Abstract
Tannerella forsythia is among the most potent triggers of periodontal diseases, and approaches to understand underlying mechanisms are currently intensively pursued. A ~22-nm-thick, 2D crystalline surface (S-) layer that completely covers Tannerella forsythia cells is crucially involved in the bacterium–host cross-talk. The S-layer is composed of two intercalating glycoproteins (TfsA-GP, TfsB-GP) that are aligned into a periodic lattice. To characterize this unique S-layer structure at the nanometer scale directly on intact T. forsythia cells, three complementary methods, i.e., small-angle X-ray scattering (SAXS), atomic force microscopy (AFM), and single-molecular force spectroscopy (SMFS), were applied. SAXS served as a difference method using signals from wild-type and S-layer-deficient cells for data evaluation, revealing two possible models for the assembly of the glycoproteins. Direct high-resolution imaging of the outer surface of T. forsythia wild-type cells by AFM revealed a p4 structure with a lattice constant of ~9.0 nm. In contrast, on mutant cells, no periodic lattice could be visualized. Additionally, SMFS was used to probe specific interaction forces between an anti-TfsA antibody coupled to the AFM tip and the S-layer as present on T. forsythia wild-type and mutant cells, displaying TfsA-GP alone. Unbinding forces between the antibody and wild-type cells were greater than with mutant cells. This indicated that the TfsA-GP is not so strongly attached to the mutant cell surface when the co-assembling TfsB-GP is missing. Altogether, the data gained from SAXS, AFM, and SMFS confirm the current model of the S-layer architecture with two intercalating S-layer glycoproteins and TfsA-GP being mainly outwardly oriented.
Keywords: Scanning probe microscopy, small-angle X-ray scattering, S-layer structure, Tannerella forsythia, force spectroscopy, antibody recognition
INTRODUCTION
Many prokaryotic cells are decorated with a 2-D crystalline cell surface (S-) layer. S-layers are natural self-assembly systems in which individual (glyco)proteins are aligned into lattices with nanometer-scale periodicity. S-layer lattices exhibit oblique (p1, p2), square (p4), or hexagonal (p3, p6) symmetry, and lattice constants range from ~10 to 25 nm (Messner et al., 2010; Sabet et al., 2003; Sleytr and Beveridge, 1999). These dimensions make the S-layer system attractive for nanobiotechnology applications where organization of matter at the nanometer scale is desired. While it is generally assumed that S-layer provides to the bacteria a selection advantage in the native habitat, different specific functions have been discussed for S-layers, such as serving as protective coats against external host or natural environmental forces or acting as a molecular sieve and ion traps (Sleytr and Beveridge, 1999; Sabet et al., 2003; Messner et al., 2010). Currently, a novel function of S-layers is evolving concerning their participation in the bacterium–host cross-talk.
In this context, the S-layer of the Gram-negative oral pathogen Tannerella forsythia is being investigated (Sabet et al., 2003; Sakakibara et al., 2007; Posch et al., 2011, 2012). T. forsythia constitutes, together with Porphyromonas gingivalis and Treponema denticola, the so-called “red-complex” of bacteria, which has been implicated in the onset and development of periodontal diseases in humans (Holt and Ebersole, 2005; Inagaki et al., 2006; Chalabi et al., 2010). Despite the clinical evidence that T. forsythia also impacts systemic health, underlying virulence mechanisms are only slowly beginning to unravel (Chalabi et al., 2010; Sekot et al., 2011; Posch et al., 2012). The T. forsythia S-layer was shown to be a virulence factor, capable of delaying the bacterium’s recognition by the innate immune system of the host and mediating adhesion to and invasion of host cells (Sakakibara et al., 2007; Posch et al., 2011, 2012; Sekot et al., 2011; Settem et al., 2013). Thus, it is an important task to characterize the mechanical structure and the biochemical nature of the T. forsythia S-layer on living cells in order to contribute to a detailed understanding of the outermost cell surface of this bacterium, which serves as the immediate contact zone between the bacterium and its environment.
T. forsythia possesses a so far unique glycosylated S-layer consisting of the two regularly arrayed glycoproteins TfsA-GP (calculated molecular mass of the protein portion, 135 kDa) and TfsB-GP (calculated molecular mass of the protein portion, 152 kDa) (Sabet et al., 2003; Lee et al., 2006; Sakakibara et al., 2007). This is a unique situation because most S-layers are composed of a single (glyco)protein species (Messner et al., 2010). Recently, the cell surface ultrastructure of T. forsythia was investigated by transmission electron microscopy (TEM) and by immune fluorescence microscopy (Sekot et al., 2012). TEM analyses of T. forsythia wild-type cells revealed a distinct square S-layer lattice with an overall lattice constant of 10.1 ± 0.7 nm, while a blurred lattice with a lattice constant of ~9.0 nm was found on some areas of T. forsythia ΔtfsA (T. forsythia devoid of the TfsA S-layer glycoprotein) and ΔtfsB (T. forsythia devoid of the TfsA S-layer glycoprotein) mutant cells (Sakakibara et al., 2007; Sekot et al., 2012). Our current model of the S-layer architecture on T. forsythia cells is a ~22-nm-thick monolayer with square lattice symmetry that is formed by co-assembly of the two intercalating, presumably “mushroom”-like glycoproteins TfsA-GP and TfsB-GP in an equimolar ratio, with the “hut” of TfsA-GP mainly outwardly oriented and that of TfsB-GP oriented into the opposite direction toward the outer membrane (Sekot et al., 2012).
Despite providing valuable information, these traditional analysis methods do not allow the investigation of the bacterial cell surface under environmental conditions and at the subcellular level. Since atomic force microscopy (AFM) allows resolving surface nanostructures in their native environments with nanometer resolution, it is also well suited for characterizing the architecture of bacterial surfaces and heterogeneities of their mechanical properties (Dupres et al., 2009; Oh et al., 2012). In fact, this approach was already shown to be applicable to T. forsythia wild-type cells in an initial experiment, yielding a periodic square lattice structure of a center-to-center spacing in the range of 9.1 ± 0.8 nm (Sekot et al., 2012), thereby demonstrating the power of this technique. An additional important feature of AFM is the possibility to explore the mechanical forces of single-molecular ligand–receptor interactions under physiological conditions at the molecular level with high precision and accuracy. Such interactions determine the function and structure of many biomolecular systems (Tang et al., 2009; Bozna et al., 2011). As a remarkable advance, the AFM-based single-molecule manipulation technique allows measuring of molecule-molecule interaction forces, mechanical tensions, molecular transitions, and ligand–receptor dissociation. Theoretical models have been established to elucidate the underlying physics and chemistry (Evans and Ritchie, 1997; Dudko et al., 2008; Hummer and Szabo, 2010). Small-angle X-ray scattering (SAXS) offers another option for determining the structure of biological macromolecules in different environments under conditions close to those in physiological settings (Svergun and Koch, 2003). Its application to S-layer research on intact bacteria was demonstrated recently (Sekot et al., 2013).
In this study, we apply SAXS and AFM to characterize the native S-layer structure on intact T. forsythia cells in physiological environment. Since the scattering volume of the S-layer is low in comparison to the scattering volume of the bacterium, for SAXS data evaluation, the difference signal of wild-type and S-layer-deficient T. forsythia cells is used. AFM allows for direct measurement of the S-layer topology on the bacterial cells. To evaluate our current model of the T. forsythia S-layer architecture with two intercalating S-layer glycoproteins being aligned into a periodic lattice, the specific interaction forces between the TfsA S-layer glycoprotein as present on wild-type and TfsB-GP-deficient mutant cells, respectively, and its corresponding antibody are investigated using the single-molecular force spectroscopy (SMFS) technique. This study allowed the elucidation of the S-layer ultrastructure and its subunit arrangement with nanometer resolution on intact T. forsythia cells.
MATERIALS AND METHODS
Bacterial strain, mutants, and cultivation
Tannerella forsythia ATCC 43037 was purchased from the American Type Culture collection (ATCC, Manassas, VA, USA). T. forsythia S-layer mutants (i.e., ΔtfsA, ΔtfsB, and ΔtfsAB) were obtained upon insertional inactivation of the respective S-layer gene(s) (Sakakibara et al., 2007). Anaerobic cultivation of T. forsythia was done in 30 g/l tryptic soy broth (Gerbu, Gaiberg, Germany), supplemented with 5 g/l yeast extract (Becton Dickinson, Heidelberg, Germany), 5 g/l phytone peptone (Becton Dickinson), 0.2 g/l cysteine (Sigma, Vienna, Austria), 20 ml/l horse serum (PAA, Linz, Austria), 2.5 μg/ml hemine (Sigma), 2 g/ml menadione (Sigma), and 10 μg/ml N-acetylmuramic acid (Sigma) at 37°C for 4 days.
Raising, purification, and labeling of polyclonal antibodies against the T. forsythia S-layer proteins were described previously (Sekot et al., 2012).
SAXS measurement
A dense suspension of T. forsythia wild-type and S-layer-deficient (ΔtfsAB) cells, respectively, was filled into quartz glass capillaries that were sealed. This setup was chosen to clearly distinguish the SAXS signals obtained from the S-layer from those evoked by the bacterial cell itself (i.e., the S-layer-deficient mutant ΔtfsAB). Subsequently, SAXS was performed with a rotating anode generator equipped with a pinhole camera (Nanostar, Bruker AXS, Karlsruhe, Germany) and CuKα radiation monochromatized and collimated from cross Goebel mirrors. X-ray patterns were measured with a 2D position-sensitive detector (Vantec 2000, Bruker AXS) for 6 h for each sample. The scattering patterns were radially averaged to obtain the scattering intensity in dependence on the scattering vector q = 4π/λ sinθ, where λ = 0.1542 nm is the X-ray wavelength and 2θ the scattering angle.
For SAXS data evaluation, the value for wild-type cells was subtracted from the value for S-layer-deficient cells. After division of the signal by the form factor for infinite plates, the intensity modulations were interpreted as the structure factor to determine the size and arrangement of the two glycoproteins TfsA-GP and TfsB-GP (Sekot et al., 2013).
Conjugation of the anti-TfsA antibody to the AFM tip
Commercially available AFM cantilevers (MSCT, Bruker, Camarillo, CA, USA) with a nominal spring constant of 0.01–0.03 N/m were functionalized with amino groups by using the 3-aminopropyltriethoxysilane (APTES) coating procedure (Ebner et al., 2007). Then, a heterobifunctional aldehyde–polyethyleneglycol (PEG) linker with a length of 6–9 nm was attached to the APTES-coated cantilever via its NHS ester followed by incubating the cantilevers for 2 h at 25°C in 500 μl of chloroform containing 3.3 mg of aldehyde–PEG–NHS and 30 μl of triethylamine. Subsequently, cantilevers were washed with chloroform and dried with nitrogen gas. Anti-TfsA antibody diluted in phosphate buffered saline (PBS) to a concentration of 0.2 mg/ml was conjugated onto the tip via the reaction between a lysine residue of the antibody and an aldehyde–PEG bound to the tip. 4 μl of a mixture containing 32 mg of NaCNBH3, 50 μl of 100 mM NaOH, and 450 μl of H2O was added to 200 μl of antibody solution, and cantilevers were immersed in this solution for 2 h. After incubation, ethanolamine (Sigma) (1 M in distilled water, pH 9.6) was added to the mixture to yield a concentration of 50 mM in order to inactivate free aldehyde groups on the cantilever tip. After 10 min, the cantilevers were washed with PBS and stored in PBS at 4°C.
Sample preparation for AFM
Cells from 1 ml of T. forsythia culture were collected by centrifugation (1500 g, 2 min), washed twice with 1 ml of PBS, and resuspended in 1 ml of PBS. Subsequently, the resuspended bacteria were immobilized by mechanical trapping on 0.8 μm polycarbonate membranes (Millipore) for noninvasive in vitro imaging by AFM (Dufrêne, 2004). After filtering of the suspension of bacteria with nitrogen gas pressure (~0.4 atm), the filters were gently rinsed with PBS and attached to the sample holder using a double-side adhesive tape. The mounted sample was measured in the AFM liquid cell (Sekot et al., 2012). Since this method for sample preparation does not involve chemical treatment or a drying process, it neither affects cell viability nor triggers undesirable biological responses.
AFM measurements
All AFM measurements were carried out in PBS using a commercial Agilent 5500 AFM (Agilent Technologies, Chandler, AZ, USA) in a fluid cell containing PBS. Magnetically coated AFM cantilevers (type VII MAC lever, Agilent Technologies) with a nominal spring constant of 0.1 N/m were used for magnetic AC mode (MAC mode) imaging. The resonance frequency of the cantilever was selected between 9 and 11 kHz in liquid, and the measurement frequency was set to 20% below the resonance frequency. Scan line speed was 0.5 Hz.
Measurement of antibody recognition forces by scanning probe microscopy
Force distance cycles were performed at 25°C using anti-TfsA antibody-modified tips with 0.01–0.03 N/m nominal spring constants. The spring constant of the cantilevers was determined by measuring the thermally driven mean-square bending of the cantilever. The deflection sensitivity was calculated from the slope of the force–distance curves recorded on bare glass substrate.
In the force spectroscopy experiments, force–distance curves were acquired by recording at least thousand force–distance cycles with vertical sweep rates between 0.5 and 10 Hz at a z-range of typically 500–1000 nm, resulting in loading rates from 100 to 5000 pN/s. The loading rates were determined by multiplying the pulling velocity with the effective spring constant prior to unbinding.
The relationship between experimentally measured unbinding forces and the interaction potential is described by kinetic models from Bell (Bell, 1978) and Evans and Ritchie (Evans and Ritchie, 1997; Merkel et al., 1999). The unbinding force F* is given as function of the loading rate r
| (1) |
where kBT is the Boltzmann thermal energy, χβ marks the thermally averaged projection of the transition state along the direction of the force, and koff is the kinetic off rate for the bond in absence of applied load.
Blocking of the specific interaction between the anti-TfsA-antibody and the TfsA-GP as present on the surface of T. forsythia cells (wild-type and mutants) was done by injecting of recombinant TfsA protein (solubilized in PBS at a concentration of 0.2 mg/ml) into the bath solution. Recombinant TfsA protein was available from a previous study (Sekot et al., 2012). All AFM and force spectroscopy experiments were repeated on at least five different preparations of the bacterial cells analyzing different cell surface areas.
RESULTS
Analysis of the S-layer structure by SAXS
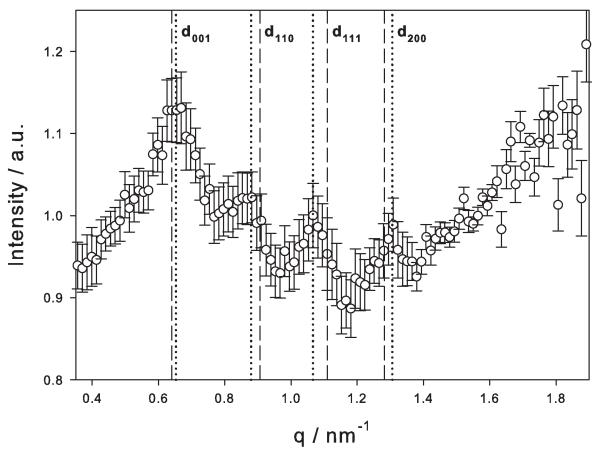
In a first approach, the cell surface of T. forsythia was characterized by SAXS. Figure 1 shows the scattering intensity of T. forsythia wild-type cells after subtracting the signal from the S-layer-deficient cells (ΔtfsAB). According to a previously elaborated strategy for measuring S-layers on intact bacteria (Sekot et al. 2013), the scattering intensity was fitted with the Fourier transform of an infinite plate with a finite thickness. As in general the scattering intensity is the product of form factor (the plate) and structure factor, division of the experimental intensity by the fitted plate function gives access to the structure factor, i.e., the ordered arrangement of the two S-layer glycoproteins TfsA-GP and TfsB-GP within the S-layer lattice. These intensity modulations are shown in Figure 2, together with the error obtained from the program GNOM (Svergun, 1992) on the basis of a Monte-Carlo technique (Rolbin et al., 1980; Svergun and Pedersen, 1994).
Figure 1.
Scattering intensity of T. forsythia wild-type cells after subtracting the signal from the S-layer-deficient cells (ΔtfsAB).
Figure 2.
SAXS data evaluation: Scattering intensity after division of the experimental data by the fit function for the Fourier transform of a plate (line in Figure 1). Dashed lines, reflections for a cubic lattice with only two layers (one shifted by half of the lattice constant toward the face diagonal); dotted lines, slightly distorted monoclinic lattice.
Although the relative intensity of the modulations is rather small, surprisingly not only peaks from pure p4 symmetry were found. This might be interpreted in two ways. Firstly, taking our current model of the T. forsythia S-layer architecture into account, implicating the presence of the two intercalating, mushroom-shaped S-layer glycoproteins TfsA-GP and TfsB-GP, with the “hut” of the former being mainly outwardly oriented, one might interpret the S-layer as a double layer of two units (of rather the same size and the same scattering contrast; i.e., TfsA-GP and TfsB-GP); an indexation of the peaks according to the structure as shown in Figure 3 would be consistent with the observed data. Reflections are only visible if h + k + 2l is even and l ≤ 1 due to the structure factor Fhkl= f1+ f2 exp(2 πi(h + k + 2l)) and nearly identical units with individual unit form factors f1 ≅ f2, respectively. The dashed lines show the positions of the reflections from a two-layer cubic lattice with in-plane lattice constant 9.8 nm and the second layer shifted by half of the lattice constant in direction of the face diagonal. An improvement of the positions of the reflections (dotted lines) would be a slight distortion of the lattice by an angle of 95° between the units, an in-plane size of 9.7 nm, and a layer-to-layer distance of 10.4 nm (see Figure 3). Secondly, another option might be that the observed modulations are partly evoked by the rough-type lipopolysaccharide (Posch et al., 2013) that is presumably evenly distributed over the outer membrane of the T. forsythia cell wall, where it is proposed to serve as an anchor for the S-layer.
Figure 3.
Cubic lattice with two layers (left image), leading to reflections shown as dashed lines in Figure 2. Slightly distorted monoclinic lattice (right image), with reflections on position of the dotted lines in Figure 2.
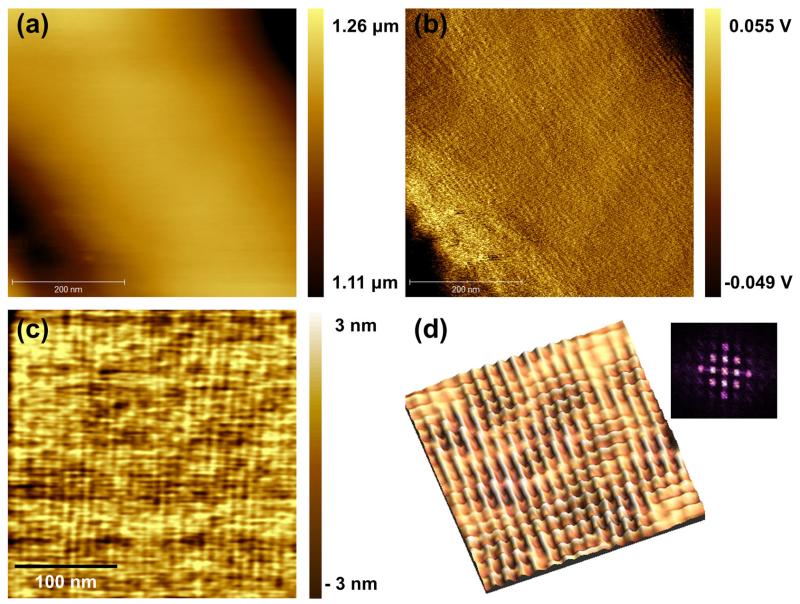
AFM analysis of the S-layer topography
Atomic force microscopy topography and amplitude images were acquired in liquid using MAC mode imaging to determine the S-layer ultrastructure as present on intact T. forsythia cells at the nanometer scale. T. forsythia wild-type bacteria are 3.5 ± 1 μm in length and 0.6 ± 0.1 μm in diameter, resulting in a large curvature of the bacterial cells (Sekot et al., 2012). Thus, the amplitude image (Figure 4b) provides higher local contrast and is more sensitive to changes in the surface topology in comparison to the topography image (Figure 4a). T. forsythia wild-type cells possess a distinct, periodic square (p4) lattice (Figure 4c), which is even more clearly revealed in the high-resolution reconverted 3D fast Fourier transform (FFT) image obtained from an enlarged topography image (Figure 4d). Peaks are due to surface periodicities, which are represented in the power spectra as the bright peaks near the origin (inset in Figure 4d). According to the AFM analyses, the p4 S-layer lattice on intact T. forsythia wild-type cells has a lattice constant of 8.74 ± 0.35 nm and an angle of 90° ± 4°; this refines previous AFM data obtained from wild-type cells of this bacterium revealing a lattice constant of 9.10 ± 0.8 nm (Sekot et al., 2012) and is also in good agreement with earlier observations made by TEM using freeze-fractured and freeze-dried preparations of bacterial cells (Sekot et al., 2012). While these data clearly confirm the presence of a closed 2D crystalline S-layer as the outermost cell surface structure of T. forsythia wild-type cells, the S-layer lattice was visible neither on the S-layer single mutants (ΔtfsA and ΔtfsB) nor on the S-layer-deficient double mutant (ΔtfsAB) that served as a negative control (data not shown).
Figure 4.
AFM images of T. forsythia wild-type bacteria: (a) topography, (b) amplitude image, (c) magnified topographic image, and (d) reconverted FFT 3D image of topography; inset, power spectrum of lattice. The p4 symmetry is clearly visible.
Determination of interaction forces between the anti-TfsA antibody and the TfsA-glycoprotein by SMFS
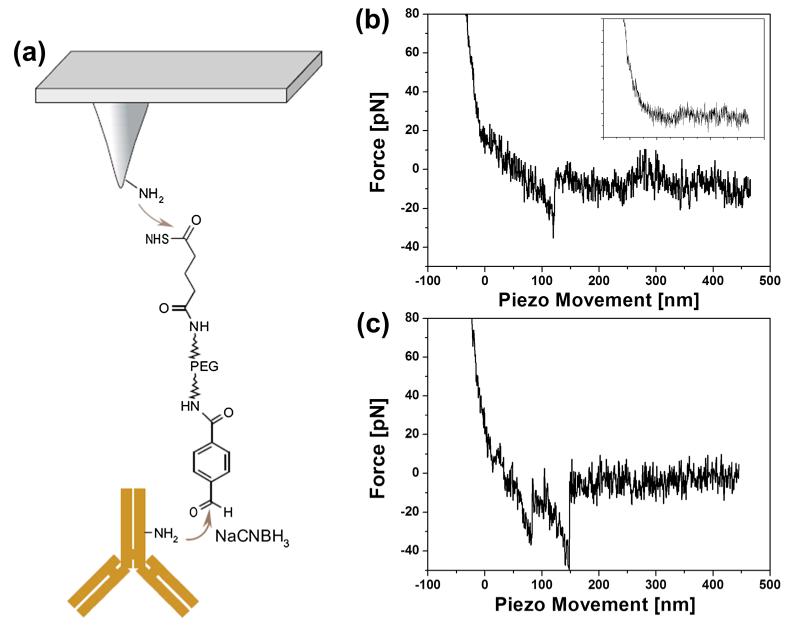
Single-molecular force spectroscopy was used to identify specific interactions between the anti-TfsA antibody and the corresponding TfsA-GP within the TfsA/TfsB S-layer glycoprotein lattice on the bacterial surface. Figure 5a shows the schematic design of the tip chemistry used for covalent antibody immobilization on the amino-functionalized tip end via a flexible PEG linker. Force–distance curves were measured by approaching the antibody-conjugated AFM tip to the surface of the intact T. forsythia wild-type cells followed by its retraction (Figure 5b). The nonlinear behavior of the force curve signal during retraction of the cantilever reflects the elastic extension of the PEG linker. The subsequently occurring unbinding event (visible as spike at about 120 nm piezo movement) corresponds to the dissociation between the anti-TfsA antibody on the tip and the TfsA-GP present within the native TfsA-GP/TfsB-GP S-layer lattice.
Figure 5.
(a) Scheme of the immobilization strategy of anti-TfsA antibody on the AFM tip via a flexible PEG linker, (b) typical unbinding event in force–distance cycles (inset, blocking of the specific interaction by injecting of recombinant TfsA protein into the bath solution), and (c) example of multiple unbinding events in force traces.
Based on the SMFS data, the anti-TfsA antibody-conjugated tip specifically recognizes the TfsA-GP as present on the bacterial wild-type cell surface (Figure 5b). In most of the measurements, single unbinding events were observed. Occasionally, multiple unbinding events were measured that were attributed to the serial rupture of two Fab fragments of the antibody bound to two TfsA-GPs (shown in Figure 5c).
The force curves were analyzed as described by Baumgartner et al. (Baumgartner et al., 2000) and Rankl et al. (Rankl et al., 2007). Each rupture event was used to calculate the distribution of specific unbinding forces, fitted with a Gaussian function, and added up to yield the empirical probability density function (pdf), as shown in Figure 6a. Most probable unbinding forces (extracted from the maxima of the pdfs) between the anti-TfsA antibody immobilized on the AFM tip were 41 pN with wild-type cells and 56 pN with TfsB-GP-deficient cells (ΔtfsB), where only the TfsA-GP is exposed on the cell surface, at a loading rate of anti-TfsA antibody of 1000 nm/s, each. In contrast, for TfsA-GP-deficient cells (ΔtfsA) and for the double knock-out mutant (data not shown), anti-TfsA antibody binding was largely absent. The rarely occurring binding events were attributed to unspecific tip surface adhesion. Differences of unbinding forces between wild-type and TfsB-deficient cells may occur due to the orientation and flexibility of the binding site of the TfsA-GP on the bacterial cell surface. The TfsA-GP on wild-type bacteria is embedded in the S-layer lattice structure (Sekot et al., 2012), whereas the TfsA-GP does not appear to be that strictly fixed on the surface of the ΔtfsB mutant where the space-filling counterpart (TfsB-GP) is missing (compare with Sekot et al., 2012).
Figure 6.
(a) Distribution of unbinding forces between anti-TfsA antibody and the bacterial cell surface (wild-type and S-layer single mutants) at a retraction velocity of 1000 nm/s, (b) comparison of binding probabilities of the anti-TfsA antibody-conjugated AFM tip to wild-type, TfsB-deficient (ΔtfsB) and TfsA-deficient (ΔtfsA) cells, as well as the blocking experiment, and (c) a plot of loading rate dependence versus unbinding force.
The binding probability, which represents the frequency of specific interaction events in force–distance cycles, is shown in Figure 6b. Wild-type and ΔtfsB cells showed 38% and 46% of binding probability to the anti-TfsA antibody conjugated to the tip, respectively. So as to prove that the measured forces were due to the specific interactions, antibody-blocking experiments were performed. After addition of recombinant TfsA protein to block the anti-TfsA antibody on the tip, the binding probability decreased to 7% for wild-type and 6% for ΔtfsB cells, respectively.
According to the theory that a single energy barrier is crossed in the thermally activated regime, a linear rise of the unbinding force with respect to a logarithmically increasing loading rate (pulling velocity × effective spring constant, cf. Materials and Methods) is expected (Sulchek et al., 2005; Rankl et al., 2008). Thus, most probable unbinding forces were determined from the maxima of pdfs (see, e.g., Figure 6a) obtained at different loading rates. They were plotted as a function of the logarithm of the loading rate for wild-type and ΔtfsB cells, respectively. Error bars account for the uncertainty in finding the most probable rupture force and in the uncertainty in determining the effective spring constant. The effective spring constant was calculated from the spring constant of the cantilever and that of the PEG-tethered cantilever, the latter of which was determined at the point rupture by fitting a worm-like chain (WLC) model (Marko and Siggia, 1995; Rankl et al., 2007).
Kinetic off-rates, koff, and the distance from the energy minimum to the transition state, χβ, were estimated by fitting the most probable unbinding forces in dependence on different loading rates (indicated with fitted line in Figure 6c), according to Equation (1). The values for koff and χβ of the interaction between the anti-TfsA antibody and the TfsA-GP as present on the cell surface of both wild-type and ΔtfsB cells are given in Table 1. For the anti-TfsA antibody/TfsA-GP interaction within the TfsA-GP/TfsB-GP S-layer lattice as present on the surface of wild-type bacteria, values of 4.46 Å for χβ and 0.751 S−1 for koff were obtained, whereas for the anti-TfsA antibody/TfsA-GP interaction on ΔtfsB cells, χβ was 5.49 Å and koff was 0.076 S−1. Differences of koff between wild-type and TfsB-deficient cells (ΔtfsA) might be explained by a less tight binding of the TfsA-GP to the anti-TfsA antibody when integrated in the native TfsA-GP/TfsB-GP S-layer lattice compared to the higher degree of spatial flexibility of the TfsA-GP on the surface of the ΔtfsB mutant.
Table 1.
Comparison of kinetic off-rates, koff, and the distance from the energy minimum to the transition state, χβ, on T. forsythia wild-type and ΔtfsB cells obtained by single-molecular force spectroscopy
| χβ [Å] | koff [S−1] | |
|---|---|---|
| Wild-type | 4.46 ± 1.33 | 0.751 ± 0.766 |
| ΔtfsB mutant | 5.49 ± 1.66 | 0.076 ± 0.13 |
The variation in the obtained kinetic parameters (Figure 6) accounted for the stochastic nature of the unbinding process and for errors of the measurements (e.g., spring constant, Gauss fitting for most probable unbinding force, and pulling speed).
DISCUSSION
In this study, we investigated the topological and mechanical structure of the S-layer as present on living T. forsythia cells using SAXS and AFM techniques in order to refine our current model of the T. forsythia S-layer architecture as a basis for a more detailed understanding of the immediate contact zone of this oral pathogen with its environment, which, in turn, is pivotal for unraveling mechanisms governing the bacterium–host cross-talk. This current T. forsythia S-layer model is based on TEM and immune fluorescence microscopy in combination with biochemical and genetic data of the constituting S-layer glycoproteins TfsA-GP and TfsB-GP (Sekot et al., 2012). It benefited from the availability of S-layer single mutants (named ΔtfsA for T. forsythia devoid of the TfsA S-layer glycoprotein, and ΔtfsB for T. forsythia devoid of the TfsB S-layer glycoprotein) and an S-layer-deficient ΔtfsAB double mutant, in addition to the wild-type bacterium (Sakakibara et al., 2007), all of which were used also in the present study. TEM analyses of T. forsythia wild-type cells revealed a distinct square S-layer lattice with an overall lattice constant of 10.1 ± 0.7 nm, while a blurred lattice with a lattice constant of 9.0 nm was found on some areas of T. forsythia ΔtfsA and ΔtfsB cells (Sekot et al., 2012). We interpreted this slight decrease in the lattice constant as a consequence of the individual S-layer glycoproteins (TfsA-GP and TfsB-GP) assuming a different conformation depending on the composition of the S-layer, i.e., monospecies-versus co-assembled S-layer lattice. Immune fluorescence microscopy experiments, in which fluorescence-labeled, polyclonal antibodies raised against the individual recombinant S-layer proteins (TfsA and TfsB) were used, allowed for an even clearer picture of the architecture of the T. forsythia S-layer. Since only for TfsA-GP as fluorescence signal could be obtained but not for TfsB-GP, we hypothesized that the antibody recognition sites of the folded TfsB-GP as present in the TfsA-GP/TfsB-GP S-layer lattice were not accessible because they might be either buried within the lattice or oriented toward the underlying outer membrane. Considering that the overall thickness of the S-layer was identical (~22 nm) on wild-type and on S-layer single mutant cells, we favored the interpretation of a T. forsythia S-layer lattice that is formed by co-assembly of the two intercalating, “mushroom”-like glycoproteins TfsA-GP and TfsB-GP in equimolar ratio, with the “hut” of the former being mainly outwardly oriented.
That model of the T. forsythia S-layer could now be supported and refined by analyzing the cell surface of intact bacteria under physiological conditions providing a more nature-like picture of the cell surface. One of the applied techniques was SAXS. The application of SAXS to S-layer research on intact bacteria was demonstrated only recently with the bacteria Geobacillus stearothermophilus ATCC 12980 and Aquaspirillum serpens MW5 (Sekot et al., 2013). It was shown in that study that SAXS and TEM are complementary methods to determine the structure of S-layers. Whereas TEM gives local information after templating, measuring in vacuum, and subsequent image processing, SAXS is able to measure the biological structure in the native environment. SAXS measurements of S-layers on intact bacteria face the difficulty that the scattering volume of the layer is small in comparison to the bacterium itself, but the applicability of the method and its accuracy were recently demonstrated (Sekot et al., 2013). In the present study, the scattering intensity of the S-layer of T. forsythia wild-type cells was obtained after subtracting the signal from the S-layer-deficient mutant cells (ΔtfsAB). SAXS data evaluation revealed a p4 S-layer lattice structure with a lattice constant of ~9.8 nm. Unexpectedly, in this experimental setup, not only peaks from pure p4 symmetry were found, but also additional modulations of weak intensities were present. This might be interpreted in two ways. Firstly, taking our current model of the T. forsythia S-layer architecture into account implicating the presence of the two intercalating, mushroom-shaped S-layer glycoproteins TfsA-GP and TfsB-GP, with the former being mainly outwardly oriented, one might interpret the S-layer as a double layer of two units (of rather the same size and the same scattering contrast, i.e., TfsA-GP and TfsB-GP). Secondly, another option might be that the observed modulations are partly evoked by the rough-type lipopolysaccharide (Posch et al., 2013) that is presumably evenly distributed over the outer membrane of the T. forsythia cell wall, where it is proposed to serve as an anchor for the S-layer. Undulations in S-layers have previously been observed when they were adsorbed on bulk lipid bilayers (Hirn et al., 1999). One has certainly to admit that the intensity modulations in Figure 1 are rather weak and only four peaks are visible. Thus, one should be cautious in the interpretation, and the presented Figure 3 is only one possible model. It is supported and derived from observations made by TEM and AFM (see below) analyses, which both gave a lattice constant in the range of 10 nm (Sekot et al., 2012).
In our AFM images, the native T. forsythia S-layer present on wild-type cells shows a distinct lattice structure with a periodicity of around 9.0 nm and an angle of about 90°. As AFM yields images of upper surfaces, these recordings represent the topography of the outermost assembly on the wild-type T. forsythia S-layer. In contrast, the cell surface of the S-layer single mutants (ΔtfsA, ΔtfsB) did not reveal any periodic structures in AFM. Thus, it appears that large-scale compact ordering requires a delicate molecular ordering of TfsA-GP and TfsB-GP. We also measured the interaction forces between the TfsA-GP on the bacterial surface and anti-TfsA antibody using SMFS. Higher unbinding forces and lower koff rates were required to dissociate the anti-TfsA antibody from the TfsA glycoprotein on ΔtfsB cells. This suggests a lack of structural flexibility or accessibility of TfsA-GP within the confined lattice structure on T. forsythia wild-type cells.
In conclusion, we demonstrated the potential of combining SAXS, AFM, and SMFS as three complementary techniques for elucidating the topography and subunit arrangement of the two-component S-layer on intact T. forsythia cells under physiological conditions. Such insights might constitute the basis for unraveling novel mechanisms governing the bacterium–host cross-talk. Especially in the context of pathogenic bacteria, such as the oral pathogen T. forsythia, this might have a direct biomedical impact.
Acknowledgements
This study was funded by a Marie Curie International Incoming Fellowship of the 7th European Community Framework Program (to Y.J.O.). Financial support came from the Austrian Science Fund projects P20605-B12 and P24317-B22 (to C.S.).
REFERENCES
- Baumgartner W, Hinterdorfer P, Schindler H. Data analysis of interaction forces measured with the atomic force microscope. Ultramicroscopy. 2000;82(1-4):85–89. doi: 10.1016/s0304-3991(99)00154-0. [DOI] [PubMed] [Google Scholar]
- Bell GI. Models for the specific adhesion of cells to cells. Science. 1978;200(4342):618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- Bozna BL, Polzella P, Rankl C, Zhu R, Salio M, Shepherd D, Duman M, Cerundolo V, Hinterdorfer P. Binding strength and dynamics of invariant natural killer cell T cell receptor/CD1d-glycosphingolipid interaction on living cells by single molecule force spectroscopy. J. Biol. Chem. 2011;286(18):15973–15979. doi: 10.1074/jbc.M110.192674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalabi M, Rezaie F, Mogharehabed A, Rezaei M, Mehraban B. Periodontopathic bacteria and herpesviruses in chronic periodontitis. Mol. Oral Microbiol. 2010;25(3):236–240. doi: 10.1111/j.2041-1014.2010.00571.x. [DOI] [PubMed] [Google Scholar]
- Dudko OK, Hummer G, Szabo A. Theory, analysis and interpretation of single–molecule force spectroscopy experiments. Proc. Natal. Acad. Sci. USA. 2008;105(41):15755–15760. doi: 10.1073/pnas.0806085105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufrêne YF. Refining our perception of bacterial surfaces with the atomic force microscope. J. Bacteriol. 2004;186(11):3283–3285. doi: 10.1128/JB.186.11.3283-3285.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupres V, Alsteens D, Pauwels K, Dufrêne YF. In vivo imaging of S-layer nanoarrays on Corynebacterium glutamicum. Langmuir. 2009;25(17):9653–9655. doi: 10.1021/la902238q. [DOI] [PubMed] [Google Scholar]
- Ebner A, Wildling L, Kamruzzahan AS, Rankl C, Wruss J, Hahn CD, Holzl M, Zhu R, Kienberger F, Blaas D, Hinterdorfer P, Gruber HJ. A new, simple method for linking of antibodies to atomic force microscopy tips. Bioconj. Chem. 2007;18(4):1176–1184. doi: 10.1021/bc070030s. [DOI] [PubMed] [Google Scholar]
- Evans E, Ritchie K. Dynamic strength of molecular adhesion bonds. Biophys. J. 1997;72(4):1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirn R, Schuster B, Sleytr UB, Bayerl TM. The effect of S-layer protein adsorption and crystallization on the collective motion of a planar lipid bilayer studied by dynamic light scattering. Biophys. J. 1999;77(4):2066–2074. doi: 10.1016/S0006-3495(99)77048-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt SC, Ebersole JL. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the ‘red complex’, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol. 2005;38(1):72–122. doi: 10.1111/j.1600-0757.2005.00113.x. 2000. [DOI] [PubMed] [Google Scholar]
- Hummer G, Szabo A. Free energy profiles from single-molecule pulling experiments. Proc. Natl. Acad. Sci. USA. 2010;107(50):21441–21446. doi: 10.1073/pnas.1015661107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki S, Onishi S, Kuramitsu HK, Sharma A. Porphyromonas gingivalis vesicles enhance attachment, and the leucine-rich repeat BspA protein is required for invation of epithelial cells by Tannerella forsythia. Infect. Immun. 2006;74(9):5023–5028. doi: 10.1128/IAI.00062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-W, Sabet M, Um H-S, Yang J, Kim HC, Zhu W. Identification and characterization of the genes encoding a unique surface (S-) layer of Tannerella forsythia. Gene. 2006;371(1):102–111. doi: 10.1016/j.gene.2005.11.027. [DOI] [PubMed] [Google Scholar]
- Marko JF, Siggia ED. Stretching DNA. Macromolecules. 1995;28(26):8759–8770. [Google Scholar]
- Merkel R, Nassoy P, Leung A, Ritchie K, Evans E. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature. 1999;397:50–53. doi: 10.1038/16219. [DOI] [PubMed] [Google Scholar]
- Messner P, Schäffer C, Egelseer EM, Sleytr UB. In: Prokaryotic Cell Wall Compounds-Structure and Biochemistry. König H, Claus H, Varma A, editors. Springer-Verlag; Berlin, Germany: 2010. pp. 53–109. [Google Scholar]
- Oh YJ, Cui Y, Kim H, Li Y, Hinterdorfer P, Park S. Characterization of Curli A production on living bacterial surfaces by scanning probe microscopy. Biophys. J. 2012;103(8):1666–1671. doi: 10.1016/j.bpj.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posch G, Pabst M, Brecker L, Altmann F, Messner P, Schäffer C. Characterization and scope of S-layer protein O-glycosylation in Tannerella forsythia. J. Biol. Chem. 2011;286(44):38714–38724. doi: 10.1074/jbc.M111.284893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posch G, Sekot G, Friedrich V, Megson ZA, Koerdt A, Messner P, Schäffer C. Glycobiology aspects of the periodontal pathogen Tannerella forsythia. Biomolecules. 2012;2(4):467–482. doi: 10.3390/biom2040467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posch G, Andrukhov O, Vinogradov E, Lindner B, Messner P, Holst O, Schäffer C. Structure and immunogenicity of the rough-type lipopolysaccharide from the periodontal pathogen Tannerella forsythia. Clin. Vaccine Immunol. 2013;20(6):945–953. doi: 10.1128/CVI.00139-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankl C, Kienberger F, Gruber H, Blass D, Hinterdorfer P. Accuracy estimation in force spectroscopy experiments. Jpn. J. Appl. Phys. 2007;46(8):5536–5539. [Google Scholar]
- Rankl C, Kienberger F, Wildling L, Wruss J, Gruber HJ, Blaas D, Hinterdorfer P. Multiple receptors involved in human rhinovirus attachment to live cells. Proc. Natl. Acad. Sci. USA. 2008;105(46):17778–17783. doi: 10.1073/pnas.0806451105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolbin A, Svergun DI, Shchedrin BM. Smoothing experimental curves of small-angle X-ray scattering. Sov. Phys. Crystallogr. 1980;25:133–137. [Google Scholar]
- Sabet M, Lee S-W, Nauman RK, Sims T, Um S-S. The surface (S-) layer is a virulence factor of Bacterioides forsythus. Microbiology. 2003;149(12):3617–3627. doi: 10.1099/mic.0.26535-0. [DOI] [PubMed] [Google Scholar]
- Sakakibara J, Nagano K, Murakami Y, Higuchi N, Nakamura H, Shimozato K, Yoshimura F. Loss of adherence ability to human gingival epithelial cells in S-layer protein-deficient mutants of Tannerella forsythensis. Microbiology. 2007;153(3):866–876. doi: 10.1099/mic.0.29275-0. [DOI] [PubMed] [Google Scholar]
- Sekot G, Posch G, Messner P, Matejka M, Rausch-Fan X, Andrukhov O, Schäffer C. Potential of the Tannerella forsythia S-layer to delay the immune response. J. Dent. Res. 2011;90(1):109–114. doi: 10.1177/0022034510384622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekot G, Posch G, Oh YJ, Zayni S, Mayer HF, Pum D, Messner P, Hinterdorfer P, Schäffer C. Analysis of the cell surface layer ultrastructure of the oral pathogen Tannerella forsythia. Arch. Microbiol. 2012;194(6):525–539. doi: 10.1007/s00203-012-0792-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekot G, Schuster D, Messner P, Pum D, Peterlik H, Schäffer C. Small-angle X-ray scattering for imaging of surface layers on intact bacteria in the native environment. J. Bacteriol. 2013;195(10):2408–2414. doi: 10.1128/JB.02164-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settem RP, Honma K, Nakajima T, Phansopa C, Roy S, Stafford GP, Sharma A. A bacterial glycan core linked to surface (S)-layer proteins modulates host immunity through Th17 suppression. Mucosal Immunol. 2013;6(2):415–426. doi: 10.1038/mi.2012.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleytr UB, Beveridge TJ. Bacterial S-layers. Trends Microbiol. 1999;7(6):253–260. doi: 10.1016/s0966-842x(99)01513-9. [DOI] [PubMed] [Google Scholar]
- Sulchek TA, Friddle RW, Langry K, Lau EY, Albrecht H, Ratto TV, DeNardo SJ, Colvin MW, Noy A. Dynamic force spectroscopy of parallel individual mucin1-antibody bonds. Proc. Natl. Acad. Sci. USA. 2005;102(46):16638–16643. doi: 10.1073/pnas.0505208102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Cryst. 1992;25:495–503. [Google Scholar]
- Svergun DI, Koch MHJ. Small-angle scattering studies of biological macromolecules in solution. Rep. Prog. Phys. 2003;66:1735–1782. [Google Scholar]
- Svergun DL, Pedersen JS. Propagating errors in small-angle scattering data treatment. J. Appl. Cryst. 1994;27(3):241–248. [Google Scholar]
- Tang J, Ebner A, Kraxberger B, Leitner M, Hykollari A, Kepplinger C, Grunwald C, Gruber HJ, Tampé R, Sleytr UB, Ilk N, Hinterdorfer P. Detection of metal binding sites on functional S-layer nanoarrays using single molecule force spectroscopy. J. Struct. Biol. 2009;168(1):217–222. doi: 10.1016/j.jsb.2009.02.003. [DOI] [PubMed] [Google Scholar]