SUMMARY
In vitro modeling of human disease has recently become feasible with induced pluripotent stem cell (iPSC) technology. Here, we established patient-derived iPSCs from a Li-Fraumeni Syndrome (LFS) family and investigated the role of mutant p53 in the development of osteosarcoma (OS). LFS iPSC-derived osteoblasts (OBs) recapitulated OS features including defective osteoblastic differentiation as well as tumorigenic ability. Systematic analyses revealed that the expression of genes enriched in LFS-derived OBs strongly correlated with decreased time to tumor recurrence and poor patient survival. Furthermore, LFS OBs exhibited impaired upregulation of the imprinted gene H19 during osteogenesis. Restoration of H19 expression in LFS OBs facilitated osteoblastic differentiation and repressed tumorigenic potential. By integrating human imprinted gene network (IGN) into functional genomic analyses, we found that H19 mediates suppression of LFS-associated OS through the IGN component DECORIN (DCN). In summary, these findings demonstrate the feasibility of studying inherited human cancer syndromes with iPSCs.
INTRODUCTION
Li-Fraumeni syndrome (LFS) is a genetically heterogeneous inherited cancer syndrome characterized by autosomal dominance and early onset of often multiple independent tumors within affected family members (Li and Fraumeni, 1969). In contrast to other inherited cancer syndromes predominantly characterized by site-specific cancers, LFS patients present with a variety of tumor types, including osteosarcoma (OS), soft tissue sarcoma, breast cancer, brain tumor, leukemia, and adrenocortical carcinoma. Germline mutations in the TP53 gene encoding the tumor suppressor p53 are responsible for LFS (Malkin et al., 1990). Mutations in p53 usually not only abolish normal p53 function but are also associated with additional oncogenic activities. Despite the prevalence of p53 mutations, the simultaneous presence of alterations in other tumor suppressors (e.g., RB1 and LKB1) and oncogenes (KRAS and HER2) makes it extremely difficult to study the specific role of p53 in cancer development. LFS provides an ideal genetic model system for investigating such a role. Although murine LFS models have been generated (Hanel et al., 2013; Lang et al., 2004; Olive et al., 2004), they do not fully recapitulate the tumor spectrum found in LFS patients. Therefore, other model systems are needed in order to further decipher mutant p53-associated pathogenesis.
Comprising almost 60% of the common histological bone sarcoma subtypes, OS is the most frequent primary non-hematological malignancy in childhood and adolescence (Tang et al., 2008). Despite advances in surgery and multi-agent chemotherapy, the survival rate has not increased in the past 40 years as much as for other malignancies. After leukemia, OS is the second leading cause of cancer mortality among children and adolescents and has been described as a cancer syndrome with a differentiation deficiency. OS exhibits osteoblast (OB)-like features and sustains undifferentiated OBs (Haydon et al., 2007). Furthermore, genetic alterations (e.g., p53 mutation and RB deletion) are strongly associated with OS development. Although the association of TP53 mutation with OS is strongly supported by the high risk of OS in LFS patients (Porter et al., 1992), the underlying mechanism by which triggers OS development is still unclear.
H19 is a maternally imprinted gene encoding a long non-coding RNA (lncRNA). Alterations in the expression of genes in the H19-IGF2 imprint locus are linked to both Beckwith-Wiedemann Syndrome (BWS) and Russell-Silver Syndrome (RSS) (Choufani et al., 2010; Eggermann, 2010). Gain of methylation of the upstream H19 imprinting center (IC1) leading to H19 inactivation and IGF2 activation, is found in 5%–10% of BWS patients and in >25% of patients with Wilms tumor, hepatoblastoma and rhabdomyosarcoma (Choufani et al., 2010). Although the H19-IGF2 imprinting mechanism has been well documented and serves as a paradigm for the study of epigenetic regulation, the functions of H19 in biological and pathological molecular regulatory processes remain nebulous. Recently, Varrault and colleagues meta-analyzed the set of strongly correlated genes in microarray data sets to infer the “Imprinted Gene Network” (IGN), of which H19 is a member. This IGN may be part of the complex regulatory system that induces rapid but controlled growth during development (Varrault et al., 2006). H19 has been suggested to regulate embryonic growth and differentiation by controlling the expression of IGF2 and several other interconnected imprinted genes; thus, fine-tuning equilibrium of growthactivation and repression (Gabory et al., 2009). These findings suggest that H19 may execute its biological functions through the IGN.
Modeling human genetic diseases has been facilitated by induced pluripotent stem cell (iPSC) methodologies (Takahashi et al., 2007; Takahashi and Yamanaka, 2006; Yu et al., 2007). Although iPSCs are widely utilized in the study of various genetic diseases with either Mendelian or complex inheritance, their application in cancer research has been much less extensively explored. In the present study, we have modeled LFS-associated OS by using OBs derived from LFS patient-specific iPSCs and were able to recapitulate disease characteristics. The LFS iPSC-derived OBs displayed a clear OS gene expression signature whose particular transcriptional spectra strongly correlate with clinical prognosis. By integrating global transcriptional and computational analyses, we demonstrated that downregulation of H19 and its associated IGN component DECORIN (DCN) is responsible for LFS-associated OS development. Restoring H19 expression facilitates OB differentiation and inhibits tumorigenesis. Downregulation of DCN impairs H19-mediated osteogenic differentiation and tumor suppression. In summary, our results suggest that p53 mutation-mediated H19 and IGN inactivation may contribute to OS development in LFS patients and that induction of H19 expression may have important implications for the future treatment or prevention of LFS-associated OS and/or OS with somatically acquired p53 mutations.
RESULTS
Generation and Characterization of LFS iPSCs
To elucidate how p53 mutation results in tumor development, we generated iPSCs from patient fibroblasts obtained from a LFS family representing three LFS patients and two unaffected individuals (Figure S1A). The three patients have a heterozygous c.734G>A mutation that causes a G245D missense substitution. This site is one of the hot-spot p53 mutations in both LFS patients and somatic tumors (Varley, 2003). These patients present with a broad spectrum of tumors, including OS, neurilemmoma and astrocytoma (Figure S1A). The fibroblast samples displayed a normal karyotype under low passage (Mirzayans et al., 2010). Genome sequencing further confirmed heterozygous G245D mutations in LFS fibroblasts (Figure S1B). Using non-integrating Sendai virus (SeV)-based delivery of the four Yamanaka reprogramming factors, OCT4, SOX2, KLF4 and c-MYC (Fusaki et al., 2009; Takahashi et al., 2007), we established a number of iPSC clones from the affected and unaffected family members. These iPSC clones all demonstrate hESC morphology, express pluripotency factors (NANOG, SOX2 and OCT4), surface markers (TRA-1-81 and SSEA4) and alkaline phosphatase (Figure 1A). The lines also show expression of pluripotency markers at levels comparable to H9 and HES2 hESCs by quantitative (q) RT-PCR and have a more open and demethylated OCT4 promoter than the original fibroblasts (Figures 1B and 1C). We verified loss of SeV and exogenous OCT4, SOX2, KLF4 and c-MYC transgenes (Figures S1C and S1D), demonstrating that these iPSCs are zerogenetic footprint. Importantly, the iPSC lines were karyotypically normal (Figure S1E) and demonstrated the capacity to differentiate into all three germ layers in vitro (data not shown) and in teratomas (Figure 1D). All characterizations of wild-type (WT) and LFS iPSCs are summarized in Table S1. Together, these data indicate that somatic cells from LFS patients can be properly reprogrammed, maintain a pluripotent state and can effectively differentiated.
Figure 1. LFS iPSCs are pluripotent.
(A) SeV-4F (OCT4, SOX2, KLF4 and c-MYC) reprogrammed LFS and wild type (WT) iPSCs derived from the same family express hESC pluripotency factors, hESC surface markers and AP activity. (B) qRT-PCR assay for expression of endogenous human NANOG, SOX2, OCT4, DPPA4, REX1 and TERT in iPSCs and parental fibroblasts. PCR reactions are normalized to GAPDH and plotted relative to expression levels in human H9 ESCs. Error bars indicate ±SEM of triplicates. (C) Bisulfite sequencing analysis of the OCT4 promoter showing CpG hypomethylation in WT and LFS iPSCs relative to the parental fibroblasts. The cell line and percentage of CpG methylation are indicated to the left of each cluster. Closed circle, methylated CpG; open circle, unmethylated CpG. (D) In vivo teratoma formation assay demonstrates LFS iPSC capacity to differentiate into the three germ layers. H&E-stained teratomas containing embryonic tissues all three germ layers, including enamel epithelium (endoderm); neural tube epithelium (ectoderm); cartilage corpuscle (mesoderm). Scale bar, 100 µm. See also Figure S1 and Table S1.
Impairment of p53 Function in LFS iPSC-Derived Mesenchymal Stem Cells (MSCs)
As mentioned previously, OS, notably featuring defective OB differentiation, is one of the major cancers affecting this LFS family. Therefore, we applied our iPSC model to study how mutant p53 interferes with OB differentiation and to investigate the molecular alterations caused by p53 mutation in OS development. Human OBs can be induced from hESC-derived multipotent MSCs that can give rise to bone, cartilage, muscle and adipose tissues. We first differentiated WT and LFS iPSCs to MSCs by treating them with FGF2 and PDGF-AB and sorting CD105+/CD24− cells (Figure S2A). These cells also expressed the MSC surface markers CD44, CD73, CD105 and CD166, and the MSC-related transcription factor SNAI1 as well as VIM (Figures 2A and S2B). The cells could be maintained for 2 months without loss of their MSC characteristics (Figure S2C) (Lian et al., 2007). In comparison with WT MSCs, LFS MSCs showed no mRNA expression differences of p53, MSC-associated transcription factors, and osteoblastic-associated factors (Figures 2B (right panel), S2D and S2E). Nevertheless, LFS MSCs showed lower mRNA expression levels of p53 targets p21 and MDM2 (Figure 2B, left and middle panels). Compared with p53(WT), p53(G245D) showed reduced binding to the p21 and MDM2 promoters by Chromatin-Immunoprecipitation (ChIP)-PCR analysis (Figure 2C), consistent with impaired transcriptional activity (Figure 2B). Upon MDM2 inhibitor Nutlin-3 treatment, expression of numerous p53 target genes (p21, MDM2, SFN, NOXA, FAS, TNFRSF10B, and GADD45A) were upregulated in WT MSCs but this effect was blunted in LFS MSCs (Figure 2D). All characteristics of WT and LFS MSCs are summarized in Table S1. These studies demonstrate that LFS MSCs not only maintain MSC characteristics identical to WT MSCs but also retain the defective p53 function of the parental fibroblasts (Barley et al., 1998).
Figure 2. Defective p53 activity in LFS MSCs.
(A) Surface antigen profiling of LFS MSCs by flow cytometry demonstrating the CD24−, CD105+, CD73+ and CD44+ fractions in differentiated MSCs. (B) qRT-PCR analysis for expression of p53 and its downstream target genes, p21 and MDM2, in the WT MSC group (11 lines) and the LFS MSC group (13 lines). The p53 mRNA levels do not show a significant difference between WT and LFS MSCs, while levels of p21 and MDM2 are significantly lower in LFS MSCs. (C) ChIP-PCR demonstrating lower p53 binding affinity to p21 and MDM2 promoter region in LFS MSCs. IgG ChIP is a negative control. Error bars indicate ±SEM of triplicates. (D) qRT-PCR for expression of p53 and its target genes after treatment of LFS and WT MSCs with the MDM2 inhibitor Nutlin-3 for 6 hr. Upregulation of the majority of p53 target genes is impaired in LFS MSCs in comparison with WT MSCs despite similar p53 expression in both MSC groups. qRT-PCR data are represented as mean ± SEM; n=3. See also Figure S2 and Table S1.
Recapitulating OS Characteristics in LFS MSC-Derived OBs
Since it was previously suggested that impairment of p53 function leads to OS (Walkley et al., 2008) and clinical OS samples are largely composed of poorly differentiated or undifferentiated OBs (Tang et al., 2008), we asked if dysregulation of p53 signaling is responsible for the observed differentiation defects. LFS MSCs were induced to the OB lineage and the differentiation process was monitored over time. Several p53 targets were gradually induced in WT but not LFS OBs during differentiation (Figure S3A). Consistently, ChIP-PCR showed significantly reduced p53 binding to the p21 and MDM2 promoters in LFS OBs (Figure S3B). The decrease in p53 transcriptional activity in late stage osteogenic differentiation (day 17) was confirmed by a p53 reporter assay (Figure S3C). These findings suggest that p53 signaling is active in WT OBs but impaired in LFS OBs. AP staining for detecting bone-associated ALPL enzyme activity (Figure 3A) and Alizarin Red S staining reflective of mineral deposition by functional OBs (Figure 3B) showed slower differentiation in LFS MSCs. Mineral precipitations were observed on the surface of Petri dishes in WT but not LFS OBs (Figure S3D). Consistently, in comparison with WT OBs, LFS OBs showed lower expression of COL1A1 and ALPL (pre-OB markers), BGLAP/Osteocalcin and PTH1R (mature OB markers) as well as FGF23 and MEPE (osteocyte markers) during osteogenesis (Figures 3C). Immunostaining confirmed that LFS OBs expressed lower BGLAP than WT OBs did (Figure S3E). Because osteogenic differentiation is controlled by several core transcriptional/epigenetic regulators, we monitored their expression levels during OB differentiation from LFS MSCs. Indeed, we found impaired upregulation of ZNF521 and ZEB1 (Figure 3D), indicating a defect in the normal OB gene regulatory network. Knockdown of p53 resulted in upregulation of osteogenic markers in LFS OBs and eliminated the osteogenic differentiation defect (Figure S3F) indicating that p53(G245D) may exert gain-of-function instead of loss-of-function effects in inhibiting osteogenic differentiation. Moreover, we noticed LFS OBs growing in randomly oriented piled-up foci rather than the two-dimensional monolayers of flattened cells (Figure S3G), a generally regarded manifestation of a transformed phenotype and an initiating step in tumorigenesis. To further investigate if LFS OBs are able to recapitulate tumorigenic potential, we performed in vitro anchorage-independent growth (AIG) assays and in vivo xenografts. AIG assays showed clonal growth in soft agar by many LFS OBs (6 out of 9) but not in WT OBs, undifferentiated LFS or WT MSCs (Figure 3E). Performing xenografts in nude mice, we found tumorigenic ability in LFS OBs but not WT OBs (Figure 3F, right panels). The tumors demonstrated immature OB characteristics including AP activity, collagen matrix deposition but not mineralization (Figure 3F, left panels). The lack of in vitro and in vivo tumorigenic ability in LFS MSCs implies that OS may originate from immature or poorly differentiated OBs rather than MSCs (Figures 3E and 3F). To examine if LFS OBs are able to gain malignancy during tumor progression, we performed serial transplantation using an in ovo chick embryo chorioallantoic membrane (CAM) model. As shown in Figure S3H, the tumor sizes of LFS OBs increased in the second transplantation in comparison with the first transplantation. These results imply that similar to OS cells, LFS OBs contain a population of potential tumor initiating cells (TICs) and these cells are enriched during in ovo primary transplantation and fuel secondary tumor growth. Interestingly, no additional increases in malignant tumor growth were seen following a tertiary transplantation. The persistence of a WT p53 allele during serial CAM transplantations (Figure S3I) provides a possible explanation for why there is no further gain of tumor growth in the tertiary transplantation. Taken together, these findings demonstrate that OS-related phenotypes (defective OB differentiation and tumorigenic ability) can be recapitulated in LFS iPSC-derived OBs.
Figure 3. LFS OBs show differentiation defects and oncogenic properties.
(A–B) Both AP (A) and Alizarin Red S (B) staining reveal the attenuation of OB differentiation in LFS OBs. (C–D) Expression of OB lineage markers (C) and transcriptional regulators (D) during the OB differentiation time-course. (E) In vitro AIG assay for tumorigenicity demonstrates colony numbers found in LFS OBs but not in WT OBs. Positive colonies after 1 month growth of differentiated OBs in either MSC or OB differentiation media are those larger than 50 µm (Scale bar, 50 µm). (F) Tumor xenograft experiments by subcutaneous injection in NU/NU mice demonstrate that LFS OBs but not MSCs recapture in vivo tumorigenic ability. The LFS2-B OB-derived tumors were examined by H&E, AP, PicroSirius Red and von Kossa stains to examine morphology, bone-associated AP expression, collagen production and mineral deposits, respectively. Error bars represent ±SEM. Scale bar, 1 cm. See also Figure S3 and Table S1.
OS Spectrum is Represented in LFS OBs
In order to gain insights into LFS-associated osteogenic defects and tumorigenesis, the global transcriptome was investigated by mRNA-seq during OB differentiation time-courses (Table S2). Expression profiles of LFS and WT time-course samples analyzed by Spearman’s rank correlation demonstrated that gene expression profiles of LFS samples clustered together but were distinct from WT samples (Figure 4A). The non-negative matrix factorization (NMF) method for extracting relevant biological correlations based on gene expression data showed that at day 0 LFS and WT MSCs clustered together, while at days 7, 14, 17 differentiating LFS OBs are distinct from their WT counterparts (Figure 4B). These results suggest that WT and LFS MSC gene expression profiles are initially similar but diverge during subsequent OB differentiation. Alignment of reads at individual gene loci and quantification by fragments per kilobase of exon per million fragments mapped (FPKM) values confirmed the gradual increase of OB marker ALPL and skeletal development regulators HOXA10, IGF2, and CLEC3B in WT but not in LFS-derived cells (Figure S4A). Gene Ontology (GO) analyses using Network2Canvas further revealed that OB differentiation in WT MSCs (day 17 versus day 0) affects biological process genes mainly involved in skeletal system development and cell motility; whereas, genes upregulated in LFS MSCs are primarily associated with an inflammatory response (Figure 4C, upper and middle panels). Expression levels of several skeletal system development-related genes were greatly increased during the WT osteogenesis time-course but not in the LFS samples (Figure 4C, bottom panel). Moreover, expression levels of genes involved in positive regulation of cell differentiation and negative regulation of cell proliferation were significantly increased in WT OBs. In contrast, genes involved in positive regulation of cell cycle and mitosis were enriched in LFS OBs (Figure 4C, upper and middle panels). Using the Mouse Gene Atlas database, genes upregulated at day 17 in WT OBs were more similar to the later-stage differentiated mouse OB gene profiles than were LFS OBs (Figure S4B). Further analyses of day 17 WT and LFS OB gene expression revealed that the expression pattern in WT OBs is similar to that of mouse OBs at day 21. In contrast, the gene expression pattern of LFS OBs is closest to that of mouse OBs at day 5 (Figure S4C). Consistent with our qRT-PCR analyses of p53 target gene expression (Figure S3A), gene set enrichment analysis (GSEA) confirmed that many known and predicted targets enriched in WT OBs relative to LFS OBs are found disproportionately in the set of significantly (>3-fold) upregulated genes in p53 transduced OS cells (Figure S4D). This supports the dysregulation of p53 function that occurs during LFS OB differentiation.
Figure 4. Genome-wide transcriptome analysis reveals that LFS OBs possess an OS signature.
(A) Correlation matrix of LFS and WT osteogenic time course mRNA-seq results. (B) Ordered tree linkage displaying sample clustering and metagenes representing the most variability associated with each differentiation transition. (C) GO biological processes associated with upregulated genes in WT and LFS OBs at day 17. FPKM values of skeletal system developmental genes are plotted as a heat map demonstrating their upregulation during WT but not LFS-derived osteogenic differentiation. (D) GSEA indicates that OS-associated genes are enriched in LFS OBs while normal OB-associated genes are enriched in WT OBs. (E) OS patients with the LFS OB signature show shorter tumor recurrence and poorer survival. (F) CREA analysis reveals chromosomal integrity of LFS OBs and tumors engrafted in nude mice. See also Figure S4 and Tables S2 and S6.
We next asked if the gene expression in LFS OBs is consistent with an oncogenic signature derived from OS cell lines. We identified both OB and OS signature genes by using GenePattern to compare the gene expression profiles between OB cells and OS lines (GSE39262). We then applied GSEA to compare enriched expressed genes between WT and LFS OBs against these signatures. As shown in Figure 4D, OS-associated genes are specifically enriched in LFS OBs; in contrast, OB-associated genes are specifically enriched in WT OBs. This finding strongly suggests that by the time of their generation, LFS OBs have already acquired OS characteristics. Furthermore, to determine whether the gene expression profiles of LFS OBs can potentially have prognostic value as measured by patient survival and tumor recurrence we performed Kaplan-Meier analyses restricted to two separate subsets of patients, those who had or did not have an enriched LFS iPSC-derived OB gene expression signature as defined by enriched genes in LFS OBs versus WT OBs at day 17. We found that the enriched LFS OB-associated gene signature was significantly correlated with more rapid tumor recurrence and poorer survival (p=0.004 and p=0.009, respectively) (Figure 4E). In summary, our LFS iPSC model not only recaptures the OS signature but can also predict clinical outcomes in patients.
Cytogenetic analyses of human OS have revealed numerous genomic alterations and rearrangements (Batanian et al., 2002; Bridge et al., 1993) that have been consistently replicated in a murine p53 conditional knockout OS model (Walkley et al., 2008). Since genomic alterations are common during cancer progression, it has been challenging to factor out their effects when attempting to interrogate the roles of tumor suppressor genes or oncogenes in tumor progression. To investigate if our LFS OBs provide a unique model to study early stages of tumor progression, we applied in silico cytogenetic region enrichment analysis (CREA) to our LFS OB samples to identify the potential presence of rearranged regions commonly found in human OS. The LFS OBs were compared with the gene expression signature of WT OBs at day 17 of OB differentiation, while the synovial sarcoma (SS) was compared with normal muscle. As expected, OS cell lines showed significant enrichment at 10 of the 18 regions with known cytogenetic alterations in OS. In contrast, human SS were generally not associated with any alterations in these regions (Figure 4F), suggesting that these chromosomal regions are a specific feature of OS rather than a common feature of other cancers. Notably, in comparison with WT OBs, both LFS OBs and tumors showed negligible enrichment in these regions (1 and 2 out of 18 regions, respectively), implying that chromosomal rearrangements barely occur in these LFS OB-derived tumors (Figure 4F). These results demonstrate that LFS iPSC-derived OBs can serve as a useful system to study the early stages of OS progression caused solely by p53 mutation without interference by secondary genomic alterations.
Impaired H19 Expression in LFS-Associated OS
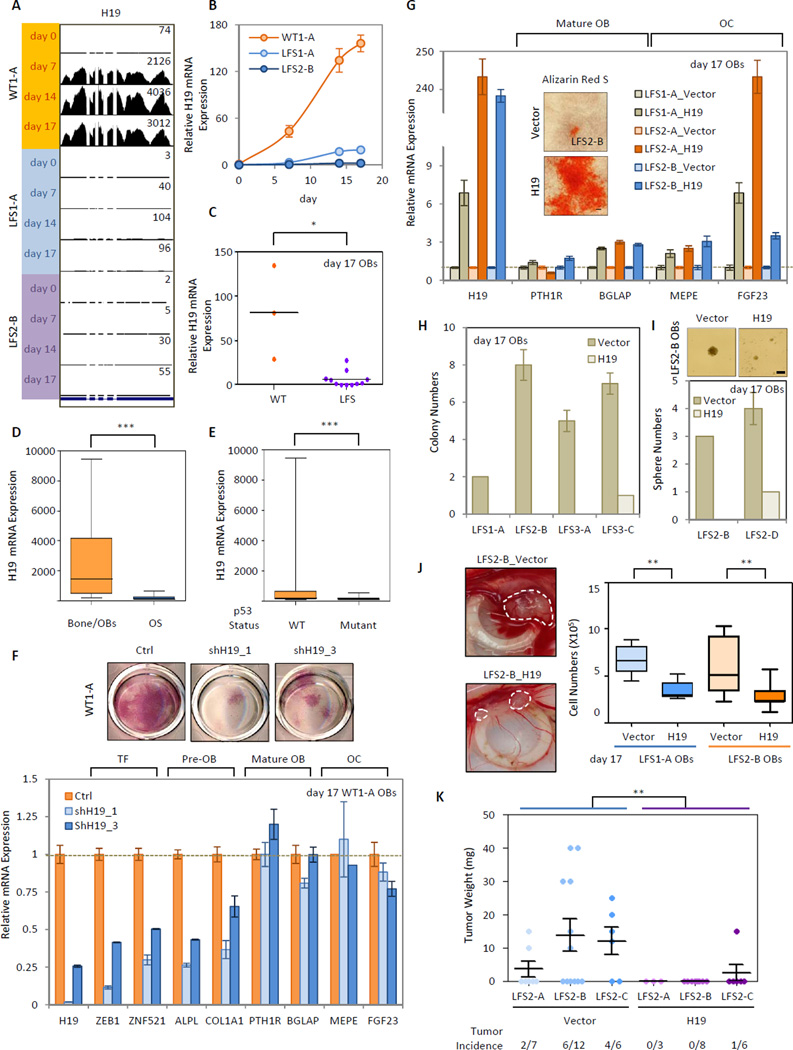
Among 421 differentially expressed genes identified in comparisons between WT and LFS OBs, H19, highly expressed in OBs but not in bone marrow or osteoclasts (Figure S5A), warranted further in-depth analyses. Alignment and quantification of reads at the H19 locus and qRT-PCR showed H19 upregulation in WT but not LFS OBs during osteogenesis (Figures 5A and 5B). The low expression of H19 in LFS OBs was further confirmed in multiple LFS iPSC-derived OBs (Figure 5C). In comparison with bone/OB tissues and p53 WT cells, H19 expression is significantly decreased in OS and p53 mutant cells, respectively (Figures 5D and 5E). These findings suggested that H19 dysregulation is a common phenomenon in OS and is correlated with p53 status. RNAi-mediated knockdown of H19 in WT OBs led to decreased expression of osteogenic factors ZEB1 and ZNF521, pre-osteoblastic makers ALPL and COL1A1 as well as AP activity (Figure 5F). Supporting the positive regulatory role of H19 in osteogenesis, ectopic expression of H19 in LFS MSCs resulted in increasing osteogenic marker expression and reactivation of OBs with consequent mineral deposition (Figure 5G). Moreover, AIG and oncosphere assays demonstrated that in vitro tumorigenic activities of LFS OBs and OS TICs were suppressed by re-expressing H19 (Figures 5H and 5I). In LFS OBs assayed in ovo with the CAM assay and in vivo by the mouse tumor xenograft model, restoration of H19 expression not only reduced the incidence of tumor development but also decreased tumor size (Figures 5J and 5K). To further investigate if restoration of H19 has any therapeutic potential for OS treatment, H19 was transduced into OS cell lines, OSA and HOS. As shown in Figure S5B, H19 reduced the incidence of OS tumor development as well as tumor size, suggesting H19 as a therapeutic target. In comparison with the original OSA tumor, the H19-transduced OSA tumor demonstrated poorly differentiated osteoblastic characteristics including positive AP activity and collagen matrix deposition but not mineralization, implying that due to heterogeneity, a small portion of OS may gain of the ability to escape H19-induced OB differentiation and tumorigenic suppression. In summary, these findings emphasize the essential role of H19 in regulating osteogenesis and the potential of re-expressed H19 to rescue defective osteogenesis and suppress tumor grow in both LFS iPSC-derived OBs and OS cell lines.
Figure 5. Involvement of H19 in LFS OB-associated defective OB differentiation and tumorigenesis.
(A) Visualization of mRNA-seq short read mapping of H19 in UCSC Genome Browser. (B) qRT-PCR shows impaired upregulated H19 in LFS during osteogenesis. Error bars indicate ±SEM of triplicates (C) Multiple LFS OBs have impaired upregulation of H19 during OB differentiation. (D–E) H19 expression is notably decreased in OS and p53 mutant cells. The analyses were performed using microarray data from GEO dataset GSE36001. (F) RNAi-mediated knockdowns of H19 in WT MSCs leads to decreased expression of osteogenic genes as well as AP activity. qRT-PCR data are represented as mean ± SEM; n=3. (G) Ectopic expression of H19 in LFS MSCs increases osteogenic gene expression and facilitates OB maturation. qRT-PCR data are represented as mean ± SEM; n=3. Scale bar, 100 µm. (H–I) AIG (H) and oncosphere (I) assays show repressed in vitro tumorigenic ability of LFS OBs upon restoration of H19. Scale bar, 100 µm. (J) In ovo CAM assay indicates H19 suppresses tumorigenic ability of LFS OBs. (K) In vivo tumor xenograft experiments indicating H19 suppression of tumorigenic ability of LFS OBs. See also Figure S5.
Since H19 expression has been shown to be suppressed by p53 (Dugimont et al., 1998), we asked if this regulation was found in LFS patients with mutant p53. Consistent with the previous findings, activation of p53 by Nutlin-3 treatment decreased H19 expression by 20–39% (Figure S5C) and RNAi-mediated p53 knockdown by two distinct RNAi molecules resulted in a 1.4–2.4 fold increase of H19 expression in WT MSCs (Figure S5D). In contrast, while suppression of H19 expression was not detected in Nutlin-3 treated LFS MSCs (Figure S5C), RNAi-mediated p53 knockdown led to a significant increase in H19 expression (7.1–19.3 folds) (Figure S5D), strongly suggesting that p53 mutants, at least p53(G245D), exert a gain-of-function effect in repressing H19. In agreement with the hypothesis that p53 does not bind the H19 promoter region and that p53-mediated H19 repression occurs through other factors (Dugimont et al., 1998), p53 ChIP showed no enrichment of p53 binding in comparison with a control IgG pulldown (Figure S5E). To further explore whether other p53 mutants could promote this regulation, we transfected multiple variants of p53 into WT MSCs and examined their effects on H19 expression. As with p53(G245D), many p53 hotspot mutants (R175H, G245S, G248W and R280K) exhibited stronger inhibition of H19 expression than did WT p53 (Figure S5F). This result not only demonstrates that the suppression of H19 expression by p53 mutants is common in LFS-associated OS but also, that this is a general mechanism found in other LFS patients with distinct p53 mutations. Since it was shown that the tumorigenic ability of several p53 mutants is at least partially accounted for by their interaction with and inhibition of p63 and/or p73 functions (Di Como et al., 1999; Gaiddon et al., 2001), we investigated whether p53(G245D) could inhibit H19 expression directly via p63 and p73. Exogenous coimmunoprecipitation (co-IP) showed that p53(G245D) and p53(R175H) but not p53(WT) can interact with p63 and p73 (Figure S5G). Depletion of p53(G245D) by siRNA increased p21mini promoter activity in two LFS-derived cells, LFS2-B (ΔWT)-1 and LFS2-B(ΔWT)-2 MSCs that lacked a functional wild-type p53 allele due to insertion of a NeoR selection marker (Lee DF and Lemischka IR, unpublished data). Both p63 and p73 were able to activate the p21mini promoter to a greater extent upon p53(G245D) knockdown (Figure S5H), confirming the dominant negative activity of p53(G245D) in regulating p63 and p73 function. However, ectopic expression of p63 and p73 does not alter H19 expression in WT MSCs (Figure S5I). These findings suggest that although one of the p53(G245D) gain-of-function effects in regulating LFS OS pathogenesis is through the suppression of normal p63/p73 function, this regulation is not involved in H19 transcriptional regulation.
Recent studies have found that p53 status may affect DNA methylation in the H19 genomic locus in a clone-specific manner in iPSCs (Yi et al., 2012). To investigate whether impaired upregulation of H19 in LFS OBs is caused by hypermethylation on the H19 locus, differentiating LFS OBs were treated with the demethylating agent 5’aza-deoxycytidine (Decitabine). As shown in Figure S5J, H19 expression in LFS OBs was slightly increased upon Decitabine treatment but remained significantly lower than in WT OBs during OB differentiation, ruling out the possibility that lower H19 expression in LFS OBs is due to H19 locus methylation. In fact, Decitabine-treated LFS OBs showed impaired OB differentiation ability (Figure S5K) and slightly increased in vitro tumorigenic potential (Figures S5L and S5M), suggesting that the clinical application of demethylating reagents, at least Decitabine, in treating LFS patients with OS is unlikely to provide much benefit.
Involvement of Human IGN in Osteogenesis and Neurogenesis
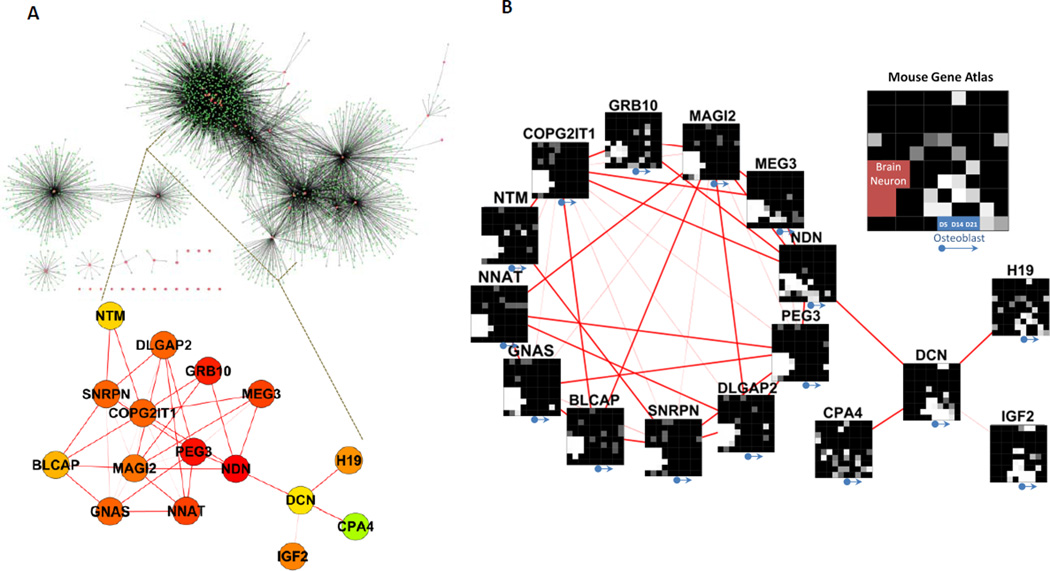
Mouse H19 controls cell growth and development by regulating the expression of several imprinted genes within the IGN (Gabory et al., 2009). Interestingly, in comparison with other bone-associated tissues, the expression of mouse imprinted genes is enriched in differentiated OBs and many of these are themselves members of the IGN (9 out of 15; i.e., H19, Ndn, Igf2, Peg3, Zac1, Sgce, Dlk1, Mest and Cdkn1c) (Figure S6A). Additionally, in comparison to normal mouse OBs, these imprinted genes, including H19, are significantly downregulated in mouse OS (Figure S6B). These results imply that the IGN may have a role in osteogenesis and that its dysregulation may promote OS in mice. We hypothesized that H19 suppresses LFS-associated OS through the imprinted gene regulatory system. Since a human IGN was not yet established, we first searched for genes frequently co-expressed with human imprinted genes and built a human IGN from a database of 79 human tissues (178 arrays) (human U133A/GNF1H Gene Atlas; GSE1133). Fifty-two imprinted genes were extracted from total of 63 known and putative human imprinted genes (http://www.geneimprint.com/). The main human IGN is composed of 16 imprinted genes and divided into 2 sub-networks, the DCN and the NDN sub-IGN (Figure 6A). The DCN sub-IGN includes 4 imprinted genes (H19, DCN, IGF2 and CPA4) and the NDN sub-IGN contains 12 imprinted genes (NDN, PEG3, NNAT, MEG3, GNAS, MAGI2, COPG2IT1, GRB10, DLGAP2, SNRPN, NTM and BLCAP). Tissue specific gene expression defines their unique biological roles and the enriched expressed genes, in general, have a role in maintaining tissue or cell specific functions. Imprinted genes have been suggested to execute their functions through modulating the expression of their coregulated genes. Accordingly, we analyzed coregulated gene expression associated with these 16 imprinted genes (Table S3) using the Mouse Gene Atlas database. Interestingly, we found that the coregulated genes associated with individual imprinted genes in the DCN sub-IGN are primarily implicated in osteogenesis. In contrast, the coregulated genes associated with the NDN sub-IGN are mainly implicated in neurogenesis (Figure 6B). These findings strongly indicate that although individual imprinted genes participate in more diverse biological functions, the main roles of the entire IGN are in osteogenesis and neurogenesis. Gene expression analyses of 318 H19 coregulated genes suggest that these may function in early OB differentiation as well as lung, lactating mammary gland and ciliary body development (Figure S6C). Interestingly, expression of these coregulated genes is mainly enriched in early but not late OB differentiation. This led us to suspect that H19 may function as an initiator of osteogenesis and those downstream molecules, including coregulated imprinted genes, may function during later osteogenic stages.
Figure 6. A network of coregulated human imprinted genes.
(A) Genes linked to human imprinted genes, including H19, identified from a set of 79 human tissues. 52 imprinted genes are extracted from total 63 known and putative human imprinted genes. The main human IGN consists of 16 imprinted genes and contains 2 sub-networks. (B) Network2Canvas analysis of these 16 imprinted coregulated genes by GO biological processes reveals that DCN-associated IGN primarily participates in osteogenesis and NDN-associated IGN in neurogenesis. See also Figure S6 and Table S3.
DCN functions downstream of H19 in LFS-Associated OS Development
We next asked whether genes in the H19-associated IGN are responsible for the observed H19-modulated OB differentiation and oncogenic repression. We noticed that the imprinted gene DCN, encoding a matrix proteoglycan, is directly linked to H19 in the IGN (Figure 6B) and that DCN-coregulated genes are relatively overexpressed in OBs in a gradually increasing pattern during osteogenesis (Figure 7A). Because we also realized that the biological functions of DCN-coregulated genes are mainly in cell adhesion and the cell matrix (Figure 7B), both of which are known to regulate OB differentiation (Sosa-Garcia et al., 2010), we pursued this candidate further. Alignment and quantification of reads at the DCN locus by FPKM values and qRT-PCR showed that DCN is gradually upregulated in WT but not LFS OBs during differentiation (Figures S7A and S7B). Decreased DCN expression in LFS OBs was further confirmed in OBs from multiple LFS iPSC lines (Figures S7C). DCN expression was significantly decreased in OS and p53 mutant cells in comparison with bone/OB tissues and p53 WT cells (Figures S7D and S7E). RNAi-mediated knockdown of DCN led to decreased expression of the osteogenic factor ZEB1, ALPL and several osteogenic differentiation-associated genes (Figure S7F), supporting an essential role for DCN in normal osteogenesis. Furthermore, knockdown of H19 downregulated DCN while ectopic expression of H19 upregulated it in WT and LFS MSCs, respectively (Figure S7G). In agreement with these results, DCN expression was directly correlated with H19 expression in both WT and LFS samples as well as in OS cell lines (Figures S7H and S7I). In contrast, knockdown of DCN did not alter H19 expression (Figure S7J). These findings demonstrate that H19 functions as an upstream regulator of DCN expression. To further elucidate whether DCN functions as a downstream regulator involved in H19-mediated OB differentiation and tumor suppression, we depleted DCN in LFS OBs expressing ectopic H19 and found that H19-mediated upregulation of the pre-osteoblastic marker ALPL was abolished upon DCN knockdown (Figure 7C) and that the in vitro and in ovo H19-mediated suppression of LFS OB tumorigenesis was inhibited (Figures 7D and 7E). These findings suggest that H19 regulation of osteogenesis and suppression of OS is, at least in part, mediated via DCN. In comparison with MSCs, GSEA of DCN-coregulated genes revealed their enrichment during OB differentiation (Figure 7F, left panel). Additionally, in comparison with WT OBs and MSC/OB tissues, DCN-coregulated genes were significantly decreased in LFS OBs and OS (Figure 7F, right panels), implying that DCN may negatively regulate OS development. Supporting its tumor suppressor function, ectopic expression of DCN reduced the incidence of OSA and HOS-initiated tumor development as well as tumor size (Figure 7G). Taken together, these findings reveal that dysregulation of the H19-DCN IGN is strongly linked to LFS-associated osteogenic defects culminating in OS development.
Figure 7. DCN functions downstream of H19 in LFS-associated OS development.
(A) GO biological processes examined by Network2Canvas indicate DCN coregulated genes are significantly upregulated during osteogenesis. (B) Panther analysis indicates DCN-coregulated genes are mainly involved in cell adhesion and cell matrix functions. (C) H19-mediated upregulation of pre-osteoblastic marker ALPL is abolished upon DCN depletion. Error bars indicate ±SEM of triplicates. (D–E) RNAi-mediated DCN knockdown impairs H19-mediated inhibition of LFS OB tumorigenic activity in vitro (D) and in ovo (E). Error bars indicate ±SEM. (F) GSEA of DCN-correlated gene expression indicates expression of DCN network genes during OB differentiation that is impaired in LFS OBs and OS cells. (G) DCN inhibits both OSA and HOS tumorigenesis. The sizes of HOS and OSA tumors were examined 1 and 2 months after subcutaneous injection, respectively. See also Figure S7.
DISCUSSION
In vitro modeling of human disease has been greatly facilitated by iPSC methodologies (Takahashi et al., 2007; Takahashi and Yamanaka, 2006; Yu et al., 2007). Characterized by their ability to self-renew indefinitely and differentiate into all cell lineages of an organism, iPSCs provide a powerful system for human disease modeling. The p53 tumor suppressor is considered a promising therapeutic target to treat tumors with p53 mutations or deletions (Freed-Pastor and Prives, 2012). However, the lack of a reliable model limits the development of useful approaches to treat cancers caused by either genetic or somatic p53 mutations. Instead of regular application of clinical patient samples, cancer cell lines and mouse models have been utilized to study p53 function. Here we demonstrate the possibility of using LFS iPSCs to turn clinical samples into cell lines and models to study the pathological mechanisms caused by mutations in p53. This model system not only serves as an alternative tool to study p53 mutation-associated disorders but also provides substantial benefits for studying the role of p53 in the early-stages of tumor development.
The role of mutant p53 in osteogenesis and OS
A series of studies have demonstrated that the p53 tumor suppressor promotes differentiation in a variety of cell types (Lee et al., 2012; Molchadsky et al., 2010). Since some types of cancer, such as OS, are considered undifferentiated, it is logical to regard the cancer to be a defect in protective differentiation that would normally suppress unchecked cell proliferation and thus, prevent tumor development. However, recent in vivo evidence from p53 knockout and conditional MDM2 deletion mice, suggesting that wild-type p53 attenuates OB differentiation and bone development (Lengner et al., 2006; Wang et al., 2006), makes the situation far more complex. In our current studies, we found that H19 promotes OB differentiation and is repressed by p53; thus, providing a possible explanation for how p53 can suppress OB differentiation.
Strikingly, the p53(G245D) mutant exerted a gain-of-function effect in repressing H19 transcription (Figures S5D and S5F), indicating that the defective OB differentiation in LFS OBs may result from inhibition of H19-mediated osteogenesis. In contrast to the p53(G245D) gain-of-function effect in downregulating H19 expression, this mutant exhibited a partial loss-of-function effect in upregulating the majority of known p53 downstream targets (e.g., MDM2, p21 and SFN). Advanced systems-level studies to characterize p53(G245D) function by identifying genome-wide differences between WT and mutant p53 will be needed to elucidate the comprehensive mechanisms involved in LFS-associated OS development.
Is H19 a tumor suppressor or an oncogene?
It has been suggested that H19 may act as a tumor suppressor (Hao et al., 1993; Yoshimizu et al., 2008). In contrast, several in vitro culture experiments have suggested a controversial oncogenic role for H19 (Lustig-Yariv et al., 1997; Verkerk et al., 1997). This discrepancy could be explained by both differences in the experimental systems and the complexity of H19 functions in developmental and physiological processes. In our studies, H19 is both significantly upregulated during osteogenesis and commonly downregulated in OS, suggesting both differentiation-promoting and tumor-suppressing roles. Moreover, ectopic expression of H19 in LFS OBs restored normal osteogenic differentiation. H19 acts not only by directly modulating downstream targets as a lncRNA but also, indirectly controls an entire group of genes via its associated IGN. Notably, many distinct cancers are associated with dysregulation of imprinted genes (Joyce and Schofield, 1998). Such dysregulation may disrupt the H19-associated IGN and additional gene networks; thus, offering a possible explanation for the distinct effects, variously as an oncogene or a tumor suppressor observed after ectopic expression of H19. Moreover, it must also be acknowledged that the H19 locus may play more complex roles than regulation of the IGN, and these unidentified functions may also play roles in its bivalent function in tumorigenesis both in different tissues and at different tumor developmental stages.
LFS iPSC disease model: an alternative system to study the early stages of OS development
One key feature of clinically apparent OS is its numerous chromosomal alterations and rearrangements (Batanian et al., 2002; Bridge et al., 1993). A high level of genomic instability, in particular, chromosomal instability, is commonly found in OS. Tumor suppressor genes are frequently lost and oncogenes are duplicated. Because genomic instability is not only a consequence of tumor progression but also an active driver of tumor evolution, it creates a heterogeneous cell population and makes it more difficult to understand the initial steps of tumorigenesis. In comparison with normal differentiated OBs, human OS cell lines and a conditional mouse OS model (Walkley et al., 2008) showed strong enrichment of certain cytogenetic rearrangement regions. Because human/mouse OS lines are only isolated after many steps of tumor evolution, using these to study the initial stage of OS tumorigenesis is challenging. In marked contrast, LFS OBs and tumors showed a negligible degree of common OS cytogenetic rearrangements in comparison with WT OBs (Figure 4F), demonstrating the existence of a relatively intact genome. This relatively undisturbed genome also helps to explain the lower rate of and weak tumorigenicity of LFS OBs in vitro and in vivo. Thus we anticipate that LFS iPSC-derived OBs will be used in future studies focused on the role of mutant p53 in early OS progression prior to development of broad genomic alterations.
In summary, LFS iPSC-derived OBs not only provide a high-fidelity model system to elucidate the pathological mechanism of p53 mutant-associated OS development but also document a path for using LFS-associated gene expression patterns to predict clinical outcomes. More generally, iPSC approaches will also facilitate the definition of inherited versus somatically acquired causal components in many cancers. Further investigations to identify the regulatory mechanism of H19-DCN IGN and to develop drugs to activate H19 and DCN may have powerful clinical implications for the treatment and/or prevention of OS in patients with either inherited or somatically acquired p53 mutations.
EXPERIMENTAL PROCEDURES
Somatic cell programming with non-integrating SeV
3 LFS patients and 2 relatives WT fibroblasts were cultured and maintained in DMEM media supplemented 10% (vol/vol) Benchmark FBS (Gemini Bio-Product) and antibiotics. These fibroblasts were reprogrammed by transducing SeV expressing the 4 reprogramming factors OCT4, SOX2, KLF4 and c-MYC (CytoTune reprogramming kit, Invitrogen) according to manufacturer protocol. The reprogramming cells were maintained in hESC media (DMEM/F12 (Cellgro, Mediatech) containing 20% (vol/vol) KnockOut Serum Replacement (Invitrogen), L-glutamine, non-essential amino acids, β-mercaptoethanol, antibiotics and bFGF). After 3–4 weeks post-induction, individual clones with hESC/iPSC morphology and positive for TRA-1-60 and TRA-1-81 live staining were picked, passaged on MEFs and examined for loss of SeV by both staining with anti-SeV specific antibody (PD029, MBL) and by quantitative RT-PCR measurement of expression of the exogenous 4 factors. The specific qPCR primers targeting exogenous OCT4, SOX2, KLF4 and c-MYC are listed in Table S4.
In vitro differentiation of iPSCs to MSCs
In vitro differentiation of WT and LFS iPSCs to MSCs was performed by a well-defined MSC differentiation protocol described previously (Lian et al., 2007). Briefly, iPSCs were seeded in gelatin-coated plates and cultured in MSC-differentiation media (DMEM supplemented with 10% Knockout serum replacement, 5 ng/ml FGF2 and 5 ng/ml PDGF-AB (PeproTech)) to induce differentiation. When differentiated cells were confluent, cells were trypsinized, split, and maintained. After 3-weeks of differentiation, the differentiated MSCs were sorted as the CD105 (eBioscience)-positive and CD24 (BD Pharmingen)-negative cells by BD AriaII in the Mount Sinai Flow Cytometry Shared Facility and expanded in MSC media (DMEM supplemented with 10% FBS). These differentiated MSCs were further examined for expression of other MSC surface markers CD44 (BD Pharmingen), CD73 (BD Pharmingen) and CD166 (BD Pharmingen) as well as MSC-associated factors SNAI and VIM by immunostaining with anti-SNAI1 (Santa Cruz) and anti-VIM (Millipore) antibodies.
In vitro osteogenic differentiation of MSCs
LFS and WT iPSC-derived MSCs were plated in 12-well plate or 6-well plate at a density of 1×104 cells or 2×104 cells per well, respectively, in osteogenic differentiation medium (α-MEM supplemented with 10% FBS, 0.1µM dexamethasone, 10mM β-glycerol phosphate, and 200µM ascorbic acid) (Barberi et al., 2005). Cells were differentiated for specific time points as noted in the main text before characterization.
In vitro AIG and oncosphere assays
LFS and WT MSCs were cultured and passaged in 6-well plate at a density of 2×104 cells per well. Cells were cultured in OB differentiation medium for 7 days, split and 1×104 cells resuspended in OB differentiation medium with 0.4–0.5% LMP agarose. The cell suspensions were then plated in 12-well plates containing solidified 0.8% agarose in OB differentiation medium. Cells were maintained in osteogenic differentiation medium for 1 month with medium changes every three days. Colony (considered to have a diameter≥50 µm) were counted under a microscope. For the oncosphere assay, 2×104 7-day differentiated osteoblasts were washed by DPBS twice, resuspended in oncosphere medium (α-MEM supplemented with 0.1µM dexamethasone, 10mM β-glycerol phosphate, 200µM ascorbic acid, B27 supplement, 5µg/ml Heparin, 20 ng/ml bFGF, and 20 ng/ml EGF) and seeded in ultra-low attachment 6-well plates (3471, Corning). The number of oncospheres (diameter ≥50 µm) was calculated after 12 days.
Xenotransplantation
All animal procedures were performed in accordance with the Mount Sinai’s Institutional Animal Care and Use Committee (IACUC). 2 ×106 Matrigel-mixed differentiated osteoblasts, 2 ×106 Matrigel-mixed HOS (ATCC CRL-1543) and 1×106 OSA (SJSA-1, ATCC CRL-2098) cells were injected subcutaneously into both right and left hind legs of 8-week-old immunocompromised nude mice (Charles River Laboratories). Tumors were excised around 6–10 weeks after injection. Tumors were weighed, fixed overnight in 10% neutral buffer formalin, embedded in paraffin, sectioned and stained with H&E, AP, PicroSirius red, and von Kossa stains to examine bone AP activity, AP expression, collagen matrix deposition and mineralization, respectively, by HistoWiz, Inc.
Statistical Analyses
Results are expressed as the mean and error bars represent SEM. Difference between two groups were examined by two-tailed unpaired t test. *, p<0.05; **, p<0.01; and ***, p<0.001.
Supplementary Material
ACKNOWLEDGEMENTS
We thank P. Yang, X. Wei, C.-K. Chou, C.F. Pereira, Y. Liu and X. Niu, for experimental assistance. We also thank E. Lopez-Rivera and M. Li for CAM assay assistance, the Mount Sinai Genomic Core Facility and Columbia Genome Center for mRNA-seq analysis, as well as Mount Sinai’s Flow Cytometry Core for sorting MSCs. We gratefully acknowledge K.A. Moore, and A. Waghray for discussions. This research was funded by grants from the National Institutes of Health (NIH) to I.R.L. (5R01GM078465), the Empire State Stem Cell Fund through New York State Department of Health (NYSTEM) to I.R.L. (C024176 and C024410), Sister Institution Fund of China Medical University and Hospital and MDACC, The National Breast Cancer Foundation, Inc. NSC 102-2321-B-039 (Taiwan) to M.-C.H., the Canadian Breast Cancer Foundation-Prairies/NWT region to R.M. and NIH Pathway to Independence Award (K99CA181496) to D.-F.L.. B.C. is a trainee in NIDCR-Interdisciplinary Training in Systems and Developmental Biology and Birth Defects (T32HD075735).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
All mRNA-seq data are listed in Table S2 and deposited in NCBI-Gene Expression Omnibus database under accession number GSE58123.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, seven figures and six tables.
REFERENCES
- 1.Barberi T, Willis LM, Socci ND, Studer L. Derivation of multipotent mesenchymal precursors from human embryonic stem cells. PLoS Med. 2005;2:e161. doi: 10.1371/journal.pmed.0020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barley RD, Enns L, Paterson MC, Mirzayans R. Aberrant p21WAF1-dependent growth arrest as the possible mechanism of abnormal resistance to ultraviolet light cytotoxicity in Li-Fraumeni syndrome fibroblast strains heterozygous for TP53 mutations. Oncogene. 1998;17:533–543. doi: 10.1038/sj.onc.1202271. [DOI] [PubMed] [Google Scholar]
- 3.Batanian JR, Cavalli LR, Aldosari NM, Ma E, Sotelo-Avila C, Ramos MB, Rone JD, Thorpe CM, Haddad BR. Evaluation of paediatric osteosarcomas by classic cytogenetic and CGH analyses. Mol Pathol. 2002;55:389–393. doi: 10.1136/mp.55.6.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bridge JA, Bhatia PS, Anderson JR, Neff JR. Biologic and clinical significance of cytogenetic and molecular cytogenetic abnormalities in benign and malignant cartilaginous lesions. Cancer Genet Cytogenet. 1993;69:79–90. doi: 10.1016/0165-4608(93)90080-6. [DOI] [PubMed] [Google Scholar]
- 5.Choufani S, Shuman C, Weksberg R. Beckwith-Wiedemann syndrome. Am J Med Genet C Semin Med Genet. 2010;154C:343–354. doi: 10.1002/ajmg.c.30267. [DOI] [PubMed] [Google Scholar]
- 6.Di Como CJ, Gaiddon C, Prives C. p73 function is inhibited by tumor-derived p53 mutants in mammalian cells. Mol Cell Biol. 1999;19:1438–1449. doi: 10.1128/mcb.19.2.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dugimont T, Montpellier C, Adriaenssens E, Lottin S, Dumont L, Iotsova V, Lagrou C, Stehelin D, Coll J, Curgy JJ. The H19 TATA-less promoter is efficiently repressed by wild-type tumor suppressor gene product p53. Oncogene. 1998;16:2395–2401. doi: 10.1038/sj.onc.1201742. [DOI] [PubMed] [Google Scholar]
- 8.Eggermann T. Russell-Silver syndrome. Am J Med Genet C Semin Med Genet. 2010;154C:355–364. doi: 10.1002/ajmg.c.30274. [DOI] [PubMed] [Google Scholar]
- 9.Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012;26:1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gabory A, Ripoche MA, Le Digarcher A, Watrin F, Ziyyat A, Forne T, Jammes H, Ainscough JF, Surani MA, Journot L, et al. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development. 2009;136:3413–3421. doi: 10.1242/dev.036061. [DOI] [PubMed] [Google Scholar]
- 12.Gaiddon C, Lokshin M, Ahn J, Zhang T, Prives C. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol Cell Biol. 2001;21:1874–1887. doi: 10.1128/MCB.21.5.1874-1887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanel W, Marchenko N, Xu S, Yu SX, Weng W, Moll U. Two hot spot mutant p53 mouse models display differential gain of function in tumorigenesis. Cell Death Differ. 2013;20:898–909. doi: 10.1038/cdd.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hao Y, Crenshaw T, Moulton T, Newcomb E, Tycko B. Tumour-suppressor activity of H19 RNA. Nature. 1993;365:764–767. doi: 10.1038/365764a0. [DOI] [PubMed] [Google Scholar]
- 15.Haydon RC, Luu HH, He TC. Osteosarcoma and osteoblastic differentiation: a new perspective on oncogenesis. Clin Orthop Relat Res. 2007;454:237–246. doi: 10.1097/BLO.0b013e31802b683c. [DOI] [PubMed] [Google Scholar]
- 16.Joyce JA, Schofield PN. Genomic imprinting and cancer. Mol Pathol. 1998;51:185–190. doi: 10.1136/mp.51.4.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, Valentin-Vega YA, Terzian T, Caldwell LC, Strong LC, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 18.Lee DF, Su J, Ang YS, Carvajal-Vergara X, Mulero-Navarro S, Pereira CF, Gingold J, Wang HL, Zhao R, Sevilla A, et al. Regulation of embryonic and induced pluripotency by aurora kinase-p53 signaling. Cell Stem Cell. 2012;11:179–194. doi: 10.1016/j.stem.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lengner CJ, Steinman HA, Gagnon J, Smith TW, Henderson JE, Kream BE, Stein GS, Lian JB, Jones SN. Osteoblast differentiation and skeletal development are regulated by Mdm2-p53 signaling. J Cell Biol. 2006;172:909–921. doi: 10.1083/jcb.200508130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li FP, Fraumeni JF., Jr Rhabdomyosarcoma in children: epidemiologic study and identification of a familial cancer syndrome. J Natl Cancer Inst. 1969;43:1365–1373. [PubMed] [Google Scholar]
- 21.Lian Q, Lye E, Suan Yeo K, Khia Way Tan E, Salto-Tellez M, Liu TM, Palanisamy N, El Oakley RM, Lee EH, Lim B, et al. Derivation of clinically compliant MSCs from CD105+, CD24- differentiated human ESCs. Stem Cells. 2007;25:425–436. doi: 10.1634/stemcells.2006-0420. [DOI] [PubMed] [Google Scholar]
- 22.Lustig-Yariv O, Schulze E, Komitowski D, Erdmann V, Schneider T, de Groot N, Hochberg A. The expression of the imprinted genes H19 and IGF-2 in choriocarcinoma cell lines. Is H19 a tumor suppressor gene? Oncogene. 1997;15:169–177. doi: 10.1038/sj.onc.1201175. [DOI] [PubMed] [Google Scholar]
- 23.Malkin D, Li FP, Strong LC, Fraumeni JF, Jr, Nelson CE, Kim DH, Kassel J, Gryka MA, Bischoff FZ, Tainsky MA, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250:1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 24.Mirzayans R, Andrais B, Scott A, Paterson MC, Murray D. Single-cell analysis of p16(INK4a) and p21(WAF1) expression suggests distinct mechanisms of senescence in normal human and Li-Fraumeni Syndrome fibroblasts. J Cell Physiol. 2010;223:57–67. doi: 10.1002/jcp.22002. [DOI] [PubMed] [Google Scholar]
- 25.Molchadsky A, Rivlin N, Brosh R, Rotter V, Sarig R. p53 is balancing development, differentiation and de-differentiation to assure cancer prevention. Carcinogenesis. 2010;31:1501–1508. doi: 10.1093/carcin/bgq101. [DOI] [PubMed] [Google Scholar]
- 26.Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, Crowley D, Jacks T. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 27.Porter DE, Holden ST, Steel CM, Cohen BB, Wallace MR, Reid R. A significant proportion of patients with osteosarcoma may belong to Li-Fraumeni cancer families. J Bone Joint Surg Br. 1992;74:883–886. doi: 10.1302/0301-620X.74B6.1447251. [DOI] [PubMed] [Google Scholar]
- 28.Sosa-Garcia B, Gunduz V, Vazquez-Rivera V, Cress WD, Wright G, Bian H, Hinds PW, Santiago-Cardona PG. A role for the retinoblastoma protein as a regulator of mouse osteoblast cell adhesion: implications for osteogenesis and osteosarcoma formation. PLoS One. 2010;5:e13954. doi: 10.1371/journal.pone.0013954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 30.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 31.Tang N, Song WX, Luo J, Haydon RC, He TC. Osteosarcoma development and stem cell differentiation. Clin Orthop Relat Res. 2008;466:2114–2130. doi: 10.1007/s11999-008-0335-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varley JM. Germline TP53 mutations and Li-Fraumeni syndrome. Hum Mutat. 2003;21:313–320. doi: 10.1002/humu.10185. [DOI] [PubMed] [Google Scholar]
- 33.Varrault A, Gueydan C, Delalbre A, Bellmann A, Houssami S, Aknin C, Severac D, Chotard L, Kahli M, Le Digarcher A, et al. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev Cell. 2006;11:711–722. doi: 10.1016/j.devcel.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 34.Verkerk AJ, Ariel I, Dekker MC, Schneider T, van Gurp RJ, de Groot N, Gillis AJ, Oosterhuis JW, Hochberg AA, Looijenga LH. Unique expression patterns of H19 in human testicular cancers of different etiology. Oncogene. 1997;14:95–107. doi: 10.1038/sj.onc.1200802. [DOI] [PubMed] [Google Scholar]
- 35.Walkley CR, Qudsi R, Sankaran VG, Perry JA, Gostissa M, Roth SI, Rodda SJ, Snay E, Dunning P, Fahey FH, et al. Conditional mouse osteosarcoma, dependent on p53 loss and potentiated by loss of Rb, mimics the human disease. Genes Dev. 2008;22:1662–1676. doi: 10.1101/gad.1656808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, Kua HY, Hu Y, Guo K, Zeng Q, Wu Q, Ng HH, Karsenty G, de Crombrugghe B, Yeh J, et al. p53 functions as a negative regulator of osteoblastogenesis, osteoblast-dependent osteoclastogenesis, and bone remodeling. J Cell Biol. 2006;172:115–125. doi: 10.1083/jcb.200507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yi L, Lu C, Hu W, Sun Y, Levine AJ. Multiple roles of p53-related pathways in somatic cell reprogramming and stem cell differentiation. Cancer Res. 2012;72:5635–5645. doi: 10.1158/0008-5472.CAN-12-1451. [DOI] [PubMed] [Google Scholar]
- 38.Yoshimizu T, Miroglio A, Ripoche MA, Gabory A, Vernucci M, Riccio A, Colnot S, Godard C, Terris B, Jammes H, et al. The H19 locus acts in vivo as a tumor suppressor. Proc Natl Acad Sci U S A. 2008;105:12417–12422. doi: 10.1073/pnas.0801540105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.