Summary
We have solved the high-resolution crystal structure of the Drosophila melanogaster alcohol-binding protein LUSH in the complexes it forms with a series of short chain n-alcohols. LUSH is the first known non-enzyme protein with a defined in vivo alcohol-binding function. The structure of LUSH reveals a set of molecular interactions that define a specific alcohol-binding site. A group of amino acids, Thr57, Ser52 and Thr48 form a network of concerted hydrogen bonds between the protein and the alcohol that provide a structural motif to increase alcohol binding affinity at this site. This motif appears to be conserved in a number of mammalian ligand-gated ion channels that are directly implicated in the pharmacological effects of alcohol. Further, these sequences are found in regions of ion-channels that are known to infer alcohol sensitivity. We suggest the alcohol-binding site in LUSH represents a general model for alcohol-binding sites in proteins.
Introduction
The biochemical effects of alcohol are linked to changes in the structure and function of a range of different proteins including N-methyl-d-aspartate (NMDA) type of glutamate receptors, γ–aminobutyric acid (GABA) receptors and glycine receptors (Gly-R)1,2 which are ligand-gated ion channels. The altered function of NMDA and GABAA receptors have been linked to alcohol tolerance, seizures during alcohol withdrawal, the development of alcohol dependency1, and Fetal Alcohol Syndrome3. Recently, evidence has emerged that alcohol-sensitive receptors possess specific sites that make them more sensitive to clinical concentrations of ethanol (reviewed in refs. 4 and 5). Single point mutations in these sites can abolish or dramatically reduce the sensitivity to ethanol6–8 or can change the length of the alcohol that affects receptor function9. It is proposed that alcohols stabilize a conformation of the receptor with altered function that persists until the alcohol is removed (reviewed in refs. 4,5 and 10). These alcohol-binding sites also overlap with binding sites for some inhaled anesthetics including isoflurane, halothane and chloroform6,11–13 however, the binding sites for anesthetics and alcohols involve different subsets of amino acids.
There is little structural information available about the nature of the alcohol-binding sites in alcohol-sensitive ion channels. The major proteins with a native alcohol-binding function are alcohol dehydrogenases, which invariably involve coordination of metal ions and other cofactors to the alcohol to perform an enzymatic reaction14–17. This mode of alcohol binding is unrelated to the non-enzymatic sites that occur in alcohol-sensitive ion-channels. Information from an analysis of protein crystal structures solved using alcohols as co-solvents revealed that H-bonds between the alcohol and protein are formed predominantly with atoms in the protein backbone18. These are non-specific interactions (i.e. no sequence dependence) and the mode of binding at these sites may be an artifact of the high concentrations of alcohol used (7–60% v/v). In contrast, pharmacological effects of alcohols are evident in the range of 5–150 mM (0.02–0.9% v/v). A blood alcohol level of 0.1%, the legal limit for driving in many US states, corresponds to a concentration of ~22 mM. Structural and biophysical studies of anesthetics bound to model proteins have defined the importance of both non-polar and polar groups in the binding sites of these molecules19–23 and binding sites for alcohols will also posses such an amphipathic nature18.
LUSH is an odorant binding protein (OBP) from Drosophila that has a specific alcohol-binding function in vivo. LUSH is one of many OBPs found exclusively in the Drosophila olfactory system, and is expressed in a small subset of the chemosensory hairs that contain the olfactory neurons. OBPs are a family of 14 kDa proteins that are essential components of the insect olfactory system that mediate the neuronal responses to airborne stimuli of pheromones and food odors. The OBP family of proteins includes the Pheromone Binding Proteins (PBP) and the General Odorant Binding Proteins (GOBP). These proteins are secreted into the sensillum lymph surrounding the olfactory neurons where they bind to odorant molecules24. The exact function of OBPs remains unclear but they may act (a) as transporters of hydrophobic odorant molecules that protect the odorant from degradation prior to activation of the receptors, (b) as essential cofactors in activating odorant receptors or (c) to remove odorants from the sensillum lymph. There are other chemosensory proteins implicated in insect olfaction including those typified by CSPMbraA6 from the moth Mamestra brassicae. These proteins share no sequence homology with the OBP family and have completely different structures25,26.
LUSH is required for the normal avoidance response to high, potentially toxic, concentrations of short chain n-alcohols27. Deletion of the lush gene results in a loss of this avoidance response only to ethanol, n-propanol and n-butanol. Wild-type and mutant flies show no difference in their response to methanol, longer n-alcohols and branched-chain alcohols such as iso-propanol. This implies that LUSH has a sterically defined alcohol-binding pocket and a LUSH-alcohol complex may be required to activate the olfactory neurons27,28.
We have solved the crystallographic structure of LUSH in the complexes with ethanol, n-propanol and n-butanol. There is only a single alcohol molecule bound to each protein, indicating the presence of a specific alcohol-binding site. Binding to alcohols occurs through a conserved mechanism, although the solution properties of the three complexes as detected by nuclear magnetic resonance (NMR) differ. Interestingly, the alcohol-binding motif in LUSH is also found in several mammalian alcohol-sensitive receptors and therefore may represent a general mode of “high-affinity” alcohol binding.
Results
Structure of LUSH-alcohol complexes
Data collection and refinement statistics for LUSH in the complexes with ethanol (1.49 Å), n-propanol (1.45 Å) and n-butanol (1.25 Å) at pH 4.6 and the n-butanol complex at pH 6.5 (2.04 Å) are presented in Table 1. The protein conformation is essentially identical in all these structures. The structure of the LUSH-butanol complex is shown (Fig. 1a). The protein structure is similar to PBP from Bombyx mori and the complex it forms with bombykol, a C-16 unsaturated alcohol29 (Fig. 1b). In summary, six α-helices (labeled α1-α6) and a short stretch of 310 helix, labeled α-5′, are arranged around a central hydrophobic cavity of ~114 Å3 in volume (Fig 1a). The alcohol-binding pocket is located at one end of this cavity.
Table 1.
Data Collection, Phasing and Refinement Statistics
| Data Collection and Phasing
| |||||||
|---|---|---|---|---|---|---|---|
| Se-Met | Native | ||||||
| λ1 (peak) | λ2 (inflection) | λ3 (remote) | Ethanol | Propanol | Butanol | pH 6.5 | |
| Resolution (Å) | 2.2 | 2.2 | 2.2 | 1.49 | 1.45 | 1.25 | 2.04 |
| Wavelength (Å) | 0.9785 | 0.9794 | 0.9611 | 0.97895 | 0.97857 | 0.97895 | 1.54180 |
| Completeness 1 (%) | 99.9 (100) | 99.9 (100) | 99.9 (100) | 98.2 (99.5) | 98.7 (98.9) | 96.7 (89.4) | 100.0 (100.0) |
| Rmerge 2 (%) | 7.9 (20.0) | 6.4 (16.8) | 7.1 (20.9) | 3.9 (19.6) | 4.1 (22.0) | 4.3 (17.2) | 5.4 (24.7) |
| I/σ (I) | 40.6 (28.6) | 52.0 (34.2) | 47.6 (27.9) | 25.4 (2.5) | 27.1 (3.2) | 21.8 (2.8) | 57.9 (10.7) |
| Redundancy | 26.8 | 26.9 | 26.7 | 3.1 | 4.2 | 3.1 | 14.9 |
| Total Reflections | 182,801 | 184,029 | 183,311 | 119,487 | 177,608 | 196,015 | 112,114 |
| Unique Reflections | 6,831 | 6,843 | 6,877 | 38,748 | 42,187 | 64,084 | 7,504 |
| Space Group | p43212 | p43212 | p43212 | p43 | p43 | p43 | p61 |
| Cell Dimensions (Å) | a = b = 46.83 | a = b = 46.94 | a = b = 46.98 | a = b = 46.94 | a = b = 55.37 | ||
| c = 111.35 | c = 111.54 | c = 111.46 | c = 111.33 | c = 66.95 | |||
| Figure of Merit | Before density modification | 0.61 | |||||
| After density modification | 0.69 | ||||||
| Refinement
| ||||
|---|---|---|---|---|
| Ethanol | Propanol | Butanol | pH 6.5 | |
| Resolution3 (Å) | 30-1.49 | 30-1.45 | 30-1.25 | 40-2.04 |
| Rcryst (%) | 17.8 (20.3) | 17.4 (19.6) | 16.0 (17.4) | 19.5 (20.9) |
| Rfree4 (%) | 21.0 (23.9) | 19.7 (21.1) | 18.5 (20.2) | 24.7 (27.1) |
| Number of Atoms (Non-Hydrogen) | 2,286 | 2,295 | 2,436 | 1,076 |
| Protein | 2,030 | 2,025 | 2,081 | 986 |
| Ligand | 6 | 8 | 10 | - |
| Water | 234 | 246 | 329 | 90 |
| Acetate ions | 16 | 16 | 16 | - |
| Average B-factor (Å2) | 20.7 | 17.3 | 14.3 | 27.8 |
| Protein | 19.5 | 16.0 | 12.7 | 27.2 |
| Ligand | 37.7 | 40.7 | 32.7 | - |
| Water | 30.6 | 26.7 | 24.0 | 34.2 |
| R.m.s.d. from ideal bond lengths (Å) | 0.016 | 0.023 | 0.011 | 0.006 |
| R.m.s.d. from ideal bond angle (°) | 1.453 | 1.678 | 1.318 | 1.172 |
| Ramachandran statistics5 | ||||
| Most favored | 95.5% | 95.5% | 95.5% | 94.5% |
| Allowed | 4.5% | 4.5% | 4.5% | 5.5% |
Numbers in parenthesis represent values for the highest resolution shell, which are 2.28-2.20 Å (Se-Met), 1.54-1.49 Å (Ethanol), 1.50-1.45 Å (Propanol), 1.29-1.25 Å (Butanol), and 2.11-2.04 Å (pH 6.5).
Rmerge = Σ | I − <I> |/ΣI, where I is the intensity of a given reflection.
Numbers in parenthesis represent values for the highest resolution shell used in refinement, which are 1.571-1.490 Å (Ethanol), 1.528-1.450 Å (Propanol), 1.318-1.250 Å (Butanol), and 2.150-2.040 Å (pH 6.5).
Rfree was calculated with 10% of reflections.
5Ramachandran values were calculated using PROCHECK.
Figure 1. Ribbon diagrams of LUSH and PBP.

(a) Diagram of the LUSH-butanol structure solved to 1.25 Å resolution. Individual helices are labeled α1-α6. α5′ is a stretch of 310-helix. Butanol is represented as a space-filling model at the center of the protein. The alkyl chain is colore in cyan and the hydroxyl group in red. (b) Diagram of the PBP-bombykol complex from B. mori. The elements of secondary structure are colored as for LUSH. Bombykol is shown in a ball-and-stick representation.
Differences between individual structures solved at pH 4.6 occur predominantly at seven surface residues and at two amino acids at the non-crystallographic dimer interface. When these nine residues are omitted from the comparison, the root mean squared deviation (RMSD) from the mean structure for all heavy atoms in the six individual proteins is 0.28 Å, and the RMSD for backbone atoms is 0.04 Å. Except where specific differences are mentioned, the following discussions are valid for all three LUSH-alcohol complexes solved at pH 4.6. Amino acids are numbered from the methionine at position 1, which is the cleavage point of the signal peptide and corresponds to residue 30 in the full-length protein. The protein construct used in these studies contains three additional amino acids at the N-terminus, Gly-Ser-His, from the thrombin protease site. These residues are generally disordered and not discussed in the text.
In the three LUSH-alcohol complexes solved at pH 4.6, the protein forms a non-crystallographic dimer, which buries 1280 Å2 of surface area at the interface. In contrast, the structure of the LUSH-butanol complex solved at pH 6.5 shows no evidence of any dimer in the crystal even though the protein conformation is maintained with a backbone RMSD of 0.55 Å. This latter complex has higher B-factors than structures solved at pH 4.6 (Table 1) and the position of the alcohol is not well defined.
Structure of the Alcohol-Binding Site
Alcohol binds in a small pocket formed by amino acids from helix-3, helix-6 and the C-terminal strand (Fig. 2a). Electron density maps for ethanol and butanol in the binding pocket are shown in Figures 2b and 2c respectively. This is the only observable alcohol-binding site in LUSH; there is no evidence of any other site. This is significant and suggests the chemistry in the observed site is unique and has evolved to bind alcohols with higher affinity and/or specificity than other sites. This is also in contrast to volatile anesthetics that often bind to multiple sites within proteins19. Within the binding pocket, Ser52 and Thr57 form a polar edge to the lip of the pocket with Phe64, Phe113 and Trp123 forming the remaining edge. Non-polar groups from Val106, Thr109 and Ala110 line the bottom of the pocket. The alcohol hydroxyl makes H-bonds with Thr57 and Ser52. H-bonds between alcohol and Thr57 Oγ are significantly shorter (dO-O 2.51 Å, s.d. 0.21 Å) than H-bonds between the Ser52 Oγ and alcohol (dO-O 3.18 Å, s.d. 0.14 Å). This is consistent with Thr57 being a weaker acid than Ser52 and forming a stronger H-bond when it acts as a H-bond donor. If Thr57 forms a H-bond to the alcohol, this reduces the likelihood of the alcohol accepting a second H-bond30 and increases the probability the alcohol acts as a H-bond donor to Ser52. Ser52 is then more likely to form H-bonds to solvent or other groups in the protein. Molecular dynamics simulations (data not shown) provide support for these hydrogen-bonding patterns and suggest the hydroxyl of Ser52 H-bonds to the main-chain carbonyl of Thr48. This H-bond pattern is diagrammed in Figure 2a.
Figure 2. Structure of the LUSH-alcohol binding pocket.
(a) Key residues in the alcohol-binding pocket formed by helix-3 (gold), helix-6 (blue) and the C-terminal strand. The network of hydrogen bonds formed by Thr48, Ser52, Thr57 and ethanol are illustrated by red dotted lines. (b) Stereo view of the electron density of residues in the binding pocket of the LUSH-ethanol complex. (c) The same view of the alcohol-binding pocket in the LUSH-butanol complex.
The conformation of the alcohols’ alkyl chain varies between structures and depends on its length. Ethanol adopts a different orientation in each of the proteins within the dimer. In one molecule, the methyl group points out of the pocket and in the other molecule it points down into the pocket. In the propanol complex, both chains point down into the pocket and contact Phe113, Trp123 and Ala110. If butanol bound in this same orientation it would introduce steric clashes with the ring of Phe113, and so instead, it extends up from the pocket and contacts Leu76, Ala55 and the methyl of Thr57. Binding of propanol may already introduce unfavorable interactions with Phe113 as some conformational heterogeneity of Phe113 is observed in this complex. In contrast, Phe113 is in a single well-defined conformation in the ethanol and butanol complexes (Figs. 2b and c).
Longer alcohols preferentially stabilize the conformation of LUSH
We examined the stability of different LUSH-alcohol complexes in solution using NMR spectroscopy (Fig. 3). In the 1H-15N heteronuclear single quantum correlation (HSQC) spectrum of LUSH, we expect to observe 118 peaks from the backbone amide groups. In the spectrum of the LUSH-ethanol complex (Fig. 3b), we observe 73 sharp peaks; the remainder are either broad or missing, indicating that regions of the protein are in intermediate exchange on the NMR timescale. In contrast, in the spectrum of the LUSH-butanol complex we observe 107 of the expected 118 peaks (Fig. 3a), which represents a dramatic improvement in the overall stability of the protein. When alcohol is removed from these samples the majority of the peaks are now broadened or no longer visible (Fig. 3c). However, a common subset of peaks is observed in all three spectra. We conclude that in the absence of alcohol, core regions of the protein appear to be well structured but other regions of the protein are in conformational exchange. Binding of alcohol stabilizes the protein structure and the degree of stabilization depends on the length of the alcohol chain.
Figure 3. 1H-15N HSQC Spectra of LUSH recorded with and without alcohol.
1H-15N HSQC spectra of (a) LUSH-butanol complex (b) LUSH-ethanol complex and (c) LUSH without alcohol. The alcohol concentration was ~ 40 mM. Without alcohol, a large number of peaks are in intermediate exchange. The presence of ethanol produces a significant improvement in the quality of the spectra but the presence of butanol produces the best appearance of the spectrum.
Discussion
We have solved the three-dimensional structure of a novel alcohol-binding protein, LUSH, bound to a series of short-chain n-alcohols. These results have implications for both odorant recognition in the invertebrate olfactory system and in understanding alcohol-binding sites in alcohol-sensitive proteins in mammalian systems.
Functional Consequences in Odorant Binding Proteins
The structure of LUSH has features in common with both the free31 and bound29 forms of PBP from B. mori (Fig. 1). The major difference between LUSH and PBP is the packing of helix-1 and the C-terminal tail. In LUSH, the C-terminal tail folds into the core and forms part of the alcohol-binding pocket, while helix-1 packs on the outside of the protein (Fig. 1a). In the PBP-bombykol complex, this configuration is reversed: helix-1 packs in the interior of the protein and forms part of the ligand-binding pocket and the C-terminal tail packs on the outside of the protein (Fig. 1b). This would indicate that members of the OBP family have at least two distinct structural isoforms. LUSH and PBP share common structural features with the protein THP-1232, which is a putative carrier of hydrophobic molecules in the beetle Tenebrio molitor. The structure of THP-12 was solved without ligand and under these conditions the N-terminal region, corresponding to α-helix-1in LUSH, is unstructured32.
Conformational changes associated with ligand binding in OBPs and other invertebrate chemosensory proteins have been suggested as a mechanism that allows activation of odorant receptors25,29,32. The conformational changes in the structure of PBP associated with pheromone binding at pH 6.5 are very small31,33. In contrast, binding of alcohol to LUSH produces a dramatic change in conformational stability and this altered conformation may be recognized by odorant receptors, or other proteins in the sensillum lymph. Such conformational changes provide a mechanism to distinguish between the bound and unbound forms of the OBP. A comparison with the structure of THP-12 would suggest that these conformational changes are likely to be associated with α-helix-1. However, preliminary NMR data suggest that changes in LUSH are associated with amino acids in αhelices 3, 4, 5 and 6 that provide the scaffold of the alcohol-binding pocket (unpublished results). The magnitude of these conformational changes is unclear and remains part of ongoing studies.
PBP undergoes a pH dependent conformational change that results in a structure that cannot bind bombykol34. At low pH the C-terminal tail of PBP forms an α-helix that folds into the core of the protein and occupies the ligand-binding site33. This does not appear to be the case for LUSH, which has the same structure in the crystal at pH 4.6 and pH 6.5 and still binds to alcohols at pH 4.6. Therefore, conformational changes in LUSH associated with ligand binding appear to be independent of local pH factors that have been suggested to mediate PBP function33.
Towards a “high-affinity” alcohol binding site
Alcohols are relatively weak hydrogen-bond donors and/or acceptors. Our studies of LUSH point to a protein structural motif that may help to increase the relative binding affinities of such molecules to proteins by the formation of a concerted network of hydrogen bonds. If this were not the case, we would expect alcohols to bind to multiple sites within the protein wherever a small hydrophobic patch is in close proximity to a H-bond acceptor or donor group. This is not the case for LUSH where we observe only a single alcohol-binding site. This motif explains the need for both H-bond donors and H-bond acceptors in this site. H-bonds to the alcohol change the polarization of the hydroxyl and make it a better H-bond donor to other groups in the protein30. This may be particularly important for short chain alcohols such as ethanol, whereas binding of long-chain alcohols may be dominated by non-polar interactions that would require fewer H-bonds to produce the same overall binding affinity as ethanol.
Molecular modeling studies of alcohol-binding sites in the GABAA and glycine receptors have identified a potential site as a hydrophobic pocket formed by amino acids from the second and third transmembrane domains6,9,35–37. The recent structure of the transmembrane domains of the Torpedo n-acetylcholine receptor (nAchR) supports this model38 and suggests the alcohol-binding site is in a pocket between the transmembrane helices. Interestingly the alcohol-binding motif in LUSH also appears to occur in GABAA-R, Gly-R, nAchR and in the alcohol-sensitive region of the Drosophila Shaw voltage sensitive potassium channel (Fig. 4), which is sensitive to ethanol concentrations of 17–170 mM39. Other members of this family of potassium channels, which have a leucine or glycine in place of the final threonine, are not sensitive to these alcohol concentrations39.
Figure 4. Comparison of the alcohol-binding site in LUSH with regions of alcohol-sensitive ion channels.
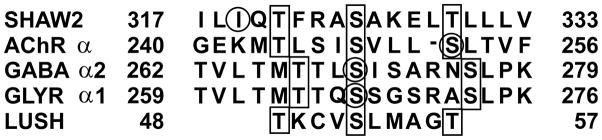
Residues that bind alcohol in LUSH and which are potentially conserved in alcohol-sensitive ion-channels are outlined in the black boxes. Residues in the ion channels that have been shown to affect alcohol sensitivity by site-directed mutagenesis are circled.
In Figure 4, the sequence alignment with LUSH is not absolutely conserved and we do not expect sequence alignments to correctly identify all alcohol-binding sites. It is more likely that conformational proximity of Serine and Threonine residues from distal regions of a protein will provide the appropriate structural motif to form a “high-affinity” alcohol-binding site. Inspection of the nAChR transmembrane domains reveals a number of sites where a pair of Serine or Threonine residues may be able to form concerted hydrogen bonds with alcohols in the manner observed in LUSH. Several of these potential sites are in close proximity to Leu257, which is equivalent to Ser270 in GABA-R and Ser267of Gly-R. However, in contrast to the effects seen with the GABA-R and Gly-R, mutation of this Leucine in the rat nAChR-α2 subunit (Leu261) does not eliminate ethanol sensitivity, but rather increases potentiation to acetylcholine40. This led to the conclusion that this Leucine is not part of an alcohol-binding site but rather is in proximity to an alcohol-binding site40. Therefore, while our studies do not fully explain the properties of all potential alcohol-binding sites in membrane-associated protein, the structure of LUSH may provide insight into the structural requirements of specific alcohol-binding sites that appear to be common to several alcohol-sensitive ion channels, including those with direct medical importance.
METHODS
Protein expression and purification
LUSH cDNA (accession # AF001621) was PCR amplified and subcloned into the NdeI/BamH1 site of pET28a (Novagen). The primers used were 5′-GACCTTTGGCATATGACCATGGAACAGTTTCTGACCTCTCTGGACATGATCCGCAGTGGC-3′ and 5′-TTAGCGCGGATCCTATTAAGGCCACATGAACTGCCCATC-3′ with the NdeI and BamH1 sites underlined, respectively.
LUSH was prepared by overexpression in E. coli strain BL21 (DE3) (Novagen). Cells were resuspended and lysed in 20 mM Tris pH 7.4, 25% sucrose, 2 mM MgCl2, and 5 mg lysozyme. After addition of an equal volume of 20 mM Tris, pH 7.5, 4 mM EDTA, 0.2M NaCl, 1% deoxycholic acid, 1% NP-40 the solution was sonicated (3x 30 s) on ice, made up to 10 mM MgCl2 and incubated with 1 mg of DNaseI (Sigma). After centrifugation, the pellet was washed twice with 20 ml 0.5% Triton X-100, 1 mM EDTA. The protein was solubilized in 5 M guanidine hydrochloride, 5 mM dithio-threitol and refolded using a cysteine/cystine redox reaction34 in the presence of 1% v/v of the n-alcohol. Soluble protein was dialyzed against 20 mM Na2HPO4/NaH2PO4 pH 6.5, 0.3% v/v alcohol and concentrated before purification by Ni2+-NTA (QIAGEN) affinity chromatography. The hexa-histidine tag was removed using the Thrombin Cleavage Capture Kit (Novagen) and the identity of the purified protein was confirmed by MALDI-TOF mass-spectrometry (m/z+ 14472)
Crystallization and data collection
LUSH crystals were grown in sodium acetate 100 mM, pH 4.0, 25–30% PEG 4000, and 0.3% alcohol at either 4 °C or 18 °C and flash frozen in liquid nitrogen. Selenomethionyl-substituted (Se-Met) LUSH was prepared by expression in E. coli strain B834 (Novagen) grown in M9 minimal media containing 100 mg L−1 Se-Met (Sigma). Multiwavelength anamolous diffraction (MAD) data was collected at three wavelengths on beamline X12C at the National Synchrotron Light Source at Brookhaven National Laboratory through the “Fed-Ex” data collection service. Data were processed using DENZO and SCALEPACK (HKL Research, Inc)41. Native data for LUSH-alcohol complexes were collected at the Advanced Photon Source at Argonne National Laboratory on Beamline SBC 19BM and processed using HKL2000.
Crystals of the LUSH-butanol complex at pH 6.5 were grown using a protein solution in 100 mM Tris pH 8.0, 0.3% n-butanol against a reservoir solution of 100 mM MOPS pH 6.5, 30% PEG 4000, 0.3% n-butanol. A complete data set for this structure was collected at UCHSC using a Rigaku Raxis-IV++ area detector and processed with DENZO and SCALEPACK.
Structure Determination and Refinement
Initial phases for the MAD data set were calculated using the program SOLVE42 which identified four of the possible nine selenium sites. After solvent flattening with RESOLVE42 models were built in O43 and the structure refined to 2.2 Å. The native data sets were used to refine the structures in the p43 space group using the programs Refmac44 and CNS45. Hydrogens were added in all three structures and anisotropic temperature factors were used for refinement of the n-butanol data. Waters, four acetate molecules and two molecules of ethanol, n-propanol, and n-butanol were added per dimer in each of the respective structures. The structure at pH 6.5 was solved using molecular replacement in CCP444. The stereochemistry of all protein models was analyzed with PROCHECK46, CNS, and Refmac.
NMR Spectroscopy
15N-labeled LUSH was prepared by cell growth in minimal media containing 15N-labeled (99 atom %) ammonium chloride. Samples for NMR were made in 90% H2O/10% D2O containing 20 mM Na2HPO4/NaH2PO4 pH 6.5 and 30 mM alcohol as described for X-ray crystallography. The protein concentration was 400–600 μM. Samples without alcohol were prepared by exhaustive dialysis into the same buffer but without the alcohol. The concentration of these samples was 100 μM. 1H-15N HSQC spectra47 were collected on a Varian Inova 500 MHz spectrometer at UCHSC.
All structural models were made using Molscript48 and electron density figures were constructed using Bobscript49 and rendered using Raster3D50.
Coordinates
Protein structures have been deposited in the Protein Data Bank with accession codes 1OOF, 1OOG, 1OOH, and 1OOI.
Acknowledgments
We thank Dr. Mair Churchill for her invaluable advice with crystallography studies and Dr. Howard Robinson for MAD data collection at BNL. X-ray crystallography facilities and NMR spectrometers were purchased with funds provided by Howard Hughes Medical Institute and their operation is supported by the University of Colorado Cancer Center Core Grant. SWK is supported by an American Heart Association Pre-doctoral Fellowship. This work is supported by an American Heart Association Development Grant and NIH/NIAAA RO1 to DNMJ.
References
- 1.Faingold CL, N’Gouemo P, Riaz A. Ethanol and neurotransmitter interactions - From molecular to integrative effects. Prog Neurobiol. 1998;55:509–535. doi: 10.1016/s0301-0082(98)00027-6. [DOI] [PubMed] [Google Scholar]
- 2.Mascia MP, Machu TK, Harris RA. Enhancement of homomeric glycine receptor function by long-chain alcohols and anaesthetics. Br J Pharmacol. 1996;119:1331–1336. doi: 10.1111/j.1476-5381.1996.tb16042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ikonomidou C, et al. Ethanol-induced apoptotic neurodegeneration and fetal alcohol syndrome. Science. 2000;287:1056–1060. doi: 10.1126/science.287.5455.1056. [DOI] [PubMed] [Google Scholar]
- 4.Lovinger DM. Alcohols and neurotransmitter gated ion channels: Past, present and future. Naunyn-Schmiedebergs Arch Pharmacol. 1997;356:267–282. doi: 10.1007/pl00005051. [DOI] [PubMed] [Google Scholar]
- 5.Harris RA. Ethanol actions on multiple ion channels: Which are important? Alcoholism (NY) 1999;23:1563–1570. [PubMed] [Google Scholar]
- 6.Mihic SJ, et al. Sites of alcohol and volatile anesthetic action on GABA(A) and glycine receptors. Nature. 1997;389:385–389. doi: 10.1038/38738. [DOI] [PubMed] [Google Scholar]
- 7.Ye Q, et al. Enhancement of glycine receptor function by ethanol is inversely correlated with molecular volume at position alpha 267. J Biol Chem. 1998;273:3314–3319. doi: 10.1074/jbc.273.6.3314. [DOI] [PubMed] [Google Scholar]
- 8.Ueno S, Wick MJ, Ye Q, Harrison NL, Harris RA. Subunit mutations affect ethanol actions on GABA(A) receptors expressed in Xenopus oocytes. Br J Pharmacol. 1999;127:377–382. doi: 10.1038/sj.bjp.0702563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wick M, et al. Mutations of gamma-aminobutyric acid and glycine receptors change alcohol cutoff: Evidence for an alcohol receptor? Proc Natl Acad Sci, USA. 1998;95:6504–6509. doi: 10.1073/pnas.95.11.6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woodward JJ. Ionotropic glutamate receptors as sites of action for ethanol in the brain. Neurochem Int. 1999;35:107–113. [PubMed] [Google Scholar]
- 11.Krasowski MD, et al. Propofol and other intravenous anesthetics have sites of action on the gamma-aminobutyric acid type a receptor distinct from that for isoflurane. Mol Pharmacol. 1998;53:530–538. doi: 10.1124/mol.53.3.530. [DOI] [PubMed] [Google Scholar]
- 12.Kash TL, Jenkins A, Harrison NL. Molecular volume determines the activity of the halogenated alkane bromoform at wild-type and mutant GABA(A) receptors. Brain Res. 2003;960:36–41. doi: 10.1016/s0006-8993(02)03748-4. [DOI] [PubMed] [Google Scholar]
- 13.Jenkins A, et al. Evidence for a common binding cavity for three general anesthetics within the GABA(A) receptor. J Neurosci. 2001;21:U7–U10. doi: 10.1523/JNEUROSCI.21-06-j0002.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li CM, Heatwole J, Soelaiman S, Shoham M. Crystal structure of a thermophilic alcohol dehydrogenase substrate complex suggests determinants of substrate specificity and thermostability. Proteins. 1999;37:619–627. doi: 10.1002/(sici)1097-0134(19991201)37:4<619::aid-prot12>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 15.Benach J, Atrian S, Gonzalez-Duarte R, Ladenstein R. The refined crystal structure of Drosophila lebanonensis alcohol dehydrogenase at 1.9 angstrom resolution. J Mol Biol. 1998;282:383–399. doi: 10.1006/jmbi.1998.2015. [DOI] [PubMed] [Google Scholar]
- 16.Xie PG, Parsons SH, Speckhard DC, Bosron WF, Hurley TD. X-ray structure of human class IV sigma sigma alcohol dehydrogenase - Structural basis for substrate specificity. J Biol Chem. 1997;272:18558–18563. doi: 10.1074/jbc.272.30.18558. [DOI] [PubMed] [Google Scholar]
- 17.Niederhut MS, Gibbons BJ, Perez-Miller S, Hurley TD. Three-dimensional structures of the three human class I alcohol dehydrogenases. Protein Sci. 2001;10:697–706. doi: 10.1110/ps.45001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dwyer DS, Bradley RJ. Chemical properties of alcohols and their protein binding sites. Cell Mol Life Sci. 2000;57:265–275. doi: 10.1007/PL00000689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eckenhoff RG, Johansson JS. Molecular interactions between inhaled anesthetics and proteins. Pharmacol Rev. 1997;49:343–367. [PubMed] [Google Scholar]
- 20.Franks NP, Jenkins A, Conti E, Lieb WR, Brick P. Structural basis for the inhibition of firefly luciferase by a general anesthetic. Biophys J. 1998;75:2205–2211. doi: 10.1016/S0006-3495(98)77664-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishizawa Y, Pidikiti R, Liebman PA, Eckenhoff RG. G protein-coupled receptors as direct targets of inhaled anesthetics. Mol Pharmacol. 2002;61:945–952. doi: 10.1124/mol.61.5.945. [DOI] [PubMed] [Google Scholar]
- 22.Johansson JS, Scharf D, Davies LA, Reddy KS, Eckenhoff RG. A designed four-alpha-helix bundle that binds the volatile general anesthetic halothane with high affinity. Biophys J. 2000;78:982–993. doi: 10.1016/S0006-3495(00)76656-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sachsenheimer W, Pai EF, Schulz GE, Schirmer RH. Halothane Binds in Adenine-Specific Niche of Crystalline Adenylate Kinase. FEBS Lett. 1977;79:310–312. doi: 10.1016/0014-5793(77)80809-0. [DOI] [PubMed] [Google Scholar]
- 24.Vogt RG, Riddiford LM. Pheromone binding and inactivation by moth antennae. Nature. 1981;293:161–163. doi: 10.1038/293161a0. [DOI] [PubMed] [Google Scholar]
- 25.Campanacci V, et al. Moth chernosensory protein exhibits drastic conformational changes and cooperativity on ligand binding. Proc Natl Acad Sci U S A. 2003;100:5069–5074. doi: 10.1073/pnas.0836654100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mosbah A, et al. Solution structure of a chemosensory protein from the moth Mamestra brassicae. Biochem J. 2003;369:39–44. doi: 10.1042/BJ20021217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim MS, Repp A, Smith DP. LUSH Odorant-binding protein mediates chemosensory responses to alcohols in Drosophila melanogaster. Genetics. 1998;150:711–721. doi: 10.1093/genetics/150.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du G, Prestwich GD. Protein structure encodes the ligand binding specificity in pheromone binding proteins. Biochemistry. 1995;34:8726–8732. doi: 10.1021/bi00027a023. [DOI] [PubMed] [Google Scholar]
- 29.Sandler BH, Nikonova L, Leal WS, Clardy J. Sexual attraction in the silkworm moth: structure of the pheromone-binding-protein-bombykol complex. Chem Biol. 2000;7:143–151. doi: 10.1016/s1074-5521(00)00078-8. [DOI] [PubMed] [Google Scholar]
- 30.Laurence C, Berthelot M. Observations on the strength of hydrogen bonding. Perspect Drug Discov Design. 2000;18:39–60. [Google Scholar]
- 31.Lee D, et al. NMR structure of the unliganded Bombyx mori pheromone-binding protein at physiological pH. FEBS Lett. 2002;531:314–318. doi: 10.1016/s0014-5793(02)03548-2. [DOI] [PubMed] [Google Scholar]
- 32.Rothemund S, Liou YC, Davies PL, Krause E, Sonnichsen FD. A new class of hexahelical insect proteins revealed as putative carriers of small hydrophobic ligands. Struct Fold Des. 1999;7:1325–1332. doi: 10.1016/s0969-2126(00)80022-2. [DOI] [PubMed] [Google Scholar]
- 33.Horst R, et al. NMR structure reveals intramolecular regulation mechanism for pheromone binding and release. Proc Natl Acad Sci U S A. 2001;98:14374–14379. doi: 10.1073/pnas.251532998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prestwich GD. Bacterial expression and photoaffinity labeling of a pheromone binding protein. Prot Sci. 1993;2:420–428. doi: 10.1002/pro.5560020314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mascia MP, Trudell JR, Harris RA. Specific binding sites for alcohols and anesthetics on ligand-gated ion channels. Proc Natl Acad Sci U S A. 2000;97:9305–9310. doi: 10.1073/pnas.160128797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamakura T, Bertaccini E, Trudell JR, Harris RA. Anesthetics and ion channels: Molecular models and sites of action. Annu Rev Pharmacol Toxicol. 2001;41:23–51. doi: 10.1146/annurev.pharmtox.41.1.23. [DOI] [PubMed] [Google Scholar]
- 37.Bertaccini E, Trudell JR. Molecular modeling of ligand-gated ion channels: Progress and challenges. Int Rev Neurobiol. 2001;48:141–166. doi: 10.1016/s0074-7742(01)48015-9. [DOI] [PubMed] [Google Scholar]
- 38.Miyazawa A, Fujiyoshi Y, Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 2003;423:949–955. doi: 10.1038/nature01748. [DOI] [PubMed] [Google Scholar]
- 39.Covarrubias M, Vyas TB, Escobar L, Wei A. Alcohols Inhibit a Cloned Potassium Channel At a Discrete Saturable Site - Insights Into the Molecular-Basis of General-Anesthesia. J Biol Chem. 1995;270:19408–19416. doi: 10.1074/jbc.270.33.19408. [DOI] [PubMed] [Google Scholar]
- 40.Borghese CM, Ali DN, Bleck V, Harris RA. Acetylcholine and alcohol sensitivity of neuronal nicotinic acetylcholine receptors: Mutations in transmembrane domains. Alcoholism (NY) 2002;26:1764–1772. doi: 10.1097/01.ALC.0000042012.58231.D9. [DOI] [PubMed] [Google Scholar]
- 41.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meths Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 42.Terwilliger TC, Berendzen J. Automated structure solution for MIR and MAD. Acta Crystallogr. 1999;D55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones TA, Zou JY, Cowan SW, Kjelgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 44.Bailey S. The CCP4 Suite - Programs for Protein Crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 45.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 46.Laskowski RA. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 47.Bodenhausen G, Ruben DJ. Natural Abundance N-15 NMR By Enhanced Heteronuclear Spectroscopy. Chem Phys Lett. 1980;69:185–189. [Google Scholar]
- 48.Kraulis PJ. Molscript - a Program to Produce Both Detailed and Schematic Plots of Protein Structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 49.Esnouf RM. Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. 1999;D55:938–940. doi: 10.1107/s0907444998017363. [DOI] [PubMed] [Google Scholar]
- 50.Merritt EA, Bacon DJ. Raster3D: Photorealistic molecular graphics. Meths Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]




