Abstract
Most eukaryotic genes are transcribed into mRNAs with alternative poly(A) sites. Emerging evidence suggests that mRNA isoforms with alternative poly(A) sites can perform critical regulatory functions in numerous biological processes. In recent years, a number of strategies utilizing high-throughput sequencing technologies have been developed to aid in the identification of genome-wide poly(A) sites. This unit describes a modified protocol of a recently published 3’READS (3’ region extraction and deep sequencing) method that accurately identifies genome-wide poly(A) sites and that can be used to quantify the relative abundance of the resulting 3’ mRNA isoforms. This approach minimizes non-specific sequence reads due to internal priming and typically yields a high percentage of sequence reads that are ideally suited for accurate poly(A) identification.
Keywords: 3’ mRNA isoforms, polyadenylation, poly(A) sites, RNA sequencing
INTRODUCTION
Alternative 3’ end formation adds an additional level of regulatory control to gene expression, and there is growing evidence for the importance of transcripts with alternative polyadenylation (poly(A)) sites in the regulation of a diverse number of biological processes (Elkon et al., 2013; Tian and Manley, 2013). These alternative poly(A) sites give rise to distinct 3’ mRNA isoforms possessing 3’ untranslated regions (UTRs) of different lengths. The individual mRNA isoforms have the potential to exhibit key biological differences, for example due to the presence or absence of sequences or structures important for their processing, export, or decay. Indeed, alternative 3’ mRNA isoforms arising from the same gene can sometimes exhibit highly different stabilities (Geisberg et al., 2014).
The prevalence of alternative poly(A) sites means that a frequent objective in transcriptome analysis is the precise identification of mRNA 3’ ends. In addition to identifying all the mRNA species arising from a particular gene, it is also often important to determine the relative abundance of the various isoforms. Common methods to identify poly(A) sites generally incorporate annealing of oligonucleotides to poly(A) tail, either by using oligo-dT priming from the poly(A) tail during the construction of DNA sequencing libraries or by direct annealing poly(A) tail to oligo-dT on the sequencing flow cell.
The basic protocol in this unit is modified from the procedure for 3’ Region Extraction And Deep Sequencing (3’READS) (Hoque et al., 2013), developed for mapping genome-wide poly(A) sites and quantifying the relative abundance of the resulting 3’ mRNA isoforms. At the heart of the procedure is the initial annealing of the mRNA poly(A) tails to a hybrid RNA/DNA oligonucleotide consisting of 5 U and 45 T nucleotides. A subsequent digestion with RNase H cleaves only the RNA-DNA portion of the resulting duplex, leaving intact the ~5 bp of U-A in RNA-RNA duplex at the beginning of the poly(A) tail. This has the effect of preserving the junction between the poly(A) site and the poly(A) tail, allowing more precise mapping of the 3’ boundary of the encoded mRNA after these fragments are amplified and sequenced. During the data analysis steps, the number of A bases at the 3’ terminus of RNA is compared to the number of As encoded in the genome, and any terminal As not encoded in the genome are considered to be added during polyadenylation process.
BASIC PROTOCOL
CONSTRUCTING AN INDEXED DNA SEQUENCING LIBRARY FROM 3’ mRNA ISOFORMS
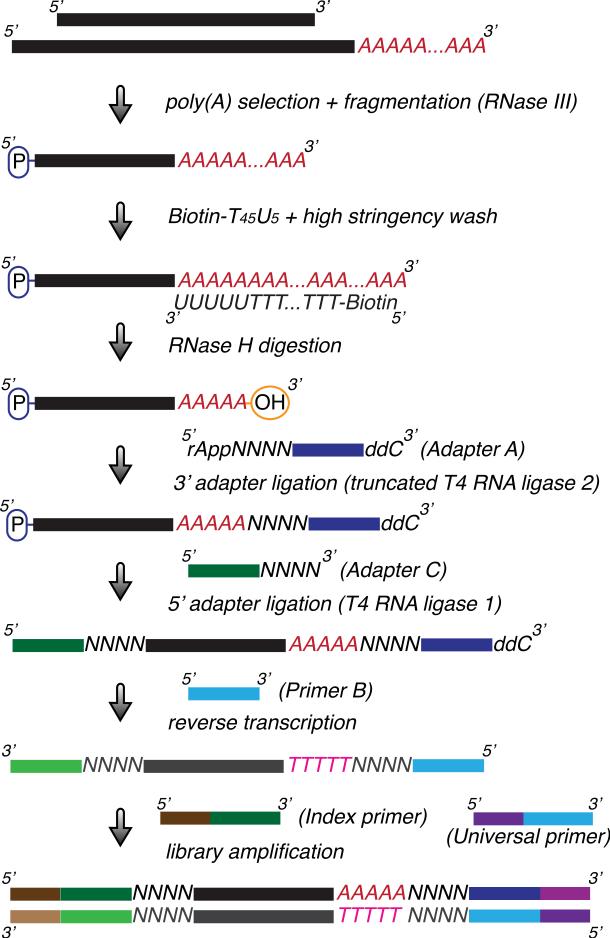
This section describes the detailed protocol for the creation of an indexed DNA library for sequencing poly(A) sites (3’ mRNA isoforms) on a genome-wide scale on the Illumina sequencing platform. This protocol is modified from the published 3’READS method (Hoque et al., 2013; Hoque et al., 2014). The procedure is outlined in Figure 1.
Figure 1.
Schematic of an indexed DNA library preparation for sequencing poly(A) sites (3’ mRNA isoforms) on a genomic scale. Poly(A)-tailed RNAs are first enriched and fragmented by RNase III digestion. 3’ polyadenylated RNA fragments captured by Dynabeads coated with a Biotin-T45U5 hybrid DNA/RNA oligo are subjected to RNase H digestion to generate RNA fragments containing 3’ termini with only a few remaining As. After ligation of linkers (Adapters A and C) to both ends, the resulting RNA fragments are reverse-transcribed to yield single-stranded cDNA fragments using a primer (Primer B) based on the sequence of Adapter A. The cDNA fragments are in turn amplified with a universal primer (Universal primer) and a second primer containing the desired index (e.g., Index primer 1) to generate a double-stranded DNA library. The quality and quantity of the purified DNA library can be examined using the Agilent Bioanalyzer and Quantitative-PCR.
Materials
25 μg purified total RNA (UNIT 4.1, 4.2, 4.3 and 4.16)
Oligonucleotides (see Table 1)
Table 1.
Oligonucleotide Sequences
| Oligonucleotides | Sequence |
|---|---|
| Biotin-T45U5* | 5′-Biotin-TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT UUUUU-3′ |
| Adapter A** | 5′-/5rApp/NNNNGATCGTCGGACTGTAGAACTCTGAAC/3ddC/-3′ |
| Primer B | 5′-GTTCAGAGTTCTACAGTCCGACGATC-3′ |
| Adapter C | 5′-CCUUGGCACCCGAGAAUUCCANNNN-3′ |
| lniversal primer | 5′-AATGATACGGCGACCACCGAGATCTACACGTTCAGAGTTCTACAGTCCGA-3′ |
| Index primer 1 (ATCACG) | 5′-CAAGCAGAAGACGGCATACGAGATCGTGATGTGACTGGAGTTCCTTGGCACCCGAGAATTCCA-3′ |
| Index primer 2 (CGATGT) | 5′-CAAGCAGAAGACGGCATACGAGATACATCGGTGACTGGAGTTCCTTGGCACCCGAGAATTCCA-3′ |
| Index primer 3 (TTAGGC) | 5′-CAAGCAGAAGACGGCATACGAGATGCCTAAGTGACTGGAGTTCCTTGGCACCCGAGAATTCCA-3′ |
| Index primer 4 (TGACCA) | 5′-CAAGCAGAAGACGGCATACGAGATTGGTCAGTGACTGGAGTTCCTTGGCACCCGAGAATTCCA-3′ |
| Index primer 5 (ACAGTG) | 5′-CAAGCAGAAGACGGCATACGAGATCACTGTGTGACTGGAGTTCCTTGGCACCCGAGAATTCCA-3′ |
| Index primer 6 (GCCAAT) | 5′-CAAGCAGAAGACGGCATACGAGATATTGGCGTGACTGGAGTTCCTTGGCACCCGAGAATTCCA-3′ |
| Index primer 7 (CAGATC) | 5′-CAAGCAGAAGACGGCATACGAGATGATCTGGTGACTGGAGTTCCTTGGCACCCGAGAATTCCA-3′ |
| Index primer 8 (ACTTGA) | 5′-CAAGCAGAAGACGGCATACGAGATTCAAGTGTGACTGGAGTTCCTTGGCACCCGAGAATTCCA-3′ |
| Index primer 9 (GATCAG) | 5′-CAAGCAGAAGACGGCATACGAGATCTGATCGTGACTGGAGTTCCTTGGCACCCGAGAATTCCA-3′ |
| Index primer 10 (TAGCTT) | 5′-CAAGCAGAAGACGGCATACGAGATAAGCTAGTGACTGGAGTTCCTTGGCACCCGAGAATTCCA-3′ |
| Index primer 11 (GGCTAC) | 5′-CAAGCAGAAGACGGCATACGAGATGTAGCCGTGACTGGAGTTCCTTGGCACCCGAGAATTCCA-3′ |
| Index primer 12 (CTTGTA) | 5′-CAAGCAGAAGACGGCATACGAGATTACAAGGTGACTGGAGTTCCTTGGCACCCGAGAATTCCA-3′ |
| Sequencing primer | 5′-CTACACGTTCAGAGTTCTACAGTCCGACGATC-3′ |
| Index sequencing primer | 5′-GGAATTCTCGGGTGCCAAGGAACTCCAGTCAC-3′ |
| Q-PCR checking primer F | 5′-CAAGCAGAAGACGGCATACGA-3′ |
| Q-PCR checking primer R | 5′-AATGATACGGCGACCACCGAGAT-3′ |
Italic letter represents an RNA nucleotide; N represents a random nucleotide.
Bold letters indicate an index region. Please note that the index sequence on the read is the reverse complement of the index sequence (in parentheses).
Biotin-T45U5 is a chimeric DNA/RNA oligonucleotide with a standard biotin modification at the 5′ end.
/5rApp/ refers to 5′ adenylation and /3ddC/ refers to dideoxycytidine, a 3′ chain terminator.
Oligo d(T)25 Magnetic Beads (NEB)
Dynabeads MyOne Streptavidin C1 (Invitrogen)
DNA SizeSelector-I (Aline)
Beads pre-wash buffer (see recipe)
2X Annealing buffer (see recipe)
2X Coupling buffer (see recipe)
2X Hybridization buffer (see recipe)
High-stringency wash buffer (see recipe)
RNase III digestion buffer (see recipe)
RNase H digestion buffer (see recipe)
Elution buffer (see recipe)
TE buffer (see recipe)
Gel extraction buffer (see recipe)
10 M NaOH (APPENDIX 2)
5 M NaCl (RNase-free; APPENDIX 2)
0.5 M EDTA (RNase-free; APPENDIX 2)
1 M Tris-Cl, pH7.5 (RNase-free; APPENDIX 2)
1 M Tris-Cl, pH8 (APPENDIX 2)
1 M Tris-Cl, pH8.3 (RNase-free; APPENDIX 2)
1 M MgCl2 (RNase-free; APPENDIX 2)
1 M DTT (RNase-free; APPENDIX 2)
3 M sodium acetate, pH 5.5 (RNase-free; APPENDIX 2)
TBE buffer (APPENDIX 2)
1 U/μl RNase III (NEB)
5 U/μl RNase H (NEB)
2 U/μl Phusion HF DNA polymerase (NEB)
20 U/μl SUPERase.In (Ambion)
10 U/μl T4 RNA ligase 1 (NEB)
200 U/μl truncated T4 RNA ligase 2 (NEB)
200 U/μl SuperScript III Reverse Transcriptase (Invitrogen)
12.5 and 25 mM dNTPs
Costar Spin-X centrifuge tube filter (0.45-μm cellulose acetate in 2-ml tube; Corning)
15 mg/ml GlycoBlue (Ambion)
Formamide, deionized (Ambion)
TWEEN 20 (Sigma)
10,000X SYBR Gold Nucleic Acid Gel Stain (Invitrogen)
Acid-Phenol:Chloroform, pH4.5 (Ambion)
100%, 80%, 70% ethanol
Nuclease-free H2O
Refrigerated centrifuge
Gel electrophoresis apparatus
8% TBE polyacrylamide gel (Invitrogen)
100 bp DNA Ladder (NEB)
PCR Marker (NEB)
6X Gel Loading Dye, Blue (NEB)
20-gauge needles
Vortex
0.5-ml thin-walled PCR tubes (non-stick, Ambion)
1.5-ml microcentrifuge tubes (non-stick, Ambion)
Thermal cycler
37°C, 55°C, 65°C heating blocks
Disposable scalpel
Transilluminator
Magnetic stand
Shaking platform
Prepare beads before the main procedure
Oligo d(T)25 Magnetic Beads preparation
Equilibrate the Oligo d(T)25 Magnetic Beads to room temperature (RT) for 20 min prior to use.
- Wash 100 μl of bead slurry with 500 μl of 1X Annealing buffer in a 1.5 ml microcentrifuge tube (per RNA sample). Collect the beads on a magnetic stand and discard the supernatant.Note: if bead slurry for multiple samples is prepared in a 1.5 ml microcentrifuge tube, use 1000 μl of 1X Annealing buffer Beads for each wash. Repeat the wash once.
Resuspend the washed beads in 200 μl of 1X Annealing buffer per RNA sample.
Biotin-T45U5 oligo-coupled Dynabeads MyOne Streptavidin C1 preparation
-
4Wash 20 μl of Dynabeads MyOne Streptavidin C1 slurry for each RNA sample with 100 μl of Beads pre-wash buffer in a 1.5 ml microcentrifuge tube. Collect the beads on a magnetic stand and discard the supernatant. Repeat the wash once.Note: if Dynabeads slurry for multiple samples (up to 7 samples) is prepared in a 1.5 ml microcentrifuge tube, use 1000 μl of Beads pre-wash buffer for each wash.
-
5Wash the beads with 100 μl of 1X coupling buffer. Collect the beads on a magnetic stand and discard the supernatant. Repeat the wash once.Note: if Dynabeads slurry for multiple samples is prepared in a 1.5 ml microcentrifuge tube, use 1000 μl of 1X coupling buffer for each wash.
-
6Add the following to the washed beads (per RNA sample) and incubate for 30 min at RT with gentle shaking. Collect the beads on a magnetic stand and discard the supernatant.
- 1 μl of 100 μM Biotin-T45U5 oligo
- 99 μl of nuclease-free H2O
- 100 μl of 2X Coupling buffer
Note: It can be scaled appropriately for the desired number of samples. -
7Wash the beads with 500 μl of 1X Hybridization buffer. Collect the beads on a magnetic stand and discard the supernatant. Repeat the wash twice.Note: if Dynabeads slurry for multiple samples is prepared in a 1.5 ml microcentrifuge tube, use 1000 μl of 1X Hybridization buffer for each wash.
-
8Resuspend the washed beads in 100 μl of 2X Hybridization buffer per RNA sample.Note: Biotin-T45U5 oligo-coupled Dynabeads MyOne Streptavidin C1 can be prepared ahead of time and are stable for up to 2 months at 4°C.
Main Procedure
Isolate 3’ end RNA fragments containing poly(A) sites
-
9Dilute 25 μg of high quality total RNA with 100 μl of nuclease-free H2O in a 1.5 ml microcentrifuge tube before adding 100 μl of 2X Annealing buffer.Note: RNA can be prepared by either a Hot-phenol extraction method or a TRIzol extraction method followed by purification using RNeasy Kit (Qiagen), including DNase I digestion, according to the manufacturer's instructions. The quality of RNA should be checked by electrophoresis or Agilent Bioanalyzer analysis (RNA 6000 Pico Kit).
-
10
Denature RNA sample from step 9 for 5 min at 65°C and then immediately cool on ice.
-
11
Pulse-spin the sample tube and mix the 200 μl of denatured RNA sample with 200 μl of washed Oligo d(T)25 Magnetic Beads from step 3. Incubate the tube for 30 min at RT with gentle shaking. Collect the beads on a magnetic stand and discard the supernatant.
-
12
Wash the beads with 200 μl of 1X Annealing buffer. Collect the beads on a magnetic stand and discard the supernatant. Repeat the wash twice.
-
13
Wash beads once in 50 μl of RNase III digestion buffer. Collect the beads on a magnetic stand and discard the supernatant.
-
14
Resuspend the RNA-bound beads in 50 μl of RNase III digestion buffer with 2.5 U of RNase III (add 2.5 μl of 1U/μl RNase III to 47.5 μl of RNase III digestion buffer). Incubate for 15 min at 37°C. Collect the beads on a magnetic stand and discard the supernatant.
-
15
Wash beads with 200 μl of 1X Annealing buffer. Collect the beads on a magnetic stand and discard the supernatant. Repeat the wash once.
-
16
Add 200 μl of TE buffer and gently resuspend the beads by pipetting. Incubate for 2 min at 55°C. Collect the beads on a magnetic stand and transfer the supernatant to a new 1.5-ml microcentrifuge tube.
-
17
Add 200 μl of acid phenol-chloroform and mix well. Centrifuge at 20,000 x g for 5 min at RT and carefully transfer 190 μl of the upper aqueous phase to a new 1.5-ml microcentrifuge tube.
-
18
Add 19 μl of 3 M sodium acetate and 1 μl of 15 mg/ml GlycoBlue to the purified RNA sample. Mix well. Add 475 μl of 100% ethanol, mix well, and incubate for at least 30 min at -20°C.
-
19
Centrifuge at 20,000 × g for 30 min at 4°C, to precipitate RNA.
-
20
Discard the supernatant and gently wash the pellet with 500 μl of 80% ice-cold ethanol. Centrifuge at 20,000 × g for 5 min at 4°C. Discard all the supernatant carefully without disturbing the pellet.
-
21Air-dry the pellet for 10 min at RT. Resuspend the air-dried RNA in 100 μl of nuclease-free H2O.Note: the size of purified RNA fragments is generally smaller than 1000 nt, and the majority should be around 100 – 300 nt. The size distribution of the RNA fragments can be examined by Agilent Bioanalyzer analysis (RNA 6000 Pico Kit). The RNA sample can be stored for up to 3 months at -80°C.
-
22
Denature RNA sample from step 21 for 5 min at 65°C and then immediately cool on ice.
-
23
Pulse-spin the sample tube and mix 100 μl of denatured RNA sample with 100 μl of prepared Biotin-T45U5 oligo-coupled Dynabeads MyOne Streptavidin C1 from step 8. Incubate for 30 min at RT with gentle shaking. Collect the beads on a magnetic stand and discard the supernatant.
-
24
Wash the beads with 500 μl of High-stringency wash buffer. Collect the beads on a magnetic stand and discard the supernatant. Repeat the wash twice.
-
25
Wash the beads with 100 μl of RNase H digestion buffer. Collect the beads on a magnetic stand and discard the supernatant.
-
26
Gently resuspend the beads in 50 μl of RNase H digestion buffer with 5 U of RNase H (add 1 μl of 5U/μl RNase H to 49 μl RNase H digestion buffer). Incubate for 30 min at 37°C. Collect the beads on a magnetic stand and transfer the supernatant to a new 1.5-ml microcentrifuge tube.
-
27
Collect any residual RNA by washing the beads with 150 μl of Elution buffer. Separate the beads on a magnetic stand and combine this supernatant with the one from step 26. The total collected volume should be 200 μl.
-
28
Add 200 μl of acid phenol-chloroform and mix well. Centrifuge at 20,000 × g for 5 min at RT and carefully transfer 190 μl of the upper aqueous phase to a new 1.5-ml microcentrifuge tube.
-
29
Add 19 μl of 3 M sodium acetate and 1 μl of 15 mg/ml GlycoBlue to the purified RNA sample. Mix well. Add 475 μl of 100% ethanol, mix well, and incubate for at least 30 min at -20°C.
-
30
Centrifuge at 20,000 × g for 30 min at 4°C, to precipitate RNA.
-
31
Discard the supernatant and gently wash the pellet with 500 μl of 80% ice-cold ethanol. Centrifuge at 20,000 × g for 5 min at 4°C. Discard all the supernatant carefully without disturbing the pellet.
-
32Air-dry the pellet thoroughly for 10 – 20 min at RT. Resuspend the air-dried RNA in 7 μl of nuclease-free H2O.Note: the quality and quantity of RNA fragments can be examined by Agilent Bioanalyzer analysis (RNA 6000 Pico Kit). The size distribution is similar to that seen in the RNA samples from step 21. The RNA sample can be stored for up to 3 months at -80°C.
Ligate linkers to RNA fragments and perform reverse transcription
Note: use a thermal cycler for all incubations
-
33
Transfer 6 μl of RNA from step 32 to a new 0.5-ml thin-walled PCR tube. Add 1 μl of 1 μM Adapter A (3’ DNA adapter) and mix by pipetting. Denature for 2 min at 70°C and then immediately cool on ice.
-
34Prepare the following ligation reaction master mix (per RNA sample) in a separate tube on ice and mix by pipetting.
- 1.5 μl of 10X T4 RNA ligase buffer (supplied with T4 RNA ligase 2, NEB)
- 1 μl of 20 U/μl SUPERase.In
- 4.5 μl of 50% PEG8000 (supplied with T4 RNA ligase 2, NEB)Note: It can be scaled appropriately for the desired number of samples. Dispense PEG8000 with a wide-bore tip to improve accuracy.
-
35
Add 7 μl of the reaction master mix from step 34 to the denatured sample from step 33. Add 1 μl of 200 U/μl truncated T4 RNA ligase 2 and mix by pipetting. Incubate for 2 hr at 25°C.
-
36
Add 1 μl of 5 μM Primer B (RT-PCR primer) to the ligation mix and mix by pipetting. Incubate the sample for 2 min at 70°C, then for 5 min at 37°C, followed by a cool down to 4°C.
-
37Prepare 5 μl of 5 μM Adapter C (5’ RNA adapter) in a new 0.5-ml thin-walled PCR tube. Denature for 2 min at 70°C and then immediately cool on ice.Note: 1 μl of Adapter C is needed for each RNA sample. We prefer to denature a larger volume of Adapter C (e.g., 5 – 10 μl) to avoid potential volume loss in denaturation process.
-
38Prepare the following ligation reaction master mix (per RNA sample) in a separate tube on ice and mix by pipetting.
- 0.5 μl of 10X T4 RNA ligase buffer (supplied with T4 RNA ligase 1, NEB)
- 1 μl of denatured Adapter C from step 37
- 1.5 μl of 10 mM ATP (supplied with T4 RNA ligase 1, NEB)Note: It can be scaled appropriately for the desired number of samples.
-
39
Add 3 μl of the reaction master mix from step 38 to the sample from step 36. Add 1 μl of 10 U/μl T4 RNA ligase 1 and mix by pipetting. Incubate for 1.5 hr at 25°C.
-
40
Add 7 μl of 5X First-Strand buffer (supplied with SuperScript III RT, Invitrogen) to the sample from step 39. Incubate for 5 min at 70°C and then immediately cool on ice.
-
41Prepare the following reverse transcription reaction master mix (per RNA sample) in a separate tube on ice and mix by pipetting.
- 1.75 μl of 12.5 mM dNTPs
- 3.5 μl of 100 mM DTT (supplied with SuperScript III RT, Invitrogen)
- 1 μl of 20 U/μl SUPERase.In
- 0.75 μl of nuclease-free H2ONote: It can be scaled appropriately for the desired number of samples.
-
42Add 7 μl of the reaction master mix from step 41 to the mixture from step 40. Add 1 μl of 200 U/μl SuperScript III reverse transcriptase and mix by pipetting. Incubate for 60 min at 50°C, then for 15 min at 70°C, followed by a cool down to 4°C.Note: If not proceeding immediately, the cDNA sample can be stored indefinitely at -20°C.
PCR amplify DNA library for sequencing
-
43Prepare the following PCR reaction master mix (per cDNA sample) for amplification in a separate tube on ice and mix by pipetting.
- 10 μl of 5X HF buffer (supplied with Phusion HF DNA polymerase, NEB)
- 0.5 μl of 25 mM dNTPs
- 1 μl of 25 μM Universal primer
- 19.5 μl of nuclease-free H2ONote: It can be scaled appropriately for the desired number of samples.
-
44
Transfer 17.5 μl of cDNA sample from step 42 to a new 0.5-ml thin-walled PCR tube. Add 1 μl of 25 μM desired Index primer and mix by pipetting.
-
45
Add 31 μl of PCR reaction master mix from step 43 to the mixture from step 44. Add 0.5 μl of 2 U/μl Phusion HF DNA polymerase and mix by pipetting.
-
46Perform PCR as follows:
- 98°C for 30 sec
- 16 cycles of 98°C for 30 sec, 60°C for 30 sec, 72°C for 30 sec
- 72°C for 10 min
- 4°C holdNote: Higher yield of DNA library can be achieved through increasing the number of PCR amplification cycles up to 18 cycles. Generally we don't find PCR reactions in this condition (16 – 18 cycles) to be saturated or to produce any spurious high molecular weight PCR products.
Purify amplified DNA library using DNA SizeSelector-I SPRI magnetic beads
-
47
Equilibrate the DNA SizeSelector-I SPRI magnetic beads to RT for 20 min.
-
48
Transfer the DNA library from step 46 to a new 1.5-ml microcentrifuge tube. Add nuclease-free H2O to adjust the final volume to 100 μl.
-
49Add 200 μl of thoroughly-vortexed SPRI bead slurry to the DNA sample from step 48 and mix by pipetting.Note: The recommended 2:1 volume ratio of SPRI bead slurry to DNA sample is specific for preferentially purifying DNA fragment larger than 200 bp (see explanation in step 54). The ratio should be empirically determined for every batch of SPRI beads using DNA size ladder as a reference (e.g., 100 bp DNA Ladder). Precise volumes of bead slurry and DNA sample are critical because even small ratio variations can significantly alter the size range of the DNA fragments retained by the SPRI beads. In order to minimize pipetting variation, we prefer to increase the DNA sample volume to 100 ul.
-
50
Incubate the mixture for 5 min at RT. Collect the beads on a magnetic stand and carefully discard the supernatant.
-
51
Wash the beads with 200 μl of 70% ethanol. Collect the beads on a magnetic stand and discard the supernatant. Repeat the wash once.
-
52Air-dry the beads on a magnetic stand for 10 min or until the beads are completely dry.Note: The dried patch of beads will show small “cracks” and other irregularities that are visible with careful inspection.
-
53Resuspend the dried beads in 22 μl of 10 mM Tris-Cl, pH 8. Collect the beads on a magnetic stand. Taking great care to avoid any carry-over of beads, transfer only 20 μl of the supernatant to a new 1.5-ml microcentrifuge tube.Note: the DNA library can be stored indefinitely at -20°C.
Evaluate DNA sequencing library
-
54Examine the quality and concentration of the purified DNA library using Agilent Bioanalyzer analysis (High-sensitivity DNA Kit).Note: A typical result from Agilent Bioanalyzer analysis is shown in Figure 2A. The concentration of the DNA library is calculated using only those DNA fragments with sizes ranging from 175 – 500 bp. A small amount of DNA smaller than 175 bp will remain in SPRI-beads-purified samples. This includes library DNAs with very short cDNA inserts (which are poor candidates for unique mapping to a reference genome), PCR primer dimers, and residual PCR primers. Gel purification is required to further extract DNA fragments with sizes only in the desired 175 – 500 bp range. If multiple uniquely indexed DNA libraries are planned to be sequenced in a single multiplexed sequencing run, the DNA libraries can be pooled according to the desired molar ratio at this stage before further gel purification. In determining the molar ratio, consider only the concentration of DNA fragments in the 175 – 500 bp range for each sample.
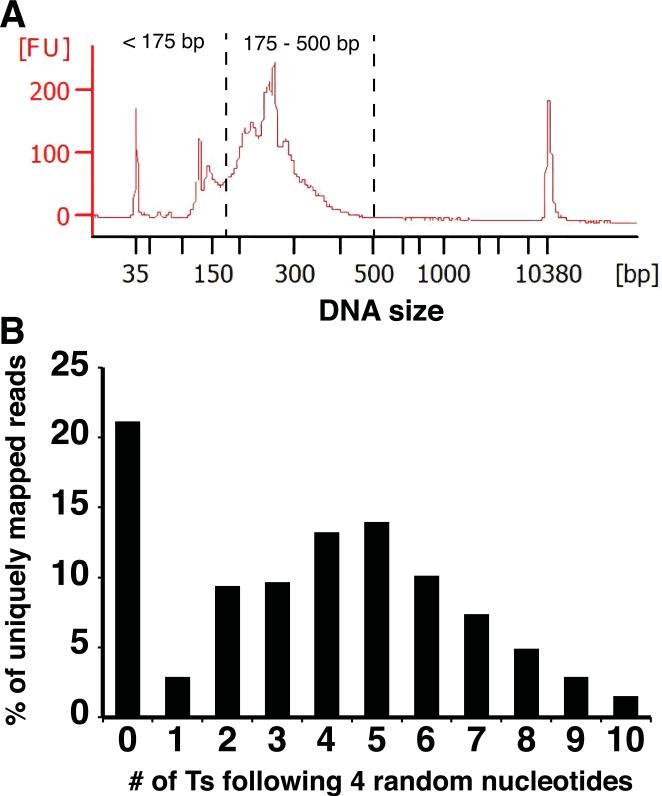
Figure 2.
(A) Agilent Bioanalyzer analysis (High-sensitivity DNA Kit) of a SPRI-beads-purified DNA library is shown. DNA fragments smaller than 175 bp include library DNAs with very short cDNA inserts, PCR primer dimers, and residual PCR primers. Gel purification is required to further extract only DNA fragments between 175 bp and 500 bp for Illumina sequencing. The DNA peaks at 35 bp and 10380 bp are high sensitivity DNA markers used in Agilent Bioanalyzer analysis. (B) Distribution of the number of consecutive Ts following four random nucleotides in uniquely mapped sequence reads. The stretch of Ts corresponds to the As remaining from poly(A) tails after RNase H digestion. Reads with no Ts are likely to be caused by RNase H over-digestion and contaminating non-3’ RNA fragments.
Purify the DNA sequencing library
-
55
Add an appropriate amount of 6X Gel Loading Dye to the DNA library and load the mixture to an 8% TBE polyacrylamide gel. Run at 180 V for 40 min.
-
56
Stain gel with SYBR Gold (1:10,000 dilution) in 30 ml TBE buffer for 3 min at RT with gentle shaking.
-
57Excise DNA fragments in the 175 – 500 bp range with a disposable scalpel on a transilluminator.Note: avoid inclusion of any smaller DNA fragments.
-
58
Transfer the excised gel piece to a 0.5-ml thin-walled PCR tube with the bottom pierced through by a 20-gauge needle. Nest the 0.5-ml PCR tube on top of a new 1.5-ml microcentrifuge tube.
-
59
Centrifuge the nested tubes at 20,000 × g for 3 min at RT, to effectively pulverize the relatively large gel piece into smaller pieces by squeezing the gel through the hole in the pierced tube.
-
60
Add 400 μl of Gel extraction buffer to resuspend the pulverized gel pieces. Incubate overnight at RT with agitation.
-
61
Transfer the whole gel/buffer mixture to the filter chamber of a Costar Spin-X centrifuge tube. Centrifuge at 20,000 × g for 2 min at RT.
-
62
Transfer 400 μl of the eluate from the collection tube to a new 1.5-ml microcentrifuge tube. Add 1 μl of 15 mg/ml GlycoBlue to the eluted DNA sample and mix well. Add 1000 μl of 100% ethanol, mix well, and incubate for at least 30 min at -20°C.
-
63
Centrifuge at 20,000 × g for 30 min at 4°C, to precipitate DNA.
-
64
Discard the supernatant and gently wash the pellet with 750 μl of 80% ice-cold ethanol. Centrifuge at 20,000 × g for 5 min at 4°C. Discard all the supernatant carefully without disturbing the pellet.
-
65Air-dry the pellet for 10 min. Resuspend the air-dried DNA in 15 μl of 10 mM Tris-Cl, pH 8.Note: the DNA sequencing library can be stored indefinitely at -20°C. The quality and concentration of the gel-purified DNA library can be examined by Agilent Bioanalyzer analysis (High-sensitivity DNA Kit). The DNA library can be readily processed on Illumina HiSeq sequencing platform for single-end sequencing using the sequencing and index sequencing primers listed in Table 1 (Both primers are those standardly used in Illumina TruSeq Small RNA sequencing runs).
SUPPORT PROTOCOL
Sequencing data analysis
The protocol here briefly describes how the raw sequencing data can be processed to identify poly(A) sites (Figure 3) and also describes typical downstream analyses.
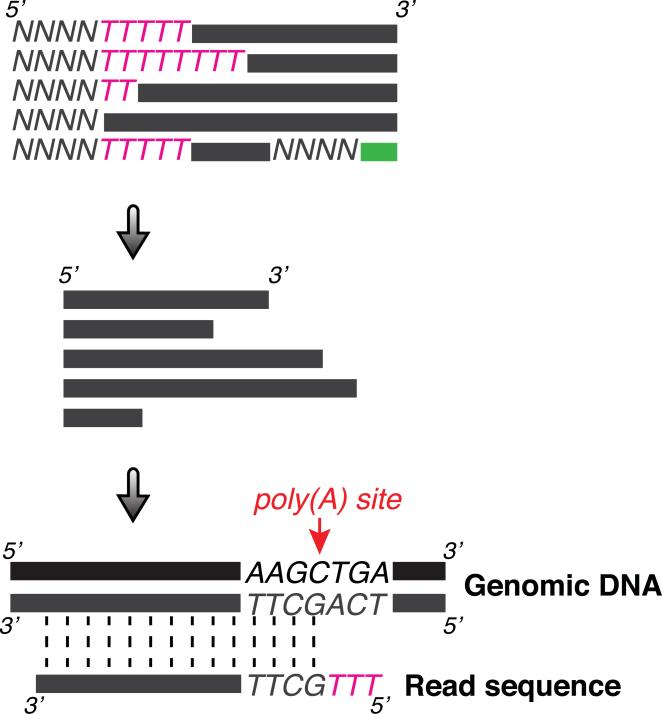
Figure 3.
Schematic of sequencing data processing to identify poly(A) sites. Raw reads are first trimmed at 5’ end (and 3’ end if necessary) and then aligned to the reference genome sequence. Poly(A) sites are defined by PASS reads, which are the uniquely mapped reads with at least two Ts not matched with genomic sequence. The last aligned position is considered as the poly(A) site.
Identify poly(A) site supporting (PASS) reads
1. Trim the first four nucleotides (corresponding to four random nucleotides from Adapter A) and consecutive Ts from 5’ end of sequence reads. Record the four nucleotides and number of Ts trimmed for later PASS reads identification. Also trim any sequences from the 3’ end of sequence reads corresponding to Adapter C.
Note: the antisense strand of the DNA library is sequenced.
2. Align the trimmed reads to reference transcriptome sequences and genomic sequences. (e.g., using Tophat command tophat –p 2 –G...)
3. Select uniquely aligned reads and compare previously trimmed T stretches from each read to 3’ end genomic sequences immediately after where that read is aligned. Reads with at least two Ts not matched with genomic sequence are considered as PASS reads. The last aligned position is considered the poly(A) site.
Note: When 3’ end of an aligned region has encoded consecutive Ts in its genomic sequence, it is impossible to determine the true poly(A) site. Arbitrarily, the poly(A) site is assigned to the last aligned non-T nucleotide. Reads with only one non-genomically encoded T can also be used for Poly(A) identification, if desired.
Cluster poly(A) sites (optional)
Note: For certain analyses, poly(A) sites located in a small window are typically clustered and treated as one representative poly(A) site. Here is one way to cluster poly(A) sites.
Cluster defined poly(A) sites based on PASS reads within 24 nt from each other. If a cluster size is ≤24 nt, the site with the highest number of PASS reads is assigned as the representative poly(A) site. If a cluster size is > 24 nt, the representative poly(A) site with the highest number of PASS reads in the cluster is assigned and PASS reads located > 24 nt from the assigned representative poly(A) site is re-clustered. Repeat this process until all representative poly(A) sites in clusters are defined.
Note: One way to increase the stringency of the poly(A) site identification and reduce false positives is to require that a poly(A) site be supported by at least two distinct PASS reads (based on a different number of nongenomic Ts and/or a different set of four nucleotides at the 5’ end of a read resulting from random nucleotides in Adapter A). Another method is to require that the number of PASS reads used to define a poly(A) site be ≥5% of all PASS reads for that particular gene.
Calculate gene expression levels and usage of alternative poly(A) sites
The gene expression level is calculated as the total number of PASS reads assigned to a gene (located in exons, introns, and 3’ UTRs), normalized to the total PASS reads in a dataset. Non-PASS reads (reads with no non-genomic Ts at the 5’ end) can also be included for gene expression analysis when comparing different datasets. However, it is not recommended to include non-PASS reads when comparing expression levels of different genes in the same dataset, because the number of non-PASS reads is correlated with A-richness of mRNA sequences.
A poly(A) site usage level is estimated as the ratio of number of PASS reads assigned to a particular poly(A) site and total number of PASS reads belonging to the gene.
REAGENTS AND SOLUTIONS
Use deionized, distilled water in all recipes and protocol steps. For common stock solutions, see APPENDIX 2; for suppliers, see APPENDIX 4.
Beads pre-wash buffer (100 ml)
1 ml 10 M NaOH (100 mM final; APPENDIX 2)
1 ml 5 M NaCl (50 mM final; APPENDIX 2)
50 μl TWEEN 20 (0.05% final)
Add up to 100 ml with nuclease-free H2O
Store indefinitely at RT
2X Annealing buffer (100 ml)
2 ml 1 M Tris-Cl, pH7.5 (20 mM final; APPENDIX 2)
2.4 ml 5 M NaCl (120 mM final; APPENDIX 2)
400 μl 0.5 M EDTA (2 mM final; APPENDIX 2)
Add up to 100 ml with nuclease-free H2O
Store indefinitely at RT
2X Coupling buffer (100 ml)
2 ml 1 M Tris-Cl, pH7.5 (20 mM final; APPENDIX 2)
40 ml 5 M NaCl (2 M final; APPENDIX 2)
400 μl 0.5 M EDTA (2 mM final; APPENDIX 2)
100 μl TWEEN 20 (0.1% final)
Add up to 100 ml with nuclease-free H2O
Store indefinitely at RT
2X Hybridization buffer (100 ml)
2 ml 1 M Tris-Cl, pH7.5 (20 mM final; APPENDIX 2)
6 ml 5 M NaCl (300 mM final; APPENDIX 2)
400 μl 0.5 M EDTA (2 mM final; APPENDIX 2)
100 μl TWEEN 20 (0.1% final)
Add up to 100 ml with nuclease-free H2O
Store indefinitely at RT
High-stringency wash buffer (100 ml)
1 ml 1 M Tris-Cl, pH7.5 (10 mM final; APPENDIX 2)
20 μl 5 M NaCl (1 mM final; APPENDIX 2)
200 μl 0.5 M EDTA (1 mM final; APPENDIX 2)
50 μl TWEEN 20 (0.05% final)
10 ml Formamide (10% final)
Add up to 100 ml with nuclease-free H2O
Store for up to 1 year at 4°C
RNase III digestion buffer (100 ml)
1 ml 1 M Tris-Cl, pH8.3 (10 mM final; APPENDIX 2)
1.2 ml 5 M NaCl (60 mM final; APPENDIX 2)
1 ml 1 M MgCl2 (10 mM final; APPENDIX 2)
100 μl 1 M DTT (1 mM final; APPENDIX 2)
Add up to 100 ml with nuclease-free H2O
Store in small aliquots for up to 2 years at -20°C
RNase H digestion buffer (100 ml)
5 ml 1 M Tris-Cl, pH8.3 (50 mM final; APPENDIX 2)
20 μl 5 M NaCl (1 mM final; APPENDIX 2)
300 μl 1 M MgCl2 (3 mM final; APPENDIX 2)
1 ml 1 M DTT (10 mM final; APPENDIX 2)
Add up to 100 ml with nuclease-free H2O
Store in small aliquots for up to 2 years at -20°C
Elution buffer (100 ml)
20 μl 5 M NaCl (1 mM final; APPENDIX 2)
200 μl 0.5 M EDTA (1 mM final; APPENDIX 2)
50 μl TWEEN 20 (0.05% final)
Add up to 100 ml with nuclease-free H2O
Store indefinitely at RT
TE buffer (100 ml)
1 ml 1 M Tris-Cl, pH7.5 (10 mM final; APPENDIX 2)
200 μl 0.5 M EDTA (1 mM final; APPENDIX 2)
Add up to 100 ml with nuclease-free H2O
Store indefinitely at RT
Gel extraction buffer (100 ml)
1 ml 1 M Tris-Cl, pH8 (10 mM final; APPENDIX 2)
6 ml 5 M NaCl (300 mM final; APPENDIX 2)
200 μl 0.5 M EDTA (1 mM final; APPENDIX 2)
Add up to 100 ml with nuclease-free H2O
Store indefinitely at RT
COMMENTARY
Background Information
Poly(A) tails at the ends of most mRNAs (and some noncoding RNAs) in eukaryotic organisms are typically generated through a cleavage and polyadenylation process on nascent RNAs (Colgan and Manley, 1997). Cis-elements and trans-factors work in concert to mediate poly(A) site selection (Mandel et al., 2008; Moqtaderi et al., 2013; Proudfoot and O'Sullivan, 2002; Proudfoot, 2011). Most eukaryotic genes are transcribed into mRNAs with alternative poly(A) sites (Tian and Manley, 2013). mRNA isoforms generated through alternative poly(A) sites have different 3’ UTRs, where many cis-elements reside for gene expression regulation (Di Giammartino et al., 2011). Previous studies have shown that mRNA isoforms might have important biological roles in a variety of processes, such as development (Ji et al., 2009; Mangone et al., 2010; Thomsen et al., 2010), cell proliferation (Sandberg et al., 2008), and cancer cell transformation (Mayr and Bartel, 2009; Singh et al., 2009).
In order to better understand the potential functions of mRNA isoforms, precise identification of all poly(A) sites on a genomic scale is of critical importance. In recent years, a number of strategies utilizing high-throughput sequencing have been proposed to identify poly(A) sites in a genome-wide manner. One popular approach is to initiate cDNA synthesis from the 3’ end of polyadenylated RNAs by using an oligo-dT primer or variant thereof (e.g., oligo-(dT)20 followed by two locking bases, a non-T nucleotide, then a random nucleotide), followed by library construction (Derti et al., 2012; Fox-Walsh et al., 2011; Shepard et al., 2011; Yoon and Brem, 2010). One obvious shortcoming of the oligo-dT based methods is that a significant fraction of the resulting sequence reads reflect internal priming from A-rich regions in the genome, which presents the challenge of distinguishing true poly(A) sites from internal priming sites.
Another oligo-dT based method is direct RNA sequencing using the Helicos single molecule sequencing technology (Ozsolak et al., 2010; Ozsolak et al., 2009). In this method, the poly(A) tail of an individual RNA is annealed to an immobilized oligo-dT primer for direct sequencing of the hybridized RNA without any additional manipulations or library construction steps. This approach largely avoids potential biases introduced by the various manipulations typically used in the generation of sequencing libraries and is particularly good at revealing less frequently utilized poly(A) sites. Unfortunately, the commercial availability of such sequencing technology is currently very limited.
The Poly(A)-position profiling by sequencing (3P-seq) method developed by the Bartel laboratory is a strategy to identify poly(A) sites independent of reverse transcription by using an oligo-dT primer (Jan et al., 2011). Through a splint-ligation of an oligonucleotide to the end of the poly(A) tail, 3P-seq captures poly(A) sites with immediate upstream RNA sequences and dramatically decreases non-specific sequence reads arising from internal priming at A-rich loci. While 3P-seq is a useful and creative method for genome-wide poly(A) identification, the protocol can be laborious, and it requires a high sequencing depth, because only a relatively small percentage of reads can be used for accurately identifying poly(A) sites.
The protocol described in this unit is modified from a recently published method named 3’READS (Hoque et al., 2013). The main advantages of 3’READS over previously proposed methods include: a) Significantly minimized internal priming artifacts relative to oligo-dT based reverse transcription; b) Reduction in sequence reads resulting from partially degraded RNAs with short oligo(A) tails, due to the stringency of the wash conditions (10% formamide); c) Preservation of a few As from the poly(A) tail, a feature essential for poly(A) identification, thanks to the use of the hybrid Biotin-T45U5 oligo and RNase H digestion; d) A high percentage of sequence reads suitable for poly(A) identification; e) Quantitative analysis of gene expression and poly(A) site usage.
Critical Parameters and Troubleshooting
RNA fragmentation by RNase III digestion
After RNase III digestion, it is important to examine the size distribution of the bound RNA fragments with poly(A) tails. An ideal digestion would result in most RNA fragments being in the range of 100 – 300 nt. Base on our experience, the proposed digestion conditions (2.5 U of RNase III, 15 min digestion at 37°C) works fairly well on RNA samples extracted from multiple human cell lines, mouse, and yeast species. However, it is worthwhile to empirically determine good digestion conditions for a particular RNA sample if a less ideal fragmentation pattern is observed using the proposed conditions.
Removal of most of but not all poly(A) tail sequence by RNase H digestion
In order to confidently identify poly(A) sites in the genome, one of the key steps in this protocol is the generation of RNA fragments that contain the poly(A) sites with a few As remaining from the poly(A) tails. Utilizing a special chimeric DNA/RNA oligonucleotide (Biotin-T45U5) in combination with appropriate digestion by RNase H (which specifically degrades the RNA in RNA/DNA hybrids), most, but not all, poly(A) tail sequence is removed (see Figure 2B for a typical distribution of the number of As from poly(A) tails left). Insufficient digestion by RNase H can leave RNA fragments with long poly(A) tails. Consequently, a large proportion of sequence reads would include a long stretch of Ts and the remaining sequence information might be insufficient for unique alignment to a reference genome. On the other hand, extreme over-digestion by RNase H can remove all poly(A) tail sequences despite the intended protection offered by the 5 uridines in the chimeric Biotin-T45U5 oligo. RNA fragments with no As left from poly(A) sequence are unable to be definitively identified as poly(A) sites. An excessive number of these fragments is indicative of over-digestion by RNase H. In our hands, both 1 U and 5 U of RNase H for 30 min at 37°C offer appropriate digestion for a wide range of samples. To ensure adequate enzyme activity despite variation in different commercial lots, we prefer to use 5 U of RNase H.
Insertion of four random nucleotides in the end of adapters
The strong sequence and structural biases in RNA ligase-mediated ssRNA/ssRNA ligation and ssRNA/ssDNA ligation can lead to distorted RNA profiles through in high-throughput sequencing results (Jayaprakash et al., 2011; Zhuang et al., 2012). In order to minimize the effects of the ligation biases of T4 RNA ligases in the sequencing library construction steps, four random nucleotides are added to the end of adapters used for ligation. Random nucleotides in adapters also contribute to higher ligation efficiency. Furthermore, over-representation of certain RNA fragments caused by any biases from the PCR amplification step in the library construction process can be accurately assessed by analyzing the distribution frequency of the first four nucleotides in selected sequence reads. Finally, the presence of four random nucleotides in Adapter A greatly help cluster-calling on the flow cells of the Illumina sequencing platform, because the Illumina cluster-calling algorithm expects a roughly equal base composition at any given position of the first few nucleotides. Overrepresentation of a stretch of Ts (the complementary reads resulting from the residual poly(A) tail As) would cause inefficient cluster-calling.
Addition of 0.05% TWEEN 20 to buffers used with Dynabeads MyOne Streptavidin C1
Dynabeads MyOne Streptavidin C1 tend to stick on to the walls of microcentrifuge tubes as well as pipette tips during washes, transfer, and incubation steps, which makes it challenging and time-consuming to fully recover the beads and bead-bound samples. Addition of a very low percentage of TWEEN 20 (0.05% final) alleviates the difficulty in collecting and transferring Dynabeads due to stickiness, while not affecting the performance of the Dynabeads MyOne Streptavidin C1 in this protocol.
Quantification of DNA sequencing library
To examine the quality and quantity of the libraries, we prefer to use SPRI magnetic beads (DNA SizeSelector-I) to purify the PCR amplified DNA sequencing libraries prior to Agilent Bioanalyzer analysis (High-sensitivity DNA Kit). Bioanalyzer analysis allows for easy quantification of the concentration of DNA fragments in the useful range for sequencing (175 – 500 bp) as well as a visual assessment of the levels of undesirable short-length byproducts such as primer dimers and small (non-mappable) fragments. Based on the concentration of DNA fragments in the 175 – 500 bp range, uniquely indexed DNA libraries can be combined according to the desired molar ratio prior to purification on 8% TBE polyacrylamide gels. Alternatively, SPRI magnetic beads can be used to further purify combined DNA libraries, in lieu of 8% TBE polyacrylamide gels. Based on our experience, it normally takes additional two rounds of purification by SPRI magnetic beads to completely remove DNA fragments smaller than 200 bp. While much faster than the gel purification, the drawback of performing two additional rounds of SPRI size selection is that a several-fold greater amount of initial material is required to offset the loss of material during the purification, in order to obtain a comparable amount of quality DNA libraries suitable for sequencing.
Alternatively, the PCR-amplified DNA sequencing library can be directly purified by electrophoresis on an 8% TBE polyacrylamide gel in order to extract DNA fragments in the range of 175 – 500 bp. One potential drawback is that it can be time consuming to gel purify each library individually if multiple libraries are planned to be combined for a single sequencing run. After purification by an 8% polyacrylamide gel, a DNA library can be quantified by Agilent Bioanalyzer analysis (High-sensitivity DNA Kit) as well as by Quantitative-PCR (Q-PCR) analysis by comparison to previously calibrated/sequenced DNA libraries using Q-PCR checking primers F and R (Table 1). In theory, quantification by Q-PCR leads to a more accurate estimation of the amount of DNA that can be effectively clustered on the surface of flow cell. In this case, one can better predict the number of expected reads for each DNA library in a multiplexed sequencing run.
No or low DNA library content after PCR amplification
As this is a relatively long protocol, there are many steps could contribute to no or low DNA library content after final amplification. However, in our opinion, it is always useful to check the activity of each enzyme used in the protocol before other troubleshooting, since a faulty enzyme is one of typical cause of failures.
Anticipated Results
The concentration of a purified DNA library is normally 5 – 10 nM. For a typical sequencing run involving such libraries, 65 – 75% of the raw reads can be uniquely aligned to a reference genome, and 60 – 65% of the uniquely mapped reads can be used to identify poly(A) sites (only uniquely mapped reads containing at least 2 non-genomic Ts are considered for identification of poly(A) sites).
Time Considerations
This protocol can be completed over 3 – 4 days. Beads preparations and isolation of 3’ end RNA fragments containing poly(A) sites can be done in 1 day. Library construction and purification take 2 – 3 days. In this protocol, there are many steps at different stages (i.e., steps 18, 21, 29, 32, 42, and 53) where the sample preparation can be temporarily stopped without adversely affecting the outcome. As such, the entire procedure can be performed over a fairly extended time period with considerable flexibility.
Acknowledgments
We thank all members of the Struhl laboratory for insightful discussions. This work was supported by grants to K.S. from the National Institutes of Health (GM 30186 and CA 107486).
Literature Cited
- Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes & development. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- Derti A, Garrett-Engele P, Macisaac KD, Stevens RC, Sriram S, Chen R, Rohl CA, Johnson JM, Babak T. A quantitative atlas of polyadenylation in five mammals. Genome research. 2012;22:1173–1183. doi: 10.1101/gr.132563.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol Cell. 2011;43:853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon R, Ugalde AP, Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nature reviews. Genetics. 2013;14:496–506. doi: 10.1038/nrg3482. [DOI] [PubMed] [Google Scholar]
- Fox-Walsh K, Davis-Turak J, Zhou Y, Li H, Fu XD. A multiplex RNA-seq strategy to profile poly(A+) RNA: application to analysis of transcription response and 3′ end formation. Genomics. 2011;98:266–271. doi: 10.1016/j.ygeno.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisberg JV, Moqtaderi Z, Fan X, Ozsolak F, Struhl K. Global analysis of mRNA isoform half-lives reveals stabilizing and destabilizing elements in yeast. Cell. 2014;156:812–824. doi: 10.1016/j.cell.2013.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque M, Ji Z, Zheng D, Luo W, Li W, You B, Park JY, Yehia G, Tian B. Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat Methods. 2013;10:133–139. doi: 10.1038/nmeth.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque M, Li W, Tian B. Accurate mapping of cleavage and polyadenylation sites by 3′ region extraction and deep sequencing. Methods in molecular biology. 2014;1125:119–129. doi: 10.1007/978-1-62703-971-0_10. [DOI] [PubMed] [Google Scholar]
- Jan CH, Friedman RC, Ruby JG, Bartel DP. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature. 2011;469:97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaprakash AD, Jabado O, Brown BD, Sachidanandam R. Identification and remediation of biases in the activity of RNA ligases in small-RNA deep sequencing. Nucleic Acids Res. 2011;39:e141. doi: 10.1093/nar/gkr693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci U S A. 2009;106:7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3′-end processing. Cellular and molecular life sciences : CMLS. 2008;65:1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangone M, Manoharan AP, Thierry-Mieg D, Thierry-Mieg J, Han T, Mackowiak SD, Mis E, Zegar C, Gutwein MR, Khivansara V, et al. The landscape of C. elegans 3′UTRs. Science. 2010;329:432–435. doi: 10.1126/science.1191244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moqtaderi Z, Geisberg JV, Jin Y, Fan X, Struhl K. Species-specific factors mediate extensive heterogeneity of mRNA 3′ ends in yeasts. Proc Natl Acad Sci U S A. 2013;110:11073–11078. doi: 10.1073/pnas.1309384110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Kapranov P, Foissac S, Kim SW, Fishilevich E, Monaghan AP, John B, Milos PM. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell. 2010;143:1018–1029. doi: 10.1016/j.cell.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Platt AR, Jones DR, Reifenberger JG, Sass LE, McInerney P, Thompson JF, Bowers J, Jarosz M, Milos PM. Direct RNA sequencing. Nature. 2009;461:814–818. doi: 10.1038/nature08390. [DOI] [PubMed] [Google Scholar]
- Proudfoot N, O'Sullivan J. Polyadenylation: a tail of two complexes. Current biology : CB. 2002;12:R855–857. doi: 10.1016/s0960-9822(02)01353-2. [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ. Ending the message: poly(A) signals then and now. Genes & development. 2011;25:1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard PJ, Choi EA, Lu J, Flanagan LA, Hertel KJ, Shi Y. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. Rna. 2011;17:761–772. doi: 10.1261/rna.2581711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Alley TL, Wright SM, Kamdar S, Schott W, Wilpan RY, Mills KD, Graber JH. Global changes in processing of mRNA 3′ untranslated regions characterize clinically distinct cancer subtypes. Cancer research. 2009;69:9422–9430. doi: 10.1158/0008-5472.CAN-09-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen S, Azzam G, Kaschula R, Williams LS, Alonso CR. Developmental RNA processing of 3′UTRs in Hox mRNAs as a context-dependent mechanism modulating visibility to microRNAs. Development. 2010;137:2951–2960. doi: 10.1242/dev.047324. [DOI] [PubMed] [Google Scholar]
- Tian B, Manley JL. Alternative cleavage and polyadenylation: the long and short of it. Trends in biochemical sciences. 2013;38:312–320. doi: 10.1016/j.tibs.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon OK, Brem RB. Noncanonical transcript forms in yeast and their regulation during environmental stress. Rna. 2010;16:1256–1267. doi: 10.1261/rna.2038810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang F, Fuchs RT, Sun Z, Zheng Y, Robb GB. Structural bias in T4 RNA ligase-mediated 3′-adapter ligation. Nucleic Acids Res. 2012;40:e54. doi: 10.1093/nar/gkr1263. [DOI] [PMC free article] [PubMed] [Google Scholar]