Abstract
Although much is known about the mechanisms of signal-mediated protein and RNA nuclear import and export, little is understood concerning the nuclear import of plasmid DNA. Plasmids between 4.2 and 14.4 kilobases were specifically labeled using a fluorescein-conjugated peptide nucleic acid clamp. The resulting substrates were capable of gene expression and nuclear localization in microinjected cells in the absence of cell division. To elucidate the requirements for plasmid nuclear import, a digitonin-permeabilized cell system was adapted to follow the nuclear localization of plasmids. Nuclear import of labeled plasmid was time- and energy-dependent, was inhibited by the lectin wheat germ agglutinin, and showed an absolute requirement for cytoplasmic extract. Addition of nuclear extract alone did not support plasmid nuclear import but in combination with cytoplasm stimulated plasmid nuclear localization. Whereas addition of purified importin α, importin β, and RAN was sufficient to support protein nuclear import, plasmid nuclear import also required the addition of nuclear extract. Finally, nuclear import of plasmid DNA was sequence-specific, requiring a region of the SV40 early promoter and enhancer. Taken together, these results confirm and extend our findings in microinjected cells and support a protein-mediated mechanism for plasmid nuclear import.
The nuclear envelope presents an effective barrier between the nuclear and cytoplasmic compartments of the cell. Although it is impermeant to large non-nuclear molecules, a multitude of macromolecules must enter and exit the nucleus across this envelope every second in order for the cell to live. All macromolecular exchange between the nucleus and the cytoplasm studied to date occurs through the nuclear pore complex (NPC),1 is signal-dependent, and utilizes a series of receptor proteins (for a review see Ref. 1). In the case of proteins destined for the nucleus, the nuclear localization signal (NLS) interacts with one of a growing number of importin family members to target the complex to the NPC. In the classical case of NLS-containing proteins, the protein binds to importin α, the NLS “receptor,” which in turn interacts with importin β. Once at the NPC, the complex interacts with the small GTP-binding protein RAN in its GDP-bound state and its accessory factor NTF2 while being translocated across the NPC. After translocation into the nucleus, the complex disassembles because of the conversion of RAN-GDP to RAN-GTP by exchange or replacement and the importins return to the cytoplasm (2). Similar scenarios of receptor proteins interacting with signals to mediate translocation across the nuclear envelope also occur for the nuclear export of proteins (e.g. exportin and the nuclear export signal) and viral mRNAs (Crm1p, HIV Rev, and the Rev response element), and the nuclear import of small nuclear RNAs that contain both protein-encoded NLSs and the trimethylguanosine cap as import signals (1, 3).
We have recently shown that the nuclear import of plasmid DNA (pDNA) also uses a signal-mediated pathway to enter the nucleus. Using a microinjection approach, we demonstrated that pDNA can enter the nucleus in the absence of cell division by a process that is consistent with transport through the NPC (4). Such nuclear import has been detected in all mammalian cells tested to date, including those of mouse, rat, monkey, human, and chicken, as well as those from all cell types, including smooth and striated muscle, fibroblasts, endothelial cells, and epithelial cells. Furthermore, this import is sequence-specific: plasmids containing as little as 80 bp of the SV40 enhancer/early promoter region are targeted to the nucleus in the absence of cell division, whereas plasmids lacking this sequence remain cytoplasmic (4).2 This sequence specificity appears unique to SV40, because several other strong viral promoter and enhancer sequences, including the immediate early promoter/enhancer of cytomegalovirus and the long terminal repeat of Rous sarcoma virus, fail to promote nuclear import.2 Based on the sequences required for plasmid nuclear import, we have proposed a working model in which pDNA import is mediated by newly synthesized transcription factors and other sequence-specific DNA-binding proteins that bind to the DNA in the cytoplasm, thus attaching NLSs to the DNA for recognition and transport by the NLS-dependent machinery. In order to further study the mechanisms of pDNA nuclear import, an in vitro system was required. To this end, we have developed an approach to label plasmid DNA that maintains its biological activity and have adapted the digitonin-permeabilized cell assay to study plasmid nuclear localization.
EXPERIMENTAL PROCEDURES
Plasmids
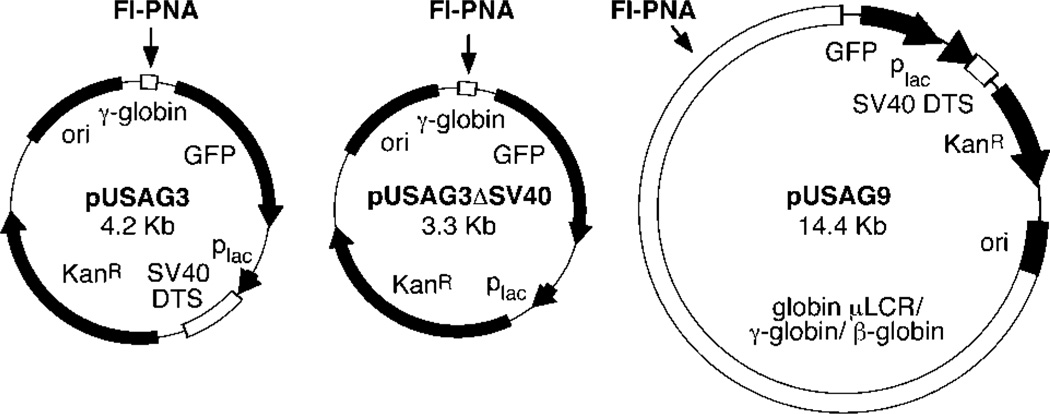
The 4.2-kb plasmid pUSAG3 contains a 60-bp sequence from the human G gamma globin promoter (−315 to −255) inserted into the SmaI site of the GFP promoter reporter vector pEGFP-1 (CLONTECH Laboratories, Palo Alto, CA) in the same orientation as the GFP gene (see Fig. 1). To create pUSAG3ΔSV40 that lacks the SV40 nuclear targeting sequence, a 913-bp fragment containing the SV40 origin and early promoter/enhancer region was removed from pUSAG3 by digestion with StuI and SspI and religation. pUSAG9 is a 14.4-kb plasmid containing the human globin locus control region and the A-γ globin and β globin genes (nucleotides 38084–43975 and 60409–65475, EMBL ID: HSHBB4R1) inserted into the SmaI site of pEGFP-1. pUSAG9 also contains the 60-bp PNA-binding site found in pUSAG3. Rhodamine-labeled pGenegrip blank vector and Rhodamine-labeled pGenegrip GFP vector were obtained from Gene Therapy Systems (San Diego, CA).
Fig. 1.
Maps of plasmids used in this study
PNA Binding
A fluorescein-labeled PNA clamp (NH2-fluorescein-OTTTTCTTCTCOOOJTJTTJTTTT-COOH; Fl-PNA) was synthesized by Perseptive Biosystems (Framingham, MA) to bind to the target GAGAAGAAA present in the 60-bp G γ globin promoter fragment (see Fig. 1). Fl-PNA (3 µ m) was reacted with the purified plasmids at a 10:1 molar ratio in TE buffer (10 mm Tris, pH 8, 1 mm EDTA) for 3 h at 37 °C. The reactions were diluted into 2 ml of TE buffer and concentrated to 50 µl in a Centricon-30 microconcentration device, diluted again to 2 ml, and reconcentrated to a final concentration of between 0.1 and 0.2 mg/ml plasmid. Approximately 30 µg of pDNA was labeled in a typical experiment. Labeled DNAs were stored at −80 °C. The pGenegrip vectors were obtained as labeled DNAs from the manufacturer.
Cells and Microinjection
HeLa cells were grown on coverslips in minimum essential medium (Life Technologies, Inc.) containing 10% fetal bovine serum and antibiotics in a humidified incubator at 37 °C with 5% CO2. Purified PNA-labeled pDNA was suspended in phosphate-buffered saline at a concentration of 0.1 mg/ml and microinjected into the cytoplasm of cells grown on etched coverslips as described (4). With a microinjection volume of approximately 3 × 10−10 ml/cell,2 about 2,000 plasmids were delivered per cell. The cells were incubated for various times, rinsed in phosphate-buffered saline, fixed for 15 min at 4 °C in 3% paraformaldehyde in phosphate-buffered saline, and mounted with 4,6-diamidino-2-phenylindole/diazobicyclo[2.2.2]octane.
Expression and Purification of His-tagged Importin α Importin β, and RAN
The human genes for importin β and RAN were polymerase chain reaction-amplified from reverse-transcribed HeLa cell RNA using oligonucleotide primers containing unique restrictions sites at the ends and cloned into the corresponding restriction sites of ptrcHisB (Invitrogen, San Diego, CA). A plasmid expressing the yeast importin α homologue, hSRP1α, was obtained from K. Weis (EMBL, Heidelberg, Germany) (5). The plasmids were transformed into Escherichia coli BL21(DE3) and expressed and purified by nickel affinity chromatography as described for the appropriate protein (5, 6).
Permeabilized Cell Assays
Permeabilized cell assays were performed as described previously using HeLa cells grown on glass coverslips (7). Briefly, cells were grown to 60% confluency, washed with wash buffer (20 mm HEPES, pH 7.3, 110 mm potassium acetate, 5 mm sodium acetate, 2 mm MgCl2, 1 mm EGTA, 2 mm dithiothreitol), and permeabilized with 40 µg/ml digitonin in wash buffer for 6–8 min on ice. The cells were rinsed for 5–10 min with several changes of cold wash buffer, and the excess buffer was removed before the assays were initiated. Reactions were carried out in a transport buffer consisting of wash buffer containing 1 mg/ml bovine serum albumin, 1 µg/ml aprotinin, leupeptin, and pepstatin, 1 mm GTP, 2 mm ATP, 10 mm phosphocreatine, and 20 units/ml of creatine phosphokinase. Rhodamine- or fluorescein-labeled NLS peptide-conjugated BSA (Fl- or Rh-BSA-NLS) were included at 25 µg/ml and fluorescein-labeled PNA plasmids were present at 10 µg/ml. Where indicated, cytoplasmic extracts prepared from HeLa cells (7) were added to between 5 and 7 mg/ml, HeLa cell nuclear extracts (in vitro transcription grade, Promega, Madison, WI) were added to 0.25 mg/ml, and affinity purified, recombinant his-tagged RAN, importin α, and importin β were used at 0.5 mg/ml each. The lectins wheat germ agglutinin (WGA) and concanavalin A were added to 0.1 mg/ml each. In reactions lacking ATP and GTP, nucleotide triphosphates, phosphocreatine, and creatine-phosphokinase were omitted from the transport buffer and ATP-depleted extracts were prepared by incubating the extracts with 10 units/ml apyrase for 30 min before addition to the cells. Other competitors were added as indicated in the text. The cells were incubated in a humidified chamber at 37 °C for 4 h, unless otherwise indicated. The reactions were terminated by washing the cells in wash buffer and fixing them at 4 °C for 15 min in 3% paraformaldehyde in wash buffer. The cells were mounted with 4,6-diamidino-2-phenylindole/diazobicyclo[2.2.2]octane, and 0.5-µm sections were viewed by confocal microscopy using an ACAS 570 laser-scanning confocal microscope.
Southern Blot
PNA-labeled and unlabeled plasmids were incubated in transport buffer containing HeLa cytoplasmic and nuclear extracts (5 and 0.25 mg/ml, respectively) for 4 h at 37 °C.At the end of the reaction, the DNA was extracted with phenol:chloroform and chloroform alone. 10 µg of tRNA were added, and the DNA was precipitated with ethanol overnight at −20 °C. The DNA was resuspended in 10 µl of TE buffer and separated on a 1% agarose gel, transferred to nylon, and immobilized by UV irradiation as described (8). The Blot was prehybridized at 42 °C for 4 h in 50% formamide containing 5× SSC, 5× Denhardt’s solution, and 2 mg/ml sheared salmon sperm DNA and hybridized with 1 × 106 cpm/ml of nick translated pUSAG3 and 1 × 106 cpm/ml of nick translated pGenegrip Blank for 18 h at 37 °C. The blot was washed at room temperature for 15 min each in 2× SSC/0.1% SDS, 0.5× SSC/0.1% SDS, 0.5× SSC/0.1% SDS, and for 30 min at 37 °C in 0.1× SSC/1% SDS, dried, and exposed to film.
RESULTS
Fluorescent PNA-labeled Plasmid DNA Is Transported into the Nuclei of Cytoplasmically Microinjected Cells
Although we have begun to make progress elucidating the mechanisms of nuclear import of plasmid DNA using microinjection and in situ hybridization, this method has its limitations because of the fact that it detects DNA in fixed samples only. A better substrate with which to follow the transport reactions in real time would be fluorescently labeled pDNA. In order to produce such substrate, we tried a variety of labeling techniques including incorporation of fluorescent nucleotide analogues (e.g. fluorescein-dUTP) by polymerase chain reaction or nick translation followed by ligation to form intact plasmid, covalent and noncovalent high affinity intercalating dyes (e.g. ethidium bromide monoazide and TOTO-1), and reaction of supercoiled plasmid with photoactivatable fluorophore conjugates. Although all of the methods produced fluorescently labeled DNA, two problems limited the use of these labeled DNAs as substrates for import reactions. First, several of the techniques produced either linear DNA or low yields of circularized plasmid, making their use impractical. More importantly, all of the labeled DNAs became inactive in both transcription of reporter genes or migration of the DNA to the nucleus in microinjected cells (not shown). We were successful, however, when we used a fluorescently labeled peptide nucleic acid (Fl-PNA) clamp; the resulting plasmids were transcriptionally active and able to localize to the nucleus as did unlabeled native plasmid.3
The PNA we used to label plasmids bound to a 10-nucleotide sequence (GAGAAGAAAA) within the G γ globin promoter. The target site was cloned upstream of the GFP gene, well removed from the SV40 nuclear targeting sequence, which was downstream of the GFP gene, 1.6 kb away (Fig. 1). One half of the PNA invaded the target site and hybridized to its complementary sequence through standard Watson-Crick base pairs, and the other half of the PNA folded back onto the PNA:DNA double helix to form a triplex structure using Hoogstein base pairs (9). Fluorescein was bound to the amino terminus of the PNA and did not interfere with binding of the PNA to the target. The resulting triplex structure is highly stable (10), and because the backbone of the PNA utilizes peptide bonds, it is resistant to nuclease digestion and even incubation in serum (11). In our hands, the PNA could not be dissociated from the bound state when incubated with a 1000-fold molar excess of target sequence at 37 °C for 8 h as followed by gel shift assays (not shown).
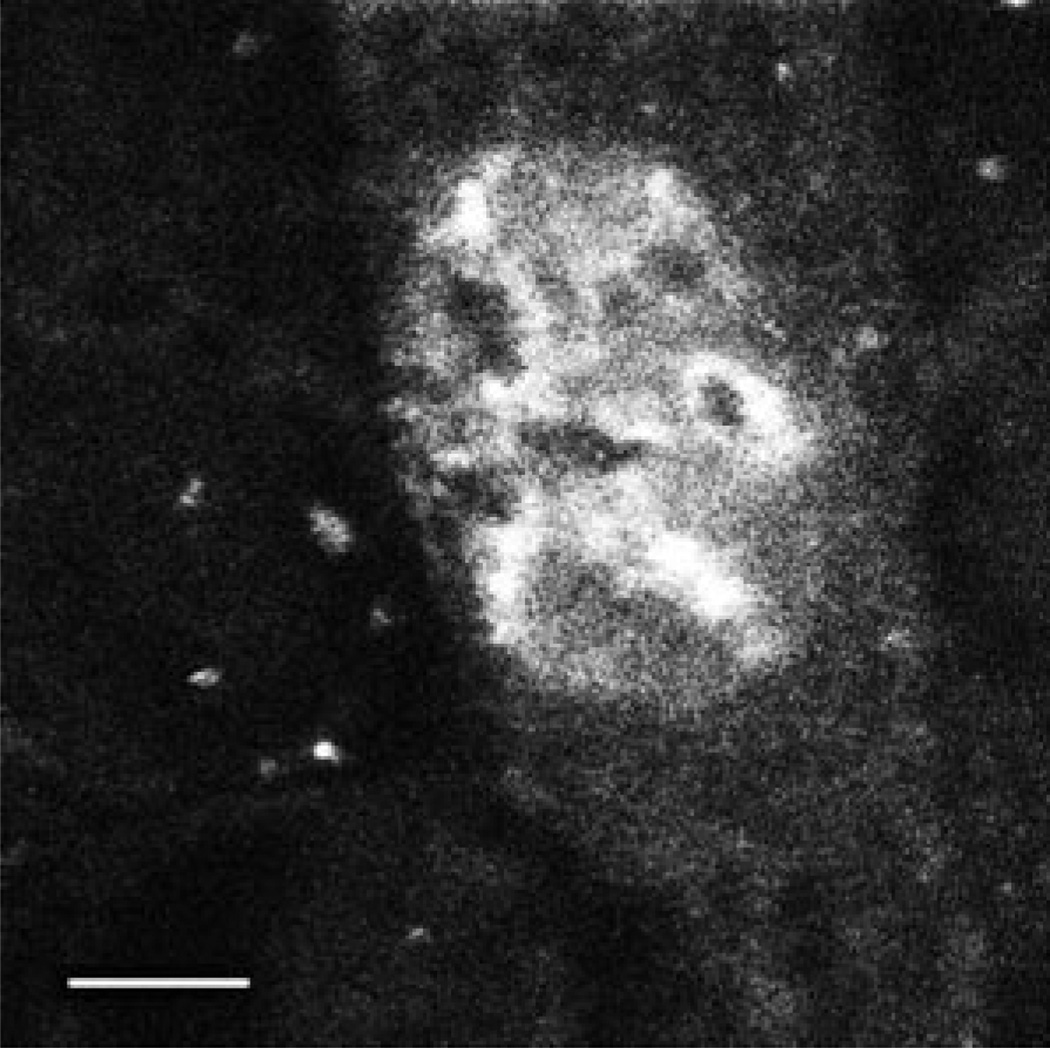
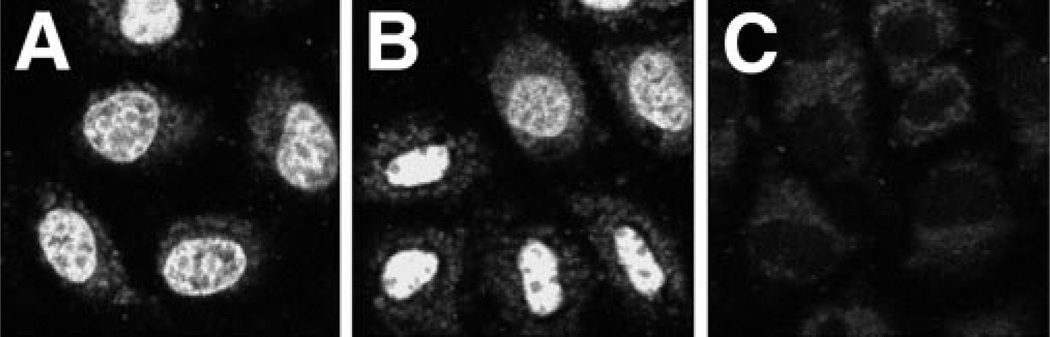
When microinjected into the cytoplasm of TC7 cells, native pDNA localizes to the nucleus within 6–8 h as detected by in situ hybridization (4). When Fl-PNA-labeled pUSAG3 was similarly microinjected into the cytoplasm, it localized to the nucleus within 6 h (Fig. 2). Further, the subnuclear distribution of the labeled pDNA and its absence in the nucleoli was very similar to that seen with native pDNA and in situ hybridization (4). Thus, because the Fl-PNA/pDNA maintained the same biological properties as unmodified plasmid, we used it as a substrate in permeabilized cell assays to dissect the transport mechanism.
Fig. 2. Nuclear localization of Fl-PNA/pDNA in microinjected cells.
TC7 cells were cytoplasmically microinjected with approximately 1000 copies/cell of F1-PNA-conjugated pUSAG3 (0.1 mg/ml). 6 h later, the cells were fixed in 4% paraformaldehyde and viewed by confocal microscopy. Bar = 10 µm.
Reconstitution of Plasmid DNA Nuclear Import in Permeabilized Cells
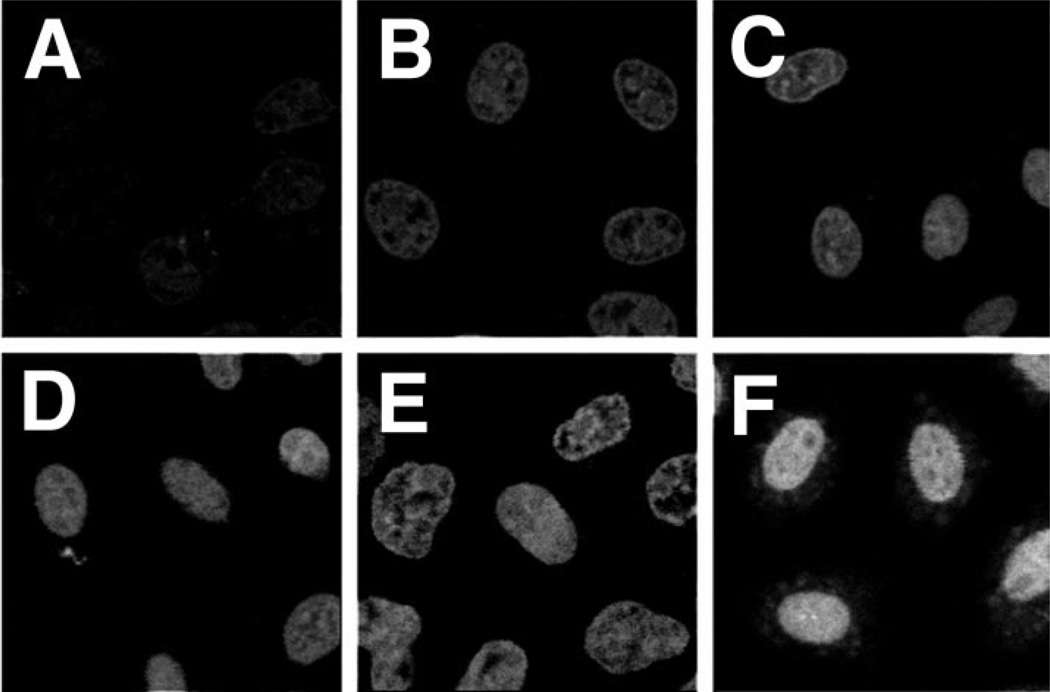
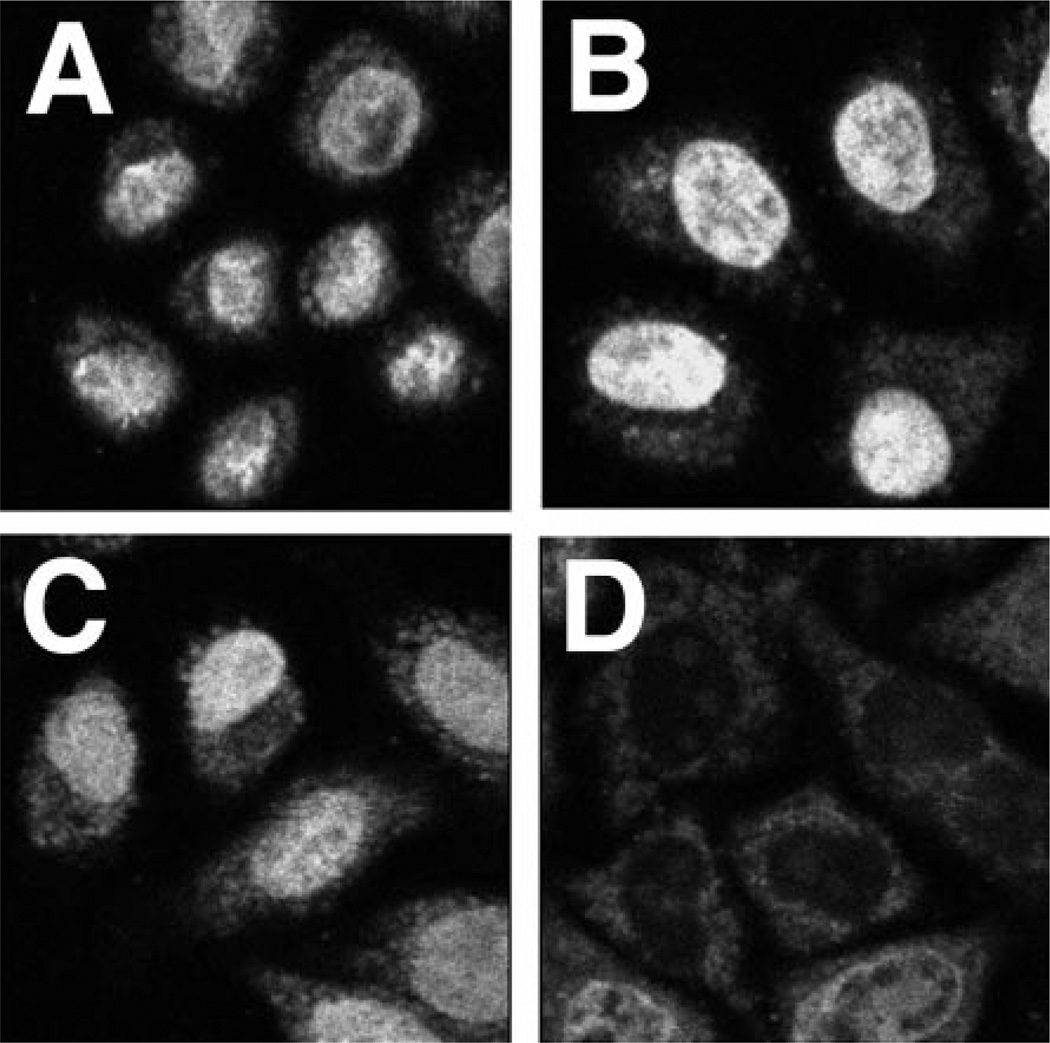
Digitonin-permeabilized HeLa cells have been used effectively to reconstitute nuclear import and export reactions of proteins, small nuclear RNA, and small linear DNA (7, 12–15). We also have used this system to study the requirements for fluorescently labeled pDNA nuclear entry. HeLa cells were permeabilized with digitonin and washed to remove cytoplasmic components as described previously (7). Because all signal-mediated nuclear import reported to date has been shown to require cytosolic proteins (e.g. importins, exportins, etc.), we reasoned that signal-mediated pDNA nuclear import would behave similarly. Thus, we added cytoplasmic extracts prepared from HeLa cells along with an ATP-regenerating system, protease inhibitors, buffer, salts, and GTP to the assays. As a positive control for nuclear import, we also followed the nuclear accumulation of rhodamine-labeled BSA conjugated to a synthetic NLS peptide (Rh-BSA-NLS) to ensure that all the components were active (not shown). As seen in Fig. 3, significant Fl-PNA/pDNA import is observed within 90 min and is maximal by 4 h. In contrast, Rh-BSA-NLS was maximally imported into the nuclei by 30 min (not shown). That the extract was still fully functional and that the nuclei were not leaky at these late times were confirmed by demonstrating that protein nuclear import occurred in an NLS-dependent manner when the substrate was added 4–8 h after the cells had been permeabilized and reacted with cytoplasmic extracts (not shown). Thus, neither the cells nor the extracts lost functional activity or selectivity over this time course. Because maximal nuclear localization was observed by 4 h, this time point was used in the remainder of the experiments.
Fig. 3. Time course of Fl-PNA/ pDNA nuclear localization in permeabilized cells.
HeLa cells were permeabilized with digitonin and washed in import buffer as described under “Experimental Procedures.” The reactions were carried out at 37 °C in transport buffer containing an ATP-regenerating system, 2 mm GTP, 5–7 mg/ml HeLa cytoplasmic extract, and 10 µg/ml Fl-PNA/pUSAG3. Reactions were stopped at 0.5 h (A), 1 h (B, 1.5 h (C), 2 h (D), 3 h (E), and 4 h (F) by washing the cells with wash buffer and fixing them at 4 °C for 15 min in 3% paraformaldehyde in wash buffer. Half-micron sections of the cells were observed and digitized by confocal microscopy.
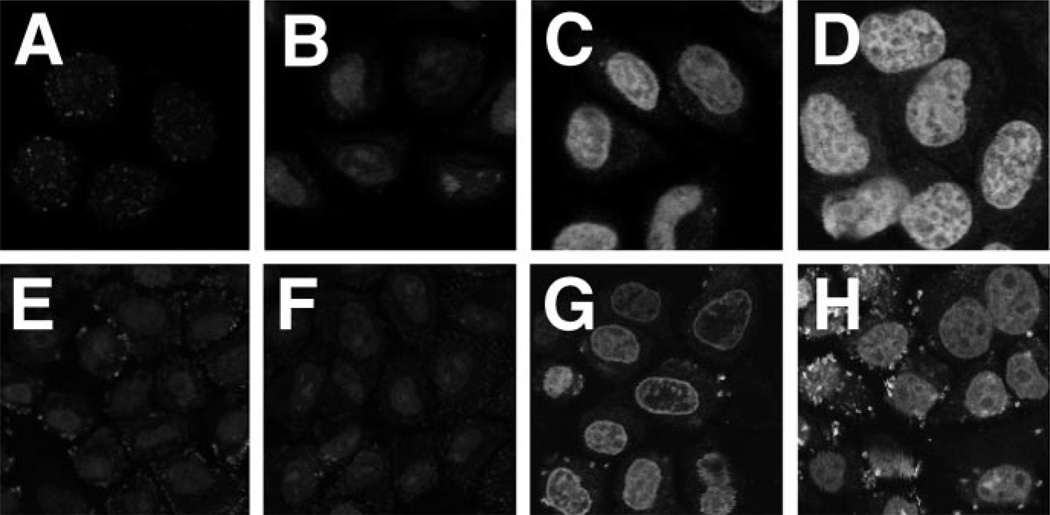
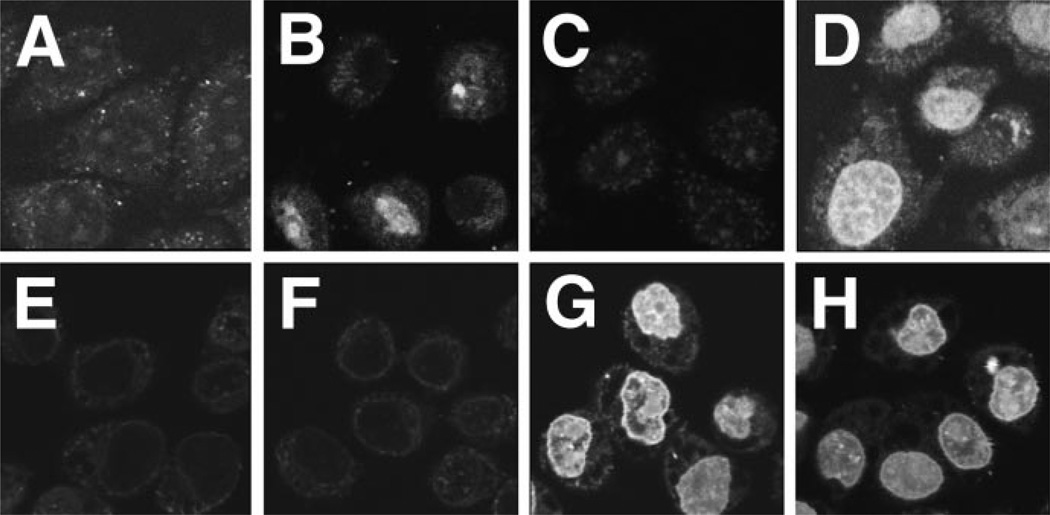
To determine whether cytoplasmic extracts were indeed required for the nuclear import of pDNA, reactions were performed with various combinations of cellular extracts (Fig. 4). Our working model for signal-mediated pDNA nuclear import postulates that sequence specificity is mediated by transcription factors and DNA-binding proteins. These proteins bind to sites within the SV40 DNA nuclear targeting sequence while the DNA and the proteins are in the cytoplasm, and the NLSs present on the transcription factors enable the pDNA-protein complex to utilize the NLS-dependent machinery for nuclear import. This model predicts that cytoplasmic extracts containing both DNA-binding proteins and the NLS-dependent import machinery are necessary for nuclear import of pDNA. In support of this model, no nuclear import was observed in the absence of cytoplasmic extract of either a 4.2-kb plasmid, pUSAG3, or a 14.4-kb plasmid, pUSAG9, both of which contain the PNA-binding site and the SV40 DNA nuclear targeting sequence (Fig. 4, A and E). In contrast, when cytoplasmic extract was provided, significant nuclear import of both plasmids was detected at 4 h (Fig. 4, C and G). Interestingly, many nuclei showed distinct rim staining of the larger plasmid (Fig. 4G). This suggests either that import was not complete at 4 h and larger plasmids take longer to enter the nucleus or that one or more factors are limiting for efficient nuclear import of the larger plasmid. Because our model predicts that transcription factors and other DNA-binding proteins mediate nuclear pDNA import, nuclear extracts were tested for their ability to support or enhance nuclear import of the plasmids. Although the addition of nuclear extract alone did not support nuclear localization of the 14.4-kb plasmid (Fig. 4F), it did allow a low level of import of the smaller plasmid (Fig. 4B). Because all components of the NLS-dependent import machinery can be found in the nucleus at levels much less than those in the cytoplasm, this is not a completely surprising result. When both nuclear and cytoplasmic extracts were provided to the cells, import of both plasmids was more robust (Fig. 4, D and H). Further, no rim staining was observed for either of the plasmids.
Fig. 4. Requirement of cellular extracts for nuclear import of pDNA.
Digitonin permeabilized HeLa cells were incubated for 4 h at 37 °C with either the Fl-PNA conjugated 4.2-kb plasmid pUSAG3 (A–D) or the Fl-PNA conjugated 14.4-kb plasmid pUSAG9 (E–H), both at 10 µg/ml, in the presence or absence of cellular extracts. BSA was added to 10 mg/ml to cells incubated in the absence of extracts (A and E). HeLa cell nuclear extract (Promega) was added to 0.5 mg/ml B and F). HeLa cell cytoplasmic extract was added to 5 mg/ml in the absence (C and G) or presence (D and H) of nuclear extract.
Nuclear Import of pDNA Requires Energy and Is Blocked by WGA
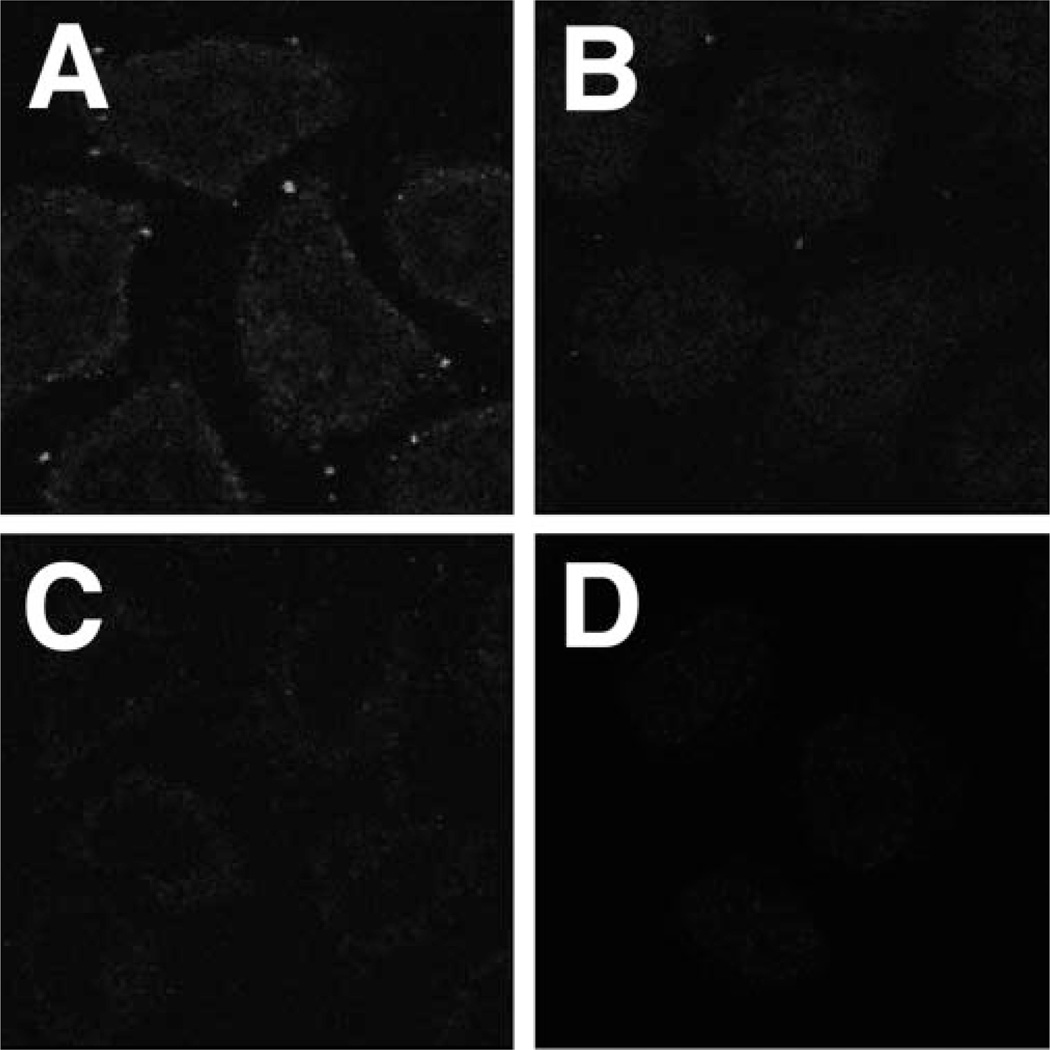
Signal-mediated nuclear import and export of proteins and RNAs has been shown to be energy-dependent (7, 12, 14, 15). Previous experiments from our laboratory using a microinjection approach have demonstrated that this is also the case for plasmid nuclear import (4). This energy dependence was confirmed in the permeabilized cell system (Fig. 5). When GTP and the ATP-regenerating system were omitted from the assays and endogenous stores of nucleotide triphosphates were depleted from the cytoplasmic and nuclear extracts by pretreatment with apyrase, no nuclear localization of pDNA was detected with any combination of extracts (Fig. 5). Similarly, these treatments also prevented nuclear accumulation of Rh-BSA-NLS (data not shown), confirming that ATP levels were sufficiently low to block NLS-mediated nuclear import.
Fig. 5. Energy dependence of pDNA nuclear import.
Digitonin permeabilized HeLa cells were incubated for 4 h at 37 °Cwith transport buffer lacking ATP, phosphocreatine, creatine phosphokinase, GTP, and Fl-PNA/pUSAG3 (10 µg/ml). The cells were then washed, fixed with 3% paraformaldehyde, and viewed by confocal microscopy. Cells incubated in the absence of cellular extracts contained 10 mg/ml BSA (A). Nuclear (B and D) and cytoplasmic (C and D) extracts were depleted of endogenous nucleotide triphosphates by preincubating the extracts with 10 units/ml apyrase at 25 °C for 30 min.
Addition of the lectin WGA that binds to N-acetylglucosamine residues present on a class of NPC proteins also inhibited nuclear accumulation of pDNA in permeabilized cells (Fig. 6). When added to cells, WGA completely inhibited the nuclear import of Fl-PNA/pUSAG3 (Fig. 6C) compared with the control reaction lacking added lectin (Fig. 6A), whereas addition of the lectin concanavalin A that has been shown not to alter transport through the NPC had no effect on pDNA import (Fig. 6B).
Fig. 6. Effects of lectins on pDNA nuclear import.
Permeabilized HeLa cells were incubated with transport buffer containing Fl-PNA/pUSAG3 (10 µg/ml) for 4 h at 37 °C. All reactions contained HeLa cytoplasm at 5 mg/ml and either no additional lectin (A), concanavalin A (0.1 mg/ml; B) or wheat germ agglutinin (0.1 mg/ml; C).
Nuclear Import of pDNA Requires the NLS Import Machinery as Well as Additional Factors
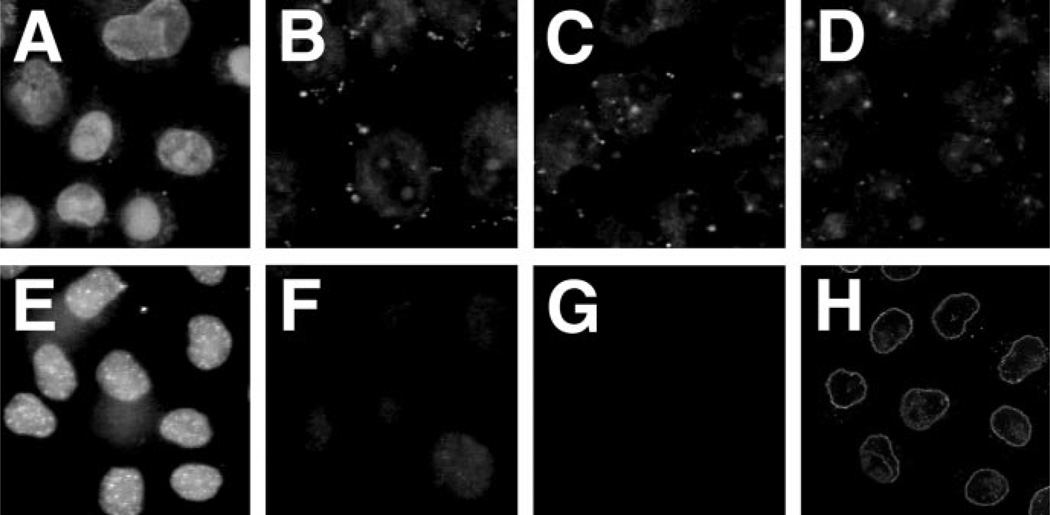
Based on the above experiments, it appeared that the need for cytoplasmic extracts for pDNA nuclear import was to supply components of the NLS-dependent transport machinery. Thus, we expressed and affinity purified histidine-tagged fusion proteins corresponding to importin α, importin β, and RAN. The proteins were judged to be >95% pure based on Coomassie Blue-stained polyacrylamide gels (data not shown). These were then added with either Rh-BSA-NLS or Fl-PNA/pUSAG3 to permeabilized cells, in the presence or absence of nuclear extract prepared from HeLa cells to provide the appropriate NLS-containing DNA-binding proteins (Fig. 7). As seen previously, in the absence of cytoplasmic extract or the importins and RAN, no nuclear import of either pDNA or Rh-BSA-NLS was observed (Fig. 7, A and E). Similarly, little or no nuclear import of either substrate was seen in the presence of nuclear extract alone (Fig. 7, B and F). Although the addition of importin α, importin β, and RAN was sufficient to drive the nuclear localization of the NLS-containing reporter protein (Fig. 7G), they were not sufficient to support import of pDNA (Fig. 7C). This suggested either that plasmid DNA does not use the importin α/β pathway for its nuclear import or that additional factors are required to allow the importins to bind to the DNA. Because neither importin α nor β bind to DNA, for them to mediate nuclear import of pDNA, an intermediary “adapter” protein would be required that binds to DNA and contains an NLS for recognition by the importins. Thus, when a nuclear extract was provided in addition to the importins and RAN, significant nuclear localization of Fl-PNA/pUSAG3 was observed (Fig. 7D). The presence of the nuclear extract had little effect on the import of Rh-BSA-NLS (Fig. 7H).
Fig. 7. Reconstitution of pDNA nuclear import using purified importins and RAN.
HeLa cells were permeabilized and incubated with transport buffer containing Fl-PNA/pUSAG3 (10 µg/ml; A–D) or Rh-BSA-NLS (25 µg/ml; E–H) at 37 °C with the indicated additions. Rh-BSA-NLS import reactions were incubated for 30 min, and pDNA import reactions proceeded for 4 h. His-tagged importin α, importin β, and RAN were purified by nickel affinity chromatography and concentrated for addition to the reactions. The reactions contained BSA alone (10 mg/ml; A and E), nuclear extract alone (0.5 mg/ml; B and F), importin α, importin β, and RAN (0.5 mg/ml each; C and G), or importin α, importin β, RAN, and nuclear extract (D and H).
To ensure that all three components of the nuclear import machinery (i.e. importin α, importin β, and RAN) were indeed required for pDNA nuclear entry, import reactions were performed in which each of the factors was left out separately (Fig. 8). As was seen in Fig. 7, significant nuclear import of both pDNA and Rh-BSA-NLS was observed in the presence of all three components and nuclear extract (Fig. 8, A and E). However, when the import reactions were performed with nuclear extract and importin α and RAN (Fig. 8, B and F), importin β and RAN (Fig. 8, C and G), or importin α and importin β (Fig. 8, D and H), nuclear import of both substrates was eliminated. The nuclear rim staining of Rh-BSA-NLS observed in the presence of the importins alone (Fig. 8H) was expected based on previous reports (16) and confirms the fidelity of the recombinant proteins and the experimental system. Thus, similar to “classical” protein nuclear import, both the importins and RAN are required for plasmid nuclear entry.
Fig. 8. Requirement of importin α, importin β, and RAN for nuclear import.
HeLa cells were permeabilized and incubated with transport buffer containing Fl-PNA/pUSAG3 (10 µg/ml; A–D) or Rh-BSA-NLS (25 µg/ml; E–H) at 37 °C with the indicated additions. Rh-BSANLS import reactions were incubated for 30 min, and pDNA import reactions proceeded for 4 h. All reactions contained HeLa cell nuclear extract (0.5 mg/ml). Affinity purified His-tagged importins and RAN were added to 0.5 mg/ml each. The reactions contained importin α, importin β, and RAN (A and E), importin α, and RAN (B and F), importin β, and RAN (C and G) or importin α and importin β (D and H).
Sequence Specificity of pDNA Nuclear Import
Competition experiments suggested that the nuclear import of Fl-PNA/ pDNA was signal-mediated. When permeabilized cells were incubated with Fl-PNA/pUSAG3 in the presence of ATP and cytoplasmic and nuclear extracts, the plasmid localized to the nuclei of cells (Fig. 9A). Addition of a 1000-fold molar excess of BSA-NLS had no effect on the nuclear localization of the plasmid (Fig. 9B). This is not entirely surprising, because the small NLS-containing protein is imported into the nuclei within minutes of addition and thus is not a true competitor for the NLS-dependent machinery, which can recycle back to the cytoplasm for import of pDNA. Striking inhibition of pDNA nuclear import was seen when a 100-fold molar excess of SV40 DNA was added to the reaction (Fig. 9D), whereas a similar excess of pUC19 had no effect on pDNA import (Fig. 9C). Because both pUSAG3 and SV40 DNA share the 360-bp SV40 promoter/enhancer region that is necessary in intact cells for nuclear targeting of pDNA, it appeared likely that the permeabilized cell system faithfully reproduced this sequence specificity. To test this directly, a 913-bp fragment containing the 360-bp SV40 DNA nuclear targeting sequence was removed from pUSAG3 to create pUSAG3ΔSV40. This plasmid was labeled with Fl-PNA and used in the permeabilized cell assay in parallel to the parent Fl-PNA/pUSAG3 (Fig. 10). As expected, neither plasmid was able to localize to the nucleus in the absence of cytoplasmic extract (Fig. 10, A and C). The plasmid lacking the SV40 sequence was also unable to localize to the nuclei in the presence of cytoplasmic extract, although faint rim staining was detected (Fig. 10D), whereas the parent pUSAG3 was imported efficiently (Fig. 10B). These experiments confirm that pDNA nuclear import in permeabilized cells faithfully represents that seen in intact cells and that nuclear uptake of plasmid DNA is sequence-specific.
Fig. 9. Competition of Fl-PNA/pDNA nuclear import by plasmid DNA and BSA-NLS.
Permeabilized HeLa cells were incubated for 4 h at 37 °C with transport buffer containing HeLa cytoplasmic extract (5 mg/ml) and Fl-PNA/pUSAG3 (10 µg/ml) in the absence (A) or presence of a 1000-fold molar excess of BSA-NLS (0.23 mg/ml; B), a 100-fold excess of pUC19 (0.7 mg/ml; C), or a 100-fold molar excess of SV40 DNA (1.3 mg/ml; D).
Fig. 10. Sequence specificity of pDNA nuclear import in permeabilized cells.
Permeabilized HeLa cells were incubated for 4 h at 37 °C in transport buffer containing BSA alone (A and C) or HeLa cytoplasmic extract at 5 mg/ml (B and D). The substrates used were either Fl-PNA/pUSAG3 (A and B) or Fl-PNA/pUSAG3DSV40 (C and D), both present at 10 µg/ml.
PNA-labeled and Unlabeled Plasmid DNA Remains Largely Intact in Cytoplasmic Extract
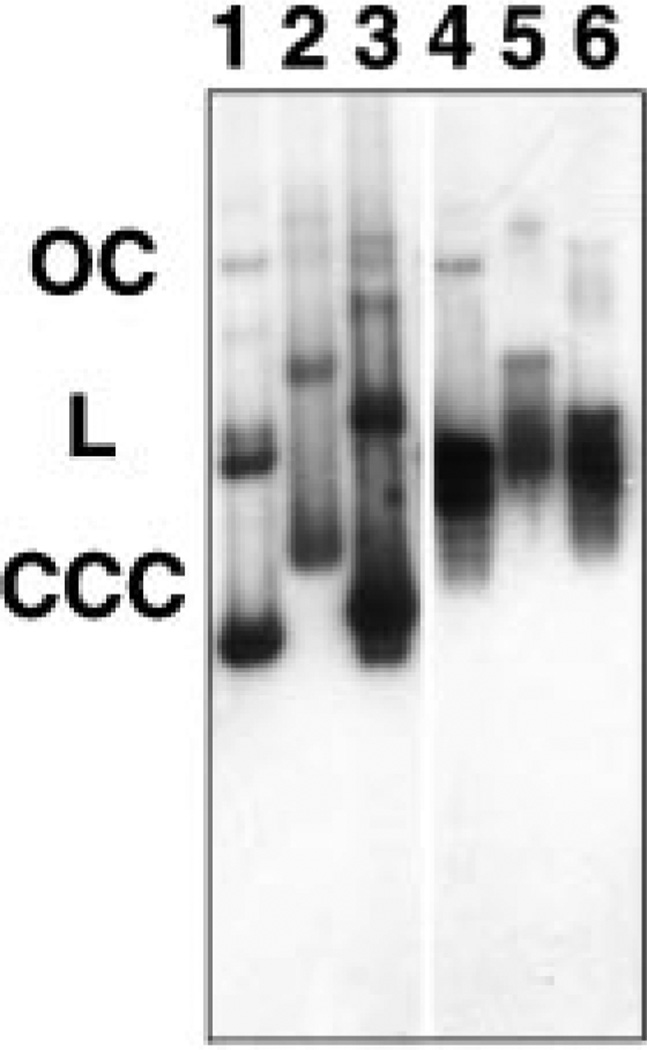
If significant degradation of pDNA is occurring in our system, the apparent nuclear import of Fl-PNA/pDNA could actually represent nuclear localization of small, labeled DNA fragments or free Fl-PNA alone liberated from the pDNA substrate upon degradation. This did not appear to be the case, because if plasmid DNA was degraded to release smaller fragments or free Fl-PNA, Fl-PNA/pUSAG3ΔSV40 should have appeared nuclear in the permeabilized cell assays as did Fl-PNA/pUSAG3 (Fig. 10). To test this directly, a Southern blot was performed on both Fl-PNA-labeled and unlabeled pDNA that had been incubated with or without cytoplasmic extract for 4 h at 37 °C, just as in our permeabilized cell assay (Fig. 11). Both Fl-PNA-labeled and unlabeled pUSAG3 were used for the Southern blot. A second rhodamine-labeled PNA-complexed plasmid, pGenegrip-GFP ((17), Gene Therapy Systems, La Jolla, CA), that expresses GFP but does not contain the SV40 DNA nuclear targeting sequence was also used in the Southern blot to determine the fate of plasmids lacking the SV40 sequence. This plasmid, like pUSAG3ΔSV40, also failed to be imported into nuclei (not shown). As can be seen in Fig. 11 (lanes 1–3), both the PNA-labeled and unlabeled plasmids used as substrate were predominantly supercoiled, with small amounts of open circular and linear DNA. After 4 h of incubation in cytoplasmic extract, all of the supercoiled form of these three plasmids had been converted to linear and open circular DNA, but relatively little of the DNA had been degraded further (Fig. 11, lanes 4–6). Thus, neither the presence of the PNA or of the SV40 sequence altered the stability of the plasmids, and it can be concluded that the observed “import” was indeed that of pDNA.
Fig. 11. Degradation of pDNA in cytoplasmic extract.
Plasmid DNA (0.1 µg) was incubated for 4 h at 37 °C with HeLa cell cytoplasmic extract (5 mg/ml) in transport buffer. Protein was removed by phenol: chloroform extraction, and the DNAs were separated by agarose gel electrophoresis, transferred to a nylon membrane, and probed using a mixture of 32P-labeled pUSAG3 and pGenegrip blank. Lanes 1–3 contain 0.1 µg of unincubated plasmid used as starting material, and lanes 4–6 contain DNA incubated with cytoplasmic extract. Three plasmids were used: Fl-PNA/pUSAG3 (lanes 1 and 4), rhodamine-labeled PNA/ pGenegrip-GFP (lanes 2 and 5), and unlabeled pUSAG3 (lanes 3 and 6).
DISCUSSION
Using the digitonin-permeabilized cell assay developed to study the nuclear import of NLS-containing proteins (7), we have shown that plasmid DNA can enter the nuclei of cells. The ability to label supercoiled plasmids at a specific site using a fluorescently labeled PNA (18) while maintaining their transcriptional (10)3 and migratory activity has allowed us to follow the nuclear import of plasmids as opposed to linear DNA fragments. That plasmids labeled in this manner can be found to enter the nucleus in microinjected cells, as do unmodified plasmids, confirms that PNA-labeled plasmids are substrates relevant for studying plasmid nuclear import in vitro. Plasmid nuclear import in digitonin permeabilized cells is energy-dependent, is inhibited by the NPC-binding lectin WGA, and requires cytoplasmic extracts. As has been found for the nuclear localization of NLS-containing proteins (1), the proteins necessary in the cytoplasmic extracts for plasmid nuclear uptake include importin α and β, and RAN. However, although addition of these proteins alone is sufficient to reconstitute BSA-NLS nuclear import, proteins present in nuclear extract are also required for pDNA nuclear entry. This, coupled with the finding that plasmid nuclear import is sequence-specific in both permeabilized cells and microinjected cells, supports a model for pDNA nuclear import in which DNA-binding proteins that contain NLSs act as intermediaries to couple the NLS import machinery to pDNA.
The development of PNA clamps as a way to specifically label pDNA at discrete sites has enabled these studies to be performed. Although multiple fluorescent labeling techniques were tested and found to produce highly modified DNA, none of them proved useful because they all destroyed the ability of the DNA to localize to the nucleus in microinjected cells. Since these techniques targeted nucleotides randomly throughout the plasmid, it is likely that they labeled sites within DNA sequences necessary for nuclear import, thus making it impossible for the appropriate DNA-binding proteins to make contact with the plasmid and mediate import. The ability to place the PNA target sequence far away from the DNA nuclear import sequence, and any desired transgene to be expressed, has allowed us to produce fluorescent pDNA that is supercoiled and able to target to the nucleus. The resulting PNA-labeled plasmids target to the nucleus with the same time course as native plasmid in microinjected cells and, further, distribute within the nucleus similarly. The exclusion of Fl-PNA/pDNA from the nucleoli and the somewhat punctate distribution within the nucleoplasm are reminiscent of the distribution of SV40 DNA in injected cells (4).
Relatively little degradation of unlabeled and Fl-PNA-labeled plasmid DNA was observed by Southern blot. Although none of the DNA remained in the supercoiled state after 4 h in cytoplasmic extract, greater than 80% of the DNA detected by hybridization was in either the open circular or linear form. Lack of cytoplasmic plasmid degradation has also been seen in microinjected cells based on the observation that as few as 10 copies of a plasmid injected into the cytoplasm can direct gene expression within 8 h.2 A similar lack of degradation of DNA by cytoplasmic extracts or in intact cells has also been seen by others (13, 19, 20). Because plasmids containing or lacking the SV40 DNA nuclear targeting sequence showed essentially the same levels of conversion to other forms and because only one of them displayed nuclear localization, it is unlikely that the nuclear import observed was due to diffusion or import of PNA alone or small DNA fragments containing the PNA-binding site bound to PNA. The energy dependence of nuclear import and inhibition by WGA demonstrate that Fl-PNA/pDNA nuclear localization is an active transport process. Although active transport could also account for nuclear import of free Fl-PNA or small DNA fragments, it is unlikely, because of the findings that small oligonucleotides appear to enter the nucleus in an energy-independent manner, consistent with diffusion (13, 21, 22). Thus, the import we observe is likely that of a mix of supercoiled, open circular, and full-length linear plasmid DNA.
The observation that proteins present in cytoplasmic extract are necessary for pDNA nuclear entry is not surprising, based on the finding that all regulated transport studied to date requires proteins found in the cytoplasm (1). Further, the ability of importin α, importin β, and RAN to mediate the nuclear import of pDNA in the presence of a nuclear extract that otherwise cannot support import unifies the import of pDNA with that of other substrates, including proteins and snRNPs. This requirement of the soluble NLS import machinery or plasmid nuclear localization appears to be in contrast to the results of Wolff and colleagues (13) who found that although nuclear import of fluorescently labeled linear DNA fragments was energy-dependent and inhibited by WGA, it occurred optimally in the absence of cytoplasmic proteins. The reason for this disparity is unclear, but a possible explanation could reside in the differences in substrates used for import. Uniformly labeled, short, linear DNA and plasmid labeled at a discrete site are two very different substrates and, as such, may utilize different pathways for entry. Further, the choice of DNA sequence may have had an effect on transport; whereas DNA fragments less than 1 kb may enter the nucleus in the absence of cell extract and a DNA nuclear targeting sequence, the inability of larger DNAs to enter could have been due to the lack of such a sequence, and consequently, the lack of cytoplasmic extract. Thus, with no DNA nuclear targeting sequence, transcription factors and other DNA-binding proteins cannot bind to provide the DNA with the NLSs needed for import by the importin pathway. Indeed, an absolute requirement for cytoplasmic extract was observed by the same group when NLS peptides were synthetically coupled to plasmid DNA (23).
The need for nuclear extract in addition to the importins and RAN for plasmid DNA nuclear import supports our model for DNA nuclear localization in which pDNA is targeted to the nucleus and translocated by an importin pathway recognizing protein NLSs that are attached to the DNA (4).2 That the SV40 DNA nuclear targeting sequence contains binding sites for multiple transcription factors also supports the model. Although we have not yet identified the protein(s) or protein complex that binds to the DNA and provides the needed NLSs, it is clear that at least one or more of these proteins can interact with importin α and importin β1 as opposed to other β homologues. However, it is also possible that additional transcription factors and proteins bind to the DNA and aid in import by utilizing other importin pathways as well (24). Several other examples in which NLS-containing proteins are thought to mediate the nuclear localization of exogenous DNA have been reported, including the matrix and Vpr proteins of HIV for the cytoplasmic reverse transcribed HIV genome (25), the virD2 and virE2 proteins of Agrobacterium tumefaciens for the pathogenic T-DNA (26, 27), the Epstein-Barr virus EBNA-1 protein for EBV oriP-containing plasmids (28), and the serum response factor for plasmids containing the smooth muscle gamma actin promoter (29). Further, many viral capsid and core proteins participate in the nuclear localization of viral genomes, apparently by similar mechanisms (30–33). Thus, such “piggy-back” transport of DNA appears a common theme, both evolutionarily and experimentally.
Import of plasmid DNA can be thought to occur in three steps: formation of the protein-DNA complex, transit to the NPC, and translocation. All three of these processes probably require significant time to occur and can account for the differences in time courses for plasmid nuclear localization (hours) and that of individual proteins or snRNPs (minutes) (4).2 The concentration of DNA-binding proteins in the cytoplasm is expected to be low, limited mainly to those proteins that have recently been translated and not yet targeted to the nucleus. Although not rate-limiting in permeabilized cells because of the excess of nuclear proteins added, the initial protein-DNA complex will form slowly in intact cells because it is a function of the concentration of the two substrates. Movement of this macromolecular complex through the cytoplasm is likely to encounter resistance because of the presence of immobile objects and cytoskeletal elements, adding to the time required for nuclear localization. Indeed, it has been shown that cytoplasmic diffusion of dextrans with hydrodynamic radii greater than 30 nm is greatly reduced compared with smaller species (34, 35). Because size estimates for condensed plasmids range from 25 to 80 nm in diameter (36–38), their diffusion through the cytoplasm is expected to be low. Finally, once at the NPC, the translocation event itself should take much longer for DNA than for proteins, based on its much larger size and shape and the fact that the rate of nuclear transport is directly correlated to the molecular weight and shape of the substrate (39, 40). It is unlikely that fully condensed plasmid is imported into the nucleus without being partially hydrated, because the upper limit to the NPC transporter appears to be 28 nm (40), the same as the lower limit for a condensed plasmid. Such similar unfolding or hydration has been suggested to participate in the nuclear translocation of similarly sized Balbiani Ring RNP particles (41).
It has recently been suggested that nuclear import of plasmid DNA may begin by one NLS or a discrete foci of NLSs interacting with the NPC machinery and being translocated and proceeds with the rest of the DNA being pulled in because of the rapid formation of higher order chromatin structures on the DNA as it enters into the nucleoplasm (42). Alternatively, based on our finding that plasmid DNA nuclear entry is dependent on transcription, it is also possible that the remainder of the plasmid is pulled into the nucleus because of transcription of plasmid DNA sequences (4). Thus, the presence of one NLS, or presumably a discrete foci of NLSs, on a plasmid should be sufficient to initiate the translocation process (42). It has also been proposed that the presence of one NLS on a plasmid will allow a plasmid to enter the nucleus through an NPC much better than would multiple NLS distributed around the plasmid (42). The reasoning is that multiple NLSs on one plasmid could interact with multiple NPCs to arrest transport in a “tug-of-war.” Based on the average density of NPCs in the nuclear envelope (43) and the persistence length of DNA, this would correspond to interactions with an NPC once every kilobase (42). Although this is an intriguing model, the demonstration that plasmids containing 70–100 NLSs distributed randomly around a plasmid are imported into the nucleus suggests that this may not be universal (23). Furthermore, the presence of DNA-binding domains facing the cytoplasm on several NPC proteins would also have the same effect of arresting transport of large DNAs, regardless of whether NLSs were present (44, 45). However, with the ability to covalently attach NLS peptides to the amino termini of PNAs and incorporate PNA target sequences at multiple sites around a plasmid, this model is easily testable.
Acknowledgments
We thank Lou Smith (GeneMedicine, The Wood-lands, TX) for advice and the gift of the pGenegrip plasmids, Karsten Weis (EMBL, Heidelberg, Germany) for the gift of pHSRPα, Mark Goulian (Institute for Physics and Biology, The Rockefeller University, New York, NY) for providing us with Fl-BSA-NLS, and Ray Hester for help using the confocal microscope. We also thank Felix Munkonge and Warren Zimmer for helpful discussions and critical reading of the manuscript.
Footnotes
This work was supported in part by Grant ALG 960006 from the Alabama Affiliate of the American Heart Association (to D. A. D.), Grant 5-FY97-0692 from the March of Dimes Foundation (to G. W.), and Grants R01 HL59956 (to D. A. D.) and 5P60 HL38639 (to D. A. D. and G. W.) from the National Institutes of Health.
The abbreviations used are: NPC, nuclear pore complex; NLS, nuclear localization signal; HIV, human immunodeficiency virus; GFP, green fluorescent protein; PNA, peptide nucleic acid; Fl, fluorescein; Rh, rhodamine; BSA, bovine serum albumin; BSA-NLS, synthetic NLS peptide-conjugated bovine serum albumin; WGA, wheat germ agglutinin; pDNA, plasmid DNA; bp, base pair(s); kb, kilobase(s).
D. A. Dean, B. S. Dean, S. Muller, and L. C. Smith, submitted for publication.
Wang, G., Xu, X., Pace, B., Dean, D. A., Glazer, P. M., Chan, P., Goodman, S. R., and Shoklenko, I. (1999) Nucleic Acids Res. 27, 2806–2813.
References
- 1.Mattaj IW, Englmeier L. Annu. Rev. Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 2.Melchior F, Gerace L. Trends Cell Biol. 1998;8:175–179. doi: 10.1016/s0962-8924(98)01252-5. [DOI] [PubMed] [Google Scholar]
- 3.Moroianu J. J. Cell. Biochem. 1998;70:231–239. [PubMed] [Google Scholar]
- 4.Dean DA. Exp. Cell Res. 1997;230:293–302. doi: 10.1006/excr.1996.3427. [DOI] [PubMed] [Google Scholar]
- 5.Weis K, Mattaj IW, Lamond AI. Science. 1995;268:1049–1053. doi: 10.1126/science.7754385. [DOI] [PubMed] [Google Scholar]
- 6.Moroianu J, Blobel G, Radu A. Proc. Natl. Acad. Sci. U. S. A. 1995;92:2008–2011. doi: 10.1073/pnas.92.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adam SA, Marr RS, Gerace L. J. Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1994. [Google Scholar]
- 9.Egholm M, Buchardt O, Christensen L, Behrens C, Freier SM, Driver DA, Berg RH, Kim SK, Norden B, Nielsen PE. Nature. 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 10.Nielsen PE, Egholm M, Buchardt O. Gene (Amst.) 1994;149:139–145. doi: 10.1016/0378-1119(94)90422-7. [DOI] [PubMed] [Google Scholar]
- 11.Demidov VV, Potaman VN, Frank-Kamenetskii MD, Egholm M, Buchardt O, Sonnichsen SH, Nielsen PE. Biochem. Pharmacol. 1994;48:1310–1313. doi: 10.1016/0006-2952(94)90171-6. [DOI] [PubMed] [Google Scholar]
- 12.Marshallsay C, Lührmann R. EMBO J. 1994;13:222–231. doi: 10.1002/j.1460-2075.1994.tb06252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagstrom JE, Ludtke JJ, Bassik MC, Sebestyén MG, Adam SA, Wolff JA. J. Cell Sci. 1997;110:2323–2331. doi: 10.1242/jcs.110.18.2323. [DOI] [PubMed] [Google Scholar]
- 14.Holaska JM, Paschal BM. Proc. Natl. Acad. Sci. U. S. A. 1998;95:14739–14744. doi: 10.1073/pnas.95.25.14739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Love DC, Sweitzer TD, Hanover JA. Proc. Natl. Acad. Sci. U. S. A. 1998;95:10608–10613. doi: 10.1073/pnas.95.18.10608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moore MS, Blobel G. Cell. 1992;69:939–950. doi: 10.1016/0092-8674(92)90613-h. [DOI] [PubMed] [Google Scholar]
- 17.Zelphati O, Liang X, Hobart P, Felgner PL. Hum. Gene Ther. 1999;10:15–24. doi: 10.1089/10430349950019156. [DOI] [PubMed] [Google Scholar]
- 18.Bentin T, Nielsen PE. Biochemistry. 1996;35:8863–8869. doi: 10.1021/bi960436k. [DOI] [PubMed] [Google Scholar]
- 19.Graessman M, Menne J, Liebler M, Graeber I, Graessman A. Nucleic Acids Res. 1989;17:6603–6612. doi: 10.1093/nar/17.16.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Escriou V, Ciolina C, Lacroix F, Byk G, Scherman D, Wils P. Biochim. Biophys. Acta. 1998;1368:276–288. doi: 10.1016/s0005-2736(97)00194-6. [DOI] [PubMed] [Google Scholar]
- 21.Beltinger C, Saragovi HU, Smith RM, LeSauteur L, Shah N, DeDionisio L, Christensen L, Raible A, Jarett L, Gewirtz AM. J. Clin. Invest. 1995;95:1814–1823. doi: 10.1172/JCI117860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartig R, Shoeman RL, Janetzko A, Grub S, Traub P. Biol. Cell. 1998;90:407–426. [PubMed] [Google Scholar]
- 23.Sebestyén MG, Ludtke JL, Bassik MC, Zhang G, Budker V, Lukhtanov EA, Hagstrom JE, Wolff JA. Nat. Biotech. 1998;16:80–85. doi: 10.1038/nbt0198-80. [DOI] [PubMed] [Google Scholar]
- 24.Albertini M, Pemberton LF, Rosenblum JS, Blobel G. J. Cell Biol. 1998;143:1447–1455. doi: 10.1083/jcb.143.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stevenson M. Trends Cell Biol. 1996;6:9–15. doi: 10.1016/0962-8924(96)81032-4. [DOI] [PubMed] [Google Scholar]
- 26.Zupan JR, Citovsky V, Zambryski P. Proc. Natl. Acad. Sci. U. S. A. 1996;93:2392–2397. doi: 10.1073/pnas.93.6.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shurvinton CE, Hodges L, Ream W. Proc. Natl. Acad. Sci. U. S. A. 1992;89:11837–11841. doi: 10.1073/pnas.89.24.11837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langle-Rouault F, Patzel V, Benavente A, Taillez M, Silvestre N, Bompard A, Sczakiel G, Jacobs E, Rittner K. J. Virol. 1998;72:6181–6185. doi: 10.1128/jvi.72.7.6181-6185.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vacik J, Dean BS, Zimmer WE, Dean DA. Gene Ther. 1999;6:1006–1014. doi: 10.1038/sj.gt.3300924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whittaker GR, Helenius A. Virology. 1998;246:1–23. doi: 10.1006/viro.1998.9165. [DOI] [PubMed] [Google Scholar]
- 31.Nakanishi A, Clever J, Yamada M, Li PL, Kasamatsu H. Proc. Natl. Acad. Sci. U. S. A. 1996;93:96–100. doi: 10.1073/pnas.93.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greber UF, Kasamatsu H. Trends Cell Biol. 1996;6:189–195. doi: 10.1016/0962-8924(96)10016-7. [DOI] [PubMed] [Google Scholar]
- 33.Kann M, Bischof A, Gerlich WH. J. Virol. 1997;71:1310–1316. doi: 10.1128/jvi.71.2.1310-1316.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luby-Phelps K, Castle PE, Taylor DL, Lanni F. Proc. Natl. Acad. Sci. U. S. A. 1987;84:4910–4913. doi: 10.1073/pnas.84.14.4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seksek O, Biwersi J, Verkman AS. J. Cell Biol. 1997;138:131–142. doi: 10.1083/jcb.138.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duguid JG, Li C, Shi M, Logan MJ, Alila H, Rolland A, Tomlinson E, Sparrow JT, Smith LC. Biophys. J. 1998;74:2802–2814. doi: 10.1016/S0006-3495(98)77987-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma C, Bloomfield VA. Biophys. J. 1994;67:1678–1681. doi: 10.1016/S0006-3495(94)80641-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blessing T, Remy JS, Behr JP. Proc. Natl. Acad. Sci. U. S. A. 1998;95:1427–1431. doi: 10.1073/pnas.95.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lanford RE, Kanda P, Kennedy RC. Cell. 1986;46:575–582. doi: 10.1016/0092-8674(86)90883-4. [DOI] [PubMed] [Google Scholar]
- 40.Dworetzky SI, Lanford RE, Feldherr CM. J. Cell Biol. 1988;107:1279–1287. doi: 10.1083/jcb.107.4.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mehlin H, Daneholt B, Skoglund U. J. Cell Biol. 1995;129:1205–1216. doi: 10.1083/jcb.129.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zanta MA, Belguise-Valladier P, Behr JP. Proc. Natl. Acad. Sci. U. S. A. 1999;96:91–96. doi: 10.1073/pnas.96.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maul GG, Deaven L. J. Cell Biol. 1977;73:748–760. doi: 10.1083/jcb.73.3.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu J, Matunis MJ, Kraemer D, Blobel G, Coutavas E. J. Biol. Chem. 1995;270:14209–14213. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]
- 45.Sukegawa J, Blobel G. Cell. 1993;72:29–38. doi: 10.1016/0092-8674(93)90047-t. [DOI] [PubMed] [Google Scholar]