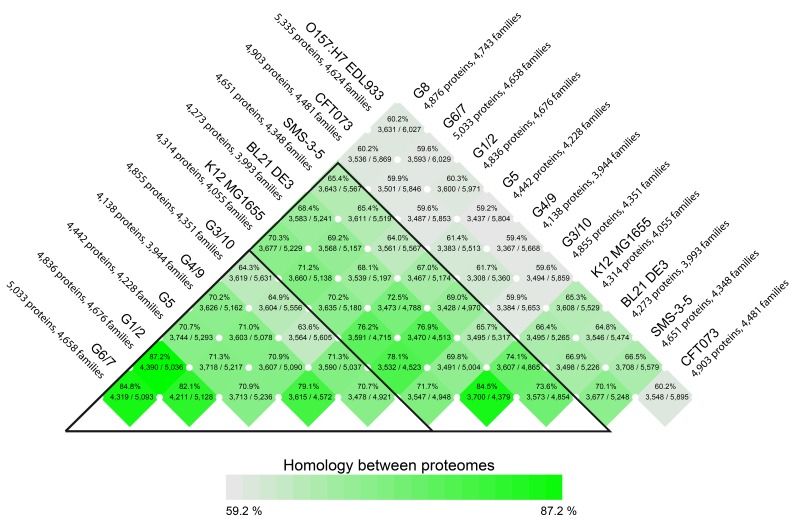
We next compared the protein coding genes of the six sequenced strains by pairwise comparison. Again, this analysis showed a high degree of similarity between the genomes, as summarized in the BLAST matrix of Fig. 1. The highest overlap (84.8%) between two proteomes was found between G6/7 and G8, whereas strain G3/10 was most dissimilar with the other probiotic strains, as indicated by the lighter colored row in the matrix. Nevertheless, that strain shares between 63.6% and 71.3% of protein genes with the other strains. Interestingly, these results, based on over 28,000 genes included in the analysis, corroborated the similarities observed in plasmid content summarized in Table 2, despite the fact that plasmid content can vary over time and is generally not a good indicator of strain similarity.
Fig. 1.
Comparison of the six probiotic E. coli and five nonrelated E. coli genomes. Protein gene similarity detected between the six Symbioflor2 E. coli strains and five other E. coli genomes is shown in a matrix, based on pairwise BLASTP comparisons of predicted coding sequences. The degree of similarity is shown by color intensity. Numbers within the cells represent the percentage identity between each compared pair, given by the ratio of their number of shared protein families and their total number of combined proteins. The probiotic genomes are more similar to each other (small black triangle) and to three other nonpathogenic strains (big black triangle) than they are to the pathogenic strains CFT073 and O157:H7
Table 2.
The six E. coli strains comprising Symbioflor2
| Features | G3/10 | G1/2 | G4/9 | G5 | G6/7 | G8 |
|---|---|---|---|---|---|---|
| G + C content (%) | 50.89 | 50.74 | 50.69 | 50.80 | 50.73 | 50.67 |
| Number of chromosomal CDS | 4780 | 4825 | 4137 | 4410 | 5022 | 4865 |
| Number of rRNAs | 22 | 22 | 22 | 22 | 22 | 22 |
| Number of tRNAs | 87 | 90 | 72 | 84 | 83 | 80 |
| Number of plasmid CDS |
60 (pSYM1) 3 (pSYM2) 1 (pSYM3) 1 (pSYM4) 4 (pSYM5) 7 (pSYM6) |
1 (pSYM10) 10 (pSYM12) |
1 (pSYM4) | 1 (pSYM3) 5 (pSYM7) 2 (pSYM8) 23 (pSYM9) 1 (pSYM11) |
1 (pSYM10) 10 (pSYM12) |
1 (pSYM10) 10 (pSYM12) |
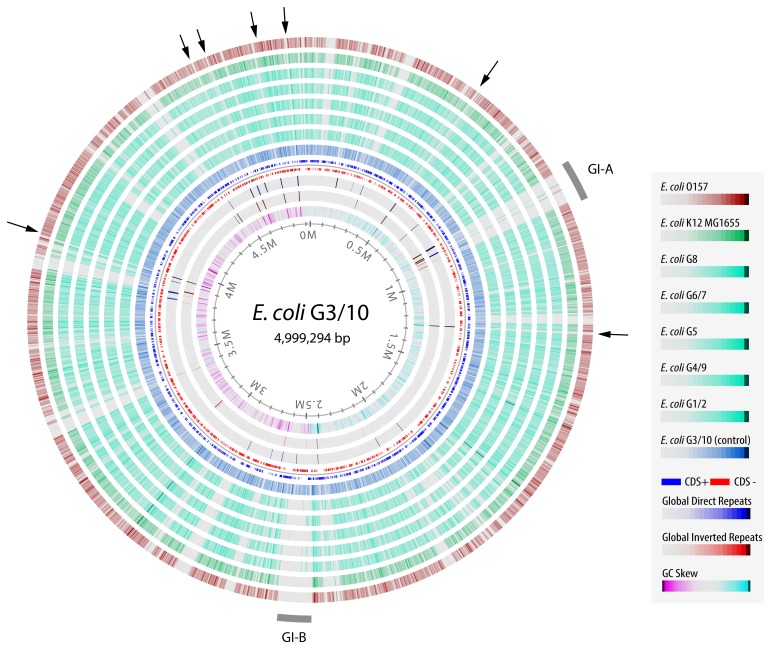
Figure 2 presents a Genome Atlas of the chromosome of strain G3/10, around which blast hits are shown for genes that detected a homolog in the other Symbioflor2 genomes. From this graphical representation, it is obvious that two genome islands can be identified that are unique to strain G3/10. This exemplifies the mosaic structure of E. coli genomes, where islands of colocated genes are often introduced or deleted by activity of mobile elements. The island named GI-A, around 0.85 MB, contains mainly transposases and other remnants of mobile DNA elements. The island GI-B, around 2.4 MB, mainly contains remnants of a prophage. It is not surprising that the variation in protein gene content between G3/10 and the other probiotic genomes is mostly due to mobile DNA, as this is frequently observed in bacteria. The two outer lanes of the atlas show the BLAST hits detected in the proteome of commensal E. coli K12 MG1655 and in a pathogenic E. coli O157 genome. The Blast lanes in which G3/10 protein genes are compared to pathogenic O157 genes does not differ much from the other blast lanes (Fig. 2). This can be explained by the relatively small genome of G3/10 compared with the much larger O157 genome: with its relatively small genome, G3/10 mostly contains proteins found in many other E. coli genomes. The genes that are typical for O157 are mostly absent in G3/10 and these are missed in this comparison (in total, there are 934 genes, comprising 782 gene families, present in strain O157:H7 EDL933 that are missing from the Symbioflor2-specific pan genome).
Fig. 2.
Blast atlas of Symbioflor2 genomes with strain G3/10 as the reference genome. Protein genes from E. coli G3/10 were compared by BlastP to other genomes, including a pathogenic strain (E. coli O157, outward lane, in red), a commensal (E. coli K12 MG 1655, dark green) and the Symbioflor2 genomes, as indicated in the legend (showing the lanes from outward, at the top, to inward, at the bottom of the legend). The dark blue Blast lane shows the positive control, blasting the protein genes of strain G3/10 against itself. Two areas with genes unique to G3/10 are indicated as GI-A and GI-B. Arrowheads indicate the position of the seven ribosomal gene loci
Comparison of Symbioflor2 E. coli with other E. coli strains
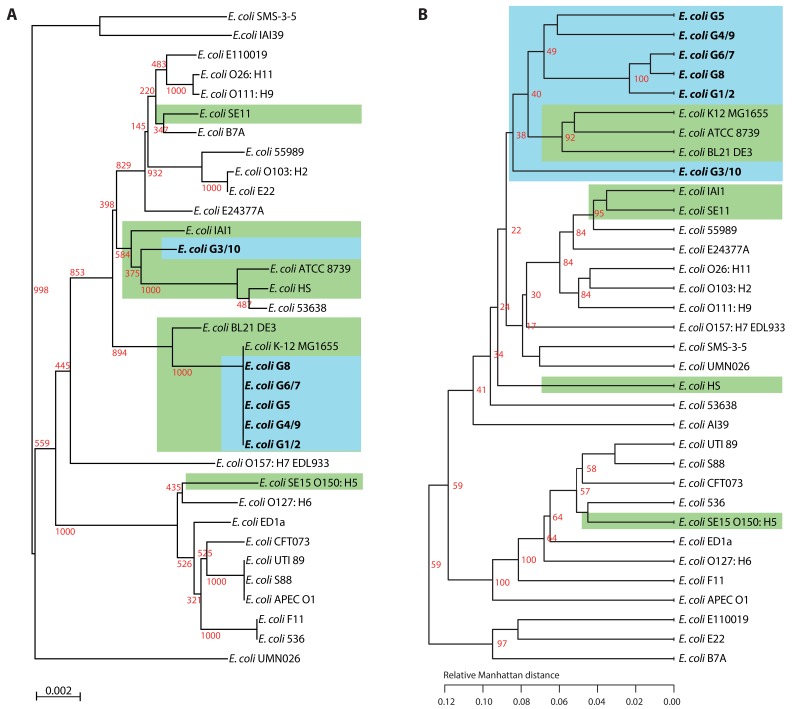
We aimed to assess further how unique the individual Symbioflor2 components were and compared their genomes with those of a selection of other sequenced E. coli strains. This selection contained both nonpathogenic (environmental) as well as pathogenic strains, of various pathotypes. In view of the large variation in gene content within this species, we applied two analyses that assessed mutually exclusive information: on the one hand, we performed a phylogenetic analysis of concatenated conserved housekeeping genes; and on the other hand, a cluster analysis of the variable gene content was carried out. For the phylogenetic analyses, seven gene fragments were selected that are frequently used for multilocus sequence typing (MLST) [17]. The results of the phylogenetic analysis are shown to the left of Fig. 3. Four of the five Symbioflor2 strains have sequence type (ST) 10, belonging to Clonal Complex 10 (http://mlst.ucc.ie/mlst/dbs/Ecoli), while strain G3/10 has different alleles for fumC, gyrB, icd, and recA, resulting in ST472. As a result, in the tree to the left of Fig. 3, strain G3/10 is separated from the ST10 Symbioflor2 strains, which cluster together with commensal E. coli K12. The cluster analysis of nonconserved genes is shown to the right of the figure where, again, G3/10 is separated from the other Symbioflor2 strains. In this analysis, a cluster of three commensal strains is placed within the cluster that contains all Symbioflor2 strains. Although the two analyses are based on different genetic information, and the topology of the trees is substantially different, in both analyses, the Symbioflor2 components are grouped together with commensal or other nonpathogenic E. coli strains. Combined with the Blast results summarized in Figs 1 and 2, it can be concluded that these probiotic E. coli strains are relatively closely related to each other.
Fig. 3.
Comparison of Symbioflor2 E. coli with commensal and pathogenic strains. In panel A, a phylogenetic tree (constructed by neighbor joining) is shown of the concatenated MLST genes adk, fumC, icd, gyrB, mdh, purA, and recA. In panel B, a cluster analysis of variable gene content is shown. Green shading indicates commensal E. coli strains (information extracted from Ref. [8] and from www.genomesonline.org). The Symbioflor2 strains are marked blue
Identification of potential virulence-associated genes
The six Symbioflor genomes were screened for presence of genes encoding potential virulence factors, since presence of such genes might be considered undesirable in probiotic organisms. A comparison of their protein-coding genes to the MvirDB database [12] resulted in between 88 and 181 hits for the different Symbioflor2 genomes. In comparison, the genome of E. coli K-12 strain MG1655 resulted in 161 hits following the same procedure, while pathogenic strain O157:H7 produced 455, and pathogenic strain CFT073 resulted in 468 hits. The other database used to identify virulence genes, VFDB [13], produced between 34 and 80 hits for Symbioflor2 genomes, 41 hits for E. coli K12 strain MG1655, and 172 and 196 hits for the pathogenic O150:H7 and CFT073 strains, respectively. Thus, although pathogens contain more genes represented in these databases than nonpathogens, the latter nevertheless reports considerable hits. These findings suggest that the used databases not only contain true virulence genes, but also genes related to colonization potential or bacterial fitness; as pointed out before, there is no clear division between these [1], and such genes can be found in pathogenic and nonpathogenic strains alike. The most important findings for the Symbioflor2 genomes are summarized in Table 4. Toxin genes were not determined, with the exception of hemolysin: hlyABCD is present in strains G1/2, G6/7, and G8, as has been described before, and these strains indeed display weak hemolytic activity in vitro, suggesting that the operon is weakly expressed [4]. Fimbriae and their adhesins can play a role in both virulence and colonization, but their biosynthesis requires multiple genes. The presence of fimbrial genes in an incomplete genetic context is frequently observed. For instance, variable csg genes, responsible for production of curli fimbriae expression, are found in Symbioflor2 genomes, and when expressed, curli fimbriae are important for biofilm formation and biosynthesis of cellulose [18]. However, none of these genomes contain the complete set (notably, csgA is missing in all). Likewise, the regulator gene mrkE, required for production of type 3 fimbriae, is missing in G3/10, though the rest of the mrk locus is present on its large plasmid (G4/9 contains an even less complete mrk locus, not shown in Table 4). Functional flagella are often associated with virulence but commensal bacteria can also be motile, for which a complete flagellar gene set is required. Although most flagellar genes are present in the Symbioflor2 genomes, fliC is missing except for G3/10 and G4/9, but these two genomes lack other crucial genes to produce functional flagella (data not shown), which explains why all Symbioflor2 strains are of the H− serotype.
Table 4.
Genes encoding proteins responsible for colonization behavior and bacterial fitness
| Gene | Description |
E. coli G1/2 |
E. coli G6/7 |
E. coli G8 |
E. coli G5 |
E. coli G4/9 |
E. coli G3/10 |
|---|---|---|---|---|---|---|---|
| hly locus | Hemolysin A (hlyABCD) | + | + | + | − | − | − |
| fim locus | Type 1 fimbriae (fimABCDEFGHI*) | − | − | − | + | + | + |
| mrk locus | Type 3 fimbriae (incomplete, mrkABCDF but not mrkE) | − | − | − | − | − | +† |
| iuc locus | Aerobactin (iucABCD, iutA) | + | + | + | − | − | − |
| ent locus | Enterobactin (entABCDEFS) | + | + | + | + | + | + |
| fec locus | Iron dicitrate system (fecABCDEIR*) | + | + | − | + | + | − |
| sigA | Serine protease autotransporter | + | + | + | − | − | − |
| upaG | Autotransporter, adhesine | − | − | − | + | + | + |
| iha | Adhesin and siderophore receptor | + | + | + | − | − | − |
| cib | Colicin IB | + | + | + | − | − | − |
| cka | Colicin IK | +† | +† | +† | − | − | − |
| cs | Colicin S4 | +† | +† | +† | − | − | − |
| gad | Glutamate decarboxylase | + | + | + | + | + | + |
| eib | Immune globulin binding, increased serum survival (eibCDEFG) | + | + | −‡ | − | + | − |
| iha | Enterobactin receptor/adhesin | + | + | − | − | − | − |
|
*Only when all genes of the locus are present,
this is recorded as + †Plasmid-encoded ‡Incomplete locus is present | |||||||
Another group of adhesins is represented by the autotransporter subgroup of proteins. In combination with type IV-secretion mechanisms, these typically form trimeric β-barrel structures that allow transport of a passenger domain across the inner and outer bacterial membrane to reach the extracellular space, where it can bind to extracellular matrix proteins and host cells [19]. Various types of autotransporters are found in the six Symbioflor2 E. coli components (Table 4). Autotransporter serin protease SigA is reported to be involved in bacterial pathogenicity of Shigella flexneri; in this organism, it encodes an exported cytopathic protease that is involved in intestinal fluid accumulation following infection [20]. Genes encoding colicins can provide a selective advantage to compete with other bacteria; three colicin genes were found present in three of the six Symbioflor2 genomes (Table 4). Such genes are only related to virulence when they promote the colonization of pathogenic strains.
Of note is the observation that genes encoding resistance to antimicrobial drugs were absent from all chromosomes and plasmids.
Colonization potential of Symbioflor2 E. coli following a single dose
In order to identify the safety of intake of Symbioflor2 under experimental conditions, a volunteer study was performed; this would also shed light on the colonization potential of the product, since we were able to identify sequences that were specific for Symbioflor2 E. coli, and these sequences were used as probes to detect the bacteria from stool. Five human volunteers, who did not have Symbioflor2 strains in their stool prior to the experiment, took a single, high oral dose of Symbioflor2 on day 1. Despite a two to ten times higher intake dose compared to the daily recommended dose of 1·108 CFU (2·109 CFU for volunteers B and E, 1·109 CFU for A and C, and 2·108 CFU for D), none of the volunteers reported any side effects; changes in stool consistency or frequency were also not reported. The stools of these volunteers were sampled from day zero onwards, to detect the total E. coli counts as well as the presence of Symbioflor2 E. coli. The results of the total E. coli counts are shown in Fig. 4. For four out of five individuals, an increase of total E. coli counts during the first week of monitoring was not apparent, with the exception of person E, who had no detectable E. coli in the stool prior to the experiment (in contrast to the other volunteers, this person had taken an antibiotic course during the 6 months prior to the experiment, though not in the 4 weeks prior to day zero). For the other volunteers between 3·104 (for A) and 6·106 (B) CFU/g, E. coli was detected in their stools prior to the experiment (geometric mean 3.57·104 CFU/g of all five and 4.91·105 CFU/g excluding E). During the examined period, the numbers of detected E. coli fluctuated considerably over time for each individual, although there was a trend towards more stable counts after 28 days following ingestion of Symbioflor2. Volunteer E had to take an antibiotic course to treat sinusitis during days 47–52 of the experiment, which resulted in a dramatic drop of E. coli counts that reversed within 8 days. Unfortunately, for a period of 17 weeks, no samples from person E were available for analysis. The sudden drop in person C, detected at day 62, could not be explained. At the end of the experiment, around day 190, the total E. coli counts varied from 1.1·105 to 8.8·106 CFU/g (geometric mean 7.5·105 CFU/g).
Fig. 4.
Total E. coli counts in the stools of five volunteers who took a high single dose of Symbioflor2. The counts at day zero, before intake of the probiotic and around day 190, at the end of the experiment, are shown as dots. The time scale beyond 84 days is changed to 3 weeks intervals, indicated by the dotted curves. The arrow indicates the time-point when volunteer E (shown in green) had to take an antibiotic. The detection level was 50 CFU E. coli per gram stool
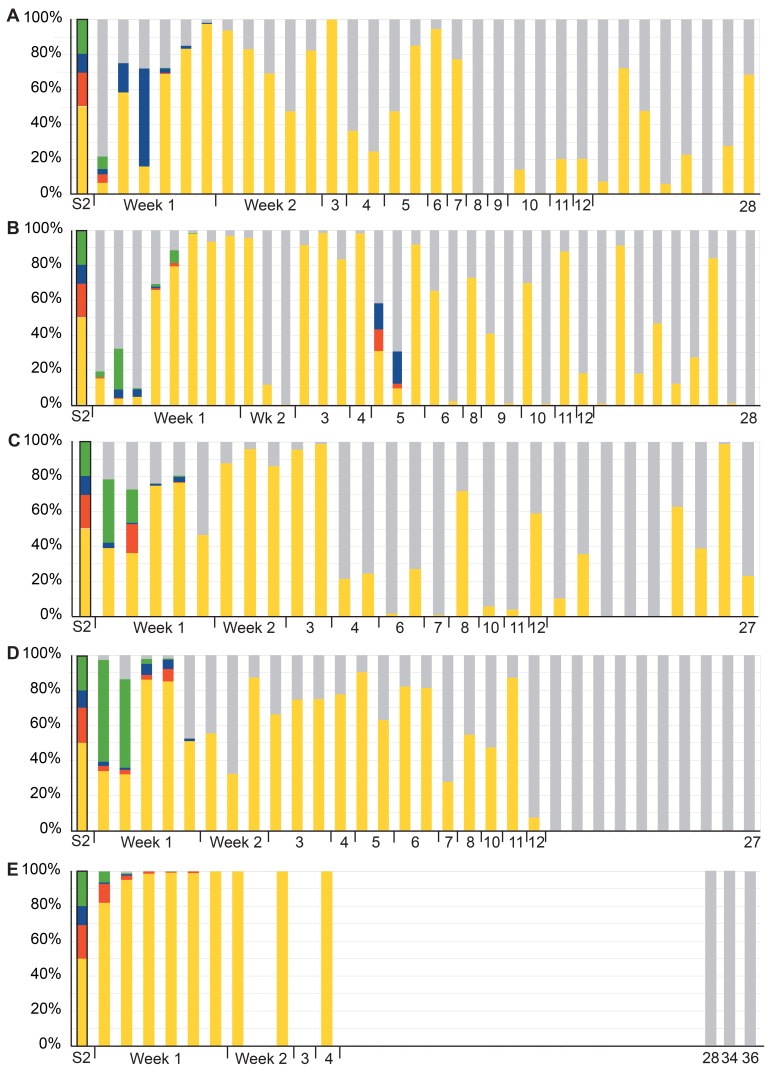
It was investigated which proportion of the detected E. coli could be attributed to the Symbioflor2 intake. Three of the individual E. coli components of Symbioflor2 could be detected in stool with the use of specific oligonucleotide probes, while the three others (G1/2, G6/7, and G8) were detected in combination. The fractions of Symbioflor2 components of E. coli detected in the stool samples are shown in Fig. 5. As can be seen, during the first week most or all components of the probiotic mixture were detected, albeit in different amounts per individual; whereas Symbioflor2 strains represented a minority of all E. coli detected in person B during the first week, for the other four individuals, these comprised 80% or more (this did not correlate to the different doses). In all five persons, strains G1/2, G6/7, and G8 combined colonized persistently; in three, colonization of these three combined components was detectable even after 12 weeks. These three strains reached a colonization maximum around 3 weeks postinoculation. For person E, who had no detectable E. coli in her stool at day zero, following intake of an antibiotic course and a period for which data are not available, host E. coli but not Symbioflor2 components could be detected in weeks 28, 34, and 36.
Fig. 5.
Distribution of Symbioflor2 strains in stool samples after a single dose. Distribution of the components of Symbioflor2 given as percentage of total E. coli determined per time point, for persons A to E (top to bottom). The distribution of G1/2, G6/7, and G8 combined (yellow), G4/9 (red), G5 (blue), and G3/10 (green) that was present in the inoculum is shown to the left of each plot, marked “S2.” Host E. coli is given in grey. For volunteer E, samples were not available for analysis between week 4 and week 28, for which reason, this person was sampled longer than the others
Safety Reports collected from over six years of Symbioflor2 sales
To evaluate the safety of the product further, the Periodic Safety Update Reports that had been collected in Europe according to BfArM and MedDRA pharmacovigilance regulations were reviewed. All safety reports, compiled in accordance to European safety regulations and covering a period of 6.6 years were compiled. The recorded symptoms reported in relation to adverse drug reactions (ADR) are summarized in Table 5.
Table 5.
Summary of all adverse drug reactions for the period June 2005 to December 2011, sorted by MedDRA system organ classes
| MedDRA system organ class | MedDRA preferred term | No. of symptoms |
Main events | Secondary events |
|---|---|---|---|---|
| Gastrointestinal disorders | 16* | 8 | 8 | |
| Abdominal discomfort | 1 | 0 | 1 | |
| Abdominal pain | 2 | 1 | 1 | |
| Abdominal pain upper | 1 | 0 | 1 | |
| Diarrhea | 3 | 1 | 2 | |
| Flatulence | 1 | 1 | 0 | |
| Gingival bleeding | 1 | 1 | 0 | |
| Glossodynia | 1 | 0 | 1 | |
| Lip swelling | 1 | 0 | 1 | |
| Nausea | 4 | 4 | 0 | |
| Swollen tongue | 1 | 0 | 1 | |
| General disorders and administration site conditions | 2 | 1 | 1 | |
| Drug intolerance | 1 | 1 | 0 | |
| Pyrexia | 1 | 0 | 1 | |
| Immune system disorders | 1 | 1 | 0 | |
| Hypersensitivity | 1 | 1 | 0 | |
| Nervous system disorders | 4 | 2 | 2 | |
| Ageusia | 1 | 0 | 1 | |
| Anosmia | 1 | 1 | 0 | |
| Burning sensation mucosal | 1 | 0 | 1 | |
| Headache | 1 | 1 | 0 | |
| Psychiatric disorders | 2 | 0 | 2 | |
| Agitation | 1 | 0 | 1 | |
| Sleep disorder | 1 | 0 | 1 | |
| Respiratory, thoracic and mediastinal disorders | 1 | 1 | 0 | |
| Dyspnoea | 1 | 1 | 0 | |
| Skin and subcutaneous tissue disorders | 4 | 4 | 0 | |
| Acne | 1 | 1 | 0 | |
| Angioedema | 1 | 1 | 0 | |
| Rash | 1 | 1 | 0 | |
| Rosacea | 1 | 1 | 0 | |
| Total | 30 | 17 | 13 | |
| *The numbers of reported adverse reactions for each MedDRA system organ class (shown in bold) are further subdivided into the MedDRA preferred terms | ||||
During the reported period over 2,125,000 treatments had been sold in Germany, Switzerland, Austria, and Hungary combined. Seventeen cases of ADR were reported, two of which were serious, according to the definitions in the International Conference on Harmonization (ICH) E2A guidelines. Temporal or spatial clustering of these 17 reported cases was not observed. The two serious cases involved: a) a 29-year old female with chronic sinusitis and allergy developed loss of smell and loss of taste after intake of Symbioflor2 for 5 months; a pharmacological relationship between the observed reactions and the administered product is unknown, and the patient’s underlying chronic sinusitis and allergy provided a possible explanation for the reduced sense of smell and taste. b) A female patient of unknown age reported Quincke’s oedema, sleep disorder, and agitation; information on the patient’s underlying disease history was not available. The woman still suffered from sleep disorder and agitation 4 weeks after discontinuation of Symbioflor2, but in lack of a mechanistic explanation, the probiotic was an unlikely cause of these symptoms. A total of 30 symptoms were reported from the 17 case reports; most frequently recorded were nausea (four times) and diarrhea (three times). Abdominal pain was recorded twice, and 21 symptoms were recorded once each. Taken together, in consideration of 2,125,000 sold treatments, the low number of 30 adverse symptoms emphasizes the favorable safety profile of Symbioflor2.
Discussion
This study emphasizes the importance of generating a genome sequence of bacterial probiotics, although some of the observations obtained from a complete sequence blueprint may at first sight appear to be disadvantageous. The advantage of having the sequences of Symbioflor2 strains available was demonstrated, as these enabled the design of strain-specific probes, which could then be used to assess the colonization capacity of the product components. Identification of strain-specific sequences would have been impossible without the genome sequence. That the Symbioflor2 components G1/2, G6/7, and G8 could not be distinguished probably reflects the close resemblance of these three strains; they may have originated from one clone, either in the host from which all Symbioflor2 components were isolated, or during continuous subculturing in the past – this could no longer be assessed.
The genome sequences further confirmed absence of antibiotic resistance genes, an absence that is desired for a probiotic strain. However, the genome sequence also revealed the presence of a number of genes that have been linked to virulence in specific pathogens. Although some of those findings were not novel (the presence of hemolysin in three of the strains had been observed before, e.g., Ref. [4]), the discovery of other genes, such as those coding for the autotransporter SigA (Table 4), was unexpected. Of course, gene presence does not guarantee gene expression. Possibly, a number of the virulence genes are not, or only weakly expressed. Moreover, the identification of virulence-associated genes that can only function in combination with other genes should be interpreted in the context of presence of those other genes; e.g., incomplete gene sets for biosynthesis of fimbriae suggest that these structures cannot be built. An automated analysis tool that could identify such incomplete gene sets would be a welcomed addition to the virulence gene database websites currently available.
The identification of putative virulence genes in the genomes of these probiotic strains is in apparent conflict to the long term clinical experience gained with Symbioflor2. Moreover, the volunteers in our study who took ten or twenty times the recommended dose did not report any symptoms or discomfort.
The finding of a complete operon for hemolysin A production in G1/2, G6/7, and G8 is particularly puzzling, although its expression in vitro is much lower than that observed in hemolytic pathogenic E. coli strains [L. Beutin (BfR, Berlin, Germany), personal communication]. Expression levels of these genes during colonization have not yet been determined. Irrespective of its in vivo expression levels, the presence of this pore-forming toxin can be considered problematic, in particular, since the product is recommended to patients suffering from irritable bowel disease. Recently, it was shown that α-hemolysin producing E. coli was frequently present in individuals suffering from ulcerative colitis (UC) or Crohn’s disease, and in a mouse model, this toxin was responsible for microlesions resulting in a “leaky gut” that triggered intestinal inflammation [21]. This is in sharp contrast with the low number of reported adverse drug reactions, collected during over 6 years of commercial use of Symbioflor2. It can be expected that many UC or Crohn’s patients were among these users, as the product is specifically recommended to treat these conditions. Lack of colonization potential of the hemolysin-producing components of Symbioflor2 cannot explain the absence of adverse effects, since precisely those components that produce hemolysin formed the highest fraction of colonizing Symbioflor2 and remained persistent for longest, following a single dose only. Thus, there is a sharp contrast between virulence prediction based on gene presence and in vivo data obtained from mouse models on the one hand and, on the other hand, the lack of adverse effects from long-term commercial use in the general population and during exposure under experimental conditions.
We conclude that the safety of the product is not questioned despite the reported findings of virulence-associated genes of Table 4. Applying the precautionary principle, the presence of such genes would deem these strains unsuitable for probiotic use, but that would be a mistake, in view of the reported ADR. In addition to a genome sequence, a volunteer exposure study that measures strain-dependent colonization levels and records possible side effects can aid to correctly evaluate absence of virulence.
An alternative explanation for the apparent conflict of presence of virulence genes and simultaneous safe use could be that the factors for which these genes encode may be involved in colonization potential or fitness and not in virulence per se. For pathogens, it is well known that colonization fitness contributes to virulence and vice versa [1, 22]. It is quite possible that similar or even identical factors that increase fitness in pathogens may improve colonization fitness in bacteria with commensal or probiotic properties. The function of “virulence” genes could be a two-sided sword. Their products may have evolved to enhance colonization in a commensal host–microbe relationship, and this is also employed by pathogens, but the evolutionary pressure may not have necessarily applied to a pathogenic relationship.
Acknowledgements
The authors thank Jochen Blom and Alexander Goesmann (Cebittec, University of Bielefeld, Germany) for assistance with genome sequencing, Michael Jarek and Maren Scharfe (Helmholtz Centre for Infection Research, Braunschweig, Germany) and Carolin Büttner (TU Dresden) for genome annotation, and Oksana Lukjancenko (DTU, Lyngby) for analysis of earlier versions of the genome sequences.
Footnotes
Possible conflicts of interest: K.Z. is employed at the research department of SymbioPharm and was involved in conception of the work and writing of the article; SymbioPharm had no influence on the analyses and interpretation or presentation of data. T.M.W. works as a consultant for SymbioPharm. The other authors have no conflict of interest to declare.
Contributor Information
Trudy M. Wassenaar, 1Molecular Microbiology and Genomics Consultants, Zotzenheim, Germany.
Anke Zschüttig, 2Institute of Medical Microbiology and Hygiene, TU Dresden, Dresden, Germany; 7GlaxoSmithKline Biologicals, Dresden, Germany.
Claudia Beimfohr, 3vermicon AG, München, Germany.
Thomas Geske, 4, Berlin, Germany.
Christian Auerbach, 2Institute of Medical Microbiology and Hygiene, TU Dresden, Dresden, Germany.
Helen Cook, 5Center for Biological Sequence Analysis, Department of Systems Biology, Technical University of Denmark, Lyngby, Denmark.
Kurt Zimmermann, 6SymbioPharm GmbH, Herborn, Germany.
Florian Gunzer, 2Institute of Medical Microbiology and Hygiene, TU Dresden, Dresden, Germany.
References
- 1.Wassenaar TM, Gaastra W. Bacterial virulence, where to draw the line? FEMS Microbiol Lett. 2001;201:1–7. doi: 10.1111/j.1574-6968.2001.tb10724.x. [DOI] [PubMed] [Google Scholar]
- 2.Wassenaar TM, Alter T. In: Beneficial microorganisms in agriculture, food and the environment. Sundh I, Goettel M, Wilckes A, editors. Vol. 201. CABI; 2012. Bacterial virulence, where to draw the line? pp. 180–196. [Google Scholar]
- 3.Wassenaar TM, Bohlin J, Binnewies TT, Ussery DW. Genome comparison of bacterial pathogens. Genome Dyn. 2009;6:1–20. doi: 10.1159/000235759. [DOI] [PubMed] [Google Scholar]
- 4.Willenbrock H, Hallin PF, Wassenaar TM, Ussery DW. Characterization of probiotic Escherichia coli isolates with a novel pan-genome microarray. Genome Biol. 2007;8:R267. doi: 10.1186/gb-2007-8-12-r267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sonnenborn U, Schulze J. The non-pathogenic Escherichia coli strain Nissle 1917 — features of a versatile probiotic. Microbial Ecology in Health and Disease. 2009;21:122–158. doi: 10.3109/08910600903444267. [DOI] [Google Scholar]
- 6.Grozdanov L, Raasch C, Schulze J, Sonnenborn U, Gottschalk G, Hacker J, Dobrindt U. Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. J Bacteriol. 2004;186:5432–5441. doi: 10.1128/JB.186.16.5432-5441.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hancock V, Vejborg RM, Klemm P. Functional genomics of probiotic Escherichia coli Nissle 1917 and 83972, and UPEC strain CFT073: comparison of transcriptomes, growth and biofilm formation. Mol Genet Genomics. 2010;284:437–454. doi: 10.1007/s00438-010-0578-8. [DOI] [PubMed] [Google Scholar]
- 8.Lukjancenko O, Wassenaar TM, Ussery DW. Comparison of 61 sequenced Escherichia coli genomes. Microb Ecol. 2010;60:708–720. doi: 10.1007/s00248-010-9717-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zschüttig, A, Auerbach, C, Meltke, S, Eichhorn, C, Brandt, M, Blom, J, Goesmann, A, Jarek, M, Scharfe, M, Zimmermann, K, Wassenaar, TM, Gunzer, F. Complete sequence of probiotic Symbioflor2 E. coli strain G3/10 and draft sequences of Symbioflor2 strains G1/2, G4/9, G5, G6/7 and G8. Genome Announcement. Submitted [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vesth T, Lagesen K, Acar O, Ussery D. CMG-Biotools, a Free Workbench for Basic Comparative Microbial Genomics. PLoS One. 2013;8:e60120. doi: 10.1371/journal.pone.0060120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Snipen L, Ussery DW. Standard operating procedure for computing pangenome trees. Stand Genomic Sci. 2010;2:135–141. doi: 10.4056/sigs.38923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou CE, Smith J, Lam M, Zemla A, Dyer MD, Slezak T. MvirDB-a microbial database of protein toxins, virulence factors and antibiotic resistance genes for bio-defence applications. Nucleic Acids Res. 2007;35(Database issue):D391–D394. doi: 10.1093/nar/gkl791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen LH, Xiong ZH, Sun LL, Yang J, Jin Q. VFDB 2012 update: toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Res. 2012;40(Database issue):D641–D645. doi: 10.1093/nar/gkr989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 15.Elvers KT, Helps CR, Wassenaar TM, Allen VM, Newell DG. Development of a strain-specific molecular method for quantitating individual campylobacter strains in mixed populations. Appl Environ Microbiol. 2008;74:2321–2331. doi: 10.1128/AEM.02269-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zschüttig A, Zimmermann K, Blom J, Goesmann A, Pöhlmann C, Gunzer F. Identification and characterization of microcin S, a new antibacterial peptide produced by probiotic Escherichia coli G3/10. PLoS One. 2012;7:33351. doi: 10.1371/journal.pone.0033351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monteiro C, Saxena I, Wang X, Kader A, Bokranz W, Simm R, Nobles D, Chromek M, Brauner A, Brown R Jr, Römling U. Characterization of cellulose production in Escherichia coli Nissle 1917 and its biological consequences. Environ Microbiol. 2009;11:1105–1116. doi: 10.1111/j.1462-2920.2008.01840.x. [DOI] [PubMed] [Google Scholar]
- 19.Linke D, Riess T, Autenrieth IB, Lupas A, Kempf VA. Trimeric autotransporter adhesins: variable structure, common function. Trends Microbiol. 2006;14:264–270. doi: 10.1016/j.tim.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 20.Al-Hasani K, Henderson IR, Sakellaris H, Rajakumar K, Grant T, Nataro JP, Robins-Browne R, Adler B. The sigA gene which is borne on the she pathogenicity island of Shigella flexneri 2a encodes an exported cytopathic protease involved in intestinal fluid accumulation. Infect Immun. 2000;68:2457–2463. doi: 10.1128/iai.68.5.2457-2463.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bücker R, Schulz E, Günzel D, Bojarski C, Lee IF, John LJ, Wiegand S, Janßen T, Wieler LH, Dobrindt U, Beutin L, Ewers C, Fromm M, Siegmund B, Troeger H, Schulzke JD. a-Haemolysin of Escherichia coli in IBD: a potentiator of inflammatory activity in the colon. Gut. 2014;63:1893–1901. doi: 10.1136/gutjnl-2013-306099. [DOI] [PubMed] [Google Scholar]
- 22.Mahan MJ, Heithoff DM, Sinsheimer RL, Low DA. Assessment of bacterial pathogenesis by analysis of gene expression in the host. Annu Rev Genet. 2000;34:139–164. doi: 10.1146/annurev.genet.34.1.139. [DOI] [PubMed] [Google Scholar]