Abstract
For nearly three decades, the sequence of the human mitochondrial genome (mtDNA) has provided a molecular framework for understanding maternally inherited diseases. However, the vast majority of human mitochondrial disorders are caused by nuclear defects, which is not surprising since the mtDNA encodes only 13 proteins. Advances in genomics, mass spectrometry, and computation have only recently made it possible to systematically identify the complement of over 1,000 proteins that comprise the mammalian mitochondrial proteome. Here, we review recent progress in characterizing the mitochondrial proteome and highlight insights into its complexity, tissue heterogeneity, evolutionary origins, and biochemical versatility. We then discuss how this proteome is being used to discover the genetic basis of respiratory chain disorders as well as to expand our definition of mitochondrial disease. Finally, we explore future prospects and challenges for using the mitochondrial proteome as a foundation for systems analysis of the organelle.
Keywords: evolution, mitochondria, organelle, respiratory chain disease, tissue diversity, oxidative phosphorylation
INTRODUCTION
The sequencing of the human mitochondrial genome (mtDNA) was an important milestone in the history of mitochondrial biology and medicine. The resulting landmark paper, published in 1981, was a technical tour de force that reported the complete sequence of this 16,569 base-pair, circular molecule (3). Companion articles in the same issue of Nature annotated its RNA and protein products (71, 81). The mtDNA genome and annotations enabled wide-ranging investigations of the organelle's evolutionary origin and biochemical function as well as explorations into human history and migration. This work also launched the race to identify the first mtDNA mutations that cause maternally inherited disorders, which were first reported in 1988 (44, 116, 117). Since then, over 150 mutations have been associated with maternally inherited syndromes (91), and ongoing work aims to distinguish pathogenic from benign variants (65, 69, 70). In addition, the mtDNA has been sequenced extensively across populations to determine its natural variation (48).
A key, early insight from the mtDNA sequence was that its limited coding capacity could account for only a small fraction of the organelle's proteome. The 13 proteins encoded by mammalian mtDNA are all components of the respiratory chain, which generates the majority of cellular ATP via oxidative phosphorylation (OXPHOS). However, the remaining ~77 respiratory chain subunits are encoded by nuclear genes, as are all proteins required for the transcription, translation, modification, and assembly of the 13 mtDNA proteins (28). All the components of numerous other mitochondrial pathways are also nuclear encoded, including the tricarboxylic acid (TCA) cycle, protein import, fatty acid and amino acid oxidation, apoptosis, and biosynthesis of ketone bodies, pyrimidines, heme, and urea. Furthermore, during the decades following the sequencing of the mtDNA, it became clear that maternally inherited mitochondrial disorders represent only ~15--20% of all inherited human mitochondrial disorders (26).
By the late 1990s, there was a growing appreciation for the role of the nuclear genome in shaping the mitochondrion in normal and disease states, yet it was not clear just how many nuclear-encoded proteins were targeted to this organelle. Quantitative two dimensional (2D) gels of highly purified mitochondria revealed approximately ~1500 distinct spots (61), providing one estimate of the size of the mitochondrial proteome. Interestingly, the genomes of alpha-proteobacteria, which are the closest living relatives of modern day mitochondria, encode a total of ~1000 distinct protein products (4). Together, such estimates suggest that the mammalian mitochondrial proteome consists of ~1000--1500 distinct proteins.
After the human genome was sequenced in 2001, ambitious campaigns were launched to systematically catalog the mammalian mitochondrial proteome. At that time, only ~350 distinct, bona fide human mitochondrial proteins were known. With the availability of well-annotated genome sequences, combined with key advances in tandem mass spectrometry and computation, it was possible to more comprehensively define this organelle's proteome. These advances have fueled progress in understanding the genetics of human mitochondrial disease. Here, we review different approaches to characterize the protein inventory of mammalian mitochondria and then discuss some remarkable biological and medical insights that have been gained. Although characterization of the mitochondrial proteome represents an extraordinarily important milestone for mitochondrial biology, really it is only the first step towards a systematic understanding of the organelle in health and disease.
DEFINING THE MITOCHONDRIAL PROTEOME
The mitochondrial proteome refers to the subset of the ~20,000 distinct mammalian proteins (20) that are primarily localized to this cellular compartment. Defining this inventory is challenging because no single targeting sequence directs import into the organelle, some proteins are of very low abundance, and other components are likely expressed only in specific developmental stages or cell types. Consequently, no single method can presently identify all mitochondrial proteins. Here we review the most fruitful strategies to date.
Targeting Signal
A purely computational strategy for identifying mitochondrial proteins is to search for their targeting sequences. There are at least five different mechanisms controlling import into the mitochondria, as described in a recent review (17). The primary pathway directs proteins to the matrix based on a cleavable peptide presequence, typically 15--50 residues long, that contains an α-helix with positively charged residues on one side and uncharged and hydrophobic residues on the other. Some of these proteins are subsequently sorted to the inner membrane and intermembrane space based on consecutive peptide sequences. A second mechanism imports outer membrane proteins via a carboxy-terminal signal. A third mechanism imports hydrophobic inner membrane proteins in a loop conformation, using a series of noncontiguous internal targeting signals (119). Fourth, some small cysteine-rich intermembrane space proteins (e.g., Tim9 and Tim10 in yeast) are imported using an internal targeting signal (8, 68). Lastly, many proteins are simply tail-anchored in the outer membrane via α-helices (17).
Many algorithms have been developed to computationally predict the cleavable, presequence targeting signal. These include TargetP (31), pTARGET (41), PSORT (77), iPSORT (9), Predotar (96), ngLoc (52), MitPred (59), MitoPred (42), and MitoProt (21), as described in a recent comparative review (40). The major limitations of these tools are that they give many false positive predictions when applied to the entire proteome, and also they cannot detect all mitochondrial proteins, as the presequence is only one of several import mechanisms. For example, the popular TargetP program detects signals in 60% of proteins with experimental evidence of mitochondrial localization, however, an estimated 69% of its predictions are false; at its most stringent setting, TargetP has a 3% false-positive rate but detects targeting signals in only 20% of mitochondrial proteins (83). Hence, due to limited sensitivity and specificity, targeting sequence prediction cannot be used in isolation to define the mitochondrial proteome.
Mass Spectrometry Based Proteomics
To date, the most successful experimental strategy for defining the mitochondrial proteome has been the application of mass spectrometry (MS) to identify proteins from highly enriched mitochondrial extracts. The ostensibly straightforward task of separating and identifying mitochondrial proteins is technically challenging given their number and dynamic range. Pioneering studies in 1998 and 2001 initially separated mitochondrial proteins by 2D gel and used peptide mass fingerprinting or MS to identify 46 proteins in human placental mitochondria (88) and 80 mitochondrial proteins from a human neuroblastoma cell line (93). Advances in separation and tandem mass spectrometry (MS/MS) techniques enabled more sensitive detection, including 615 mitochondrial proteins from human heart (103) and 399 from mouse brain, heart, kidney, and liver (72); however, careful analyses revealed that these studies detected only abundant molecules (72). Newer-generation MS/MS technology is far more sensitive and enables detection of proteins across six orders of magnitude of abundance. For example, Kislinger and colleagues identified 2533 proteins in mitochondrial extracts from mouse brain, heart, kidney, liver, lung, and placenta as part of a large study on organellar proteomics (54). A series of additional studies have identified 1130 mitochondrial proteins from adipocyte 3T3-L1 cells (1); 1162 from rat brain, liver, heart, and kidney (51); 689 proteins from rat muscle, heart and liver (34); and 297 from mouse liver (36). Pagliarini et al. recently analyzed mitochondrial extracts from 14 diverse mouse tissues and detected a total of 3881 proteins (83).
It is important to note that not all proteins detected in such mitochondrial extracts actually represent genuine mitochondrial-localized proteins. Due to the high sensitivity of these newer generation instruments, many of these detected proteins are copurifying contaminants, which represent up to 75% of proteins detected by the most sensitive MS/MS experiments. One rigorous experimental approach to distinguish contaminants is protein correlation profiling (PCP), which first separates organelles using a sucrose gradient and then compares profiles of peptides across gradient fractions to profiles of marker proteins for each organelle. Foster et al. used PCP to assign 1404 proteins to distinct organelles, including 297 mitochondrial proteins (36). In an alternate computational approach, Kislinger et al. compared MS/MS abundance measurements of each protein across four cellular compartments to confidently assign mitochondrial localization to 334 of the 2533 detected proteins (54). Pagliarini et al. recently developed a subtractive proteomics experimental approach whereby both crude and highly purified mitochondria were analyzed by MS/MS, and proteins with greater abundance in purified mitochondria received higher scores of mitochondrial localization (83).
Microscopy
A complementary experimental approach relies on microscopy to establish mitochondrial localization. This can be accomplished by immunofluorescence of native proteins using antibodies, tagging endogenous proteins with epitopes such as GFP, or transfecting cells with exogenous tagged genes. Unfortunately, the availability of high-quality antibodies limits the first approach. Tagging of endogenous proteins, which has been applied successfully to the yeast proteome (46, 57), has been extended to mouse but is not yet a high-throughput method (82). Large-scale efforts to visualize tagged exogenous proteins in mammals include the ongoing lifeDB project (http://www.lifedb.de) (66) and the completed MitoCarta project (http://www.broadinstitute.org/pubs/MitoCarta) (83). Currently, there are a total of 321 human proteins and 166 mouse mitochondrial proteins evidenced through microscopy, according to the MitoMiner database (97). Although microscopy provides excellent proof of subcellular localization, this technique is hampered in mammals by the time-consuming process, the inability to tag all relevant splice forms, the lack of necessary chaperones/modifiers in cellular models, the potential interference of the epitope tag with the import machinery, and the resolution limit of light microscopy.
Other Methods
Additional clues of mitochondrial localization can be garnered from other experimental and computational genomic methods. Sequence homology is extremely helpful in extrapolating subcellular localization data from model organisms to humans. One of the best methods for prediction of human mitochondrial proteins relies on the presence of protein domains that are found exclusively in eukaryotic SwissProt proteins known to localize to mitochondria (42, 83). In yeast, experimental genetic approaches have successfully detected hundreds of mitochondrial proteins through the observation of a petit phenotype or reduced growth when the yeast deletion strain is grown on nonfermentable substrates (25, 29, 99). Additional prediction methods include guilt-by-association of profiles of mRNA transcript levels across diverse tissues (15, 72, 73, 79, 83), detection of mRNA transcripts that are specifically induced during mitochondrial biogenesis (75), and presence of mitochondrial-specific cis-regulatory motifs in gene promoters (15, 73).
Integrative Approaches
While the individual approaches all provide useful clues to mitochondrial localization, ensemble methods that combine various techniques have yielded the most comprehensive and accurate mitochondrial protein inventories. These include ad hoc approaches, such as MitoPred (42), and supervised machine learning techniques such as linear classification in yeast (87) and Bayesian classification in human (15). The most recent and comprehensive mitochondrial inventory combines literature curation, large-scale green fluorescent protein (GFP)-tagging/microscopy, and a Bayesian integration of seven additional data sources: subtractive proteomics of mitochondria from 14 mouse tissues, presence of mitochondrial-specific protein domains, mRNA coexpression, targeting signal prediction, transcriptional induction during mitochondrial proliferation, homology to yeast mitochondrial proteins, and protein homology to Rickettsia, the relative of the mitochondrion's bacterial ancestor. This catalog, named MitoCarta (http://www.broadinstitute.org/pubs/MitoCarta/), contains 1098 distinct gene loci with an estimated 10% false-positives (83).
Collectively, the complementary techniques described here have laid the foundation for defining the inventory of mitochondrial proteins. Several online databases serve up these datasets (Table 1).
Table 1.
Online databases of mitochondrial proteins
| Resource | Species | Database description |
|---|---|---|
| MitoP2 (30) | Human, mouse, Arabidopsis thaliana, yeast, Neurospora | Integrates target signal prediction, homology, MS/MS studies, mutant screening, expression profiling, protein--protein interaction and cellular sublocalization Pros: mitochondrial reference set; localization scores for all proteins; regularly updated; good search capabilities Cons: no Entrez or RefSeq integration URL: ihg.gsf.de/mitop2 |
| MitoCarta (83) | Human, mouse | Integrates literature curation, GFP-tagging/microscopy, MS/MS proteomics across 14 tissues, targeting signal, yeast homology, coexpression, protein domains Pros: mitochondrial reference set; localization scores for all proteins Cons: not updated regularly; no SwissProt or Ensembl integration URL: www.broadinstitute.org/pubs/MitoCarta |
| MitoMiner (97) | Human,mouse, rat, cow, fruitfly, yeast, Plasmodium falciparum | Integrates HomoloGene, Gene Ontology, MS/MS data, GFP data, KEGG, OMIM Pros: sophisticated searches; user-defined lists and workspaces; GFP images; updated regularly; useful lists of functionally related genes Cons: no single mitochondrial reference set; complex user interface; difficult to view all evidence related to a single gene URL: mitominer.mrc-mbu.cam.ac.uk |
| Mito Proteome (22) | Human | Integrates literature curation, MS/MS studies, Entrez, KEGG, OMIM, MINT, DIP, PFAM, InterPro, PRINTS Pros: mitochondrial reference set; search capabilities Cons: incomplete mitochondrial gene set URL: http://www.mitoproteome.org |
| HMPDb | Human | Integrates Protein Data Bank (PDB), Entrez, OMIM, mtDB, MitoMap, Neuromuscular disease center, Human 2D PAGE databases, sublocalization Pros: detailed protein data and images, comparisons of mtDNA sequences and polymorphisms, clinical symptom search Cons: no evidence provided for mitochondrial reference set URL: http://bioinfo.nist.gov |
| Mito Phenome (92) | Human | Integrates genes, clinical features and disease diagnoses (from the literature), OMIM, literature citations, Entrez, Ensembl, SwissProt, and subcellular localization Pros: only database containing phenotype information Cons: incomplete mitochondrial gene set; not updated regularly URL: http://mitophenome.org |
Abbreviations: Mito, mitochondrial; HMPDb, Human Mitochondrial Protein Database; MS/MS, tandem mass spectrometry; GFP, green fluorescent protein; Kyoto Encyclopedia of Genes and Genomes, KEGG; Online Mendelian Inheritance in Man, OMIM; Molecular Interactions Database, MINT; Database of Interacting Proteins (DIP); Protein families, PFAM; Human Mitochondrial Genome Database, mtDB; Note that HMPDb, KEGG OMIM, MINT, DIP, PFAM, InterPro, PRINTS, mtDB, Entrez, RefSeq, SwissProt, and Ensembl are databases or datasets available online.
PROPERTIES OF THE MITOCHONDRIAL PROTEOME
The near-comprehensive inventory of mitochondrial proteins has enabled systems-level investigation of the complexity of this organelle and its diverse roles in the cell. Below, we discuss some of the interesting insights that have emerged on the size and complexity of the mitochondrial proteome and on the extent of dual localization, tissue diversity, biochemical capacities, and evolutionary conservation.
Size and Dynamic Range
The most complete mitochondrial catalog contains ~1100 gene loci that encode mitochondrial proteins (83). This MitoCarta catalog is estimated to be 85% complete, based on a Bayesian probabilistic model that incorporates training data and a prior probability that 7% of all mammalian genes are mitochondrial. Allowing the prior probability to range between 5% and 12%, the Bayesian model predicts that 1050--1400 genes encode mitochondrial proteins. Of course, each gene locus can encode multiple splice forms, and these proteins may have multiple posttranslational modifications; thus, the repertoire of unique protein forms may exceed the number of gene loci by up to tenfold. At present our best estimates suggest that a total of 1100--1400 distinct gene loci encode mitochondrial proteins.
Mitochondrial proteins span a broad dynamic range of abundance. Mitochondrial proteins that span five or six orders of magnitude of abundance have been previously reported (36, 54). Based on 2D gels, the two most abundant inner and outer membrane proteins are the adenine nucleotide translocator (ANT1) and voltage-dependent anion-selective channel (VDAC), respectively (112). Recent MS/MS analysis supports the high abundance of these proteins. Based on rough estimates of protein abundance across 14 mouse tissues (83), the five most abundant mitochondrial proteins are ATP5A1, ATP5B, ACO2, ANT1, and ANT2; VDAC1 ranks 33rd in abundance.
Dual Localization
It has long been known that proteins can be dual localized to serve similar functions in different compartments with distinct regulatory properties. This redundancy allows the ~20,000 mammalian gene loci (20) to encode proteins that are exquisitely regulated both spatially and temporally. Although gene duplications can create paralogous proteins that may have specific locations (e.g., HMGCS1 and HMGCS2), dual localization typically refers to the protein products from a single gene locus that are present in multiple compartments. This dual localization can be accomplished through several mechanisms, including alternative splicing or alternative start sites to produce two distinct proteins, one of which contains a mitochondrial targeting sequence [e.g., ISCU (107) and LRPPRC (74)], or proteins that change cellular locations upon stimulus, such as the proapoptotic protein BID that translocates from the cytosol to the mitochondrion following a death stimulus (62).
Several large-scale studies have systematically investigated dual localization of cellular proteins. Foster et al. localized 1404 mouse proteins using PCP (described above) and confidently detected multiple locations for 39% of all proteins and 16% of mitochondrial proteins (36). In yeast, Kumar et al. determined localization of 2744 proteins by tagging and microscopy and found evidence of dual localization for 11% of all proteins and 15% of proteins with mitochondrial staining (57). Collectively, these studies suggest that ~15% of mitochondrial proteins are dual-localized.
Pathways
The mitochondrion houses not only pathways for energy metabolism, such as OXPHOS and the TCA cycle, but dozens of additional pathways, such as heme biosynthesis, fatty acid/amino acid oxidation, pyrimidine biosynthesis, calcium homeostasis, and apoptosis. Of the ~1100 proteins in the mitochondrial proteome, approximately 300 have no known function and an additional 300 have only domain annotations based on sequence similarity (83). These proteins could be new components of well-studied pathways or, alternatively, represent components of pathways not previously appreciated even to reside in mitochondria. Nilsson et al. coupled the MitoCarta inventory with large-scale coexpression analysis to identify novel mitochondrial transporters and chaperones required for heme biosynthesis (79). Huynen et al. performed a functional network analysis of the mammalian mitochondrion (47), building on elegant studies in yeast (87), and highlighted uncharacterized proteins tightly connected to modules of known function now shown to reside in the mitochondrion. There have been some surprises from these initial analyses of mitochondrial pathways. For example, it's now quite clear that mammalian mitochondria house nearly all of the protein components for type II fatty acid synthesis (43), which may be important for the biosynthesis of lipoic acid, and possibly for fatty acids that are incorporated into the mitochondrial lipids. These large-scale proteomic surveys have also revealed a large number of proteins that appear to be involved in reversible phosphorylation and acetylation (83), suggesting the existence of a large signaling network within the organelle.
Tissue Diversity
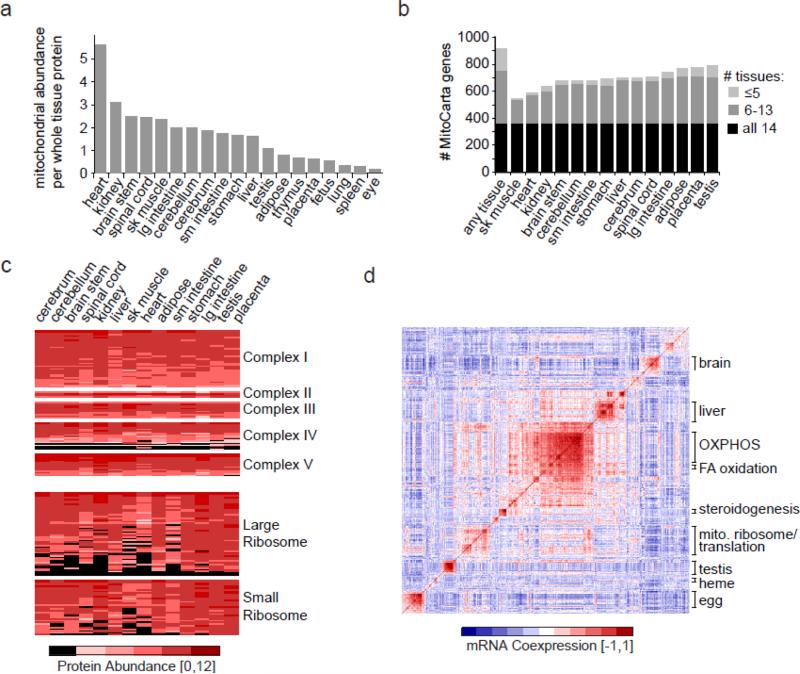
Decades of biochemical and ultrastructural studies have shown that mitochondrial function and structure varies across cell types. Large-scale proteomic surveys have provided valuable molecular insights into this tissue diversity. The heart, for example, contains over twofold more mitochondria than brain regions (Figure 1a). There are qualitative differences in these mitochondria, too. In the first proteomic comparison across tissues, Mootha et al. found that mitochondria obtained from distinct organs shared approximately 75% of their proteins (72). This general model of protein sharing has been confirmed with more extensive analyses of the organelle. Specifically, of the ~1100 mitochondrial proteins, it appears that almost half are core components found in virtually all tissues, whereas the remaining half are distributed in a tissue-specific manner (Figure 1b) (83). Mitochondria from developmentally related organs tend to share more proteins. Interestingly, a recent investigation of the mitochondrial proteome of brown fat is consistent with the recent discovery that brown fat derives from muscle (35).
Figure 1.
Tissue diversity of the mitochondrial proteome. (a) Estimate of mitochondrial abundance across tissues using cytochrome c as a proxy, reproduced with permission from Pagliarini et al. (83). (b) Number of distinct proteins detected by tandem mass spectrometry (MS/MS) within 14 mouse tissues (83). (c) Protein expression of protein complexes across tissues, where black indicates lack of detection and red indicates abundance levels if detected (83). (d) Mitochondrial mRNA coexpression, shown as a pairwise correlation matrix based on GNF mouse tissue expression (100), where red represents strong positive correlation. Prominent clusters of coexpressed genes are annotated based on their pathway or primary tissue of expression. Abbreviation: FA, fatty acid; OXPHOS, oxidative phosphorylation system
Some core pathways show rather surprising patterns of tissue diversity. As expected, complexes I, II, III, and V appear to be found in high abundance in all tissues surveyed (Figure 1c). However, complex IV is a curious outlier and appears to have a fair number of subunits that are expressed in a tissue-dependent manner (Figure 1c), consistent with previous reports (16). The mitochondrial ribosome may be the most surprising of all large macromolecular complexes. Although it is required for translating the 13 mtDNA-encoded proteins in all tissues, many of its subunits appear to be expressed in a tissue-specific manner (Figure 1c). It is tempting to speculate that perhaps these are regulatory subunits responsible for tissue-specific translational control, which may have important implications for the tissue-specific pathology observed in mtDNA disease.
Recently, Balaban et al. have not only characterized tissue-specific mitochondrial proteomes in rats (51) but have also combined these inventories with biochemical pathway maps to model the biosynthetic capacities of different organs (50). In addition, Palsson et al. have used proteomic inventories to constrain their flux balance analysis models of heart-specific mitochondrial metabolism (104, 113). These innovative studies represent some of the very first attempts to use the mitochondrial proteome as a framework for systems physiology.
Evolutionary Origins
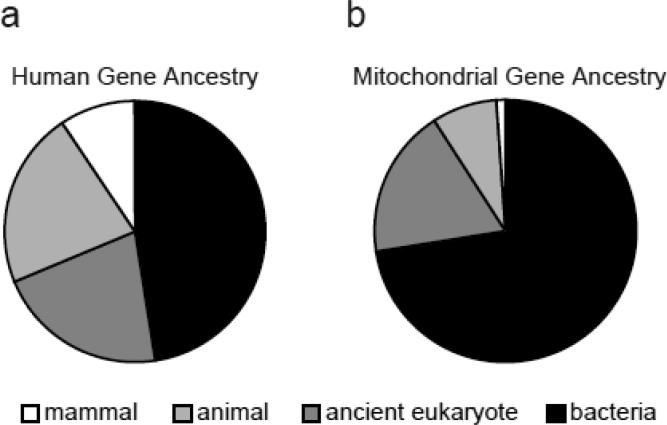
Evidence suggests that modern day mitochondria derive from an α-proteobacterial endosymbiont (4). The endosymbiont lost or transferred most of its DNA to the host nucleus and, additionally, many host proteins acquired targeting signals to the organelle. Interestingly, only about 15--20% of the current human mitochondrial proteome is estimated to be derived from the original endosymbiont DNA (38). Because the evolutionary origins of mitochondria have been recently reviewed (32, 47, 60), we focus here on some insights gained from the nearly comprehensive characterization of mammalian mitochondrial proteomes (83).
Mammalian mitochondrial proteins exhibit far more evolutionary conservation compared to other cellular components. Nearly 75% of mitochondrial proteins have clear bacterial ancestry, compared to only 48% of all mammalian proteins (Figure 2). Even though most of the mitochondrial components have ancient origin, 9% apparently have homologues only in metazoans, including 10 complex I subunits, 8 complex IV subunits, and 18 mitochondrial ribosomal proteins, suggesting recent innovations in these complexes. Some mitochondrial proteins have evolved specifically in the vertebrate lineage, including several factors required for apoptosis. Phylogenetic profiling, a method that relates proteins to each other based on shared evolutionary histories (64, 86), has been very useful in elucidating the function of several mitochondrial proteins (39, 80, 83, 101).
Figure 2.
Evolutionary history of all 24,000 human protein-encoding genes (a) and of 1100 protein-encoding mitochondrial genes (b), based on sequence similarity to proteins in 500 fully sequenced species (83).
MENDELIAN MITOCHONDRIAL DISEASES
The availability of the mitochondrial proteome has enabled systematic approaches to understanding mitochondrial diseases. In this section, we review how this protein inventory, coupled with new genomic methods, has assisted in the discovery of mitochondrial disease genes as well as in expanding our definition of these disorders.
Respiratory Chain Diseases
Respiratory chain diseases (RCD) represent a large subset of mitochondrial disorders and are biochemically characterized by defective oxidative phosphorylation. They occur at an estimated prevalence of 1 in 5000 live births and are collectively the most common inborn error of metabolism (27). The mitochondrial respiratory chain (RC) is composed of five macromolecular complexes, encoded by 13 mtDNA and ~77 nuclear genes (28), which together generate the vast majority of cellular ATP. Defects in this machinery can cause diseases that range in severity from neonatal lethality to adult-onset neurodegeneration. Clinical presentation can be highly variable, but involvement of multiple organ systems is common. Clinical features can include skeletal muscle myopathy, cardiomyopathy, seizures, strokes, ataxia, peripheral neuropathy, blindness, deafness, GI dysmotility, liver failure, bone marrow dysfunction, and pancreatic exocrine and endocrine dysfunction (27). These disorders are also genetically heterogeneous and can exhibit maternal, autosomal dominant, recessive, or X-linked inheritance patterns, although in many cases they are sporadic. Estimates suggest that ~15--20% of RCD are due to mtDNA mutations, while the rest are likely caused by nuclear defects (18, 26). Due to their clinical and genetic heterogeneity, and the pleoitropy of known disease loci, RCD diagnosis is extremely difficult (11, 76, 115).
Discovery of nuclear genes underlying RCD has accelerated dramatically over the past few years due to advances in genomics and mitochondrial proteomics. Typically, RCD genes have been identified through candidate gene approaches or through mapping techniques such as linkage analysis, homozygosity mapping, or chromosomal transfer. However mapping methods usually implicate large chromosomal intervals containing many genes, and it can be difficult to pinpoint the causal mutation. Since genes encoding mitochondrial proteins are strong candidates for RCD, crossing an implicated region with the list of mitochondrial genes can quickly focus the search to a handful of high-confidence candidates that can be resequenced.
The first genetics study to integrate large-scale genomic and proteomic information successfully discovered mutations in LRPPRC underlying Leigh Syndrome French Canadian variant (74). The same underlying approach was subsequently used to identify ETHE1 as the causal gene in ethylmalonic encephalopathy (105). Additional genomic clues were combined into a Bayesian predictor called Maestro that could be applied to predict candidate disease genes within any genetic interval (15). Maestro and related methods have been used to identify mutations in MPV17 (98), TMEM70 (19), and TACO (118). However, for very large genetic intervals, additional methods are necessary to further prioritize the mitochondrial protein-encoding genes. One innovative approach that has been used to identify genes contributing to RC complex I deficiency is phylogenetic profiling. This method relies on the observation that complex I has been lost multiple times in eukaryotic evolution and thus proteins sharing the same pattern of evolutionary loss may be functionally related. This method was first applied to pinpoint causal mutations in NDUFAF2 (80) and later applied to discover mutations in C8orf38 (83) and C20orf7 (101).
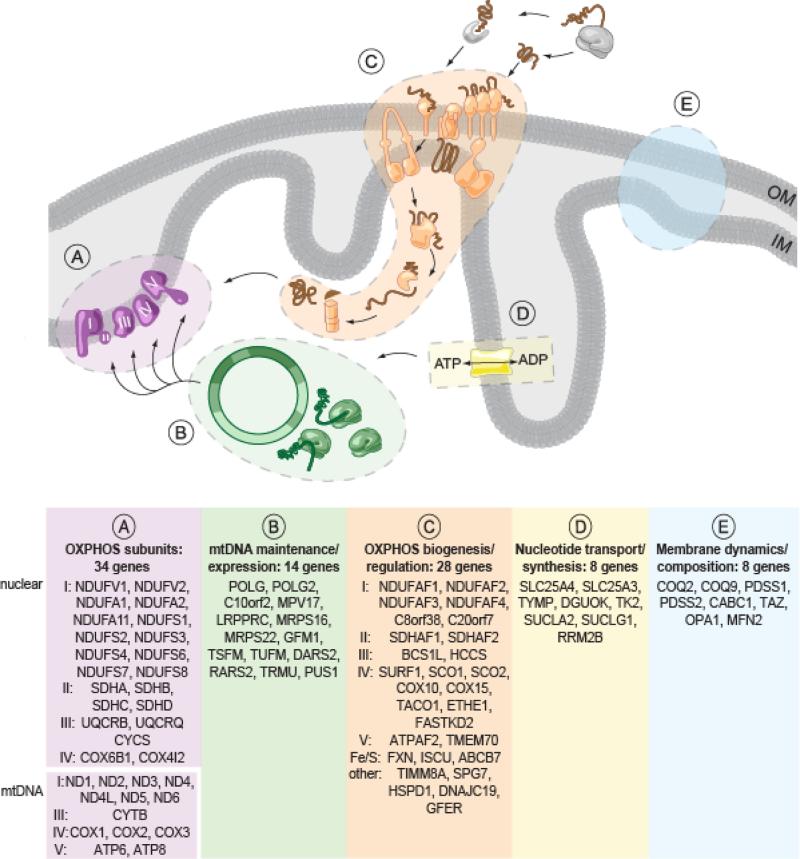
The growing catalog of genes underlying RCD (123) provides valuable insights into molecular pathogenesis. To date, 92 protein-encoding genes have been identified whose mutations underlie RCD. These 92 proteins span five pathways (Figure 3), as described by Kirby et al. (53). First, mutations in any of the 13 mtDNA protein-encoding genes (or in any of its tRNAs or rRNAs) can underlie RCDs (28, 85, 102, 110). Second, mutations in many of the nuclear genes encoding these subunits can cause RCD. Third, a large class of RCD genes encode proteins that are essential for the proper import, maturation, and assembly of the OXPHOS complexes. Fourth, membrane dynamics are required for proper import. Finally, the mtDNA requires a balanced pool of nucleotides for proper replication and transcription, and it's now clear that mutations in this pathway can give rise to OXPHOS disease secondary to loss of mtDNA homeostasis. Some genes are not readily classified, because the gene function is not well characterized (e.g., MPV17 and FASTKD2) or because the method of pathogenesis is more indirect, as is the case with ETHE1 mutations that cause complex IV inhibition via H2S toxicity (106). As additional RCD genes are identified, more pathways essential to respiratory chain function are likely to be uncovered, and human genetics may help us to understand how cells assemble their mitochondria.
Figure 3.
Biological pathways underlying respiratory chain disease (RCD), adapted from Kirby et al. (53). Bottom panel lists 92 protein-encoding genes that have been shown to cause RCD, including 79 genes encoded in the nucleus and 13 encoded by the mtDNA. Noted are genes associated with specific complexes (I--V) or iron/sulfur cluster biogenesis (Fe/S). Not listed are the 24 noncoding mtDNA genes that can also underlie RCD (2 rRNA and 22 tRNA genes). Abbreviation: OXPHOS, oxidative phosphorylation
Expanding the Definition of Mitochondrial Disease
Traditionally, mitochondrial disease has referred primarily to disorders of oxidative ATP production, as discussed above. However the breadth of the mitochondrial proteome now implicates a large number of additional phenotypes, such as soft tissue tumors (paragangliomas) and diabetes mellitus (27). The discovery of new disease genes will further expand the clinical phenotypes associated with mitochondrial defects. For example, recently discovered mutations in mitochondrial protein PYCR1 cause a syndrome with advanced aging features and cutis laxa (89), and mutations in mitochondrial protein DHODH underlie Miller syndrome, characterized by micrognathia, cleft lip, and limb hypoplasia (78A). If we now define disorders of the mitochondrial proteome as the set of diseases caused by mutations in genes encoding mitochondria-localized proteins, then over 150 different disorders may be encompassed (92).
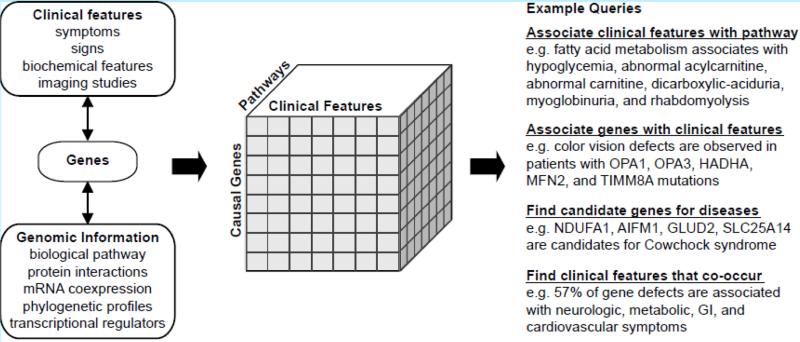
How can we begin to systematically understand the remarkable biochemical and clinical diversity of disorders stemming from defects in this organelle? A useful approach may lie in connecting disease genes, clinical features, and biological pathways (Figure 4). Using the wealth of genomic information that is now available, it is possible to identify biological pathways centered on a given disease gene. By connecting genes with clinical features, this framework can facilitate discovery of specific pathways underlying pathogenesis and can implicate functionally related genes as novel disease candidates.
Figure 4.
Exploring the relationship between disease genes, biological pathways, and clinical features. Causal genes provide a link between clinical features and biological pathways (left), enabling the creation of multidimensional datasets (middle) that can be used to address biological queries (right). Examples are derived from the MitoPhenome database integrated with Kyoto Encyclopedia of Genes and Genomes (KEGG) biological pathways. Disease gene candidates, reported by Scharfe et al. are derived from known disease genes with similar phenotypic consequences as well as functionally related genes (92).
A prerequisite for such integration is a careful phenotypic annotation of the diseases. Scharfe et al. recently made important progress in this direction by performing an extensive literature review of clinical phenotypes associated with defects in mitochondrial genes. These investigators created a database of 502 hierarchical clinical features associated with defects in 174 mitochondrial proteins (http://www.mitophenome.org) (92). This dataset currently is both incomplete (missing approximately half of known RCD genes) and includes many proteins that may not reside primarily in the organelle, such as p53 (63, 67) and WFS1 (33). Despite these caveats, the data set can be used as an initial framework for linking genes and pathways to clinical phenotypes. First, it becomes easy to quantify the co-occurrence of symptoms. For example, the integrated database can be used to learn that 89% of mitochondrial disease genes cause neurologic symptoms whereas 57% cause combined neurologic, metabolic, gastrointestinal and cardiovascular symptoms. Second, it becomes possible to look for biological connections between genes that cause specific phenotypes in patients. By querying this database, for example, it is clear that color vision defects (dyschromatopsia or monochromatic vision) can accompany mutations in any of five different mitochondrial genes (90, 108, 109, 122, 124) (Figure 4). Third, by linking genes to known biological processes, it becomes straightforward to identify clinical features associated with particular biological pathways, such as fatty acid metabolism (Figure 4). Such integration also makes it possible to identify novel disease gene candidates, as Scharfe et al. did by building gene networks based on similar clinical features (92) (Figure 4). In a comparable vein, Huynen et al. highlighted similar disease phenotypes within gene modules clustered on STRING function interactions (47). Although these databases of clinical phenotypes are still in their infancy and suffer from significant ascertainment bias, they represent important steps in the systematic understanding of mitochondrial disease.
FUTURE PROSPECTS AND CHALLENGES
Mitochondrial biology has been studied extensively for decades using traditional biochemical and molecular approaches. The comprehensive characterization of its protein inventory opens up exciting new opportunities for the systems-level analysis of this organelle in health and disease. Some exciting prospects and challenges lie ahead.
What can we anticipate in the fast-moving area of mitochondrial proteomics? In the coming years, the remaining few hundred mitochondrial proteins not present in current catalogs will likely be identified through a combination of high-throughput and traditional biochemical strategies to yield a finished mitochondrial proteome. It will then be important to understand the extent of splice variation and post-translational modifications of all of these proteins (5, 6, 13), as well as their specific localization within the mitochondrion. Progress in this latter area has already been charted for the inner mitochondrial membrane and the mtDNA nucleoid (12, 23) and may benefit from emerging superresolution imaging technology (45). Another major challenge lies in understanding how mitochondrial proteins function together in pathways and complexes. Hundreds of currently uncharacterized proteins will likely be linked to functions using high-throughput approaches such as RNAi, protein--protein interaction mapping, and computational predictions (10, 37, 49, 79, 80, 83). Physical and functional interaction maps will be useful not only for understanding biological pathways within the organelle but also for linking the mitochondrion to other organelles, as illustrated recently by an elegant synthetic biology approach in yeast (56). As the protein inventory and the complexes of the mitochondria become refined, it will be important to characterize its diversity across tissues, developmental states, and diseases. Initial efforts in this area have already begun (51, 54, 58, 78, 83, 84, 94).
In the coming decade, perhaps the most exciting physiological and clinical insights will come from genetic studies of rare and common variation in the mitochondrial proteome. One of the most famous examples of common variation in the mitochondrial proteome is a missense polymorphism in the aldehyde dehydrogenase 2 (ALDH2) gene which in East Asians confers sensitivity to alcohol (121). Many other such examples may be uncovered thanks to next-generation sequencing technology. Large-scale projects, such as the 1000 Genomes Project, will soon catalog the spectrum of normal genomic variation (http://www.1000genomes.org). Resequencing of individuals with extreme mitochondrial phenotypes may yield a slew of additional highly penetrant variants. The challenge will be to establish causal links between genetic variants and specific phenotypes. Such challenges have been faced for decades by mitochondrial researchers, as evidenced by lively debates on how to distinguish pathogenic from benign variants in mtDNA (65, 69, 70) and by the expanding number of presumed monogenic diseases that are actually caused by multiple loci (14, 95, 111, 114).
Understanding rare disorders of the mitochondrial proteome will also shed mechanistic insights into common disease. This has already been evident for neurodegenerative disease and diabetes (27), but it could extend to many other common disorders. For example, Vogelstein et al. recently discovered frequent mutations in cytosolic isocitrate dehydrogenase 1 (IDH1) that underlie a variety of cancers, including brain tumors (7, 120). Recently an innovative metabolomics study demonstrated that the mutation is a gain-of-function variant causing the enzyme to produce hydroxyglutarate (24). Interestingly, hydroxyglutarate is known to be elevated in inborn errors of mitochondrial metabolism and is believed to be involved in the development of brain tumors (2, 55). Together, these studies suggest that hydroxyglutarate is important in the etiology of both rare and common tumors. Hence, the rare mitochondrial disorders will no doubt offer a valuable lens into the pathogenesis of a variety of common human diseases.
The sequencing the human mtDNA in 1981 represented a landmark achievement that sparked the genetic era of mitochondrial medicine. This sequence has served as a framework for understanding molecular biology of the mtDNA and maternally inherited diseases. Now, the initial characterization of the mitochondrial proteome represents perhaps an even more important milestone for mitochondrial biology and medicine. Not only will it serve as a molecular foundation for systems-level analysis of mitochondria, but it will bring us one step closer to a genomic nosology of a large and challenging class of human disorders.
SUMMARY POINTS.
The human mitochondrial proteome consists of an estimated 1100--1400 distinct proteins, of which 13 are encoded by the mitochondrial DNA (mtDNA).
Approximately 1100 of these proteins have been identified to date, mainly through large-scale proteomics, microscopy, and computation.
Approximately 15% of mitochondrial proteins are dual-localized.
Mitochondrial protein composition varies across tissues: Approximately half of mitochondrial proteins are ubiquitous and found across all organs surveyed, whereas the remainder show tissue-specific expression.
The mitochondrial ribosome shows surprising diversity across tissues.
Seventy-five percent of the mammalian mitochondrial proteome shows homology to bacterial proteins, far exceeding the 48% rate across all mammalian proteins.
The list of disorders of the mitochondrial proteome is rapidly growing thanks to advances in proteomics and genetics.
The nearly complete protein characterization of mitochondria, combined with its tractability for experimental studies, makes the organelle an excellent test bed for clinical systems biology.
FUTURE ISSUES.
Approximately 15% of the mitochondrial proteome awaits identification.
Further work is needed to characterize the mitochondrial proteome at the level of suborganellar localization, macromolecular complexes, and posttranslational modifications.
Additional studies are required to uncover how the mitochondrial proteome differs across developmental and disease states, and to unravel the molecular basis of these differences.
The molecular basis of tissue-specific differences in the mitochondrial proteome remains elusive.
Advances in sequencing technology will help uncover links between inherited variation in the mitochondrial proteome and biochemical and clinical phenotypes.
ACKNOWLEDGMENTS
The authors thank Scott B. Vafai, Steve Hershman, Joshua M. Baughman, David R. Thorburn, and members of the Mootha Laboratory for valuable feedback on this review, and Sigrid Hart for assistance in preparing figures. This work was supported by grant no. R01GM077465 from the National Institutes of Health.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Adachi J, Kumar C, Zhang Y, Mann M. In-depth analysis of the adipocyte proteome by mass spectrometry and bioinformatics. Mol. Cell Proteomics. 2007;6:1257–-73. doi: 10.1074/mcp.M600476-MCP200. [DOI] [PubMed] [Google Scholar]
- 2.Aghili M, Zahedi F, Rafiee E. Hydroxyglutaric aciduria and malignant brain tumor: a case report and literature review. J. Neurooncol. 2009;91:233–-36. doi: 10.1007/s11060-008-9706-2. [DOI] [PubMed] [Google Scholar]
- 3.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–-65. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 4.Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Ponten T, Alsmark UC, et al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–-40. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 5.Aponte AM, Phillips D, Harris RA, Blinova K, French S, et al. 32P labeling of protein phosphorylation and metabolite association in the mitochondria matrix. Methods Enzymol. 2009;457:63–-80. doi: 10.1016/S0076-6879(09)05004-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aponte AM, Phillips D, Hopper RK, Johnson DT, Harris RA, et al. Use of (32)P to study dynamics of the mitochondrial phosphoproteome. J. Proteome Res. 2009;8:2679–-95. doi: 10.1021/pr800913j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116:597–-602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 8.Banci L, Bertini I, Cefaro C, Ciofi-Baffoni S, Gallo A, et al. MIA40 is an oxidoreductase that catalyzes oxidative protein folding in mitochondria. Nat. Struct. Mol. Biol. 2009;16:198–-206. doi: 10.1038/nsmb.1553. [DOI] [PubMed] [Google Scholar]
- 9.Bannai H, Tamada Y, Maruyama O, Nakai K, Miyano S. Extensive feature detection of N-terminal protein sorting signals. Bioinformatics. 2002;18:298–-305. doi: 10.1093/bioinformatics/18.2.298. [DOI] [PubMed] [Google Scholar]
- 10.Baughman JM, Nilsson R, Gohil VM, Arlow DH, Gauhar Z, Mootha VK. A computational screen for regulators of oxidative phosphorylation implicates SLIRP in mitochondrial RNA homeostasis. PLoS Genet. 2009;5:e1000590. doi: 10.1371/journal.pgen.1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernier FP, Boneh A, Dennett X, Chow CW, Cleary MA, Thorburn DR. Diagnostic criteria for respiratory chain disorders in adults and children. Neurology. 2002;59:1406–-11. doi: 10.1212/01.wnl.0000033795.17156.00. [DOI] [PubMed] [Google Scholar]
- 12.Bogenhagen DF, Rousseau D, Burke S. The layered structure of human mitochondrial DNA nucleoids. J. Biol. Chem. 2008;283:3665–-75. doi: 10.1074/jbc.M708444200. [DOI] [PubMed] [Google Scholar]
- 13.Boja ES, Phillips D, French SA, Harris RA, Balaban RS. Quantitative mitochondrial phosphoproteomics using iTRAQ on an LTQ-Orbitrap with high energy collision dissociation. J. Proteome Res. 2009;8(10):4665–4675. doi: 10.1021/pr900387b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bugiani M, Invernizzi F, Alberio S, Briem E, Lamantea E, et al. Clinical and molecular findings in children with complex I deficiency. Biochim. Biophys. Acta. 2004;1659:136–-47. doi: 10.1016/j.bbabio.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 15.Calvo S, Jain M, Xie X, Sheth SA, Chang B, et al. Systematic identification of human mitochondrial disease genes through integrative genomics. Nat. Genet. 2006;38:576–-82. doi: 10.1038/ng1776. [DOI] [PubMed] [Google Scholar]
- 16.Capaldi RA, Halphen DG, Zhang YZ, Yanamura W. Complexity and tissue specificity of the mitochondrial respiratory chain. J. Bioenerg. Biomembr. 1988;20:291–-311. doi: 10.1007/BF00769634. [DOI] [PubMed] [Google Scholar]
- 17.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–-44. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chinnery PF. Searching for nuclear-mitochondrial genes. Trends Genet. 2003;19:60–-62. doi: 10.1016/s0168-9525(02)00030-6. [DOI] [PubMed] [Google Scholar]
- 19.Cizkova A, Stranecky V, Mayr JA, Tesarova M, Havlickova V, et al. TMEM70 mutations cause isolated ATP synthase deficiency and neonatal mitochondrial encephalocardiomyopathy. Nat. Genet. 2008;40:1288–-90. doi: 10.1038/ng.246. [DOI] [PubMed] [Google Scholar]
- 20.Clamp M, Fry B, Kamal M, Xie X, Cuff J, et al. Distinguishing protein-coding and noncoding genes in the human genome. Proc. Natl. Acad. Sci. USA. 2007;104:19428–-33. doi: 10.1073/pnas.0709013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 1996;241:779–-86. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 22.Cotter D, Guda P, Fahy E, Subramaniam S. MitoProteome: mitochondrial protein sequence database and annotation system. Nucleic Acids Res. 2004;32:D463–67. doi: 10.1093/nar/gkh048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Da Cruz S, Xenarios I, Langridge J, Vilbois F, Parone PA, Martinou JC. Proteomic analysis of the mouse liver mitochondrial inner membrane. J. Biol. Chem. 2003;278:41566–-71. doi: 10.1074/jbc.M304940200. [DOI] [PubMed] [Google Scholar]
- 24.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–-86. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 26.DiMauro S, Davidzon G. Mitochondrial DNA and disease. Ann. Med. 2005;37:222–-32. doi: 10.1080/07853890510007368. [DOI] [PubMed] [Google Scholar]
- 27.DiMauro S, Hirano M, Schon EA. Mitochondrial Medicine. Informa Healthcare; New York: 2006. p. 348. [Google Scholar]
- 28.DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. N. Engl. J. Med. 2003;348:2656–-68. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- 29.Dimmer KS, Fritz S, Fuchs F, Messerschmitt M, Weinbach N, et al. Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002;13:847–-53. doi: 10.1091/mbc.01-12-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elstner M, Andreoli C, Ahting U, Tetko I, Klopstock T, et al. MitoP2: an integrative tool for the analysis of the mitochondrial proteome. Mol. Biotechnol. 2008;40:306–-15. doi: 10.1007/s12033-008-9100-5. [DOI] [PubMed] [Google Scholar]
- 31.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300:1005–-16. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 32.Embley TM, van der Giezen M, Horner DS, Dyal PL, Bell S, Foster PG. Hydrogenosomes, mitochondria and early eukaryotic evolution. IUBMB Life. 2003;55:387–-95. doi: 10.1080/15216540310001592834. [DOI] [PubMed] [Google Scholar]
- 33.Fonseca SG, Fukuma M, Lipson KL, Nguyen LX, Allen JR, et al. WFS1 is a novel component of the unfolded protein response and maintains homeostasis of the endoplasmic reticulum in pancreatic beta-cells. J. Biol. Chem. 2005;280:39609–-15. doi: 10.1074/jbc.M507426200. [DOI] [PubMed] [Google Scholar]
- 34.Forner F, Foster LJ, Campanaro S, Valle G, Mann M. Quantitative proteomic comparison of rat mitochondria from muscle, heart, and liver. Mol. Cell Proteomics. 2006;5:608–-19. doi: 10.1074/mcp.M500298-MCP200. [DOI] [PubMed] [Google Scholar]
- 35.Forner F, Kumar C, Luber CA, Fromme T, Klingenspor M, Mann M. Proteome differences between brown and white fat mitochondria reveal specialized metabolic functions. Cell Metab. 2009;10:324–-35. doi: 10.1016/j.cmet.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 36.Foster LJ, de Hoog CL, Zhang Y, Zhang Y, Xie X, et al. A mammalian organelle map by protein correlation profiling. Cell. 2006;125:187–-99. doi: 10.1016/j.cell.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 37.Friedberg I. Automated protein function prediction---the genomic challenge. Brief Bioinform. 2006;7:225–-42. doi: 10.1093/bib/bbl004. [DOI] [PubMed] [Google Scholar]
- 38.Gabaldon T, Huynen MA. From endosymbiont to host-controlled organelle: the hijacking of mitochondrial protein synthesis and metabolism. PLoS Comput. Biol. 2007;3:e219. doi: 10.1371/journal.pcbi.0030219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gabaldon T, Rainey D, Huynen MA. Tracing the evolution of a large protein complex in the eukaryotes, NADH:ubiquinone oxidoreductase (complex I). J. Mol. Biol. 2005;348:857–-70. doi: 10.1016/j.jmb.2005.02.067. [DOI] [PubMed] [Google Scholar]
- 40.Gaston D, Tsaousis AD, Roger AJ. Predicting proteomes of mitochondria and related organelles from genomic and expressed sequence tag data. Methods Enzymol. 2009;457:21–-47. doi: 10.1016/S0076-6879(09)05002-2. [DOI] [PubMed] [Google Scholar]
- 41.Guda C. pTARGET: a web server for predicting protein subcellular localization. Nucleic Acids Res. 2006;34:W210–-13. doi: 10.1093/nar/gkl093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guda C, Fahy E, Subramaniam S. MITOPRED: a genome-scale method for prediction of nucleus-encoded mitochondrial proteins. Bioinformatics. 2004;20:1785–-94. doi: 10.1093/bioinformatics/bth171. [DOI] [PubMed] [Google Scholar]
- 43.Hiltunen JK, Schonauer MS, Autio KJ, Mittelmeier TM, Kastaniotis AJ, Dieckmann CL. Mitochondrial fatty acid synthesis type II: more than just fatty acids. J. Biol. Chem. 2009;284:9011–-15. doi: 10.1074/jbc.R800068200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holt IJ, Harding AE, Morgan-Hughes JA. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature. 1988;331:717–-19. doi: 10.1038/331717a0. [DOI] [PubMed] [Google Scholar]
- 45.Huang B, Bates M, Zhuang X. Super-resolution fluorescence microscopy. Annu. Rev. Biochem. 2009;78:993–-1016. doi: 10.1146/annurev.biochem.77.061906.092014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–-91. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 47.Huynen MA, de Hollander M, Szklarczyk R. Mitochondrial proteome evolution and genetic disease. Biochim. Biophys. Acta. 2009;1792(12):1122–9. doi: 10.1016/j.bbadis.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 48.Ingman M, Gyllensten U. mtDB: Human Mitochondrial Genome Database, a resource for population genetics and medical sciences. Nucleic Acids Res. 2006;34:D749–-51. doi: 10.1093/nar/gkj010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, et al. STRING 8---a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–-16. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson DT, Harris RA, Blair PV, Balaban RS. Functional consequences of mitochondrial proteome heterogeneity. Am. J. Physiol. Cell Physiol. 2007;292:C698–-707. doi: 10.1152/ajpcell.00109.2006. [DOI] [PubMed] [Google Scholar]
- 51.Johnson DT, Harris RA, French S, Blair PV, You J, et al. Tissue heterogeneity of the mammalian mitochondrial proteome. Am. J. Physiol. Cell Physiol. 2007;292:C689–-97. doi: 10.1152/ajpcell.00108.2006. [DOI] [PubMed] [Google Scholar]
- 52.King BR, Guda C. ngLOC: an n-gram-based Bayesian method for estimating the subcellular proteomes of eukaryotes. Genome Biol. 2007;8:R68. doi: 10.1186/gb-2007-8-5-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kirby DM, Thorburn DR. Approaches to finding the molecular basis of mitochondrial oxidative phosphorylation disorders. Twin Res. Hum. Genet. 2008;11:395–-411. doi: 10.1375/twin.11.4.395. [DOI] [PubMed] [Google Scholar]
- 54.Kislinger T, Cox B, Kannan A, Chung C, Hu P, et al. Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell. 2006;125:173–-86. doi: 10.1016/j.cell.2006.01.044. [DOI] [PubMed] [Google Scholar]
- 55.Kolker S, Mayatepek E, Hoffmann GF. White matter disease in cerebral organic acid disorders: clinical implications and suggested pathomechanisms. Neuropediatrics. 2002;33:225–-31. doi: 10.1055/s-2002-36741. [DOI] [PubMed] [Google Scholar]
- 56.Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, et al. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–-81. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumar A, Agarwal S, Heyman JA, Matson S, Heidtman M, et al. Subcellular localization of the yeast proteome. Genes Dev. 2002;16:707–-19. doi: 10.1101/gad.970902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kumar GK, Prabhakar NR. Post-translational modification of proteins during intermittent hypoxia. Respir. Physiol. Neurobiol. 2008;164:272–-76. doi: 10.1016/j.resp.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kumar M, Verma R, Raghava GP. Prediction of mitochondrial proteins using support vector machine and hidden Markov model. J. Biol. Chem. 2006;281:5357–-63. doi: 10.1074/jbc.M511061200. [DOI] [PubMed] [Google Scholar]
- 60.Kutik S, Stroud DA, Wiedemann N, Pfanner N. Evolution of mitochondrial protein biogenesis. Biochim. Biophys. Acta. 2009;1790:409–-15. doi: 10.1016/j.bbagen.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 61.Lopez MF, Kristal BS, Chernokalskaya E, Lazarev A, Shestopalov AI, et al. High-throughput profiling of the mitochondrial proteome using affinity fractionation and automation. Electrophoresis. 2000;21:3427–-40. doi: 10.1002/1522-2683(20001001)21:16<3427::AID-ELPS3427>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 62.Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 1998;94:481–-90. doi: 10.1016/s0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- 63.Marchenko ND, Zaika A, Moll UM. Death signal--induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J. Biol. Chem. 2000;275:16202–-12. doi: 10.1074/jbc.275.21.16202. [DOI] [PubMed] [Google Scholar]
- 64.Marcotte EM, Pellegrini M, Thompson MJ, Yeates TO, Eisenberg D. A combined algorithm for genome-wide prediction of protein function. Nature. 1999;402:83–-86. doi: 10.1038/47048. [DOI] [PubMed] [Google Scholar]
- 65.McFarland R, Elson JL, Taylor RW, Howell N, Turnbull DM. Assigning pathogenicity to mitochondrial tRNA mutations: when “definitely maybe” is not good enough. Trends Genet. 2004;20:591–-96. doi: 10.1016/j.tig.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 66.Mehrle A, Rosenfelder H, Schupp I, del Val C, Arlt D, et al. The LIFEdb database in 2006. Nucleic Acids Res. 2006;34:D415–-18. doi: 10.1093/nar/gkj139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, et al. p53 has a direct apoptogenic role at the mitochondria. Mol. Cell. 2003;11:577–-90. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 68.Milenkovic D, Ramming T, Muller JM, Wenz LS, Gebert N, et al. Identification of the signal directing Tim9 and Tim10 into the intermembrane space of mitochondria. Mol. Biol. Cell. 2009;20:2530–-39. doi: 10.1091/mbc.E08-11-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mitchell AL, Elson JL, Howell N, Taylor RW, Turnbull DM. Sequence variation in mitochondrial complex I genes: Mutation or polymorphism? J. Med. Genet. 2006;43:175–-9. doi: 10.1136/jmg.2005.032474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Montoya J, Lopez-Gallardo E, Diez-Sanchez C, Lopez-Perez MJ, Ruiz-Pesini E. 20 years of human mtDNA pathologic point mutations: carefully reading the pathogenicity criteria. Biochim. Biophys. Acta. 2009;1787:476–-83. doi: 10.1016/j.bbabio.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 71.Montoya J, Ojala D, Attardi G. Distinctive features of the 5'-terminal sequences of the human mitochondrial mRNAs. Nature. 1981;290:465–-70. doi: 10.1038/290465a0. [DOI] [PubMed] [Google Scholar]
- 72.Mootha VK, Bunkenborg J, Olsen JV, Hjerrild M, Wisniewski JR, et al. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115:629–-40. doi: 10.1016/s0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- 73.Mootha VK, Handschin C, Arlow D, Xie X, St Pierre J, et al. Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc. Natl. Acad. Sci. USA. 2004;101:6570–-75. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mootha VK, Lepage P, Miller K, Bunkenborg J, Reich M, et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc. Natl. Acad. Sci. USA. 2003;100:605–-10. doi: 10.1073/pnas.242716699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–-73. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 76.Morava E, van den Heuvel L, Hol F, de Vries MC, Hogeveen M, et al. Mitochondrial disease criteria: diagnostic applications in children. Neurology. 2006;67:1823–-6. doi: 10.1212/01.wnl.0000244435.27645.54. [DOI] [PubMed] [Google Scholar]
- 77.Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 1999;24:34–-36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 78.Nanduri J, Yuan G, Kumar GK, Semenza GL, Prabhakar NR. Transcriptional responses to intermittent hypoxia. Respir. Physiol. Neurobiol. 2008;164:277–81. doi: 10.1016/j.resp.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ng A, S.B., et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2009;42(1):30–5. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nilsson R, Schultz IJ, Pierce EL, Soltis KA, Naranuntarat A, et al. Discovery of genes essential for heme biosynthesis through large-scale gene expression analysis. Cell Metab. 2009;10:119–-30. doi: 10.1016/j.cmet.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ogilvie I, Kennaway NG, Shoubridge EA. A molecular chaperone for mitochondrial complex I assembly is mutated in a progressive encephalopathy. J. Clin. Invest. 2005;115:2784–-92. doi: 10.1172/JCI26020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–-74. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 82.Ozawa T, Sako Y, Sato M, Kitamura T, Umezawa Y. A genetic approach to identifying mitochondrial proteins. Nat. Biotechnol. 2003;21:287–-93. doi: 10.1038/nbt791. [DOI] [PubMed] [Google Scholar]
- 83.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–-23. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Palmfeldt J, Vang S, Stenbroen V, Pedersen CB, Christensen JH, et al. Mitochondrial proteomics on human fibroblasts for identification of metabolic imbalance and cellular stress. Proteome Sci. 2009;7:20. doi: 10.1186/1477-5956-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pancrudo J, Shanske S, Coku J, Lu J, Mardach R, et al. Mitochondrial myopathy associated with a novel mutation in mtDNA. Neuromuscul. Disord. 2007;17:651–-54. doi: 10.1016/j.nmd.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl. Acad. Sci. USA. 1999;96:4285–-88. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Perocchi F, Jensen LJ, Gagneur J, Ahting U, von Mering C, et al. Assessing systems properties of yeast mitochondria through an interaction map of the organelle. PLoS Genet. 2006;2:e170. doi: 10.1371/journal.pgen.0020170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rabilloud T, Kieffer S, Procaccio V, Louwagie M, Courchesne PL, et al. Two-dimensional electrophoresis of human placental mitochondria and protein identification by mass spectrometry: toward a human mitochondrial proteome. Electrophoresis. 1998;19:1006–-14. doi: 10.1002/elps.1150190616. [DOI] [PubMed] [Google Scholar]
- 89.Reversade B, Escande-Beillard N, Dimopoulou A, Fischer B, Chng SC, et al. Mutations in PYCR1 cause cutis laxa with progeroid features. Nat. Genet. 2009;41:1016–-21. doi: 10.1038/ng.413. [DOI] [PubMed] [Google Scholar]
- 90.Reynier P, Amati-Bonneau P, Verny C, Olichon A, Simard G, et al. OPA3 gene mutations responsible for autosomal dominant optic atrophy and cataract. J. Med. Genet. 2004;41:e110. doi: 10.1136/jmg.2003.016576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ruiz-Pesini E, Lott MT, Procaccio V, Poole JC, Brandon MC, et al. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 2007;35:D823–-28. doi: 10.1093/nar/gkl927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Scharfe C, Lu HH, Neuenburg JK, Allen EA, Li GC, et al. Mapping gene associations in human mitochondria using clinical disease phenotypes. PLoS Comput. Biol. 2009;5:e1000374. doi: 10.1371/journal.pcbi.1000374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Scheffler NK, Miller SW, Carroll AK, Anderson C, Davis RE, et al. Two-dimensional electrophoresis and mass spectrometric identification of mitochondrial proteins from an SH-SY5Y neuroblastoma cell line. Mitochondrion. 2001;1:161–-79. doi: 10.1016/s1567-7249(01)00007-1. [DOI] [PubMed] [Google Scholar]
- 94.Schilling B, Yoo CB, Collins CJ, Gibson BW. Determining cysteine oxidation status using differential alkylation. Int. J. Mass Spectrom. 2004;236:117–-27. [Google Scholar]
- 95.Schuler AM, Gower BA, Matern D, Rinaldo P, Vockley J, Wood PA. Synergistic heterozygosity in mice with inherited enzyme deficiencies of mitochondrial fatty acid beta-oxidation. Mol. Genet. Metab. 2005;85:7–-11. doi: 10.1016/j.ymgme.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 96.Small I, Peeters N, Legeai F, Lurin C. Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics. 2004;4:1581–-90. doi: 10.1002/pmic.200300776. [DOI] [PubMed] [Google Scholar]
- 97.Smith AC, Robinson AJ. MitoMiner, an integrated database for the storage and analysis of mitochondrial proteomics data. Mol. Cell Proteomics. 2009;8:1324–-37. doi: 10.1074/mcp.M800373-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Spinazzola A, Viscomi C, Fernandez-Vizarra E, Carrara F, D'Adamo P, et al. MPV17 encodes an inner mitochondrial membrane protein and is mutated in infantile hepatic mitochondrial DNA depletion. Nat. Genet. 2006;38:570–-75. doi: 10.1038/ng1765. [DOI] [PubMed] [Google Scholar]
- 99.Steinmetz LM, Scharfe C, Deutschbauer AM, Mokranjac D, Herman ZS, et al. Systematic screen for human disease genes in yeast. Nat. Genet. 2002;31:400–-4. doi: 10.1038/ng929. [DOI] [PubMed] [Google Scholar]
- 100.Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, et al. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. USA. 2002;99:4465–-70. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sugiana C, Pagliarini DJ, McKenzie M, Kirby DM, Salemi R, et al. Mutation of C20orf7 disrupts complex I assembly and causes lethal neonatal mitochondrial disease. Am. J. Hum. Genet. 2008;83:468–-78. doi: 10.1016/j.ajhg.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Taylor RW, Schaefer AM, McDonnell MT, Petty RK, Thomas AM, et al. Catastrophic presentation of mitochondrial disease due to a mutation in the tRNA(His) gene. Neurology. 2004;62:1420–-23. doi: 10.1212/01.wnl.0000120667.77372.46. [DOI] [PubMed] [Google Scholar]
- 103.Taylor SW, Fahy E, Zhang B, Glenn GM, Warnock DE, et al. Characterization of the human heart mitochondrial proteome. Nat. Biotechnol. 2003;21:281–-6. doi: 10.1038/nbt793. [DOI] [PubMed] [Google Scholar]
- 104.Thiele I, Price ND, Vo TD, Palsson BO. Candidate metabolic network states in human mitochondria. Impact of diabetes, ischemia, and diet. J. Biol. Chem. 2005;280:11683–-95. doi: 10.1074/jbc.M409072200. [DOI] [PubMed] [Google Scholar]
- 105.Tiranti V, D'Adamo P, Briem E, Ferrari G, Mineri R, et al. Ethylmalonic encephalopathy is caused by mutations in ETHE1, a gene encoding a mitochondrial matrix protein. Am. J. Hum. Genet. 2004;74:239–-52. doi: 10.1086/381653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tiranti V, Viscomi C, Hildebrandt T, Di Meo I, Mineri R, et al. Loss of ETHE1, a mitochondrial dioxygenase, causes fatal sulfide toxicity in ethylmalonic encephalopathy. Nat. Med. 2009;15:200–-5. doi: 10.1038/nm.1907. [DOI] [PubMed] [Google Scholar]
- 107.Tong WH, Rouault T. Distinct iron-sulfur cluster assembly complexes exist in the cytosol and mitochondria of human cells. EMBO J. 2000;19:5692–-700. doi: 10.1093/emboj/19.21.5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tranebjaerg L, Jensen PK, Van Ghelue M, Vnencak-Jones CL, Sund S, et al. Neuronal cell death in the visual cortex is a prominent feature of the X-linked recessive mitochondrial deafness--dystonia syndrome caused by mutations in the TIMM8a gene. Ophthalmic. Genet. 2001;22:207–-23. doi: 10.1076/opge.22.4.207.2220. [DOI] [PubMed] [Google Scholar]
- 109.Tyni T, Kivela T, Lappi M, Summanen P, Nikoskelainen E, Pihko H. Ophthalmologic findings in long-chain 3-hydroxyacyl-CoA dehydrogenase deficiency caused by the G1528C mutation: a new type of hereditary metabolic chorioretinopathy. Ophthalmology. 1998;105:810–-24. doi: 10.1016/S0161-6420(98)95019-9. [DOI] [PubMed] [Google Scholar]
- 110.Uusimaa J, Finnila S, Remes AM, Rantala H, Vainionpaa L, et al. Molecular epidemiology of childhood mitochondrial encephalomyopathies in a Finnish population: sequence analysis of entire mtDNA of 17 children reveals heteroplasmic mutations in tRNAArg, tRNAGlu, and tRNALeu(UUR) genes. Pediatrics. 2004;114:443–-50. doi: 10.1542/peds.114.2.443. [DOI] [PubMed] [Google Scholar]
- 111.Van Goethem G, Lofgren A, Dermaut B, Ceuterick C, Martin JJ, Van Broeckhoven C. Digenic progressive external ophthalmoplegia in a sporadic patient: recessive mutations in POLG and C10orf2/Twinkle. Hum. Mutat. 2003;22:175–-76. doi: 10.1002/humu.10246. [DOI] [PubMed] [Google Scholar]
- 112.Vieira HL, Haouzi D, El Hamel C, Jacotot E, Belzacq AS, et al. Permeabilization of the mitochondrial inner membrane during apoptosis: impact of the adenine nucleotide translocator. Cell Death Differ. 2000;7:1146–-54. doi: 10.1038/sj.cdd.4400778. [DOI] [PubMed] [Google Scholar]
- 113.Vo TD, Greenberg HJ, Palsson BO. Reconstruction and functional characterization of the human mitochondrial metabolic network based on proteomic and biochemical data. J. Biol. Chem. 2004;279:39532–-40. doi: 10.1074/jbc.M403782200. [DOI] [PubMed] [Google Scholar]
- 114.Vockley J. Metabolism as a complex genetic trait, a systems biology approach: implications for inborn errors of metabolism and clinical diseases. J. Inherit. Metab. Dis. 2008;31:619–-29. doi: 10.1007/s10545-008-1005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Walker UA, Collins S, Byrne E. Respiratory chain encephalomyopathies: a diagnostic classification. Eur. Neurol. 1996;36:260–-67. doi: 10.1159/000117269. [DOI] [PubMed] [Google Scholar]
- 116.Wallace DC, Singh G, Lott MT, Hodge JA, Schurr TG, et al. Mitochondrial DNA mutation associated with Leber's hereditary optic neuropathy. Science. 1988;242:1427–-30. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- 117.Wallace DC, Zheng XX, Lott MT, Shoffner JM, Hodge JA, et al. Familial mitochondrial encephalomyopathy (MERRF): genetic, pathophysiological, and biochemical characterization of a mitochondrial DNA disease. Cell. 1988;55:601–-10. doi: 10.1016/0092-8674(88)90218-8. [DOI] [PubMed] [Google Scholar]
- 118.Weraarpachai W, Antonicka H, Sasarman F, Seeger J, Schrank B, et al. Mutation in TACO1, encoding a translational activator of COX I, results in cytochrome c oxidase deficiency and late-onset Leigh syndrome. Nat. Genet. 2009;41:833–-37. doi: 10.1038/ng.390. [DOI] [PubMed] [Google Scholar]
- 119.Wiedemann N, Pfanner N, Ryan MT. The three modules of ADP/ATP carrier cooperate in receptor recruitment and translocation into mitochondria. EMBO J. 2001;20:951–-60. doi: 10.1093/emboj/20.5.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009;360:765–-73. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yoshida A. Molecular genetics of human aldehyde dehydrogenase. Pharmacogenetics. 1992;2:139–-47. doi: 10.1097/00008571-199208000-00001. [DOI] [PubMed] [Google Scholar]
- 122.Yu-Wai-Man P, Griffiths PG, Hudson G, Chinnery PF. Inherited mitochondrial optic neuropathies. J. Med. Genet. 2009;46:145–-58. doi: 10.1136/jmg.2007.054270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhu X, Peng X, Guan MX, Yan Q. Pathogenic mutations of nuclear genes associated with mitochondrial disorders. Acta Biochim. Biophys. Sin. (Shanghai) 2009;41:179–-87. doi: 10.1093/abbs/gmn021. [DOI] [PubMed] [Google Scholar]
- 124.Zuchner S, De Jonghe P, Jordanova A, Claeys KG, Guergueltcheva V, et al. Axonal neuropathy with optic atrophy is caused by mutations in mitofusin 2. Ann. Neurol. 2006;59:276–-81. doi: 10.1002/ana.20797. [DOI] [PubMed] [Google Scholar]