Abstract
Using transgenic animals harboring a targeted LacZ insertion, we studied the expression pattern of the C9ORF72 mouse ortholog. Unlike most genes mutated in ALS, which are ubiquitously expressed, the C9ORF72-ortholog was most highly transcribed in the neuronal populations sensitive to degeneration in ALS and FTD. Thus, our study provides a potential explanation for the cell type specificity of neuronal degeneration caused by C9ORF72 mutations.
Keywords: Amyotrophic lateral sclerosis (ALS), frontotemporal dementia (FTD), C9ORF72, motor neuron, cortical neuron, hippocampus, astrocyte, microglial
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disorder that primarily affects motor neurons in the spinal cord, brain stem, and cerebral cortex 1. Frontotemporal dementia (FTD) is the second most common cause of presenile dementia. FTD is characterized by degeneration of the frontal and temporal lobes of the brain resulting in progressive changes in personality, behavior and language, with relative preservation of perception and memory2, 3. Recently, expansion of a noncoding hexanucleotide repeat in C9ORF72 was identified4, 5 as a common cause of both ALS and FTD6.
Several potential mechanisms by which the C9ORF72 mutation might lead to neuronal degeneration have been proposed. Significant reduction in transcript abundance has been observed in patients carrying expanded repeats5, 7, suggesting that haploinsufficiency could play a role in disease. The repeat expansion is associated with formation of nuclear RNA foci, potentially indicating alterations in RNA metabolism4, 8. Finally, ubiquitinated inclusions containing dipeptide proteins non-canonically translated from the GGGGCC repeats have been found upon autopsy of C9ORF72 patients 9, 10. However, whether one or more of these mechanisms are the cause of neuronal degeneration has not been resolved. Regardless of which molecular mechanism, or mechanisms, are responsible for the mutation's negative effects, it remains to be determined whether this mutation acts primary in the neural subtypes subject to degeneration or through non-neuronal cell types, as has been suggested by studies of mutant SOD1. Moreover, the normal physiological function of C9ORF72 and its expression pattern in the developing and adult nervous system have not been explored. As a first step towards understanding the function of C9ORF72 in development, normal brain function and disease, we produced animals harboring a LacZ reporter gene targeted to the mouse ortholog and used them to study the gene's expression pattern 11.
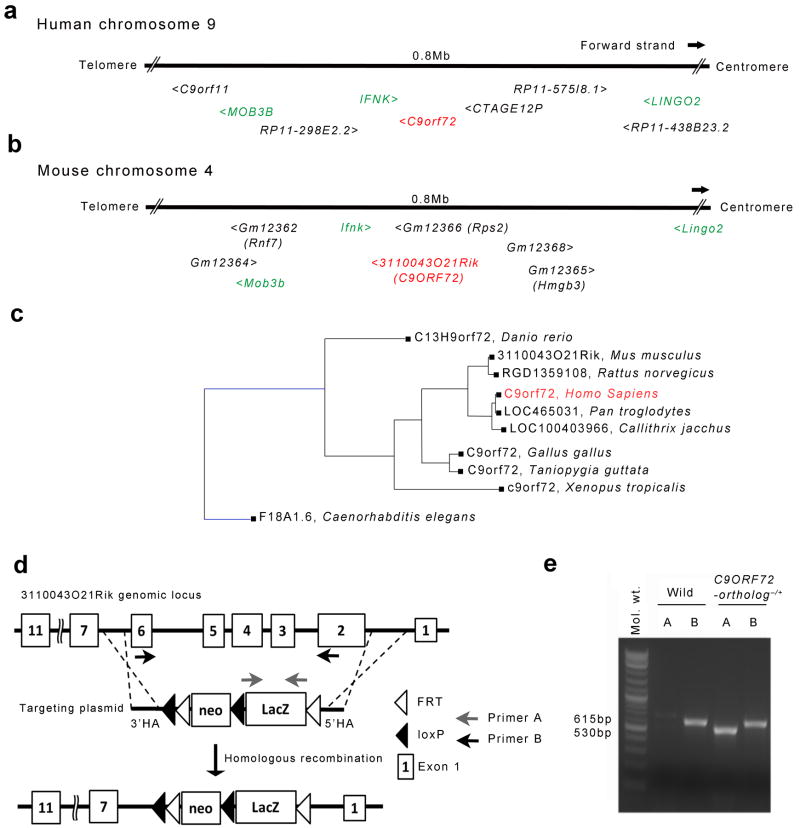
The C9ORF72 gene is located on the reverse strand of chromosome 9 (Fig. 1a). We found that the mouse 3110043O21Rik gene was located on the reverse strand of chromosome 4 in a syntenic position, centromeric to Mob3b as well as Ifnk and telomeric to Lingo2 (Fig. 1b). BlastN revealed >90% identity between the predicted human C9ORF72 and the mouse 311043O21Rik protein. Non-human primates, other mammals, Xenopus and Zebrafish also possess apparent orthologs with 66 - 98 % amino acid identity (Fig. 1c and Supplementary Fig. 1). Only 9 amino acids differ between the predicted protein sequences encoded by the mouse and human genes (Supplementary Fig. 2). In light of these findings, we will refer to the 311004O21Rik gene as the mouse C9ORF72-ortholog.
Figure 1. Mouse 3110043O21Rik gene is the ortholog of human C9ORF72.
Human C9ORF72 gene is located on the reverse strand of chromosome 9 (27,546,544-27,573,864) (a), while mouse 3110043O21Rik gene is located in a region of apparent synteny on the reverse strand chromosome 4 (35,138,536-35,173,129) (b). (c) Homology of C9ORF72 gene and its apparent orthologs. Black line represents ×1 branch length, while blue line represents ×10 branch length. F18A1.6 in C elegans has 23 % identity with human C9ORF72. (d) Construction of the C9ORF72 LacZ “knock-in” mice. Boxes represent exons. Frt sites (open triangle) and Loxp sites (closed triangle) are shown. Exon 2 through exon 6 are deleted and substituted with LacZ sequence. Grey and black arrows represent the locations of primers used in Fig. 1e respectively. (e) LacZ genotyping result. Primer-set A detects LacZ sequence (530 bp). Primer-set B detects exon 4 and 5 of C9ORF72 ortholog (615 bp). Mol. Wt = Molecular Weight.
Analysis of predicted transcripts and expressed sequence tags (ESTs) demonstrated that the mouse and human orthologs also share similar predicted intron-exon structures (Supplementary Fig. 3). In humans, predicted isoform 1 and a rare EST did not include the repeat expansion sequence, while isoforms 2, 3 and several rare ESTs did. Predicted transcript isoform 1 and 2 in mouse did not contain the location of the human repeat while isoform 3 and relatively rare ESTs did. To experimentally determine the relative abundance of these isoforms, we performed RNA sequencing on mouse cortex and purified mouse as well as human embryonic stem cell-derived motor neurons. Among these transcripts, mouse isoforms 1 and 2 were most highly expressed in cortex and motor neurons, whereas isoform 3 and the predicted ESTs were far less abundant. In human motor neurons, isoform 1 was most highly expressed, while isoform 2, 3 and ESTs were only present at more modest levels (Supplementary Fig. 3c-e).
Comparative sequence analysis at the site of human repeat expansion revealed that the unexpanded GGGGCC repeat sequence was conserved in chimpanzees (Supplementary Fig. 4). This precise repeat sequence was not completely conserved in mouse. However, the CpG island in which the human repeat resides was highly conserved between Chimpanzee and Human (98% identity), and well as between Human and Mouse (58.3 % identity). Thus, although the hexanucleotide repeat itself is not conserved outside of primates, it resides in region that it is well conserved to the rodent and may therefore have a regulatory function.
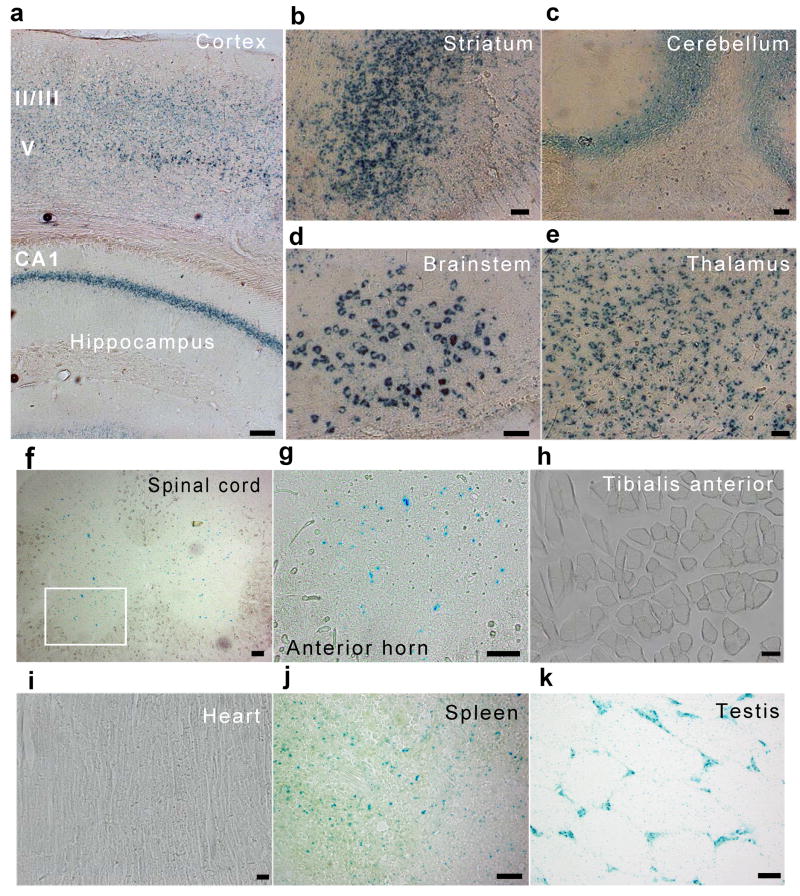
Review of the Knockout Mouse Project database revealed that mouse ES cells harboring a LacZ insertion replacing exons 2 through 6 of the C9ORF72-ortholog had been produced by gene targeting (Fig. 1d)11. Via standard methods, we produced chimeric mice that transmitted this mutant allele through the germ-line to offspring (Fig. 1d and e). Using 7 to 9 week old heterozygous mice, we studied the expression pattern of the C9ORF72-ortholog using X-gal staining (Supplementary table 1, Supplementary Fig. 5). In the brain, we found X-gal activity in the hippocampus, dentate gyrus, striatum, thalamus, brainstem nucleus, cerebellum and throughout the cortex, (Fig. 2a-e). We did not detect X-gal staining in white matter regions such as the corpus callosum (Supplementary Fig. 5). In the spinal cord, X-gal activity was distributed throughout the grey matter, with highest levels found in the ventral horn (Fig. 2f, g).
Figure 2. Distribution of X-gal in heterozygous animals with C9ORF72-ortholog LacZ insertions.
(a-e). X-gal staining of the brain of C9ORF72 ortholog knock-in mice. Expression of X-gal in cortex and hippocampus (a), striatum (b), cerebellum (c), brainstem nucleus (d), thalamus (e). X-gal expression distributes mainly in grey matter of the spinal cord in C9ORF72 ortholog knock-in mice (f, g). (g) Shows higher magnification image of white rectangle in (f). X-gal staining of non-nervous system organs (h, k). X-gal staining was undetected in tibialis anterior muscle (h) and heart (i). Note that peripheral of germinal center in spleen (j) and testis (k) were positive for X-gal staining. Bar, 200 μm (a), 100 μm (f), 50 μm (b-e, g-k).
Outside the CNS, the tibialis anterior muscle, heart, lung, liver, and kidney were X-gal negative (Fig2h, i Supplementary Fig. 6). The testis and germinal centers in the spleen were X-gal positive (Fig. 2j, k).
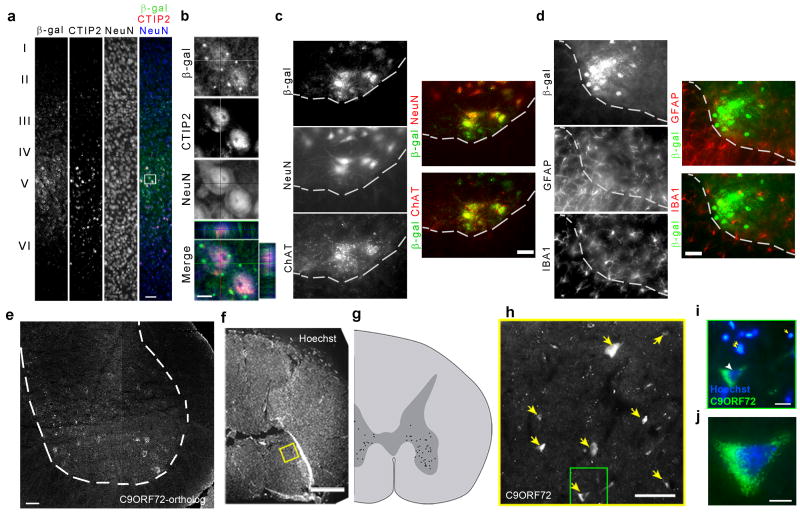
To determine the identity of X-gal positive cells in the CNS, we performed co-imunnostaining with anti-β-gal antibodies and antibodies that labeled relevant classes of neuronal and non-neuronal CNS cell-type (Fig. 3). We found that 128/130 β-gal+ cells in layer V of the cortex expressed NeuN (98%) and that 120/195 (62%) of these cells further co-stained with antibodies specific to CTIP2, a transcription factor selectively expressed in cortical spinal motor neurons and other projection neurons of layers V and VI (Fig. 3a and b). In cortical layers II and III 112/114 β-gal+ cells expressed NeuN (98%), with 107/112 (96%) of these NeuN+ cells further expressing CUX1, a transcription factor found in callosal projection neurons (Supplementary Fig. 7). Throughout the spinal cord, cells expressing β-gal uniformly expressed NeuN (111/115, 97%), with a fraction in the ventral horn further co-labeling with anti-ChAT antibodies, indicating that many were spinal motor neurons (65/115, 57%) (Fig. 3c and Supplementary Fig. 8). In striking contrast, spinal cord microglia as identified by IBAI staining, and astrocytes identified by GFAP expression, were largely and entirely β-gal negative respectively (Iba1: 7/172, 4% and GFAP: 0/172, 0%) (Fig. 3d and Supplementary Fig. 9).
Figure 3. Characterization of the cells expressing β-gal under control of the C9ORF72-ortholog promoter.
(a). Co-localization of β-gal, CTIP2 (layer V) and NeuN in cortex. Green-β-gal, red-CTIP2, blue-NeuN. (b) shows Z-stack and orthogonal images of white rectangle in (a). (c) Co-localization of β-gal with ChAT, and β-gal with NeuN in ventral horn of the spinal cord are shown. Dashed line represents the border between grey and white matter. (d) β-gal positive cells are not colocalized with GFAP or Iba1 in ventral horn of the spinal cord. (e-j) Spliced C9ORF72 mRNA is specifically expressed in motor neuron-like cells in the mouse (e) and human spinal cord (f-j). (f) Composite photomicrograph of a coronal section of a human spinal cord stained with Hoechst to visualize DNA. (g) Map of C9ORF72-expressing cells (black dots) observed in the spinal cord section show in panels (f-j). Labeled cells were sparse and were confined to cell bodies distributed in the ventral and lateral horns of the spinal cord. The wedge-shaped tear on the left dorsolateral edge of the spinal cord is a histological artifact. (h) Photomicrograph of the yellow-boxed region in f, showing the in situ hybridization signal of DIG-labeled antisense riboprobe against spliced human C9ORF72 mRNA (yellow arrows). Hybridization was strong and specific. (i) High power photomicrograph of the green boxed region in (h), illustrating a representative labeled cell (green), whose nucleus (arrowhead) is considerably larger than those of surrounding cells (yellow arrows). (i and j) Representative C9ORF72-expressing cells are large and pyramidal, and strongly reminiscent of spinal cord motor neurons. Bar, 50 μm (a, e) and 20 μm (b, c, d, h), 2mm (f), 100μm (i), and 10μm (j).
Through in situ hybridization using probes targeting exons 2 through 6 of the C9ORF72 gene and its ortholog, we found that many cells with a neuronal morphology were labeled in both the human and mouse spinal cord (Fig. 3e-j). Labeled cells were predominantly observed in the ventral and lateral horns of the mouse and human spinal cord grey matter and absent from the white matter, a distribution identical to β-gal+ cells observed in heterozygous animals.
Expression data compiled from the Allen Brain Atlas confirmed the expression pattern for the C9ORF72-ortholog (Supplementary Fig. 10-12) 12. We also carried out a developmental survey of X-gal activity and found that transcription of the C9ORF72-ortholog was undetectable during prenatal stages and became activated in an expression pattern similar to that found in the adult over the first two weeks of post-natal life (Supplementary Table 2).
Here we show that 3110043O21Rik is the mouse ortholog of human C9ORF72, which we found to be highly conserved between vertebrate species. Using mice we produced carrying a LacZ reporter at the C9ORF72 ortholog, we found that transcription was most abundant in neural types known to degenerate in ALS /FTD. In contrast, the C9ORF72 ortholog was largely absent or undetectable in microglia and astrocytes. Although the results from our reporter analyses are clear, it is important to note that one limitation of this approach is that post transcriptional regulation of the C9ORF72 ortholog could alter the relative localization of the protein it encodes. While our findings do not rule out low levels of ortholog expression in these non-neuronal cell-types, our results do seem to argue against the notion that the C9ORF72 mutations act predominantly through them to mediate neural degeneration. Regardless of whether C9ORF72 repeat expansions act in disease through a loss of function or gain of function mechanism, our studies of the mouse ortholog provide a potential explanation for the cell-type selectivity of neural degeneration in individuals harboring this mutation: The neuronal types most sensitive to ALS and FTD transcribe the highest levels of this gene.
Methods
Methods and any associated references are available in the online version of the paper.
Online Methods
Bioinformatics
We referred spacial expression pattern of C9ORF72-ortholog of mice brain and spinal cord from Allen Brain Atlas database13. Probes are made from transcripts of exon 4 through exon 11.
Generation of C9ORF72 Knock-in Mice
Target vector was designed in the National Institutes of Health Knockout Mouse (KOMP) project14. Briefly, Electroporation of C57BL/6N mouse ES cells with linearized plasmid DNA was carried out in electroporation cuvettes. Stable clones were selected in medium containing Geneticin. The ES cells were injected into C57BL/6 blastocysts to create chimeric mice, which were bred with C57BL/6 mice to generate heterozygous C9ORF72-ortholog knock-in mice. Postnatal day 60 male mice are used for the experiments otherwise particulary mentioned. Up to 5 animals are housed in the same cage. All of the experimental protocols and procedures were approved by the Animal Committee of the Harvard University.
Genotyping
Genomic DNA from ear biopsies was lysed in lysis buffer with proteinase K at 60°C for 12 h. The PCR contained primer set A (5′- ATCACGACGCGCTGTATC-3′ and 5′- ACATCGGGGAAATAATATCG-3′) which detect LacZ sequence and primer set B (5′- CCATGCTTACTGGGGAAGTC-3′ and 5′- AAGAAAGCCTTCGTGACAGC -3′), which detect deleted exon 4 and 5. Genomic DNA and primers (50 nM each) were placed in standard Taq buffer supplemented with 1.25 units of Taq polymerase for 10 min at 94 °C. After enzymatic amplification for 35 cycles, the PCR products were resolved on 2% agarose gel in Tris acetate-EDTA buffer.
X-gal Staining of Tissues
Mice were anesthetized with avertin and perfusion-fixed with 4% paraformaldehyde (PFA). Tissues were harvested and post-fixed in 4% PFA overnight at 4 °C, washed with PBS, immersed in 30% sucrose for 24 h at 4 °C, and frozen in optimal cutting temperature compound for sectioning using cryostat. Some of the specimen from CNS are washed with PBS and cut using vibratome. X-galactosidase (X-gal) activity was assessed by incubating 10-50 μm thick sections with LacZ staining solution (1.0 mg/ml of X-gal, 5mM potassium ferrocyanide, 5 mM potassium ferricyanide, 2 mM MgCl2) for 30 min-12 h at 37 °C. The sections were examined and photographed with a Zeiss AX10. Antibodies. The following antibodies were used: β-gal (1:500, CGAL-45A-Z, ICL), ChAT (1:100, AB144P, Millipore), CTIP2 (1:200, 25B6, Abcam), CuxI (1:200, sc13024, Santa Cruz), NeuN (1:200, MAB377, Millipore), IbaI (1:200, 019-19741, Wako), and GFAP (1:500, G3893, Sigma).
Immunohistochemistry15, 16
Cryosections of tissue (10-50 μm thick) were cut from the brain, spinal cord and other organs, placed on poly-L-lysine-coated slides, air-dried, and pre-incubated in phosphate-buffered saline (PBS) containing 5 % goat serum for 30 min at room temperature. The primary antibodies were applied overnight at 4 °C. Following incubation with the appropriate secondary antibodies, the mounted sections were observed using Zeiss AX10 microscope or Zeiss LSM 700 laser scanning confocal microscope (Carl Zeiss Micro-Imaging GmbH). Images were processed using ZEN 2010 software. Stainings were performed at least three times and representative figures were shown.
Motor neuron differentiation from mouse and human ES cells (ESCs)
Mouse and Human derived ES cells with HB9GFP reporter are differentiated and fluorescence activated cell sorting (FACS) purified as described previously16, 17. Briefly, human ESCs are incubated in accutase and resuspended in low-attachment flasks. ESCs are differentiated into motor neurons with dual-SMAD inhibition, and then retinoic acid (RA) and smoothened antagonist (SAG). Mouse ESCs were differentiated into motor neurons according to methods previously described16. For differentiation to motor neurons, mouse ESCs are cultured in medium supplemented with RA and SAG for 7 days. After motor neuron differentiation, GFP positive motor neurons are collected using FACS.
RNA sequence
Following harvesting of RNA from indicated sources, RNA quality was determined using bioAnalyzer (Aligent). RNA integrity numbers (RIN) above 7.5 were deemed sufficiently high quality to proceed with library preparation. In brief, RNA sequencing libraries were generated from ∼250ng total RNA using the illumina TruSeq RNA kit v2, according to the manufacturers' directions. Libraries were sequenced at the Harvard Bauer Core Sequencing facility on a HiSeq 2000. Libraries were generated from at least 2 independent biological replicates and 20-40 million, 100 base-pair, paired end reads were obtained for each sample. Reference files of the genome build hg19 (human) and mm9 (mouse) as well as ensembl transcript annotations were obtained from iGenomes (http://support.illumina.com/sequencing/sequencing_software/igenome.ilmn). Reads were aligned to the genome using the split read aligner Tophat (v2.0.7) and Bowtie2 (v2.0.5)18. Transcript assembly and isoform specific quantitation was performed using Cufflinks (v2.1.1). Abundance of individual isoforms is reported as Fragments per Kilobase of transcript per Million mapped reads (FPKM). Computations were performed on the Odyssey cluster supported by the FAS Science Division Research Computing Group at Harvard University. No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those generally employed in the field.
In situ hybridization
A 658 base pair fragment spanning exons 2-6 of spliced human C9ORF72 was amplified from a human whole brain cDNA library. Primers used were TCTCCAGCTGTTGCCAAGAC, AGTGTGAGCTGATGGCATTGA. For mouse, primers used were TTGGCGGCTACCTTTGCTTA, GTCTGCAGGTGTGAGCTGAT. The amplicon was purified, A-tailed, cloned into the pGEM-T expression vector (Promega), and sequenced. Digoxigenin (DIG)-labeled antisense riboprobe was transcribed in vitro from the linearized C9ORF72 cDNA clone (Roche), purified, and hybridized to acetylated human spinal cord sections overnight at 65 C. After stringent washes in 0.2× SSC at 65 C, sections were probed with anti-DIG antibody directly coupled with horseradish peroxidase (Roche) for 1 hour, washed, and developed for 20 minutes with Cy3 tyramide (Perkin Elmer) in the presence of 10% dextran sulfate and 2 mM of 4-iodophenol (Sigma Aldrich). Stained tissue was then washed with PBST and mounted with Aqua Polymount (Polysciences).
Human sample
Adult human spinal cord (70 year old, male) was derived from Upstate New York Transplant Services (UNYTS). The protocol was approved by Harvard IRB.
Supplementary Material
Supplementary Figure 1. Gene tree of C9ORF72 gene and its orthologs. Note that C9ORF72 orthologs are highly conserved among several species, especially in vertebrata. Black line represents ×1 branch length, while blue line represents ×10 branch length. Green box represents alignment match, while white box represents alignment gap. F18A1.6 in C elegans has 23 % identity with human C9ORF72.
Supplementary Figure 2. Amino acid sequence of human C9ORF72 and mouse 3110043O21Rik gene. Upper and lower lane represent human and mouse ortholog respectively.
Supplementary Figure 3. Comparison of splicing isoforms between human C9ORF72 (a) and mouse 3110043O21Rik gene (b). Black boxes represent translated exons and grey boxes represent predicted non-coding exons. Blue font represents translated isoforms. Red arrowhead and dashed line represent the location of hexanucleotide repeat expansion in human patients with ALS/FTD (a) and analogous site of patient C9ORF72 hexanucleotide repeat (b). RNA sequence data (c-e). Isoform 1 is highly expressed in mouse cortex (c), mouse purified ES derived motor neuron (d), and human purified ES derived motor neuron (e). FPKM, Fragments Per Kilobase of exon per Million mapped fragments. Error bars indicate s.d. n=2 for (c), n=3 for (d) and (e).
Supplementary Figure 4. Comparison of sequence between human C9ORF72, mouse 3110043O21Rik and chimpanzee LOC465031 genes around the sequence of hexanucleotide repeats in human. 58.3 % identities are found between human and mouse. Note that GC rich region in both sequences.
Supplementary Figure 5. X-gal staining in the brain and several organs of C9ORF72-ortholog knockin mice. (a) 0.6 mm from the midline. A, anterior; P, posterior; OB, olfactory bulb; cc, corpus callosum; CPu, caudate putamen (striatum); DC, granular dentate gyrus; thal, thalamus; cereb, cerebellum. Lung (b), liver (c), and kidney (d) are negative for X-gal staining. Testis (e) is shown as positive control. Bar, 1 mm (a) and 50 μm (b-e).
Supplementary Figure 6. Co-localization of β-gal, Cux1, and NeuN is shown (a). β-gal positive cells are also located in superficial layers (b). (b) shows Z-stack and orthogonal images of white rectangle in (a). Bar, 50 μm (a) and 20 μm (b).
Supplementary Figure 7. Co-localization of β-gal, ChAT, and NeuN in ventral horn of spinal cord. (b-d) shows Z-stack and orthogonal images of white rectangle in (a). Arrowheads show ChAT positive motor neurons and arrows are ChAT negative cells. Bar, 50 μm (a) and 20 μm (b-d).
Supplementary Figure 8. β-gal positive cell does not co-localize with IbaI and GFAP in anterior horn of spinal cord (a-f). Images are taken from the same slice. Z-stack images are also shown. Green-β-gal, red-IbaI (e) or GFAP (f) and blue-DAPI. Bar, 50 μm (a-f).
Supplementary Figure 9. Brain expression pattern of C9ORF72 ortholog from Allen Brain Atlas (a-l) Expression pattern of C9ORF72-ortholog of brain from Allen Brain Atlas database. Allen Brain Atlas used probes from exon 4 through exon 11. In situ hybridization of C9ORF72-ortholog in sagittal section of mice brain at P56: Nissl staining (a) and expression image (b) from similar slices. Higher magnification of cortex (c, d), hippocampus (e, f), brainstem (g, h), cerebellum (i, j), and thalamus (k, l) from similar slices. Nissl staining (a, c, e, g, i, k) and expression of C9ORF72-ortholog (b, d, f, h, j, l). Note that hippocampus has higher expression. Bar, 1 mm (a, b) and 300 μm (c-l). Note that each image is a distinct section.
Supplementary Figure 10. Spinal cord expression pattern of C9ORF72 ortholog from Allen Brain Atlas (a-g) Expression pattern of C9ORF72-ortholog in spinal cord from Allen Brain Atlas database. Allen Brain Atlas used probes from exon 4 through exon 11. In situ hybridization of C9ORF72-ortholog in sagittal section of mouse brain at P56: Nissl staining (a) and expression image (b-g) from similar slices. Lower (a-d) and higher (e-g) magnification are presented. Expression pattern of C9ORF72-ortholog (d, e), ChAT (b, f) and GFAP (c, g) are shown. Note that C9ORF72-ortholog transcript distributes mainly in the grey matter and displays a pattern similar to, but modestly broader than ChAT. White line shows the border between grey and white matter (e-g). Bar, 200 μm (a-d), 100 μm (e-g). note that each image is from a distinct section.
Supplementary Table 1. Distribution of C9ORF72-ortholog in CNS
Supplementary Table 2. X-gal expression during embryonic and postnatal stages in CNS. N/A, not applicable.
Acknowledgments
The authors wish to thank all K.E. lab members for their technical support and helpful discussions. We also thank HSCRB Histology Core. This work was supported by HHMI, Project ALS, p2ALS, Target ALS and NINDS grant #164520. N.S. was supported by 2011 Lilly Scientific Fellowship Program.
Footnotes
Note: Supplementary information is available in the online version of the paper.
Author Contributions: K.E. conceived the project; K.E., N.S. and A.M. designed all the experiments. K.K. supported animal experiments. A.M., A. I. and N.S. supported staining. F.M. and I.A. support in situ hybridization. R.M. and B. D. support RNA sequenceing data analysis. N.S., A.M., and K. E. wrote the manuscript. K. E. supervised the project.
Competeing Financial Interests: The authors declare no competing financial interests.
References
- 1.Ludolph AC, Brettschneider J, Weishaupt JH. Amyotrophic lateral sclerosis. Current opinion in neurology. 2012;25:530–535. doi: 10.1097/WCO.0b013e328356d328. [DOI] [PubMed] [Google Scholar]
- 2.Rademakers R, Neumann M, Mackenzie IR. Advances in understanding the molecular basis of frontotemporal dementia. Nature reviews Neurology. 2012;8:423–434. doi: 10.1038/nrneurol.2012.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mackenzie IR, et al. Nomenclature for neuropathologic subtypes of frontotemporal lobar degeneration: consensus recommendations. Acta neuropathologica. 2009;117:15–18. doi: 10.1007/s00401-008-0460-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeJesus-Hernandez M, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Renton AE, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Majounie E, et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet neurology. 2012;11:323–330. doi: 10.1016/S1474-4422(12)70043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gijselinck I, et al. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet neurology. 2012;11:54–65. doi: 10.1016/S1474-4422(11)70261-7. [DOI] [PubMed] [Google Scholar]
- 8.Reddy K, Zamiri B, Stanley SY, Macgregor RB, Pearson CE. The disease-associated r(GGGGCC)n repeat from the C9ORF72 gene forms tract length-dependent uni- and multi-molecular RNA G-quadruplex structures. The Journal of biological chemistry. 2013 doi: 10.1074/jbc.C113.452532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mori K, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 10.Ash PE, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skarnes WC, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henry AM, Hohmann JG. High-resolution gene expression atlases for adult and developing mouse brain and spinal cord. Mammalian genome : official journal of the International Mammalian Genome Society. 2012;23:539–549. doi: 10.1007/s00335-012-9406-2. [DOI] [PubMed] [Google Scholar]
- 13.Henry AM, Hohmann JG. High-resolution gene expression atlases for adult and developing mouse brain and spinal cord. Mammalian genome : official journal of the International Mammalian Genome Society. 2012;23:539–549. doi: 10.1007/s00335-012-9406-2. [DOI] [PubMed] [Google Scholar]
- 14.Skarnes WC, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arlotta P, Molyneaux BJ, Jabaudon D, Yoshida Y, Macklis JD. Ctip2 controls the differentiation of medium spiny neurons and the establishment of the cellular architecture of the striatum. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:622–632. doi: 10.1523/JNEUROSCI.2986-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Giorgio FP, Carrasco MA, Siao MC, Maniatis T, Eggan K. Non-cell autonomous effect of glia on motor neurons in an embryonic stem cell-based ALS model. Nature neuroscience. 2007;10:608–614. doi: 10.1038/nn1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Giorgio FP, Boulting GL, Bobrowicz S, Eggan KC. Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying an ALS-causing mutation. Cell stem cell. 2008;3:637–648. doi: 10.1016/j.stem.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 18.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Gene tree of C9ORF72 gene and its orthologs. Note that C9ORF72 orthologs are highly conserved among several species, especially in vertebrata. Black line represents ×1 branch length, while blue line represents ×10 branch length. Green box represents alignment match, while white box represents alignment gap. F18A1.6 in C elegans has 23 % identity with human C9ORF72.
Supplementary Figure 2. Amino acid sequence of human C9ORF72 and mouse 3110043O21Rik gene. Upper and lower lane represent human and mouse ortholog respectively.
Supplementary Figure 3. Comparison of splicing isoforms between human C9ORF72 (a) and mouse 3110043O21Rik gene (b). Black boxes represent translated exons and grey boxes represent predicted non-coding exons. Blue font represents translated isoforms. Red arrowhead and dashed line represent the location of hexanucleotide repeat expansion in human patients with ALS/FTD (a) and analogous site of patient C9ORF72 hexanucleotide repeat (b). RNA sequence data (c-e). Isoform 1 is highly expressed in mouse cortex (c), mouse purified ES derived motor neuron (d), and human purified ES derived motor neuron (e). FPKM, Fragments Per Kilobase of exon per Million mapped fragments. Error bars indicate s.d. n=2 for (c), n=3 for (d) and (e).
Supplementary Figure 4. Comparison of sequence between human C9ORF72, mouse 3110043O21Rik and chimpanzee LOC465031 genes around the sequence of hexanucleotide repeats in human. 58.3 % identities are found between human and mouse. Note that GC rich region in both sequences.
Supplementary Figure 5. X-gal staining in the brain and several organs of C9ORF72-ortholog knockin mice. (a) 0.6 mm from the midline. A, anterior; P, posterior; OB, olfactory bulb; cc, corpus callosum; CPu, caudate putamen (striatum); DC, granular dentate gyrus; thal, thalamus; cereb, cerebellum. Lung (b), liver (c), and kidney (d) are negative for X-gal staining. Testis (e) is shown as positive control. Bar, 1 mm (a) and 50 μm (b-e).
Supplementary Figure 6. Co-localization of β-gal, Cux1, and NeuN is shown (a). β-gal positive cells are also located in superficial layers (b). (b) shows Z-stack and orthogonal images of white rectangle in (a). Bar, 50 μm (a) and 20 μm (b).
Supplementary Figure 7. Co-localization of β-gal, ChAT, and NeuN in ventral horn of spinal cord. (b-d) shows Z-stack and orthogonal images of white rectangle in (a). Arrowheads show ChAT positive motor neurons and arrows are ChAT negative cells. Bar, 50 μm (a) and 20 μm (b-d).
Supplementary Figure 8. β-gal positive cell does not co-localize with IbaI and GFAP in anterior horn of spinal cord (a-f). Images are taken from the same slice. Z-stack images are also shown. Green-β-gal, red-IbaI (e) or GFAP (f) and blue-DAPI. Bar, 50 μm (a-f).
Supplementary Figure 9. Brain expression pattern of C9ORF72 ortholog from Allen Brain Atlas (a-l) Expression pattern of C9ORF72-ortholog of brain from Allen Brain Atlas database. Allen Brain Atlas used probes from exon 4 through exon 11. In situ hybridization of C9ORF72-ortholog in sagittal section of mice brain at P56: Nissl staining (a) and expression image (b) from similar slices. Higher magnification of cortex (c, d), hippocampus (e, f), brainstem (g, h), cerebellum (i, j), and thalamus (k, l) from similar slices. Nissl staining (a, c, e, g, i, k) and expression of C9ORF72-ortholog (b, d, f, h, j, l). Note that hippocampus has higher expression. Bar, 1 mm (a, b) and 300 μm (c-l). Note that each image is a distinct section.
Supplementary Figure 10. Spinal cord expression pattern of C9ORF72 ortholog from Allen Brain Atlas (a-g) Expression pattern of C9ORF72-ortholog in spinal cord from Allen Brain Atlas database. Allen Brain Atlas used probes from exon 4 through exon 11. In situ hybridization of C9ORF72-ortholog in sagittal section of mouse brain at P56: Nissl staining (a) and expression image (b-g) from similar slices. Lower (a-d) and higher (e-g) magnification are presented. Expression pattern of C9ORF72-ortholog (d, e), ChAT (b, f) and GFAP (c, g) are shown. Note that C9ORF72-ortholog transcript distributes mainly in the grey matter and displays a pattern similar to, but modestly broader than ChAT. White line shows the border between grey and white matter (e-g). Bar, 200 μm (a-d), 100 μm (e-g). note that each image is from a distinct section.
Supplementary Table 1. Distribution of C9ORF72-ortholog in CNS
Supplementary Table 2. X-gal expression during embryonic and postnatal stages in CNS. N/A, not applicable.